Summary
Injured peripheral neurons successfully activate a pro-regenerative transcriptional program to enable axon regeneration and functional recovery. How transcriptional regulators co-ordinate the expression of such program remains unclear. Here we show that hypoxia-inducible factor 1α (HIF-1α) controls multiple injury-induced genes in sensory neurons and contribute to the preconditioning lesion effect. Knockdown of HIF-1α in vitro or conditional knock out in vivo impairs sensory axon regeneration. The HIF-1α target gene Vascular Endothelial Growth Factor A (VEGFA) is expressed in injured neurons and contributes to stimulate axon regeneration. Induction of HIF-1α using hypoxia enhances axon regeneration in vitro and in vivo in sensory neurons. Hypoxia also stimulates motor neuron regeneration and accelerates neuromuscular junction re-innervation. This study demonstrates that HIF-1α represents a critical transcriptional regulator in regenerating neurons and suggests hypoxia as a tool to stimulate axon regeneration.
Introduction
Permanent disabilities following central nervous system (CNS) injuries result from the failure of injured axons to regenerate and rebuild functional connections. The poor intrinsic regenerative capacity of mature CNS neurons is a major contributor to the regeneration failure (Di Giovanni, 2009; Fagoe et al., 2014; Liu et al., 2011; Lu et al., 2014). In contrast to CNS neurons, peripheral nervous system (PNS) neurons successfully activate intrinsic signaling pathways to enable axon regeneration (Bradke et al., 2012; Hoffman, 2010; Liu et al., 2011). Defining how injured PNS neurons transition to a pro-regenerative state may suggest therapeutic approaches to improve neuronal recovery following axon injury.
Sensory neurons with cell body in dorsal root ganglia (DRG) provide a useful model system to study intrinsic regenerative pathways. DRG neurons are pseudounipolar and in addition to their peripherally projecting branch, they extend a second branch through the dorsal root to innervate targets in the spinal cord or brainstem. Whereas injury to the peripheral branch increases DRG neurons’ intrinsic growth capacity and promote expression of pro-regenerative genes such as c-Jun and STAT3, injury to the dorsal root only minimally simulates axon growth capacity and pro-regenerative gene expression (Broude et al., 1997; Schwaiger et al., 2000; Smith and Skene, 1997). Similarly, injury to the peripheral branch, but not the central branch elicits changes in the chromatin that contribute to regulation of the transcriptional response (Cho et al., 2013; Finelli et al., 2013; Puttagunta et al., 2014). Activation of such a pro-regenerative gene expression program following peripheral injury is illustrated by the conditioning injury paradigm, in which a sensory neuron exposed to a prior peripheral lesion exhibits a dramatic improvement in axon regeneration compared to that of a naive neuron (McQuarrie and Grafstein, 1973; Richardson and Issa, 1984; Smith and Skene, 1997). This paradigm also reveals that activation of a pro-regenerative program can partially overcome the inhibitory environment of the CNS (Neumann and Woolf, 1999).
Activation of a pro-regenerative program in peripheral neurons relies on the expression of multiple regeneration-associated genes (RAGs) (Blackmore, 2012). Although many genes have been identified for their pro-regenerative influence, individual gene based approaches have yielded limited success in axon regeneration (Blackmore, 2012; Fagoe et al., 2014), illustrating that manipulation of individual RAGs is unlikely to be sufficient to stimulate robust and meaningful long-distance axon regeneration in the injured CNS. Hence, understanding how a large ensemble of RAGs can be simultaneously activated after injury could reveal strategies to initiate the transcriptional pro-regenerative program.
Many transcriptional profiling studies have demonstrated the differential gene expression patterns in regenerating vs. non-regenerating neurons (Blackmore, 2012; Fagoe et al., 2014; Moore and Goldberg, 2011). Because of the complexity and magnitude of the gene changes detected, several studies have also focused on searching the data set for transcription factor binding sites (Michaelevski et al., 2010). A comparison between four lists of transcription factors and transcriptional modulators predicted to be active in regenerating DRG identified c-Jun as the only transcription factor common to four profiling studies (Blackmore, 2012). ATF3 and members of the KLF and SMAD families, which play important roles in axon regeneration (Moore et al., 2009; Moore and Goldberg, 2011; Zou et al., 2009) were common in three profiling studies (Blackmore, 2012). Another interesting transcription factor identified in this comparative study is the Hypoxia-Inducible Factor (HIF) (Blackmore, 2012). In C. elegans, hif-1 mutants show reduced regeneration (Nix et al., 2014), but whether HIF-1 functions in mammal to regulate axon regeneration has not been examined in detail.
HIF-1 is a transcriptional mediator of the cellular response to hypoxia, a form of cellular stress. HIF-1 is a heterodimeric transcription factor consisting of two subunits, HIF-1α and HIF-1β (Dunwoodie, 2009; Yee Koh et al., 2008). Whereas HIF-1β is constitutively expressed, HIF-1α expression and activity are regulated by cellular oxygen concentration. Under normal oxygen levels HIF-1α hydroxylation targets it for ubiquitylation and proteasomal degradation (Dunwoodie, 2009; Yee Koh et al., 2008). In addition, HIF-1α hydroxylation blocks its binding to the transcriptional co-activator CBP/p300 (Kallio et al., 1998; Lisy and Peet, 2008). In low oxygen conditions, the rate of hydroxylation is reduced, resulting in HIF-1α accumulation and translocation to the nucleus, where it binds HIF binding sites within hypoxia response elements on HIF target gene promoters or enhancers (Pawlus and Hu, 2013). Activation of HIF-1α to a fully competent transcriptional regulatory protein complex involves control of protein stability, subcellular localization, DNA-binding and interaction with transcriptional co-regulators (Pawlus and Hu, 2013; Ruas and Poellinger, 2005). Some of these transcription co-regulators harbor histone acetyltransferase activity, such as p300/CBP (Arany et al., 1996) or SRC-1 (Carrero et al., 2000) or histone deacetylase activity, such as class II HDACs (Kato et al., 2004; Seo et al., 2009). Other transcriptional co-regulators include chromatin-remodeling complexes (Dekanty et al., 2010) and transcription factors (Pawlus and Hu, 2013) such as c-Jun and STAT3 (Pawlus and Hu, 2013), HIF-1α thus controls gene expression by recruiting co-activators and modifying the chromatin structure.
Here we reveal that a large proportion of the known (Benita et al., 2009) or predicted (Ortiz-Barahona et al., 2010) HIF-1α target genes are up-regulated by axon injury in cultured DRG neurons. In the absence of HIF-1α, in vitro and in vivo regeneration of sensory axons is impaired. Consistent with a role of HIF-1α in pro-regenerative gene expression, we found that sciatic nerve injury, but not dorsal root injury leads to the accumulation of nuclear HIF-1α in DRG neurons. We further show that HIF-1α is required to fully activate the intrinsic regenerative program that mediates the conditioning injury paradigm. The HIF-1α target gene VEGFA is expressed in injured neurons and contribute to stimulate axon regeneration. We also demonstrate that acute intermittent hypoxia enhances sensory axon regeneration in a HIF-1α- dependent manner, as well as axon regeneration in motor neurons. This study identifies HIF-1α as an important transcriptional regulator participating in the activation of a pro-regenerative program and suggests a role for hypoxia as a non-invasive tool to stimulate axon regeneration.
Results
A large proportion of HIF-1α target genes are induced following injury
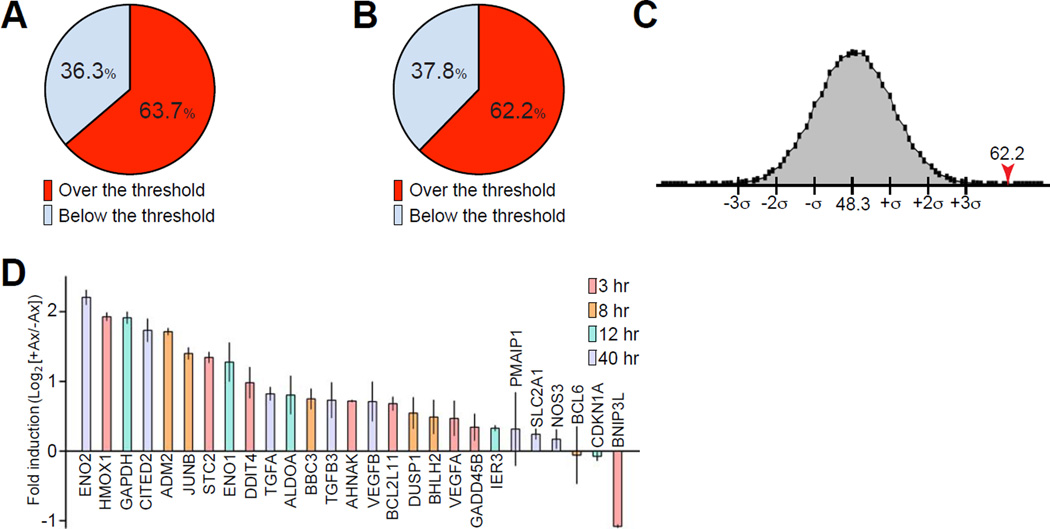
To determine whether HIF-1α participates in the activation of a pro-regenerative program, we generated a list of genes activated in injured cultured DRG neurons from our previous microarray data (Cho et al., 2013) and compared injury-induced genes with identified (Benita et al., 2009) or predicted (Ortiz-Barahona et al., 2010) HIF-1α target genes. We found that 63.7% (58 out of 91) of genes known as HIF-1α targets were up-regulated after axon injury over the 1.2 fold threshold (Figure 1A and Table S1), and 62.2% (61 out of 98) of predicted HIF-1α target genes were up-regulated after injury (Figure 1B and Table S2). To test the probability of obtaining these percentages within a given random group of genes, we generated 220,000 groups each containing 189 genes (with detection p value below 0.01) randomly selected from a total of 15,310 genes detected in our microarray analysis (Cho et al., 2013). The size of the group (189 genes) was selected to represent the size of the group of predicted and known HIF-1α target genes. The percentage of up-regulated genes over the threshold in every group was calculated and plotted, with a mean value of 48.3% (Figure 1C). This result indicates that for any random group of 189 genes detected within the array, 48.3% are up-regulated after injury. The probability of obtaining 62.2% of up-regulated genes calculated in Figure 1B is below 0.005%, revealing that transcription of a significant proportion of HIF-1α target genes occurs after injury. We next performed qPCR analysis and found that 22 out of 27 HIF-1α target genes tested were up-regulated at different time points after axon injury in cultured DRG neurons (Figure 1D). 5 out of 27 candidate genes tested were not different between injured and uninjured conditions, and one gene was down-regulated by injury (Figure 1D).
Figure 1. Statistical analysis of genes activated by axon injury.
(A) Percentage of up-regulated genes after in vitro axon injury that are known HIF-1α target genes. The expression of 91 known HIF-1α target genes was compared to the genes up-regulated in injured cultured DRG neurons. 58 genes out of 91 (63.7%) were up-regulated over the 1.2 threshold at 3, 8, 12 or 40 h after in vitro axotomy. (B) As in A but comparison to predicted HIF-1α target genes. 61 genes out of 98 (62.2%) were up-regulated over the threshold at 3, 8, 12 or 40 h after in vitro axotomy. (C) Distribution of percentages of up-regulated genes over the threshold in 220,000 randomly selected groups of 189 genes from a total of 15310 genes detected in the microarray analysis (with detection p value below 0.01). The percentage of up-regulated genes over the threshold in each group was calculated and plotted (mean, 48.3%; SD, 3.4). The probability of obtaining 62.2% of up-regulated genes in a group of 189 genes is below 0.005%. (D) Quantitative PCR analysis of mRNA prepared from mouse DRG cultures at 0, 3, 8, 12 and 40 h after axotomy (n=3; mean±SEM). For each gene, the log2-transformed expression fold change at the time after axotomy at which maximum fold change was detected is indicated.
To validate that these genes are indeed HIF-1α transcriptional target genes, we measured the fold induction 24 h following in vitro axotomy of a selected group of genes in control DRG neurons or in DRG neurons in which HIF-1α was knocked down. qPCR for HIF-1α confirmed the efficiency of the knock down (Figure S1). Compared to house keeping genes (SF3B5 and MRLP10), whose expression was not affected by injury or HIF-1α knock down, we observed that the induction of seven genes following in vitro axotomy was reduced by HIF-1α knock down (Figure S1). These results indicate that HIF-1α contributes to the expression of injury-responsive genes in DRG neurons.
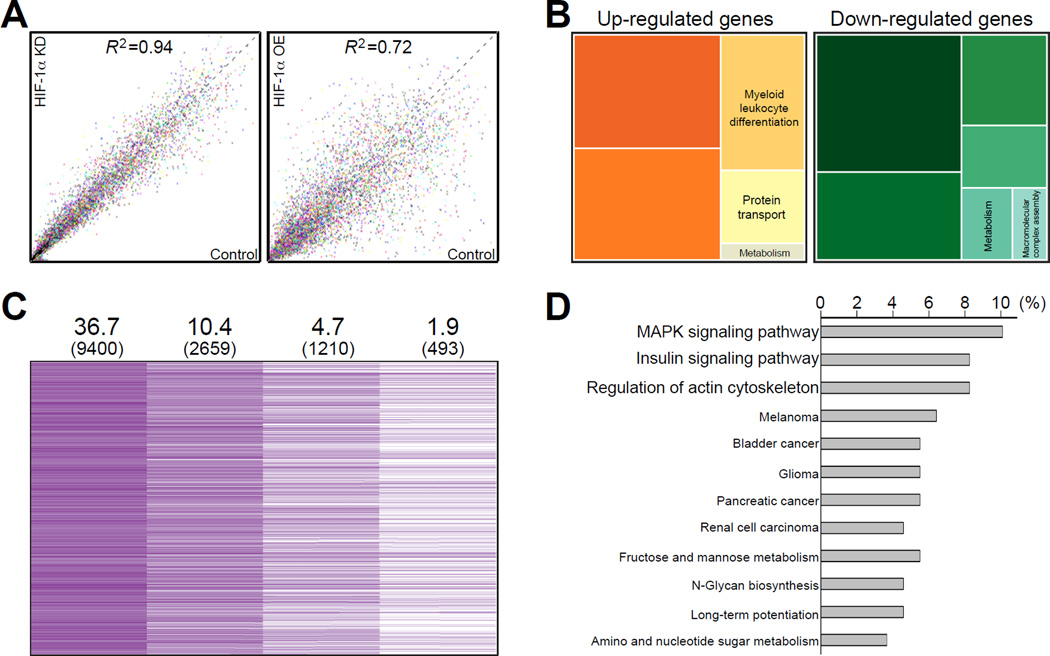
To further study the genes regulated by HIF-1α in DRG neurons, we examined changes in gene expression in cultured DRG by microarray analysis, comparing DRG control condition to DRG overexpressing HIF-1α or to DRG in which HIF-1α was knocked down, in uninjured conditions as well as 3 and 12 h after in vitro axotomy. 9,400 probes (36.7% of total probes) were detected with significant detection p value. In uninjured neurons HIF-1α knock down did not significantly change the gene expression profile, whereas gene expression was dramatically altered by constitutive over-expression of HIF-1α (Figure 2A). Over-expression of HIF-1α not only induced expression of some genes but also suppressed other genes (Figure 2A, 2B, Tables S3 and S4). Biological functions of genes down-regulated by HIF-1α over-expression were mainly involved in RNA processing and vesicle transport (Figure 2B and Table S4). These results imply that the overall gene expression profile in DRG neurons is modestly affected by HIF-1α knock down but more significantly affected by constitutive over-expression of HIF-1α.
Figure 2. Microarray analysis of genes regulated by HIF-1α in DRG neurons.
(A) Microarray analysis of gene expression profiles from RNA samples of control DRG cultures, HIF-1α-knock down (KD) or HIF-1α-overexpression (OE) cultures (X-axis, arbitrary units of normalized expression levels of control; Y-axis, arbitrary units of normalized expression levels of HIF-1α-knockdown (left) or HIF-1α-overexpression (right); R2, coefficient of determination). (B) Tree map of functional ontology from genes up-regulated by HIF-1α-overexpression (left) or down-regulated by HIF-1α-overexpression (right). (C) Schematic diagram of the screen identifying injury-responsive genes regulated by HIF-1α. (D) Functional gene ontology of HIF-1α-regulated injury-responsive genes.
Among the 9,400 probes detected, 2659 (10.4%) were up-regulated above the threshold of 1.2 fold at 3 or 12 h after in vitro axotomy in DRG control condition. To screen for injury--induced HIF-1α-dependency, a differential expression slope value was calculated (see supplemental experimental procedure section for details). Only those genes with expression level at 3 or 12 h post-axotomy higher in the control set than in HIF-1α knock down set were included, leading to 1210 (4.7%) injury-responsive HIF-1α-target genes. Finally, the 1210 genes were further screened for HIF-1α-dependency by comparing their basal expression level at 0 h in control and in HIF-1α-overexpression set. This final filtering lead to 493 genes (1.9%) defined as genes induced by axon injury and regulated by HIF-1α (Figure 2C and Table S5). Gene ontology showed that most of these genes were involved in MAPK signaling pathway, insulin signaling pathway and regulation of actin cytoskeleton pathway (Figure 2D).
HIF-1α and HIF-1α dependent genes regulate axon regeneration in DRG neurons
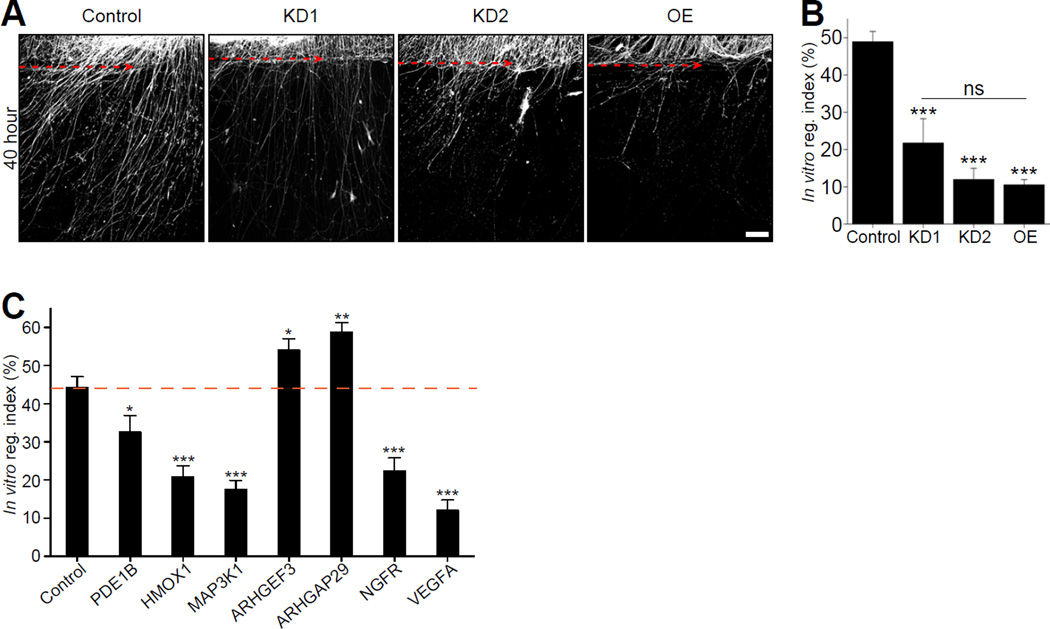
Since HIF-1α appears to control the expression of numerous genes after axon injury, we used an in vitro axotomy assay (Cho and Cavalli, 2012; Cho et al., 2013) to test whether modulating HIF-1α levels changes the regenerative capacity of cultured DRG neurons. Spot-cultured DRG neurons were infected with control shRNA or two different shRNA targeting HIF-1α. Both shRNA constructs efficiently reduced the levels of HIF-1α to 22.6±5.7% and 18.4±3.3% (Figure S2). GFP- HIF-1α protein expression was ~3 fold over endogenous HIF-1α levels (Figure S2). DRG were axotomized and immunostained for SCG10, a marker of regenerating axons (Shin et al., 2014) 40 h after injury. To assess the regenerative capacity of injured axons, the ratio of SCG10 fluorescence intensity proximal and distal to the axotomy line was measured, as described (Cho et al., 2013). This measure reflects both the length and number of regenerating axons. HIF-1α knock down significantly impaired axon regeneration in vitro (Figure 3A and 3B). We found that the constitutive overexpression of HIF-1α also inhibited axon regeneration to an extent similar to HIF-1α knock down (Figure 3A and 3B). These results indicate that regulation of HIF-1α levels following axon injury plays an important role in axon regeneration.
Figure 3. HIF-1α is required for axon regeneration in vitro.
(A) DRG spot-cultured neurons were infected with control shRNA (control), two different shRNA targeting HIF-1α (KD1, KD2) or HIF-1α–overexpressing lentivirus (OE), fixed and immunostained with SCG10 antibody 40 h after axotomy. Scale bar, 200µm. Dotted line indicates the axotomy site. (B) In vitro regeneration index was calculated from images in (A) (n=9, 14 and 16 for each condition; ***p<0.001 by one-way ANOVA with Tukey test; mean±SEM; ns, not significant). (C) In vitro regeneration assay was performed with the knockdown of selected candidate HIF-1α target genes (n=11, 12, 12, 17, 13, 15, 13 and 11 for Control, PDE1B, HMOX1, MAP3K1, ARHGEF3, ARHGAP29, NGFR and VEGFA; ***p<0.001, **p<0.01, *p<0.05 by one-way ANOVA with Tukey test; mean±SEM).
To examine whether HIF-1α-target genes affect regenerative capacity, we examined the role of a small group of candidate HIF-1α-dependent genes in the in vitro axotomy assay. We selected the candidate genes (PDE1B, HMOX1, MAP3K1, ARHGEF3, ARHGAP29, NGFR and VEGFA) based on their high basal expression and their high fold change following injury (Figure S5). HMOX1 and VEGFA are also known HIF-1α-dependent genes (Figures 1D and S1), whereas all others genes were selected from our screen for injury-induced, HIF-1α-dependent genes (Figure 2C and Table S5). Knock down of PDE1B, HMOX1, MAP3K1, NGFR and VEGFA significantly inhibited axon regeneration, demonstrating that HIF-1α-dependent genes can positively control axon regeneration (Figure 3C). However, knocking down ARHGEF3 and ARHGAP29, two genes which are involved in cytoskeleton re-modeling through small GTPase Rho (Saras et al., 1997) showed enhanced regeneration, suggesting that some of the HIF-1α-dependent genes can negatively impact axon regeneration (Figure 3C). These results suggest that HIF-1α-dependent genes can either promote or impair axon regeneration.
HIF-1α regulates axon regeneration in DRG neurons in vivo
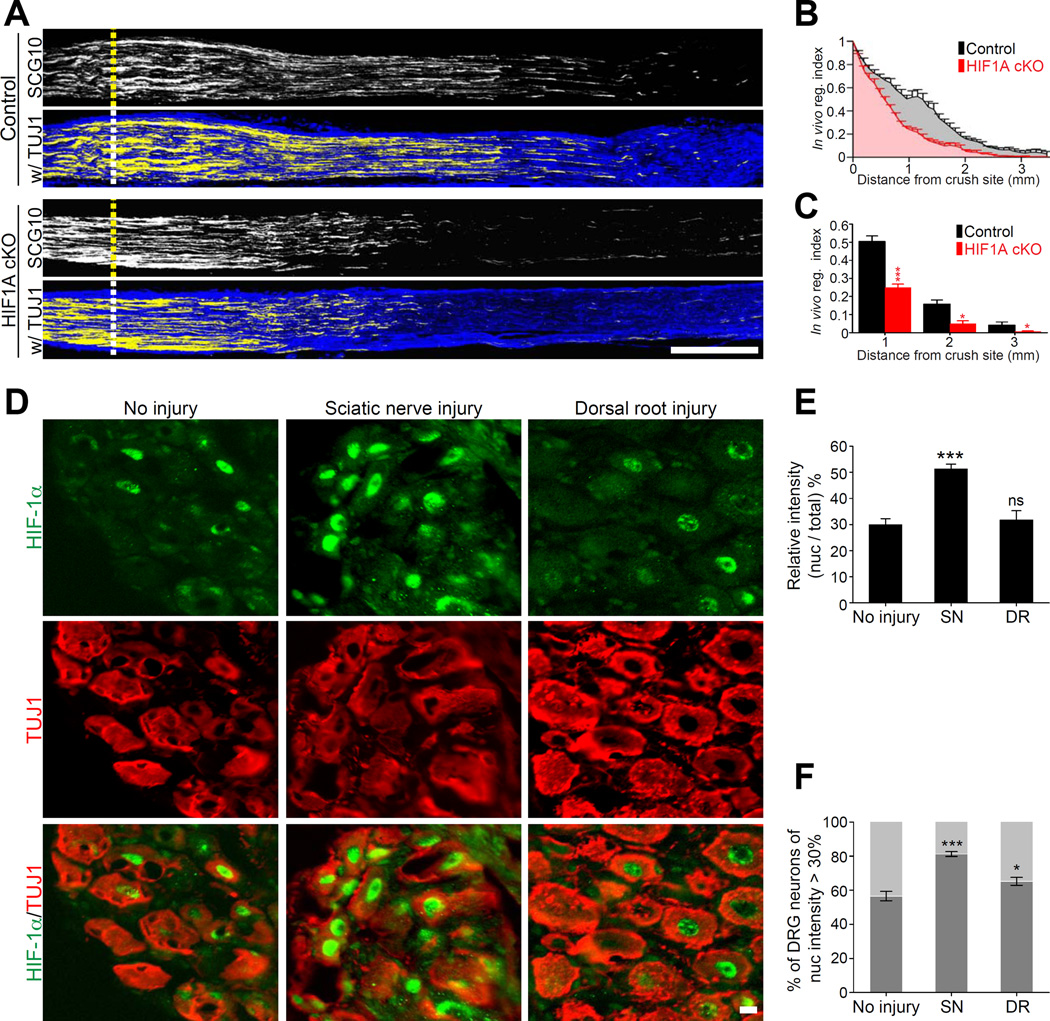
To investigate the contribution of HIF-1α to axon regeneration in vivo, we crossed HIF1Aflox/flox mice to mice expressing Cre under the control of the Advillin promoter, which is expressed almost exclusively in peripheral sensory neurons (Hasegawa et al., 2007; Zhou et al., 2010). HIF1Aflox/flox;AdvillinCre/+ mice are hereafter referred to as HIF1AcKO and HIF1Aflox/flox;Advillin+/+ as littermate control. The efficiency of Advillin-Cre mediated HIF1A knock out in DRG neurons was verified by western blot and immunofluorescence (Figure S3A–D). Sciatic nerves of control or HIF1AcKO mice were crushed and regenerating axons were labeled by SCG10 three days later, as described (Cho et al., 2013; Shin et al., 2014). To quantify axon regeneration, SCG10 fluorescence intensity was measured along the length of the nerve. A regeneration index was calculated by normalizing the average SCG10 intensity at distances away from the crush site to the SCG10 intensity at the crush site. This measure takes into account both the length and the number of regenerating axons past the crush site. Absence of HIF-1α in sensory neurons reduced axon regeneration past the crush site (Figures 4A, 4B and 4C), indicating that HIF-1α contributes to axon regeneration in vivo.
Figure 4. HIF-1α is required for axon regeneration in vivo.
(A) Representative longitudinal sections of sciatic nerve from control or HIF1AcKO mice three days after crush injury stained with SCG10 and βIII tubulin (TUJ1) antibody. Scale bar, 500µm. Dotted lines indicate the crush site, identified as the maximal SCG10 intensity. (B and C) In vivo regeneration index was calculated from images in (A). SCG10 intensity was measured from the crush site towards the distal end and normalized to the intensity at the crush site. SCG10 intensity was plotted as a function of the distance from the crush site (n=7 and 10 for control and HIF1AcKO, respectively; ***p<0.001, *p<0.05 by one-way ANOVA with Tukey test; mean±SEM). (D) Representative longitudinal sections of mouse L4 DRGs dissected 24 h after sciatic nerve or dorsal root injury, immunostained with HIF-1α and βIII tubulin. Scale bar, 20µm. (E) Average of relative intensity of nuclear HIF-1α over total HIF-1α from images in (D) (n=20, 22 and 18 for no injury, sciatic nerve injury (SN) and dorsal root injury (DR), respectively; ***p<0.001 by one-way ANOVA with Tukey test; mean±SEM; ns, not significant). (F) Average percentage of DRG neurons displaying nuclear HIF-1α intensity over 30% (n=10, 12 and 12 for no injury, sciatic nerve injury (SN) and dorsal root injury (DR), respectively; ***p<0.001, *p<0.05 by one-way ANOVA with Tukey test; mean±SEM).
Activation of HIF-1α to a competent transcriptional activator involves control of protein stability and nuclear localization. The cellular level of HIF-1α protein is regulated by oxygen level and also by various signal transduction pathways (Yee Koh et al., 2008), including calcium, which enhances HIF-1α transcriptional activity (Mottet et al., 2003) and protein levels (Hui et al., 2006; Liu et al., 2007). We thus tested whether axon injury elicits changes in HIF-1α levels in DRG neurons. Western blot analysis showed a 1.2 fold increase in HIF-1α protein levels in DRG 24 h after sciatic nerve injury (Figure S3E,F). Quantitative PCR analysis also showed a 1.5 fold increase in HIF-1α mRNA levels following in vitro axotomy (Figure S3G). We next tested whether the increase in HIF-1α protein levels in injured neurons requires the back-propagating calcium wave elicited by axon injury (Cho and Cavalli, 2012; Cho et al., 2013). We found that calcium-chelation with BAPTA at the site of injury prevented the increase in HIF-1α protein levels in DRG soma (Figure S3H,I).
We next tested whether the increase in HIF-1α protein levels correlate with an increase in nuclear localization, and whether nuclear localization would be differently affected by sciatic nerve injury or dorsal root injury. Mouse L4 DRG were dissected 24 h after sciatic nerve injury or after L4 dorsal root injury, immunostained for HIF-1α and βIII tubulin (TUJ1), a neuron specific marker, and compared to uninjured controls. Sciatic nerve injury induced nuclear accumulation of HIF-1α in DRG neurons (Figure 4D and 4E). The proportion of DRG neurons having nuclear HIF-1α was also increased by sciatic nerve injury (Figure 4F). In contrast, dorsal root injury did not affect the average relative intensity of nuclear HIF-1α (Figure 4E) and only modestly increased the proportion of neurons having nuclear HIF-1α (Figure 4F). These experiments indicate that sciatic nerve injury, but not dorsal root injury, leads to the nuclear accumulation of HIF-1α in a calcium-dependent manner.
HIF-1α is involved in regulating pre-conditioning lesion effects
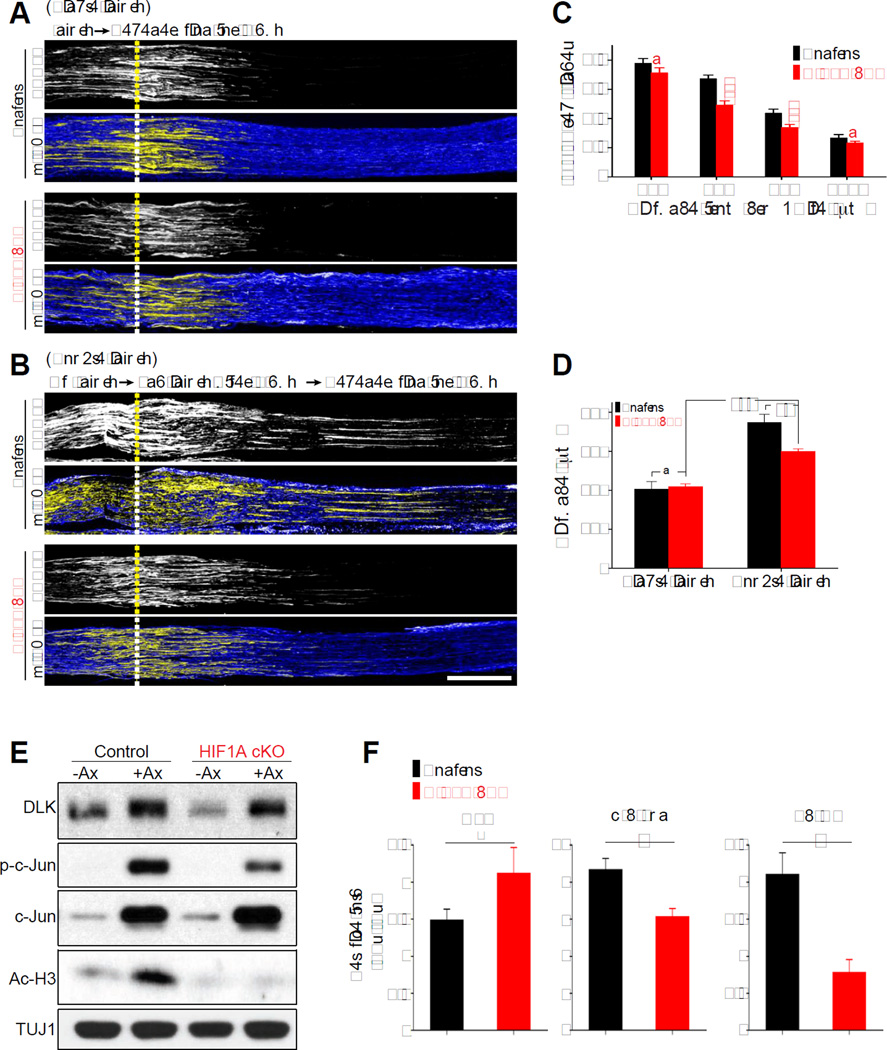
Injured PNS axons form new growth cones that lead to axon extension. Axon regeneration is subsequently promoted by the up-regulation of pro-regenerative genes. Given the transcriptional role of HIF-1α, we next tested whether HIF-1α stabilization in injured DRG neurons contributes to the induction of the regeneration program rather than the local and initial axon extension. To test this hypothesis, we took advantage of the pre-conditioning lesion paradigm, in which DRG neurons primed by a prior injury exhibit improved regeneration after a second injury, compared to that after the first injury (McQuarrie and Grafstein, 1973; Richardson and Issa, 1984; Shin et al., 2012; Smith and Skene, 1997). To first examine the early phase of axon regeneration, the sciatic nerves of control or HIF1AcKO were subjected to a crush lesion and allowed to re-grow for one day: at this early time point induction of the transcriptional response is not fully involved (Shin et al., 2012). We found that regeneration of SCG10-positive axons after one day was not affected by absence of HIF-1α (Figure 5A and 5D), demonstrating that HIF-1α is not required for the early phase of axon regeneration. When we performed a preconditioning injury, in which we crushed the sciatic nerve three days prior to a second crush to allow for induction of the pro-regenerative program (Shin et al., 2012; Smith and Skene, 1997), we observed an ~2 fold increase in axon regeneration in control mice, which was significantly reduced by conditional knock out of HIF-1α (Figure 5B, 5C and 5D). These experiments indicate that HIF-1α contributes to the activation of a pro-regenerative program that primes DRG neurons for accelerated growth after injury, consistent with the role of HIF-1α as a transcriptional regulator.
Figure 5. HIF-1α contributes to pre-conditioning lesion effects.
(A) Representative longitudinal section images of sciatic nerve dissected 1 day after single injury (“single injury”). Sections from control or HIF1AcKO mice were immunostained with SCG10 and βIII tubulin. Scale bar, 500µm. Dotted lines indicate the crush site. (B) Representative longitudinal section images of mouse sciatic nerve 1 day after a second injury, given 3 days after a pre-conditioning injury (“double injury”). Sections from control or HIF1AcKO were immunostained with SCG10 and βIII tubulin. Scale bar, 500µm. Dotted lines indicate the crush site. (C) In vivo regeneration index was calculated from images in (B). SCG10 intensity was measured from the crush site towards the distal end and normalized to the intensity at the crush site. SCG10 intensity was plotted as a function of the distance from the crush site (n=6 for each condition; ***p<0.001 by one-way ANOVA with Tukey test; mean±SEM; ns, not significant). (D) The distance at which regenerating axons display 50% of the SCG10 intensity at the crush site was calculated (n=6 for each condition; ***p<0.001, **p<0.01 by one-way ANOVA with Tukey test; mean±SEM; ns, not significant). (E) Western blot of dissected L4 and L5 DRGs from control or HIF1AcKO mice that receveid (+Ax) or not (−Ax) a sciatic nerve axotomy 3 days earlier. (F) Average fold changes in intensity (+Ax/−Ax) (n=3 for each condition; #p>0.05, *p<0.05 by t- test; mean±SEM).
Because the dual leucine zipper kinase (DLK) is essential for the activation of the pro-regenerative program (Shin et al., 2012), we next tested whether HIF-1α regulates the preconditioning effect by modulating the DLK pathway. We investigated whether absence of HIF-1α impacts the elevation of DLK levels by injury (Huntwork-Rodriguez et al., 2013) or the activation of the transcription factor c-Jun, a well-characterized downstream effector of DLK (Shin et al., 2012). We found that three days after sciatic nerve injury, DLK protein levels were increased in DRG in both control and HIF1AcKO (Figure 5E and 5F), with a more pronounced increase in DLK expression in HIF1AcKO compared to control. In contrast, phosphorylation levels of c-Jun were reduced in HIF1AcKO compared to control (Figure 5E and 5F), consistent with the observation that HIF-1α-dependent genes are involved in MAPK signaling pathway (Figure 2D). To further examine the link between the DLK and the HIF-1α pathways, we determined the levels of HIF-1α after nerve injury in mice lacking DLK in sensory neurons (DLKcKO) (Shin et al., 2012). The levels of nuclear HIF-1α increased to a similar extent in both control and DLKcKO mice (Figure S4). These results indicate that the injury-induced increase in the protein levels of DLK and HIF-1α does not require the function of each other. Instead, both the DLK and HIF-1α pathways impact the activation of c-Jun and the preconditioning program, with a more prominent role for DLK (Shin et al., 2012) compared to HIF-1α.
Because HIF-1α is known to interact with histone acetyl transferases (Arany et al., 1996; Carrero et al., 2000) and increased histone acetylation levels correlates with regenerative capacity (Cho et al., 2013; Finelli et al., 2013; Puttagunta et al., 2014), we examined the levels of acetylated histone H3. We found that the injury-induced increase in acetylated histone H3 at lysine 9 and 14 was impaired in HIF1AcKO (Figure 5E and 5F). These results suggest that HIF-1α in injured neurons contribute to the activation of c-Jun and to the increase in histone H3 acetylation levels, which support the activation of the pro-regenerative gene expression program.
The HIF-1α target gene VEGFA enhances axon regeneration
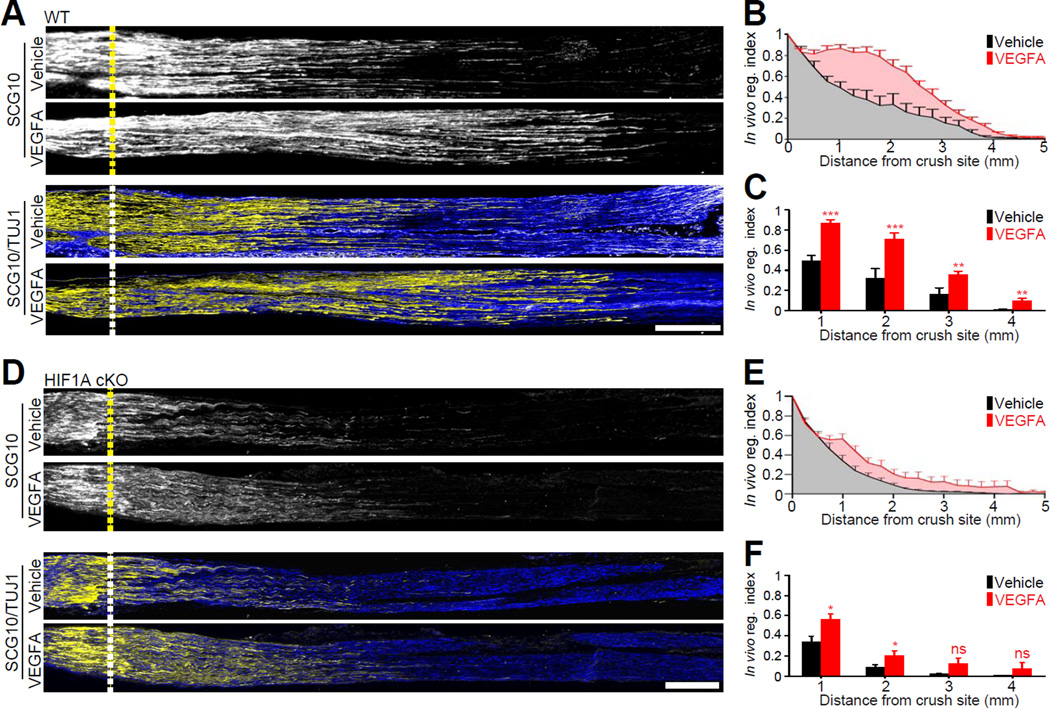
Vascular endothelial growth factor A (VEGFA) is a well-established HIF-1α target gene (Arany et al., 1996; Forsythe et al., 1996) which is mostly known for its role in blood vessel growth. However, VEGFA also promotes a wide range of neuronal functions, including neuronal survival and axon guidance (Mackenzie and Ruhrberg, 2012). We found that VEGFA is an injury-induced HIF-1α target gene (Figure S1) and previous studies showed that the presence of VEGFA within acellular conduits (Hobson et al., 2000) or VEGFA gene therapy (Pereira Lopes et al., 2011) increased vascularization and enhanced nerve regeneration. In addition, we verified that the levels of VEGFA are reduced in DRG of HIF1AcKO mice compared to controls (Figure S5). We thus asked whether the direct delivery of VEGFA to injured sciatic nerve in control mice increases axon regeneration and whether it would restore axon regeneration in HIF1AcKO mice. Because VEGF receptors are present on axons and growth cones (Sondell et al., 1999; Sondell et al., 2000) and VEGF stimulates axonal outgrowth by acting both on the growing axons and cell bodies (Sondell et al., 2000), we administered VEGFA at the site of injury. Delivery of VEGFA to the injury site enhanced axon regeneration in control mice (Figure 6A, 6B and 6C). In HIF1AcKO mice, VEGFA applied to the injured sciatic nerve also enhanced axon regeneration (Figure 6D, 6E and 6F) but did not fully restore axon regeneration to the level of control mice. These results indicate that VEGFA is part of the ensemble of genes regulated by HIF-1α in injured neurons, but that expression of other gene is also required to stimulate axon regeneration.
Figure 6. The HIF-1α target gene VEGFA enhances axon regeneration.
(A) Representative longitudinal sections of sciatic nerves from wild type mice treated with vehicle or recombinant mouse VEGFA, dissected 3 days after injury and immunostained for SCG10 and βIII tubulin. Scale bar, 500nm. Dotted lines indicate the crush site. (B and C) In vivo regeneration index calculated from (A) (n=6 and 8 for vehicle and VEGFA; ***p<0.001, **p<0.01 by t-test; mean±SEM). (D) Representative longitudinal sections of sciatic nerves from HIF1AcKO mice treated with vehicle or recombinant mouse VEGFA, dissected 3 days after injury and immunostained for SCG10 and βIII tubulin. Scale bar, 500^m. Dotted lines indicate the crush site. (E and F) In vivo regeneration index calculated from (D) (n=12 for each condition; *p<0.05 by t-test; mean±SEM; ns, not significant).
Hypoxia increases HIF-1α levels and stimulates axon regeneration in vitro
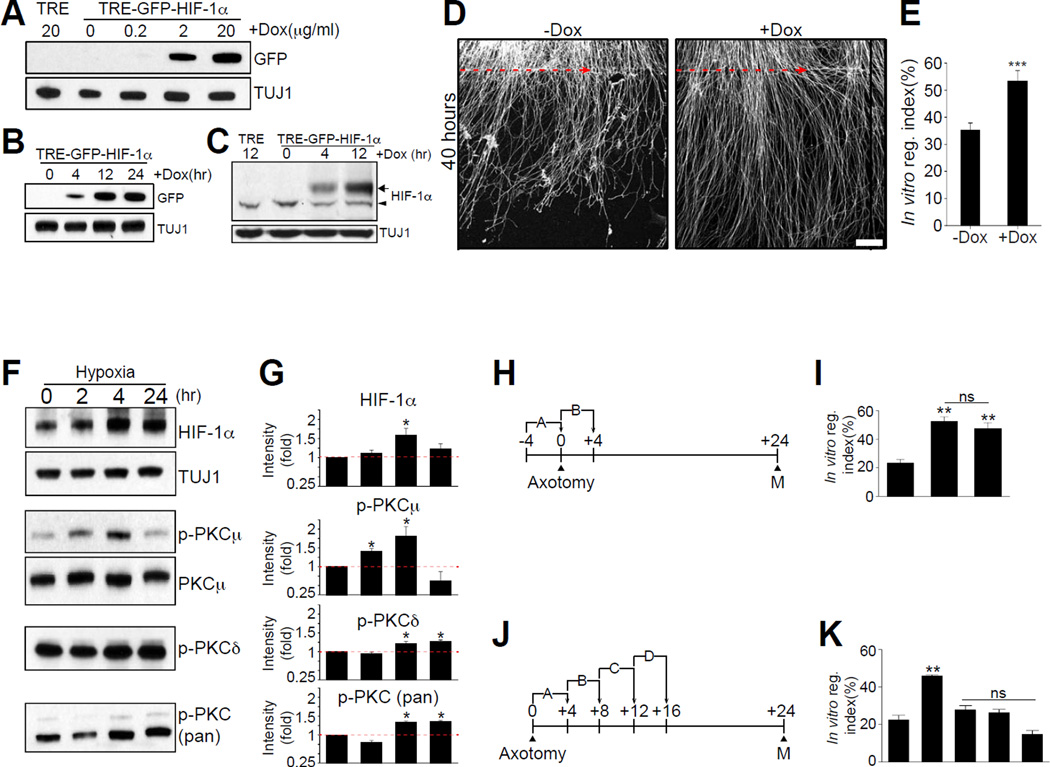
Inducing HIF-1α levels in DRG neurons may increase regeneration capacity by activating a group of HIF-1α-dependent genes that contribute to axon regeneration. Since we observed that constitutive over-expression of HIF-1α suppresses axon regeneration (Figure 3A and 3B), we tested whether a temporally controlled induction of HIF-1α levels in DRG neurons would promote axon regeneration, hoping that this may more closely mimic HIF-1α stabilization induced by injury in vivo. To achieve this, we first used an inducible tetracyclin-on HIF-1α lentiviral expression vector and determined the dose of doxycycline and the duration of application needed to reach HIF-1α protein levels similar to those observed following sciatic nerve injury. We found that GFP-HIF-1α protein was minimally induced with a treatment of 2µg/ml of Doxycyclin for 4 h (Figure 7A, 7B, 7C and S6). We then used these conditions in an in vitro axon regeneration assay and observed that induction of HIF-1α with Doxycycline (+Dox) enhanced axon regeneration by 17% compared to control (−Dox) (Figure 7D and 7E). These experiments demonstrate that a temporally controlled increase in HIF-1α levels, in contrast to constitutive over-expression (Figure 3A and 3B), enhanced axon regeneration.
Figure 7. Temporally controlled HIF-1α induction enhances axon regeneration in vitro.
(A and B) Western blot analysis of tetracyclin-inducible tre-GFP-HIF-1α expression in cultured DRG neurons with different doxycycline concentrations (A) or different induction time (B). (C) Western blot analysis of tre-GFP-HIF-1α expression by doxycycline induction. The arrow indicates the exogenous GFP-HIF-1α and the arrowhead indicates the endogenous HIF-1α. (D) Representative images of in vitro regeneration of tre-GFP-HIF-1α-transduced DRG neurons without (−Dox) or with doxycycline induction (+Dox). Scale bar, 100^m. (E) In vitro regeneration index calculated from (H) (n=16 for each condition; ***p<0.001 by t-test; mean±SEM). (F) Western blot analysis of cultured DRG neurons treated with hypoxia for the indicated amount of time. (G) Quantification of (F) (n=4; *p<0.05 by one-way ANOVA with Tukey analysis; mean±SEM). (H and J) Experimental schemes for the in vitro regeneration assay with different hypoxia conditions. Numbers indicate hour at which hypoxia condition was started. In vitro axotomy was given at the 0 h, A refers to a 4 h hypoxia treatment before axotomy, B, refers to a 4 h of hypoxia treatment after axotomy. B, C and D refer to hypoxia starting 4, 8 or 12 h after axotomy, respectively. (I and K) In vitro regeneration index from the experimental schemes in C and E. N: normoxia. (n=15, 14 and 18 for N, A and B of (C); n=15, 18, 12, 12 and 16 for N, A, B, C and D of (E); **p<0.01 by one-way ANOVA with Tukey test; ns, not significant; mean±SEM).
Since oxygen controls HIF-1α levels, we next sought to test whether hypoxia increases HIF-1α levels in cultured DRG neurons. Hypoxia is defined as the reduction or lack of oxygen in tissues or cells. To generate hypoxic conditions in cultured DRG neurons, we used a mixture of 5% CO2 balanced with 95% N2, as described (Wu and Yotnda, 2011). DRG spot-cultured neurons were kept in hypoxic conditions for 2, 4 and 24 h. We observed a 1.5-fold increase in HIF-1α levels after a period of 4 h of continuous hypoxia (Figure 7F and 7G). We also observed an increase in phosphorylation levels of protein kinase C (PKC) (Figure 7F and 7G), which is known to be activated by hypoxia (Goldberg et al., 1997; Lee et al., 2007). We next tested whether 4 h of continuous hypoxia enhanced in vitro axon regeneration (Figure 7H). 4 h of continuous hypoxia before or immediately after injury enhanced axon regeneration by ~2 fold (Figure 7I). However, if hypoxia treatment was started with a delay of 4, 8 or 12 h after injury, no differences in axon regeneration were observed (Figure 7J and 7K). These results demonstrate that in vitro axon regeneration can be enhanced with a hypoxia treatment administered immediately before or after axon injury.
Acute intermittent hypoxia enhances axon regeneration in vivo
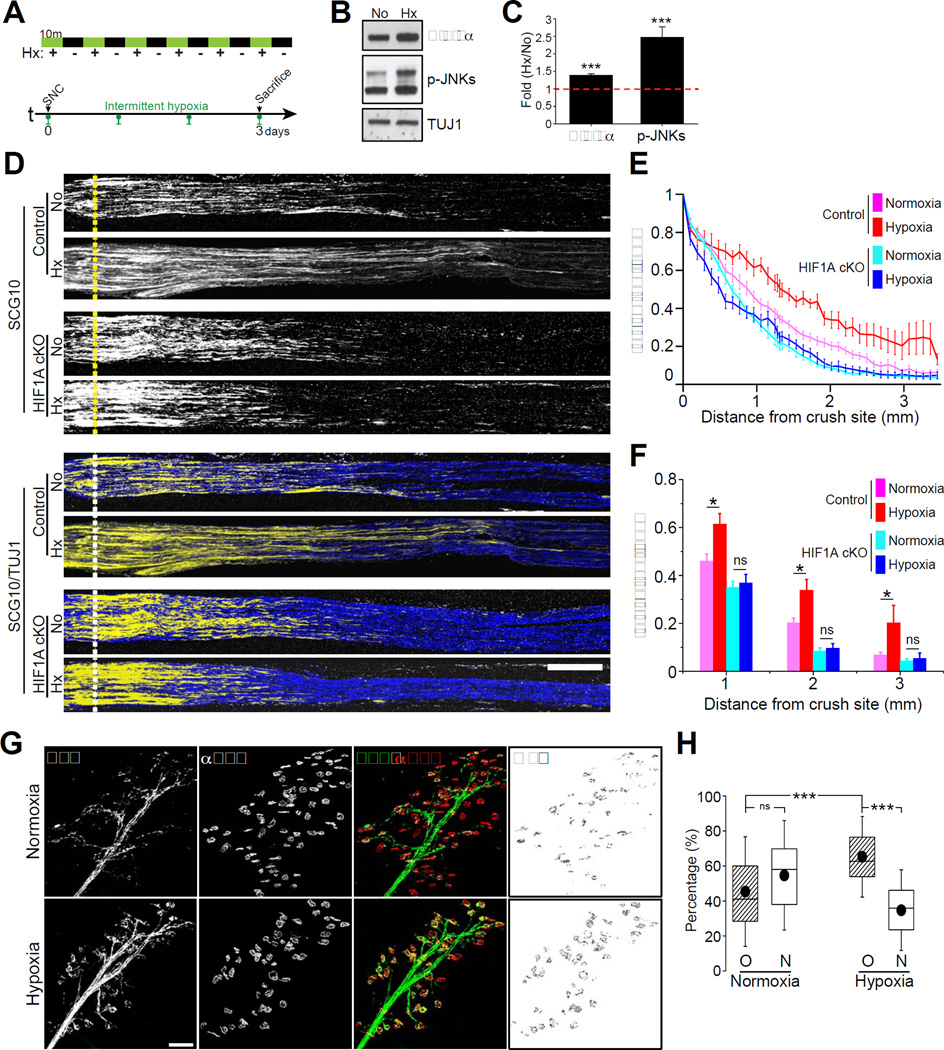
Next we asked whether hypoxia also stimulates regenerative responses in injured sensory neurons in vivo. We chose Acute Intermittent Hypoxia (AIH), which has been widely used for its beneficial effects in multiple physiological systems (Navarrete-Opazo and Mitchell, 2014). We thus assessed whether axon regeneration can be enhanced by AIH in vivo. To test this possibility, mice were submitted to AIH (10 min episodes of 8% O2 balanced with N2 with 10 min intervals of normoxia for 2 h; Figure 8A). This 2 h AIH treatment was effective in increasing HIF-1α levels and in activating the hypoxia-induced JNK phosphorylation in DRGs (Gozal et al., 1999) (Figure 8B and 8C). Immediately following this 2 h AIH treatment, we also observed that the expression of a subset of HIF-1α-dependent genes, including VEGFA were up-regulated in DRGs (Figure S7A). Since VEGFA is a well-established HIF-1α target gene (Figure S1 and see (Arany et al., 1996; Forsythe et al., 1996)), these results further suggest that the increased protein levels of HIF-1α correlate with a transcriptionally active HIF-1α. We then administered daily AIH or normoxia for three days, starting 2 h after sciatic nerve injury in control and HIF1AcKO mice. After three days mice were sacrificed, sciatic nerve harvested and regenerating axons quantified as in Figure 4A. Compared to normoxia, AIH increased axon regeneration past the crush site in control mice (Figure 8D, 8E and 8F). In contrast, we observed that AIH failed to stimulate axon regeneration in HIF1AcKO mice compared to control mice (Figures 8D, 8E and 8F). These results indicate that AIH acts mainly through HIF-1α to stimulate axon regeneration (Figure S7B).
Figure 8. Acute intermittent hypoxia (AIH) enhances axon regeneration in vivo.
(A) Experimental scheme for in vivo AIH: 6 10min hypoxic episodes (8% oxygen) with equivalent 10min normoxic intervals. Green bar indicates 10 min hypoxic condition and black bar indicates 10 min normoxic condition (AIH total duration=120 min). Control mice followed the same 120 min regime under normoxia. SNC: sciatic nerve crush. (B) Western blot analysis of mouse L4 and L5 DRGs treated for 120 min of normoxia (No) or hypoxia (Hx) using the AIH protocol. (C) Quantifications of protein level of HIF-1α and p-JNKs (n=3 for each condition; ***p<0.001, **p<0.01 by t-test; mean±SEM). (D) Representative longitudinal sections of sciatic nerves from control or HIF1AcKO mice, dissected 3 days after injury with or without daily AIH and immunostained for SCG10 and βIII tubulin. Scale bar, 500µm. (E and F) In vivo regeneration index calculated from (D) (n=7 and 10 for normoxia and hypoxia from control mice, n=10 and 8 for normoxia and hypoxia from HIF1AcKO mice; *p<0.05 by one-way ANOVA with Tukey test; mean±SEM; ns, not significant). (G) Motor axons reinnervation assays. EHL muscles of thy1-YFP-16 mice dissected 12 days after sciatic nerve injury with or without daily AIH were stained with Alexa Fluor 647-conjugated α-Bungarotoxin (αBTX). Scale bar, 100µm. (H) Quantification of percentage of axon-non-occupied (N) and axon-re-occupied NMJ end plates (O) (n=15 for each condition; box, 25 to 75%; whisker, standard deviation; closed circle, mean; ***p<0.001 by one-way ANOVA with Tukey test; ns, not significant).
To test whether AIH also stimulates long-range axon regeneration in motor neurons, we measured reinnervation of the neuromuscular junction in the extensor hallucis longus muscle 12 days after sciatic nerve injury. AIH was administered daily for 7 days starting 1 h after sciatic nerve injury in YFP-16 mice (Feng et al., 2000), which express YFP in motor neurons. Reinnervation was quantified 5 days later by measuring the percentage of NMJ boutons that co-localized with YFP, as described (Cho et al., 2013). We observed accelerated regeneration promoted by AIH treatment in motor axons reinnervating the NMJ (Figure 8G and 8H). The observed accelerated motor neuron regeneration may result from non-cell-autonomous or secondary effects, but together with the observation that daily AIH induces functional improvement in respiratory and non-respiratory motor pathways after chronic and acute cervical spinal injury (Lovett-Barr et al., 2012; Navarrete-Opazo et al., 2015), this result suggests that the HIF-1α-dependent mechanisms we describe in DRG neurons may contribute to increase the growth capacity of motor neurons.
Together, these results identify HIF-1α as an important transcriptional activator in axon regeneration and suggest a role for hypoxia as a non-invasive tool to stimulate axon regeneration.
Discussion
Injured sensory neurons successfully regain axon growth ability via activation of a pro-regenerative transcriptional program that enables axon regeneration and functional recovery. Here we demonstrate that HIF-1α contributes to the injury-induced transcriptional response and that modulating HIF-1α levels with acute intermittent hypoxia increases axon regeneration in sensory neurons, representing a non-invasive yet effective way to activate the pro-regenerative program.
Injury to the peripheral axon branch of sensory neurons increases their intrinsic growth capacity and also promotes some level of regeneration of their centrally projecting axons (Neumann and Woolf, 1999; Richardson and Issa, 1984). This pre-conditioning paradigm requires transcriptional events (Smith and Skene, 1997) and reveals that activation of a pro-regenerative program can overcome the inhibitory environment of the CNS. Here we show that HIF-1α represents an important transcriptional regulator of the pre-conditioning effect, with reduced levels of HIF-1α impairing the accelerated growth promoted by a prior injury. In the absence of HIF-1α in DRG, we observed reduced activation of c-Jun, although not to the same extent as in mice lacking DLK (Shin et al., 2012). Hence our results suggest that both the DLK and HIF-1α pathways impact the activation of the preconditioning program, with a more prominent role for DLK (Shin et al., 2012) compared to HIF-1α. Whether HIF-1α operates in parallel or together with DLK in the pre-conditioning effect remains to be tested. Although we saw a comparable increase in HIF-1α levels in both control and DLKcKO mice, whether HIF-1α in DLKcKO represents a fully competent transcriptional complex remains to be determined. Indeed, other injury-induced co-factors may interact with HIF-1α at the promoter of HIF-1α-target genes (Pawlus and Hu, 2013). In particular, two transcription factors known to be activated by axon injury, c-Jun and STAT3 (Broude et al., 1997; Schwaiger et al., 2000) functionally co-operate HIF-1α in hypoxia-induced gene transcription (Alfranca et al., 2002; Gray et al., 2005; Oh et al., 2011). Since the levels of c-Jun and STAT3 activation are reduced in DLKcKO mice (Shin et al., 2012), HIF-1α might not represent a fully competent transcriptional activator in the absence of DLK signaling. Future studies are needed to compare gene expression in both HIF1cKO and DLKcKO to establish the extent of overlap between DLK and HIF-1α -dependent genes.
Interestingly, we found that HIF-1α is also required for the injury-induced increase in histone acetylation, which promotes axon regeneration in peripheral (Cho et al., 2013) and central axons (Finelli et al., 2013; Puttagunta et al., 2014). Failure to increase histone acetylation in response to axon injury in the absence of HIF-1α may result from a decreased recruitment of transcriptional co-regulators that harbor histone acetyltransferase activity, such as p300/CBP (Arany et al., 1996) or SRC-1 (Carrero et al., 2000). Stabilization and nuclear accumulation of HIF-1α following axon injury may thus contribute to control the chromatin structure in injured sensory neurons, since HIF-1α is known to be involved in chromatin changes in other systems (Perez-Perri et al., 2011).
What are the signaling mechanisms promoting HIF-1α stabilization and nuclear accumulation after injury in DRG neurons? HIF-1α is usually hydroxylated by prolyl hydroxylases (PHD1-PHD3) and recognized by von Hippel Lindau (VHL) for proteasomal degradation (Yee Koh et al., 2008). It will be interesting to determine whether the activity of PHD and VHL is regulated following axon injury. HIF-1α is also regulated by oxygen-independent mechanisms (Yee Koh et al., 2008). Our data suggest that a potential mechanism could involve the elevated calcium levels we previously observed in DRG soma after injury (Cho et al., 2013) and the receptor of activated protein kinase C (RACK1). Indeed, RACK1 homodimerization recruits components of the E3 ligase complex to HIF-1α, leading to HIF-1α ubiquitination and degradation (Yee Koh et al., 2008). In the presence of calcium, RACK1 dimerization is blocked by the calcium-dependent phosphatase calcineurin A, preventing RACK-1-mediated HIF-1α degradation (Liu et al., 2007). Future studies should determine the precise molecular pathways controlling HIF-1α levels and localization in injured neurons.
Our results indicate that HIF-1α regulates a large ensemble of genes following axon injury. VEGFA expression downstream of HIF-1α represents one molecular mechanism enhancing axon regeneration. We found that neurons upregulate the expression of VEGFA in response to axon injury, suggesting an autocrine function. The observation that VEGFA receptors are also expressed in neurons support the evidence that VEGFA can directly affect neurons (Mackenzie and Ruhrberg, 2012). Earlier studies have shown that VEGFA added to explants culture promotes survival, axonal outgrowth and Schwann cell proliferation (Sondell et al., 1999). An engineered HIF-1α to activate VEGFA was shown to enhance recovery after spinal cord injury as well as attenuate axonal degradation (Figley et al., 2014; Liu et al., 2010), but these studies did not directly examine the impact on axon regeneration. Hence, HIF-1α-dependent VEGFA expression after injury together with injury-induced expression of the VEGF receptor (KDR) may enhance vascularization as well as directly stimulate axon regeneration, similarly to the common role of VEGFA during development of the nervous and vascular systems (Carmeliet and Ruiz de Almodovar, 2013; Ruiz de Almodovar et al., 2009).
Our demonstration that HIF-1α promotes axon regeneration by regulating an ensemble of genes suggests that regulating HIF-1α levels with AIH is a promising therapeutic strategy to promote functional recovery. AIH has been extensively studied for its therapeutic potential (Navarrete-Opazo and Mitchell, 2014). Our results suggest that in addition to plasticity and functional improvement in spared respiratory and non-respiratory motor pathways after chronic cervical spinal injury (Baker-Herman et al., 2004; Baker-Herman and Mitchell, 2002; Lovett-Barr et al., 2012), AIH enhances axon regeneration via the activation of HIF-1α-dependent pro-regenerative genes. When administered to human patients with incomplete spinal cord injury, AIH increases ankle strength (Trumbower et al., 2012) as well as walking speed and endurance (Hayes et al., 2014). The precise molecular mechanisms underlying these improvements in human patients remain unknown, but carefully controlled rAIH appears a safe and non-invasive modality that can be paired with other neurorehabilitative strategies (Gonzalez-Rothi et al., 2015). In rat models, AIH is believed to enhance respiratory and forelimb motor capacities through hypoxia induced increased release of spinal serotonin (Baker-Herman and Mitchell, 2002; MacFarlane and Mitchell, 2009) and expression of BDNF and activation of its receptor TRKB (Baker-Herman et al., 2004; Lovett-Barr et al., 2012; Satriotomo et al., 2012). Given the role of BDNF and its receptor in axon regeneration (Hoyng et al., 2014), AIH might also stimulate expression of BDNF to promote axon regeneration in sensory neurons.
AIH can trigger both beneficial and detrimental effects in multiple physiological systems (Gonzalez-Rothi et al., 2015; Navarrete-Opazo and Mitchell, 2014). Whereas acute intermittent hypoxia elicits a form of neuroplasticity termed phrenic long-term facilitation (Baker-Herman and Mitchell, 2002), prolonged intermittent hypoxia impairs this important form of respiratory plasticity (Huxtable et al., 2015). Prolonged hypoxia can also promote apoptotic neuronal death (Banasiak et al., 2000). One underlying cause for such divergent effects could be the magnitude and duration over which HIF-1α expression is modulated in response to a given AIH stimulus in a given tissue or cell type. Indeed, we found that the duration of HIF-1α expression in sensory neurons is critical for the beneficial effects in axon regeneration, with over-expression of HIF-1α having negative effects on axon regeneration and temporally controlled HIF-1α having beneficial effects. We also observed that not all HIF-1α dependent genes have pro-regenerative functions. Some HIF-1α dependent genes have anti-regenerative properties, suggesting that the degree of HIF-1α induction may impact the balance of pro- vs. anti-regenerative genes. Similarly, following hypoxia in the brain, both genes that promote and inhibit apoptosis are activated (Banasiak et al., 2000). AIH might also elicit different responses in different cell types that could influence regenerative responses and axon integrity. For example HIF-1α in oligodendrocyte precursor cells couples postnatal white matter angiogenesis and axon integrity with the onset of myelination (Yuen et al., 2014). HIF-1α target genes in neurons may also differ from target genes in other neuronal cell types. Indeed, although VEGFA is expressed by neurons, astrocytes and microglia in response to hypoxia (Mackenzie and Ruhrberg, 2012; Rosenstein et al., 2010), VEGFA is not induced in oligodendrocyte precursor cells in which HIF-1α is stabilized (Yuen et al., 2014). Given the well-known cell-specificity of hypoxia-driven responses, and their dependency on the magnitude, duration, and frequency of the hypoxic challenge, whether the AIH protocol we used to stimulate axon regeneration in sensory neurons would also stimulate axon regeneration in neurons within the CNS remains to be tested.
In conclusion, our study unveils HIF-1α as an important transcriptional regulator participating in the activation of a pro-regenerative program. This study also emphasizes that sensory neurons have harnessed a general feature of HIF-1α, which is to protect organisms against hypoxic stress. Axon injury in itself is a form of cellular stress that mobilizes HIF-1α as a part of the cellular stress response to drive an injured neuron into a pro-regenerative state. Our study also highlights that hypoxic stress represents a non-invasive therapeutic avenue to improve axon regeneration and importantly, could be applied after injury, in a post-conditioninglike fashion.
Experimental Procedures
RNA preparations, qPCR and microarray analyses
Total RNA was extracted from DRG spot cultures at 0, 3, 8, 12 and 40 h after in vitro axotomy using PureLink RNA mini kit (Life Technologies). To perform quantitative PCR, Fast SYBR Green Master Mix was used (Life Technologies) with validated primer sets from PrimerBank. To study HIF-1α-dependent gene regulation, a microarray analysis was performed to compare control DRG neurons to DRG neurons in which HIF1A was knocked down or in which human HIF1A was overexpressed. Detailed methods about microarray analysis can be found in Supplemental Experimental Procedures.
Animals
HIF1Aflox/flox mice originally generated by Dr. Randall S. Johnson (Baranova et al., 2007) were kindly provided from Dr. Gidday and Dr. Beebe at Washington University. HIF1Aflox/flox female mice were crossed to AdvillinCre/Cre male mice (Zhou et al., 2010) to generate HIF1Aflox/flox;AdvillinCre/+ conditional knock out mice and HIF1Aflox/flox;Advillin+/+ littermate controls. Genotype was tested by genomic tail DNA PCR at weaning age. DLKcKO (DLKflox/D; AdvillinCre/+) were obtained by crossing DLKflox/flox to DLK+/D;AdvillinCre/+ mice (Miller et al., 2009; Shin et al., 2012). DLKflox/+ were used as littermate controls. We used CD-1 timed pregnant mice for DRG culture. All in vivo experiments were done using 8 to 12 weeks-old littermate mice.
Surgeries and tissue preparations
All surgical procedures were performed under isofluorane anesthesia and approved by Washington University in St. Louis, School of Medicine Animal Studies Committee. Sciatic nerve injury experiments were performed as described (Cho and Cavalli, 2012). For double crush injury experiment, we followed previous protocols (Shin et al., 2012). For mouse L4 dorsal root nerve crush surgery, a small laminectomy of the caudal portion of the L2 vertebra was performed and fine forceps were used to crush the right L4 dorsal root.
For western blot analysis, mouse DRG tissues were dissected at the indicated time after nerve injury and homogenized in lysis buffer (Cell Signaling) as described (Cho and Cavalli, 2012). For immunohistochemistry, mouse DRG tissues were prepared as described (Cho and Cavalli, 2012). To measure the intensity of nuclear HIF-1α, region-of-interest (ROI) of DAPI-positive area from TUJ-1-positive cells was selected and HIF-1α-fluorescence intensity was measured using ImageJ.
Embryonic DRG neuron spot culture and in vitro regeneration assay
Embryonic DRG neurons were cultured as previously described (Cho and Cavalli, 2012). Knock down or overexpresson was achieved by lentivral infection at DIV4. In vitro regeneration assay was performed as previously described (Cho and Cavalli, 2012). DRG spot cultures were axotomized with a blade, immunostained 40 h later for SCG10. A regeneration index was calculated from the images acquired by measuring the fluorescence intensity of a square area (2.7×0.1mm) at 0.1 mm distal to the axotomy line and normalizing this intensity to the similar area 0.1 mm proximal to the axotomy line.
In vivo regeneration assays
Sciatic nerves were dissected at the indicated time after a crush injury. Longitudinal sciatic nerves sections were stained with SCG10 and TUJ1, as described in (Shin et al., 2012; Shin et al., 2014). SCG10 fluorescence intensity was measured along the length of the nerve using a line scan macro in ImageJ. A regeneration index was calculated by measuring the average SCG10 intensity at several distances away from the crush site, which is defined by the position along the nerve length with maximal SCG10 intensity, correlating with TUJ1 labeling where deformation of the nerve and disruption of axons are identified (Shin et al., 2014). Reinnervation of the neuromuscular junction after sciatic nerve injury was performed as described in (Cho et al., 2013). The number of NMJ endplates re-occupied (O) or non-occupied (N) by YFP signal was counted and normalized to the total number of endplates to calculate the percentage.
Hypoxia treatment
For in vitro hypoxia, established protocols were followed (Wu and Yotnda, 2011). Briefly, embryonic DRG cultures were placed in a gas-tight flushed for 10 minutes with 5% CO2 balanced with nitrogen at a flow rate of 20 L/min. The chamber was then placed back in the cell culture incubator for the indicated amount of time. Control cultures underwent the same procedure but 5% CO2 balanced with normal air was flushed.
All in vivo AIH procedures were approved by Washington University in St. Louis, School of Medicine Animal Studies Committee and followed established protocol (Zhang et al., 2004) with minor modifications. Mice were placed in a chamber flushed with oxygen balanced with nitrogen: 10 min episodes of 8% O2; 10 min intervals with normal air, 6 hypoxic episodes, total 120min. For testing the effect of AIH on axon regeneration, 120 m AIH protocol began 2 h after sciatic nerve injury and was applied once daily for 3 days. For muscle reinnervation, AIH was administered daily for 7 days starting 1 h after sciatic nerve injury and mice were returned to their cages for 5 days
Statistics
Student’s t-test or ANOVA (for multiple comparisons) followed by Tukey post-hoc tests were used to determine statistical significance, with p<0.05 considered statistically significant.
Supplementary Material
Acknowledgements
We thank Dr Gidday for helpful discussion and comments on hypoxic conditioning. We thank Dr DiAntonio for critical reading of the manuscript and sharing DLK knockout mouse line. We thank Marcus Mahar for assistance with bioinformatics. This work was supported in part by National Institutes of Health Grants DE022000 and NS082446, the Hope Center for Neurological Disorders Just In-Time Award, the Hope Center Viral Vectors Core at Washington University School of Medicine and the University of Missouri Spinal Cord Injuries Research Program (to V.C.), grants by the National Research Foundation of Korea (NRF-2012R1A6A3A03039290 to Y.C.) and by the Wings for Life (WFL-US-002/13 to J.E.S.). The authors declare that they have no conflict of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contribution
Conceptualization: Y.C., J.E.S., E.E.E, Y.M.O, W.P-T and V.C.; Investigation: Y.C., J.E.S, E.E.E. and Y.M.O; Writing-original Draft: Y.C., W.P-T, J.E.S. and V.C.; Writing-Review Editing: Y.C. and V.C. Visualization: Y.C.; Funding Acquisition: Y. C and V.C.
Accession Numbers
The NCBI GEO accession number for the array data reported in this paper is GSE73415.
References
- Alfranca A, Gutierrez MD, Vara A, Aragones J, Vidal F, Landazuri MO. c-Jun and hypoxia-inducible factor 1 functionally cooperate in hypoxia-induced gene transcription. Mol Cell Biol. 2002;22:12–22. doi: 10.1128/MCB.22.1.12-22.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany Z, Huang LE, Eckner R, Bhattacharya S, Jiang C, Goldberg MA, Bunn HF, Livingston DM. An essential role for p300/CBP in the cellular response to hypoxia. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker-Herman TL, Fuller DD, Bavis RW, Zabka AG, Golder FJ, Doperalski NJ, Johnson RA, Watters JJ, Mitchell GS. BDNF is necessary and sufficient for spinal respiratory plasticity following intermittent hypoxia. Nature neuroscience. 2004;7:48–55. doi: 10.1038/nn1166. [DOI] [PubMed] [Google Scholar]
- Baker-Herman TL, Mitchell GS. Phrenic long-term facilitation requires spinal serotonin receptor activation and protein synthesis. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2002;22:6239–6246. doi: 10.1523/JNEUROSCI.22-14-06239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banasiak KJ, Xia Y, Haddad GG. Mechanisms underlying hypoxia-induced neuronal apoptosis. Progress in neurobiology. 2000;62:215–249. doi: 10.1016/s0301-0082(00)00011-3. [DOI] [PubMed] [Google Scholar]
- Baranova O, Miranda LF, Pichiule P, Dragatsis I, Johnson RS, Chavez JC. Neuron-specific inactivation of the hypoxia inducible factor 1 alpha increases brain injury in a mouse model of transient focal cerebral ischemia. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:6320–6332. doi: 10.1523/JNEUROSCI.0449-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benita Y, Kikuchi H, Smith AD, Zhang MQ, Chung DC, Xavier RJ. An integrative genomics approach identifies Hypoxia Inducible Factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic acids research. 2009;37:4587–4602. doi: 10.1093/nar/gkp425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackmore MG. Molecular control of axon growth: insights from comparative gene profiling and high-throughput screening. Int Rev Neurobiol. 2012;105:39–70. doi: 10.1016/B978-0-12-398309-1.00004-4. [DOI] [PubMed] [Google Scholar]
- Bradke F, Fawcett JW, Spira ME. Assembly of a new growth cone after axotomy: the precursor to axon regeneration. Nature reviews Neuroscience. 2012;13:183–193. doi: 10.1038/nrn3176. [DOI] [PubMed] [Google Scholar]
- Broude E, McAtee M, Kelley MS, Bregman BS. c-Jun expression in adult rat dorsal root ganglion neurons: differential response after central or peripheral axotomy. Experimental neurology. 1997;148:367–377. doi: 10.1006/exnr.1997.6665. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Ruiz de Almodovar C. VEGF ligands and receptors: implications in neurodevelopment and neurodegeneration. Cellular and molecular life sciences : CMLS. 2013;70:1763–1778. doi: 10.1007/s00018-013-1283-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrero P, Okamoto K, Coumailleau P, O'Brien S, Tanaka H, Poellinger L. Redox-regulated recruitment of the transcriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1alpha. Mol Cell Biol. 2000;20:402–415. doi: 10.1128/mcb.20.1.402-415.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Cavalli V. HDAC5 is a novel injury-regulated tubulin deacetylase controlling axon regeneration. The EMBO journal. 2012;31:3063–3078. doi: 10.1038/emboj.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Sloutsky R, Naegle KM, Cavalli V. Injury-Induced HDAC5 Nuclear Export Is Essential for Axon Regeneration. Cell. 2013;155:894–908. doi: 10.1016/j.cell.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekanty A, Romero NM, Bertolin AP, Thomas MG, Leishman CC, Perez-Perri JI, Boccaccio GL, Wappner P. Drosophila genome-wide RNAi screen identifies multiple regulators of HIF-dependent transcription in hypoxia. PLoS genetics. 2010;6:e1000994. doi: 10.1371/journal.pgen.1000994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giovanni S. Molecular targets for axon regeneration: focus on the intrinsic pathways. Expert Opin Ther Targets. 2009;13:1387–1398. doi: 10.1517/14728220903307517. [DOI] [PubMed] [Google Scholar]
- Dunwoodie SL. The role of hypoxia in development of the Mammalian embryo. Developmental cell. 2009;17:755–773. doi: 10.1016/j.devcel.2009.11.008. [DOI] [PubMed] [Google Scholar]
- Fagoe ND, van Heest J, Verhaagen J. Spinal cord injury and the neuron-intrinsic regeneration-associated gene program. Neuromolecular medicine. 2014;16:799–813. doi: 10.1007/s12017-014-8329-3. [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Figley SA, Liu Y, Karadimas SK, Satkunendrarajah K, Fettes P, Spratt SK, Lee G, Ando D, Surosky R, Giedlin M, Fehlings MG. Delayed administration of a bio-engineered zinc-finger VEGF-A gene therapy is neuroprotective and attenuates allodynia following traumatic spinal cord injury. PloS one. 2014;9:e96137. doi: 10.1371/journal.pone.0096137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finelli MJ, Wong JK, Zou H. Epigenetic regulation of sensory axon regeneration after spinal cord injury. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:19664–19676. doi: 10.1523/JNEUROSCI.0589-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M, Zhang HL, Steinberg SF. Hypoxia alters the subcellular distribution of protein kinase C isoforms in neonatal rat ventricular myocytes. The Journal of clinical investigation. 1997;99:55–61. doi: 10.1172/JCI119133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rothi EJ, Lee KZ, Dale EA, Reier PJ, Mitchell GS, Fuller DD. Intermittent Hypoxia and Neurorehabilitation. Journal of applied physiology, jap 00235 02015. 2015 doi: 10.1152/japplphysiol.00235.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozal E, Simakajornboon N, Dausman JD, Xue YD, Corti M, El-Dahr SS, Gozal D. Hypoxia induces selective SAPK/JNK-2-AP-1 pathway activation in the nucleus tractus solitarii of the conscious rat. Journal of neurochemistry. 1999;73:665–674. doi: 10.1046/j.1471-4159.1999.0730665.x. [DOI] [PubMed] [Google Scholar]
- Gray MJ, Zhang J, Ellis LM, Semenza GL, Evans DB, Watowich SS, Gallick GE. HIF-1αlpha, STAT3, CBP/p300 and Ref-1/APE are components of a transcriptional complex that regulates Src-dependent hypoxia-induced expression of VEGF in pancreatic and prostate carcinomas. Oncogene. 2005;24:3110–3120. doi: 10.1038/sj.onc.1208513. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Abbott S, Han BX, Qi Y, Wang F. Analyzing somatosensory axon projections with the sensory neuron-specific Advillin gene. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:14404–14414. doi: 10.1523/JNEUROSCI.4908-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes HB, Jayaraman A, Herrmann M, Mitchell GS, Rymer WZ, Trumbower RD. Daily intermittent hypoxia enhances walking after chronic spinal cord injury: a randomized trial. Neurology. 2014;82:104–113. doi: 10.1212/01.WNL.0000437416.34298.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson MI, Green CJ, Terenghi G. VEGF enhances intraneural angiogenesis and improves nerve regeneration after axotomy. Journal of anatomy 197 Pt. 2000;4:591–605. doi: 10.1046/j.1469-7580.2000.19740591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman PN. A conditioning lesion induces changes in gene expression and axonal transport that enhance regeneration by increasing the intrinsic growth state of axons. Experimental neurology. 2010;223:11–18. doi: 10.1016/j.expneurol.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Hoyng SA, De Winter F, Gnavi S, de Boer R, Boon LI, Korvers LM, Tannemaat MR, Malessy MJ, Verhaagen J. A comparative morphological, electrophysiological and functional analysis of axon regeneration through peripheral nerve autografts genetically modified to overexpress BDNF, CNTF, GDNF, NGF, NT3 or VEGF. Experimental neurology. 2014;261:578–593. doi: 10.1016/j.expneurol.2014.08.002. [DOI] [PubMed] [Google Scholar]
- Hui AS, Bauer AL, Striet JB, Schnell PO, Czyzyk-Krzeska MF. Calcium signaling stimulates translation of HIF-alpha during hypoxia. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2006;20:466–475. doi: 10.1096/fj.05-5086com. [DOI] [PubMed] [Google Scholar]
- Huntwork-Rodriguez S, Wang B, Watkins T, Ghosh AS, Pozniak CD, Bustos D, Newton K, Kirkpatrick DS, Lewcock JW. JNK-mediated phosphorylation of DLK suppresses its ubiquitination to promote neuronal apoptosis. The Journal of cell biology. 2013;202:747–763. doi: 10.1083/jcb.201303066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxtable AG, Smith SM, Peterson TJ, Watters JJ, Mitchell GS. Intermittent Hypoxia-Induced Spinal Inflammation Impairs Respiratory Motor Plasticity by a Spinal p38 MAP Kinase-Dependent Mechanism. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2015;35:6871–6880. doi: 10.1523/JNEUROSCI.4539-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio PJ, Okamoto K, O'Brien S, Carrero P, Makino Y, Tanaka H, Poellinger L. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1alpha. The EMBO journal. 1998;17:6573–6586. doi: 10.1093/emboj/17.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Tamamizu-Kato S, Shibasaki F. Histone deacetylase 7 associates with hypoxia-inducible factor 1alpha and increases transcriptional activity. The Journal of biological chemistry. 2004;279:41966–41974. doi: 10.1074/jbc.M406320200. [DOI] [PubMed] [Google Scholar]
- Lee JW, Park JA, Kim SH, Seo JH, Lim KJ, Jeong JW, Jeong CH, Chun KH, Lee SK, Kwon YG, Kim KW. Protein kinase C-delta regulates the stability of hypoxia-inducible factor-1 alpha under hypoxia. Cancer science. 2007;98:1476–1481. doi: 10.1111/j.1349-7006.2007.00535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisy K, Peet DJ. Turn me on: regulating HIF transcriptional activity. Cell death and differentiation. 2008;15:642–649. doi: 10.1038/sj.cdd.4402315. [DOI] [PubMed] [Google Scholar]
- Liu K, Tedeschi A, Park KK, He Z. Neuronal Intrinsic Mechanisms of Axon Regeneration. Annu Rev Neurosci. 2011 doi: 10.1146/annurev-neuro-061010-113723. [DOI] [PubMed] [Google Scholar]
- Liu Y, Figley S, Spratt SK, Lee G, Ando D, Surosky R, Fehlings MG. An engineered transcription factor which activates VEGF-A enhances recovery after spinal cord injury. Neurobiology of disease. 2010;37:384–393. doi: 10.1016/j.nbd.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Liu YV, Hubbi ME, Pan F, McDonald KR, Mansharamani M, Cole RN, Liu JO, Semenza GL. Calcineurin promotes hypoxia-inducible factor 1alpha expression by dephosphorylating RACK1 and blocking RACK1 dimerization. The Journal of biological chemistry. 2007;282:37064–37073. doi: 10.1074/jbc.M705015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett-Barr MR, Satriotomo I, Muir GD, Wilkerson JE, Hoffman MS, Vinit S, Mitchell GS. Repetitive intermittent hypoxia induces respiratory and somatic motor recovery after chronic cervical spinal injury. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:3591–3600. doi: 10.1523/JNEUROSCI.2908-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Belin S, He Z. Signaling regulations of neuronal regenerative ability. Current opinion in neurobiology. 2014;27:135–142. doi: 10.1016/j.conb.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane PM, Mitchell GS. Episodic spinal serotonin receptor activation elicits long-lasting phrenic motor facilitation by an NADPH oxidase-dependent mechanism. The Journal of physiology. 2009;587:5469–5481. doi: 10.1113/jphysiol.2009.176982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie F, Ruhrberg C. Diverse roles for VEGF-A in the nervous system. Development. 2012;139:1371–1380. doi: 10.1242/dev.072348. [DOI] [PubMed] [Google Scholar]
- McQuarrie IG, Grafstein B. Axon outgrowth enhanced by a previous nerve injury. Arch Neurol. 1973;29:53–55. doi: 10.1001/archneur.1973.00490250071008. [DOI] [PubMed] [Google Scholar]
- Michaelevski I, Segal-Ruder Y, Rozenbaum M, Medzihradszky KF, Shalem O, Coppola G, Horn-Saban S, Ben-Yaakov K, Dagan SY, Rishal I, et al. Signaling to transcription networks in the neuronal retrograde injury response. Sci Signal. 2010;3:ra53. doi: 10.1126/scisignal.2000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BR, Press C, Daniels RW, Sasaki Y, Milbrandt J, DiAntonio A. A dual leucine kinase-dependent axon self-destruction program promotes Wallerian degeneration. Nature neuroscience. 2009;12:387–389. doi: 10.1038/nn.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DL, Blackmore MG, Hu Y, Kaestner KH, Bixby JL, Lemmon VP, Goldberg JL. KLF family members regulate intrinsic axon regeneration ability. Science. 2009;326:298–301. doi: 10.1126/science.1175737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DL, Goldberg JL. Multiple transcription factor families regulate axon growth and regeneration. Dev Neurobiol. 2011;71:1186–1211. doi: 10.1002/dneu.20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottet D, Michel G, Renard P, Ninane N, Raes M, Michiels C. Role of ERK and calcium in the hypoxia-induced activation of HIF-1. Journal of cellular physiology. 2003;194:30–44. doi: 10.1002/jcp.10176. [DOI] [PubMed] [Google Scholar]
- Navarrete-Opazo A, Mitchell GS. Therapeutic potential of intermittent hypoxia: a matter of dose. American journal of physiology Regulatory, integrative and comparative physiology. 2014;307:R1181–R1197. doi: 10.1152/ajpregu.00208.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarrete-Opazo A, Vinit S, Dougherty BJ, Mitchell GS. Daily acute intermittent hypoxia elicits functional recovery of diaphragm and inspiratory intercostal muscle activity after acute cervical spinal injury. Experimental neurology. 2015;266:1–10. doi: 10.1016/j.expneurol.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann S, Woolf CJ. Regeneration of dorsal column fibers into and beyond the lesion site following adult spinal cord injury. Neuron. 1999;23:83–91. doi: 10.1016/s0896-6273(00)80755-2. [DOI] [PubMed] [Google Scholar]
- Nix P, Hammarlund M, Hauth L, Lachnit M, Jorgensen EM, Bastiani M. Axon regeneration genes identified by RNAi screening in C. elegans. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:629–645. doi: 10.1523/JNEUROSCI.3859-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh MK, Park HJ, Kim NH, Park SJ, Park IY, Kim IS. Hypoxia-inducible factor-1alpha enhances haptoglobin gene expression by improving binding of STAT3 to the promoter. The Journal of biological chemistry. 2011;286:8857–8865. doi: 10.1074/jbc.M110.150557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Barahona A, Villar D, Pescador N, Amigo J, del Peso L. Genome-wide identification of hypoxia-inducible factor binding sites and target genes by a probabilistic model integrating transcription-profiling data and in silico binding site prediction. Nucleic acids research. 2010;38:2332–2345. doi: 10.1093/nar/gkp1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlus MR, Hu CJ. Enhanceosomes as integrators of hypoxia inducible factor (HIF) and other transcription factors in the hypoxic transcriptional response. Cellular signalling. 2013;25:1895–1903. doi: 10.1016/j.cellsig.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira Lopes FR, Lisboa BC, Frattini F, Almeida FM, Tomaz MA, Matsumoto PK, Langone F, Lora S, Melo PA, Borojevic R, et al. Enhancement of sciatic nerve regeneration after vascular endothelial growth factor (VEGF) gene therapy. Neuropathology and applied neurobiology. 2011;37:600–612. doi: 10.1111/j.1365-2990.2011.01159.x. [DOI] [PubMed] [Google Scholar]
- Perez-Perri JI, Acevedo JM, Wappner P. Epigenetics: new questions on the response to hypoxia. International journal of molecular sciences. 2011;12:4705–4721. doi: 10.3390/ijms12074705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttagunta R, Tedeschi A, Soria MG, Hervera A, Lindner R, Rathore KI, Gaub P, Joshi Y, Nguyen T, Schmandke A, et al. PCAF-dependent epigenetic changes promote axonal regeneration in the central nervous system. Nat Commun. 2014;5:3527. doi: 10.1038/ncomms4527. [DOI] [PubMed] [Google Scholar]
- Richardson PM, Issa VM. Peripheral injury enhances central regeneration of primary sensory neurones. Nature. 1984;309:791–793. doi: 10.1038/309791a0. [DOI] [PubMed] [Google Scholar]
- Rosenstein JM, Krum JM, Ruhrberg C. VEGF in the nervous system. Organogenesis. 2010;6:107–114. doi: 10.4161/org.6.2.11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruas JL, Poellinger L. Hypoxia-dependent activation of HIF into a transcriptional regulator. Seminars in cell & developmental biology. 2005;16:514–522. doi: 10.1016/j.semcdb.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Ruiz de Almodovar C, Lambrechts D, Mazzone M, Carmeliet P. Role and therapeutic potential of VEGF in the nervous system. Physiological reviews. 2009;89:607–648. doi: 10.1152/physrev.00031.2008. [DOI] [PubMed] [Google Scholar]
- Saras J, Franzen P, Aspenstrom P, Hellman U, Gonez LJ, Heldin CH. A novel GTPase-activating protein for Rho interacts with a PDZ domain of the protein-tyrosine phosphatase PTPL1. The Journal of biological chemistry. 1997;272:24333–24338. doi: 10.1074/jbc.272.39.24333. [DOI] [PubMed] [Google Scholar]
- Satriotomo I, Dale EA, Dahlberg JM, Mitchell GS. Repetitive acute intermittent hypoxia increases expression of proteins associated with plasticity in the phrenic motor nucleus. Experimental neurology. 2012;237:103–115. doi: 10.1016/j.expneurol.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger FW, Hager G, Schmitt AB, Horvat A, Hager G, Streif R, Spitzer C, Gamal S, Breuer S, Brook GA, et al. Peripheral but not central axotomy induces changes in Janus kinases (JAK) and signal transducers and activators of transcription (STAT) The European journal of neuroscience. 2000;12:1165–1176. doi: 10.1046/j.1460-9568.2000.00005.x. [DOI] [PubMed] [Google Scholar]
- Seo HW, Kim EJ, Na H, Lee MO. Transcriptional activation of hypoxia-inducible factor-1alpha by HDAC4 and HDAC5 involves differential recruitment of p300 and FIH-1. FEBS Lett. 2009;583:55–60. doi: 10.1016/j.febslet.2008.11.044. [DOI] [PubMed] [Google Scholar]
- Shin J, Cho Y, Beirowski B, Milbrandt J, Cavalli V, DiAntonio A. Dual leucine zipper kinase is required for retrograde injury signaling and axonal regeneration. Neuron. 2012;74:1015–1022. doi: 10.1016/j.neuron.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JE, Geisler S, DiAntonio A. Dynamic regulation of SCG10 in regenerating axons after injury. Experimental neurology. 2014;252:1–11. doi: 10.1016/j.expneurol.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DS, Skene JH. A transcription-dependent switch controls competence of adult neurons for distinct modes of axon growth. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1997;17:646–658. doi: 10.1523/JNEUROSCI.17-02-00646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondell M, Lundborg G, Kanje M. Vascular endothelial growth factor stimulates Schwann cell invasion and neovascularization of acellular nerve grafts. Brain research. 1999;846:219–228. doi: 10.1016/s0006-8993(99)02056-9. [DOI] [PubMed] [Google Scholar]
- Sondell M, Sundler F, Kanje M. Vascular endothelial growth factor is a neurotrophic factor which stimulates axonal outgrowth through the flk-1 receptor. The European journal of neuroscience. 2000;12:4243–4254. doi: 10.1046/j.0953-816x.2000.01326.x. [DOI] [PubMed] [Google Scholar]
- Trumbower RD, Jayaraman A, Mitchell GS, Rymer WZ. Exposure to acute intermittent hypoxia augments somatic motor function in humans with incomplete spinal cord injury. Neurorehabilitation and neural repair. 2012;26:163–172. doi: 10.1177/1545968311412055. [DOI] [PubMed] [Google Scholar]
- Wu D, Yotnda P. Induction and testing of hypoxia in cell culture. Journal of visualized experiments : JoVE. 2011 doi: 10.3791/2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee Koh M, Spivak-Kroizman TR, Powis G. HIF-1 regulation: not so easy come, easy go. Trends in biochemical sciences. 2008;33:526–534. doi: 10.1016/j.tibs.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Yuen TJ, Silbereis JC, Griveau A, Chang SM, Daneman R, Fancy SP, Zahed H, Maltepe E, Rowitch DH. Oligodendrocyte-encoded HIF function couples postnatal myelination and white matter angiogenesis. Cell. 2014;158:383–396. doi: 10.1016/j.cell.2014.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SX, Miller JJ, Gozal D, Wang Y. Whole-body hypoxic preconditioning protects mice against acute hypoxia by improving lung function. Journal of applied physiology. 2004;96:392–397. doi: 10.1152/japplphysiol.00829.2003. [DOI] [PubMed] [Google Scholar]
- Zhou X, Wang L, Hasegawa H, Amin P, Han BX, Kaneko S, He Y, Wang F. Deletion of PIK3C3/Vps34 in sensory neurons causes rapid neurodegeneration by disrupting the endosomal but not the autophagic pathway. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9424–9429. doi: 10.1073/pnas.0914725107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Ho C, Wong K, Tessier-Lavigne M. Axotomy-induced Smad1 activation promotes axonal growth in adult sensory neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:7116–7123. doi: 10.1523/JNEUROSCI.5397-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.