Abstract
Interleukin-17 is an ancient cytokine implicated in a variety of immune defense reactions. We have indentified five members of the sea lamprey IL-17 family (IL-17D.1, IL-17D.2, IL-17E, IL-17B and IL-17C) and six IL-17 receptor genes (IL-17RA.1, IL-17RA.2, IL-17RA.3, IL-17RF, IL-17RE/RC and IL-17RD), determined their relationship with mammalian orthologues, and examined their expression patterns and potential interactions in order to explore their roles in innate and adaptive immunity. The most highly expressed IL-17 family member is IL-17D.1 (mammalian IL-17D like), which was found to be preferentially expressed by epithelial cells of skin, intestine and gills and by the two types of lamprey T-like cells. IL-17D.1 binding to recombinant IL-17RA.1 and to the surface of IL-17RA.1-expressing B-like cells and monocytes of lamprey larvae was demonstrated, and treatment of lamprey blood cells with recombinant IL-17D.1 protein enhanced transcription of genes expressed by the B-like cells. These findings suggest a potential role for IL-17 in coordinating the interactions between T-like cells and other cells of the adaptive and innate immune systems in jawless vertebrates.
Keywords: Lamprey, Interleukin-17, Interleukin-17 receptor
Introduction
Interleukin-17 (IL-17) is an ancient cytokine, orthologues of which have been identified in invertebrates that employ innate immune responses for protection against pathogens (1–5). In mammals, six members of the IL-17 family (IL-17A, IL-17B, IL-17C, IL-17D, IL-17E and IL-17F) have been characterized, where they play important roles in a variety of immune responses. IL-17A and IL-17F are essential for adaptive immune responses against extracellular bacteria and fungi in humans, and they contribute to the pathogenesis of inflammatory diseases, such as rheumatoid arthritis, inflammatory bowel disease, and multiple sclerosis (6). IL-17B and IL-17C are shown to enhance TNF-α and IL-1β production, both important immune regulators, by a human leukemic monocytic cell line (7). With an extended C-terminal domain, the IL-17D protein is the largest IL-17 family member; it is expressed at relatively high levels in skeletal muscle, brain, adipose, heart, lung, and pancreas tissues, and at lower levels by resting CD4+ T cells and B cells in humans (8). IL-17D suppresses myeloid progenitor cell proliferation, but enhances endothelial cell production of IL-6 and IL-8. IL-17D also promotes tumor rejection through recruitment of natural killer cells (9). IL-17E, also called IL-25, promotes production of Th2 cell cytokines and recruitment of eosinophils to provide protection against helminth infection and regulate proallergic responses (10, 11).
The protein structure of IL-17 family members is distinguished by the characteristic cystine-knot with four C-terminal cysteines forming two intramolecular disulfide bonds as indicated by the crystal structure of human IL-17F (12, 13). This cystine-knot motif, originally identified in studies on the nerve growth factors (14), is highly conserved. Orthologues of mammalian IL-17A and IL-17F have been described in zebrafish, rainbow trout and Japanese pufferfish (also known as fugu) (15–17). IL-17C orthologues have been reported in zebrafish, fugu, and rainbow trout (15, 17, 18). IL-17D orthologues are identified in zebrafish, fugu, Atlantic salmon, grass carp, channel catfish and Japanese lamprey (1, 15, 17, 19–22). Studies on the expression patterns of IL-17 orthologues indicate their tissue specificity. By real-time PCR analysis, IL-17D expression is evident in zebrafish spleen and gills (15); fugu IL-17D is expressed at low levels in skin, brain, head kidney and the intestine (17); Japanese lamprey IL-17D expression is most prominent in skin, gills, and intestine (22). In Atlantic salmon and grass carp, expression of IL-17D is detectable in skin and intestine by real-time PCR analysis (19, 20). A novel IL-17 ligand named IL-17N has only been identified in bony fish including zebrafish, medaka, fugu and stickleback (17, 23). In elephant shark Callorhinchus milii, three IL-17A-like sequences (GenBank accession no. NP_001279507, XP_007883573 and XP_007883503) and two IL-17C-like sequences (GenBank accession no. XP_007906115 and XP_007904597) have been identified by NCBI Eukaryotic Genome Annotation Pipeline (http://www.ncbi.nlm.nih.gov/genome/annotation_euk/Callorhinchus_milii/100). Putative IL-17 genes have also been identified in invertebrates, including vase tunicate, pearl oyster, Pacific oyster and Californian abalone (1, 2, 4, 24, 25). Several IL-17-like genes are found in other invertebrates, such as amphioxus (Branchiostoma floridae), sea urchin (Strongylocentrotus purpuratus), lottia (Lottia gigantea) and Caenorhabditis elegans, although full-length sequences for these genes are not yet available (3, 5, 23, 24).
The mammalian IL-17 receptor (IL-17R) family consists of five members (IL-17RA to IL-17RE) which share limited sequence similarity with other cytokine receptors. Members of the IL-17R family have extracellular fibronectin III-like domains, a single transmembrane domain and a conserved cytoplasmic domain called “similar expression to fibroblast growth factor genes and IL-17R” (SEFIR) that resembles the Toll/IL-1 receptor (TIR) domain. IL-17RA is the common receptor subunit in that it pairs with most other IL-17R members for the binding of IL-17 family members. In humans, IL-17RA forms a heteromeric complex with IL-17RC for IL-17A or IL-17F signaling (26–28). IL-17RA has higher affinity for IL-17A, while both IL-17A and IL-17F bind to IL-17RC with similar affinities. IL-17RA is expressed on both hematopoietic cells, including B cells and T cells, and non-hematopoietic cells such as fibroblasts, endothelial cells, and epithelial cells (29). In IL-17RA knockout mice, the host defense against infectious Klebsiella pneumoniae and Candida albicans is compromised, thereby suggesting important roles of IL-17RA in immune defense (30–32). IL-17RB binds IL-17B and can also pair with IL-17RA to bind IL-17E (33). The IL-17RA and IL-17RE receptor complex is required for IL-17C signaling (34–36). The receptor for IL-17D has not yet been identified; IL-17RD is an orphan receptor without an identified ligand. IL-17R orthologues have been found in the invertebrates such as sea urchin, amphioxus and vase tunicate (3, 4, 37, 38). IL-17RA and IL-17RD orthologues have been identified in the amphioxus genome sequences (38), but the cloning and characterization of these invertebrate IL-17R genes have not yet been reported. Several incomplete sequences of IL-17Rs including IL-17RA, IL-17RB, IL-17RC and IL-17RD have been identified in the genome sequence of the cartilaginous fish, elephant shark (38), and IL-17RE (GenBank accession no. XP_007909458) has also been identified in this cartilaginous fish by NCBI Eukaryotic Genome Annotation Pipeline. In most teleost fish, IL-17RA and IL-17RD orthologues have been identified. IL-17RA sequences have been reported in fugu, Atlantic salmon, rainbow trout, stickleback, medaka and zebrafish (16, 23, 38). IL-17RD orthologues are found in zebrafish, fugu, green puffer, stickleback and medaka (23, 38, 39). The presence of IL-17RB, IL-17RC and IL-17RE orthologues has been confirmed in the genome sequences of stickleback and zebrafish (23). A group of vertebrate IL-17R-like proteins, which lack the intracellular conserved SEFIR domain but resemble the extracellular domain of IL-17RE, have been identified in zebrafish, fugu and stickleback (38) and named IL-17RE-like (IL-17REL) proteins.
Here, we focus on the IL-17 ligand and receptor family in lampreys. These jawless vertebrates have an alternative adaptive immune system in which leucine-rich repeat (LRR)-based proteins named variable lymphocyte receptors (VLRs) are used for antigen recognition (40–43). Three VLR genes have been identified; the VLRA and VLRC genes are expressed by T-like VLRA+ and VLRC+ cells respectively, and VLRB genes are expressed by B-like VLRB+ cells (44). The present investigations were stimulated by our earlier findings on the reciprocal expression patterns of IL-17 ligand and receptor. One IL-17 orthologue was identified in lampreys (22, 41) and shown to be preferentially expressed by the VLRA+ lymphocytes. By contrast, IL-17R transcripts were predominantly expressed by the VLRB+ lymphocytes (41). The reciprocal expression of an IL-17 cytokine and cytokine receptor pair suggested their participation in crosstalk between different T-like and B-like populations in response to the antigen stimulation. In the present studies, we have defined the members of the IL-17 and IL-17R families in lampreys, determined their expression patterns and examined their potential interactions.
Materials and Methods
Animal maintenance
Petromyzon marinus larvae (outbred, 8–15 cm long and 2–4 years of age) and adult sea lampreys (outbred, 50–80 cm long and 8–14 years of age) were from Great Lakes of North America. Lampetra planeri larvae (outbred, about 10cm long) were obtained from the tributaries of the Rhine river in Germany. Larvae were maintained in sand-lined aquariums at 18 °C and were fed brewer’s yeast. Adult sea lampreys were maintained in temperature-controlled tanks (16–18 °C). Animals were sacrificed in 1g/1 MS-222 and 1.4g/l sodium bicarbonate followed by exsanguination. Peripheral blood of larvae was collected in 0.66×PBS/30mM EDTA, layered on top of 55% percoll and subjected to density centrifugation (400g, 20 min, no brake). Adult lamprey blood was collected by cutting the tail and leukocytes were separated by Lymphoprep. Subsequently, the lamprey lymphocytes were collected for following studies. Cells from kidney, intestine and gills were isolated by disrupting tissues between frosted glass slides and 400g centrifugation. The cell pellets were lysed in 1% NP-40 lysis buffer (with 5µg/ml leupeptin, 1µg/ml pepstatin, 5µg/ml aprotinin, 10µg/ml soybean trypsin inhibitor, 40µg/ml PMSF). The tissue lysates were used for immunoblot assays. All studies have been reviewed and approved by Institutional Animal Care and Use Committee at Emory University.
Transcriptome assembly of lampreys
Total RNA was extracted from the kidney, gills, blood and intestines (containing typhlosole) of lampreys, treated with DNase and Illumina Hi-Seq compatible, mRNA enriched, stranded cDNA libraries were generated using Illumina TruSeq RNA (stranded) kit, following the manufacturer’s instructions. Transcriptomes were assembled from 100bp paired-end reads (generated on the x sequencer) using the Trinity suite of programs (45) and a kmer size of 25bp. Reads were assembled in a strand-specific manner with assembly time reduced through in-silico normalization of reads to a maximum of 30x coverage using the in-built Trinity parameter. Open reading frames (ORFs) were extracted using Trinity’s Transdecoder option to select the best open reading-frame for each transcript in a strand-specific manner, with identical ORFs being collapsed into a single sequence.
Identification of lamprey IL-17 and IL-17R sequences
Known mammalian IL-17 and IL-17R sequences were used as queries for tBLASTn search against sea lamprey (http://www.ensembl.org/Petromyzon_marinus/Info/Index) and Japanese lamprey genome sequences (http://jlampreygenome.imcb.a-star.edu.sg). Unique ORF collections from European brook lamprey transcriptomes were interrogated using IL-17 and IL-17R protein sequences of various species. The signal peptide, transmembrane and putative domains were predicted by SMART software (http://smart.embl-heidelberg.de) and conserved domain database (CDD) of NCBI. The conserved IL-17 domain of IL-17s and the conserved SEFIR domain of IL-17Rs were used in the phylogenetic analysis. Multiple protein sequence alignments were conducted using the ClustalW program (46). The phylogenetic trees were constructed by (i) neighbor-joining (NJ) and (ii) maximum likelihood (ML) methods using the MEGA5.0 program (48). The evolutionary distances were computed by the JTT matrix-based method (48). The reliability of the tree was assessed by bootstrap resampling with a minimum of 1000 replications.
Cloning and sequencing sea lamprey (Petromyzon marinus) IL-17D.1 and IL-17R
Total RNA was extracted from lymphoid tissues of lamprey larvae. First-strand cDNA was synthesized with oligo(dT) primer by Superscript III (Invitrogen). PCR primers were designed according to the sea lamprey genome sequences and nested PCR was performed to clone the IL-17D.1 gene. The 5’ and 3’ end of IL-17R genes were obtained by SMARTer rapid amplification of cDNA ends (RACE) cDNA Amplification Kit (Clontech). 5’ RACE cDNA was synthesized by touchdown PCR using a modified oligo(dT) primer and SMARTer II A oligonucleotides and 3’ RACE cDNA synthesis was carried out using a special oligo(dT) primer. The 5’ and 3’ RACE PCR products were cloned into pBluescript (Stratagene) vector and sequenced. According to the complete sequence information, PCR primers were then designed and nested PCR was performed to clone the IL-17R genes. Primers used for RACE and nested PCR are all listed in Table 1.
Table 1.
List of primers and probe used in the study
| Primer name | Primer sequence (5’→3’) | Use |
|---|---|---|
| qRT_IL-17D.2_F4 | gttcccgtcacccaggccg | real-time PCR |
| qRT_IL-17D.2_R4 | cacggcgatgttgtgcgtgtc | real-time PCR |
| qRT_IL-17B_F2 | gtctccgtgcccgtcat | real-time PCR |
| qRT_IL-17B_R2 | acggcgatctcctccca | real-time PCR |
| qRT_IL-17E_F | gagcgctccctcgtctcc | real-time PCR |
| qRT_IL-17E_R | cagctccggggctatctt | real-time PCR |
| qRT_IL-17D.1_F | caagattccttgcgttaccc | real-time PCR |
| qRT_IL-17D.1_R | tgtctctaggtccggcttgt | real-time PCR |
| qRT_IL-17C_F1 | acaggatcgtggagaacc | real-time PCR |
| qRT_IL-17C_R1 | gttgacctcggtcgtctc | real-time PCR |
| qRT_IL-17RA.1_F1 | cgaggctgctgtatgtgaaa | real-time PCR |
| qRT_IL-17RA.1_R1 | ctggaacggtgtgcatattg | real-time PCR |
| qRT_IL-17RA.2_F | cagcagttggatcaaagcaa | real-time PCR |
| qRT_IL-17RA.2_R | tcttcccgaaattccacttg | real-time PCR |
| qRT_IL-17RA.3_F | gctctctcgcctcgtaccta | real-time PCR |
| qRT_IL-17RA.3_R | cctgcagccagttcatgtta | real-time PCR |
| qRT_IL-17RF_F | gccactgatatgtgccgact | real-time PCR |
| qRT_IL-17RF_R | aacgtcggggatgtctttct | real-time PCR |
| qRT_IL-17RE/RC_F2 | cttcacgtactcgtgtttct | real-time PCR |
| qRT_IL-17 RE/RC_R2 | actctggcacggtgtagt | real-time PCR |
| qRT_IL-17RD_F6 | caagacggtgaacgtgac | real-time PCR |
| qRT_IL-17RD_R6 | cgtacaccacgtacgactc | real-time PCR |
| qRT_IL-8_F | cacagagctccaactgcaag | real-time PCR |
| qRT_IL-8_R | aatgtggctgatcaccttcc | real-time PCR |
| E2A_F1 | ccaagctgggaatccttcac | real-time PCR |
| E2A_R1 | cagctttcggattcaagttcct | real-time PCR |
| Pax5_F1 | gacaatgtttgcctgggagatc | real-time PCR |
| Pax5_R1 | ttccgttccgtgtgattctg | real-time PCR |
| Blimp1_F1 | cctaccagtgtcaggtgtgc | real-time PCR |
| Blimp1_R1 | ggagcttcaggtggatgaac | real-time PCR |
| Bcl6_F1 | acacgggagagaagccctat | real-time PCR |
| Bcl6_R1 | accttggtgtttgtgatggc | real-time PCR |
| Syk_F1 | caagccctacccgaagatgaaag | real-time PCR |
| Syk_R1 | cgggcaatgttccggttt | real-time PCR |
| BCAP_F1 | gcccagcgtgtaaacccata | real-time PCR |
| BCAP_R1 | acggcaacatgcacagtacct | real-time PCR |
| GAPDH_F | ttgaggatgggaagctgttga | real-time PCR |
| GAPDH_R | gggtcacgctccgagtagac | real-time PCR |
| 3RACE_IL-17RA.1_F | tggtccttgacctctggcaagtgaa | RACE |
| 5RACE_IL-17RA.1_R | cgactttgcgtcctccttgagtgtg | RACE |
| 3RACE_IL-17RA.2_F | gacagggaggatgcagtggctgatg | RACE |
| 5RACE_IL-17RA.2_R | ctgagggcggggctgtacatgtct | RACE |
| 3RACE_IL-17RA.3_F | ggaaggagaggcctccagcagagaa | RACE |
| 5RACE_IL-17RA.3_R | gcagcgccatgcgtaatatctcagg | RACE |
| clone IL-17RA.1_F3 | gctgctgttgaatgagaacg | cloning |
| clone IL-17RA.1_R1 | gcatcttgatattcgtgcattg | cloning |
| clone_IL-17RA.2_F1 | tggtcttcgtggggtgtaga | cloning |
| clone_IL-17RA.2_F2 | tagagggcctgcgtttatgg | cloning |
| clone_IL-17RA.2_R1 | tgtgtgcttacgatggtacagttg | cloning |
| clone_IL-17RA.2_R2 | tggacacgtagttttgggagttc | cloning |
| clone_IL-17RA.3_F1 | atttcccgactgctggagtt | cloning |
| clone_IL-17RA.3_F2 | actcgcgcagaggatagagg | cloning |
| clone_IL-17RA.3_R1 | cccatgcaaaagaagttcaaca | cloning |
| clone_IL-17RA.3_R2 | gctgatctcgatttcggtcac | cloning |
| clone IL-17D.1_F2 | acggcaggagagatacagga | cloning |
| clone IL-17D.1_F3 | agctggcacgttcttttgat | cloning |
| clone IL-17D.1_Rout | cccacaggtccttctctcct | cloning |
| clone IL-17D.1_R | ggggagaatggcaatcca | cloning |
| IL-17D.1 sense probe | accession no. KR059941 | in-situ hybridization |
Real-time PCR
Tissues (skin, kidneys, intestine, gills and blood) from lamprey larvae were extracted for RNA isolation. RNA was purified from lamprey tissues using RNeasy kits with on-column DNA digestion by DNase I (QIAGEN). First-strand cDNA was synthesized with random hexamer primers by Superscript III (Invitrogen). Quantitative real-time PCR was performed using SYBR Green on 7900HT ABI Prism (Applied Biosystems). All samples were run in three replicates. Cycling conditions were 1 cycle at 50°C for 2 min, 1 cycle at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min. The values for the IL-17D.1 and IL-17R genes were normalized to the expression of GAPDH. Primers used in this experiment are listed in Table 1.
In situ hybridization analysis
RNA in situ hybridization was performed with digoxigenin (DIG)-labelled RNA riboprobes as described previously (49). The probe sequence for IL-17D.1 is listed in Table 1.
IL-17D.1 and IL-17R expression in HEK-293T cells
HEK-293T cells were maintained in Dulbecco's Modified Eagle Medium supplemented with 10% FBS, 2mM L-glutamine, 50U/ml penicillin, and 50µg/ml streptomycin at 37°C in 5% CO2. IL-17D.1 cDNA was cloned into a fusion vector made from pIRESpuro2 (Invitrogen, Grand Island, NY) harboring a puromycin resistance gene to produce a fusion protein IL-17D.1/IgG1-Fc which is composed of IL-17D.1 and the Fc region of human IgG1 (IgG1-Fc) (Supplemental Fig. 1A). The IgG1-Fc portion facilitated detection of the fusion protein with anti-IgG antibodies and its purification by protein A chromatography. The coding sequence of IL-17RA.1, IL-17RA.2 or IL-17RA.3 was subcloned into pDisplay vector (Invotrogen) in-frame with HA tag, myc tag and FLAG tag respectively. The plasmids were transfected into HEK-293T cells using linear polyethylenimine (PEI), MW 25,000 (Polysciences, Inc.) at a 3:1 (PEI:DNA) ratio. After 60 hours, cells were harvested by centrifugation at 300g for 3 minutes. The supernatant were collected. The cell pellets were washed once with PBS and lysed in 1% NP-40 lysis buffer (with 5µg/ml leupeptin, 1µg/ml pepstatin, 5µg/ml aprotinin, 10µg/ml soybean trypsin inhibitor, 40µg/ml PMSF). The whole cell lysates and the supernatants were used for immunoblot assays.
Purification of recombinant IL-17D.1/IgG1-Fc proteins
Recombinant IL-17D.1/IgG1-Fc proteins were purified from supernatants via the human IgG1-Fc portion using protein A-agarose column (GE Healthcare). 500mL cell culture supernatants were collected from stably transfected HEK-293T cells under selection of 2µg/ml puromycin (Sigma). The supernatants were then loaded onto the protein A-agarose column, and washed with 10 column volumes of PBS. The non-specific proteins were removed by 15 column volumes of 50mM citrate at pH4.5 and recombinant 17D/IgG1-Fc proteins were eluted with 15 column volumes of 50mM citrate, 50mM NaCl at pH3. The eluate was dialyzed in PBS and concentrated by centrifugation using an Amicon Ultra-15 30K Centrifugal Filter Device (Millipore).
Production of anti-IL-17D.1, anti-monocyte and anti-granulocyte monoclonal antibodies (mAbs)
BALB/c mice were hyper-immunized with the IL-17D.1/IgG1-Fc fusion proteins and the lymphocytes were isolated from the draining lymph nodes of these mice and subsequently fused with the Ag8.653 myeloma cell line using PEG-1500 (Roche). The hybridomas were screened by ELISA and Western blot and the positive clones were subcloned by serial dilutions. The ELISA and Western blot assay (Supplemental Fig. 1B) of a representative mouse anti-IL-17D.1 mAb (clone 114–178, IgG2a) was shown. Anti-monocyte mouse mAb (8A1, IgG2b) and anti-granulocyte mAb (2D4, IgG2a) were produced by immunization of BALB/c mice with fresh peripheral blood leukocytes from the adult lamprey and screened by flow cytometry using FSC/SSC. Two distinct populations of leukocytes were stained by 8A1 and 2D4 mAbs (Supplemental Fig. 1C). FSC/SSC profile and Wright-Giemsa staining of sorted monocytes and granulocytes (Supplemental Fig. 1D) are shown.
Immunoblotting
Samples treated with 2-mercaptoethanol were separated on 10% SDS-PAGE gels and then transferred onto nitrocellulose membranes (Millipore) by semi-dry transfer method. Membranes were blocked with 7% skimmed milk for an hour and incubated with primary antibodies overnight at 4 degree. The primary antibodies used in this study include HRP conjugated goat anti-human Fc (1:5000, Southern Biotech), mouse anti-HA mAb (HA.11, 1:1000, Covance), mouse anti-myc mAb (9B11, 1:1000, Cell signaling), mouse anti-FLAG M2 mAb (1:1000, Sigma), rabbit anti-γ-Tubulin (AK-15) antibody (1:1000, Sigma). After three washes with TBS-1% Tween-20, membranes were incubated with secondary antibodies including HRP conjugated goat anti-mouse Ig (1:10000, Dako), HRP conjugated goat anti-rabbit Ig (1:10000, Dako). After three washes, blots were developed using SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) for 5 minutes.
Flow cytometric analysis and sorting
Leukocytes isolated from blood were stained for flow cytometry as described (41). Briefly, leukocytes from blood were stained with primary antibodies including anti-VLRA rabbit polyclonal serum (R110), anti-VLRB mouse mAb (4C4), anti-VLRC mouse mAb (3A5), anti-monocyte mouse mAb (8A1), or anti-granulocyte mouse mAb (2D4). The cells were costained with recombinant IL-17D.1/IgG1-Fc or control proteins. Secondary antibodies were matched. Staining and washes were in 0.67×PBS with 1% BSA. Cells were gated using forward scatter-A (FSC-A) vs side scatter-A (SSC-A) (lymphocytes), FSC-A vs FSC-H (singlets), and negative propidium iodide (Sigma P4864) staining (live cells). Flow cytometric analysis was performed on a CyAn ADP (Dako) flow cytometer. VLRA+, VLRB+, VLRC+, VLR triple-negative cells, monocytes, and granulocytes were sorted on BD FACS Aria II (BD Bioscience) for real-time PCR analysis. The purity of the sorted cells was over 90%.
Immunoprecipitation
The IL-17R expressing plasmids were transfected into HEK-293T cells using PEI and the whole cell lysates were used for immunoprecipitation assays. Samples were pre-cleared by incubating with 30µl of a protein G-agarose suspension for 30 min followed by centrifugation at 12,000g for 1 min to remove the beads. The pre-cleared lysates were incubated with 30µl of the protein G-agarose suspension and IL-17D.1/IgG1-Fc or control protein on a rotator for 2 hours at 4 degree. Protein G-agarose beads were centrifuged at 12,000g for 12 seconds and washed 4 times with 1% NP-40 lysis buffer. Samples treated with 2-mercaptoethanol were separated on 10% SDS–PAGE gels and then transferred onto nitrocellulose membranes (Millipore) and immunoblotted with corresponding antibodies.
Cell culture with IL-17D.1 recombinant protein
Buffy coat leukocytes were collected, washed twice with and resuspended in 2/3 IMDM medium (HyClone, GE Healthcare) supplemented with 100IU/ml of penicillin sulfate and 100µg/ml of streptomycin. For each lamprey larvae (n=5), blood leukocytes were separated into three groups (saline, IL-17D.1/IgG1-Fc, control Fc) and seeded in a sterile 24-well plate at a density of 1.0×106 cells/ml in 1.0 ml medium. IL-17D.1/IgG1-Fc and control Fc were added to the blood leukocytes at a final concentration of 2µg/ml and equal volume of 0.67×PBS was also added as the saline control. Cells were cultured at 20°C for 16 h and then collected for RNA extraction and real-time PCR analysis.
Statistical analysis
All data are expressed as the mean ± SEM (n = 3 or more). Real-time PCR data were analyzed using one-way repeated measures ANOVA performed with GraphPad Prism Version 5.0 (GraphPad Software). If a significant difference between multiple means was reported, ANOVAs were followed by Tukey's post-hoc test. A p value of <0.05 was considered significant. The significant difference is denoted with * (p < 0.05), ** (p < 0.01), or *** (p < 0.001).
Results
Phylogenetic relationships of IL-17 sequences identified in lampreys
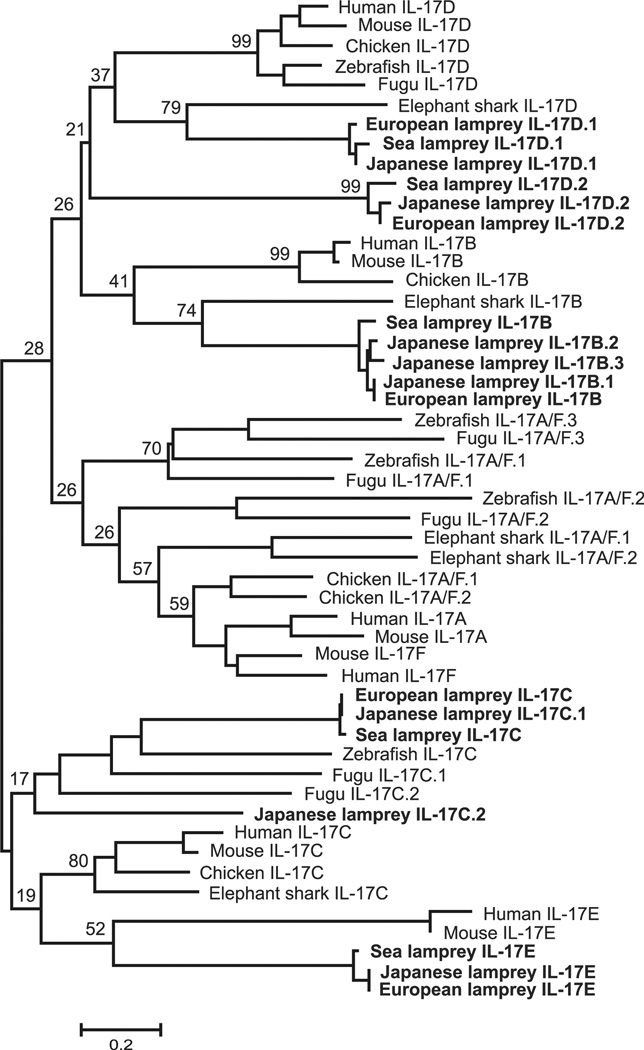
By conducting similarity searches and homology inferences from the currently available genome and transcriptome data, we identified five different IL-17 genes in the draft genome and transcriptome of sea lamprey (Petromyzon marinus), eight IL-17 genes in Japanese lamprey (Lethenteron japonicum) genome and five different IL-17 sequences in transcriptomes of European brook lamprey (Lampetra planeri) (Fig. 1, Supplemental Table 1). Although full-length sequences of the different IL-17s are not available for all three lamprey species, our phylogenetic analysis of the derived IL-17 C-terminal amino acid sequences suggests that IL-17s in lampreys can be classified into five groups designated as IL-17D.2, IL-17B, IL-17E, IL-17D.1 and IL-17C (Fig. 1). Among the eight sequences identified in Japanese lamprey, IL-17D.2, IL-17E and IL-17D.1 have one to one orthologous relationship with the corresponding sequences in sea lamprey and European brook lamprey. Three additional IL-17 sequences of the Japanese lamprey (IL-17B.1, IL-17B.2, IL-17B.3) group with IL-17B sequences; two other IL-17 sequences (IL-17C.1, IL-17C.2) are related to IL-17C of sea lamprey and European brook lamprey (Fig. 1, Supplemental Fig. 2). As the sea lamprey genome sequence is incomplete and the European brook lamprey genome sequence is not available, it is not possible to arrive at a definitive conclusion about possible duplication or deletion events affecting the IL-17 gene family in each lineage after divergence of these three lamprey species. Compatible with the presumed cystine-knot structure of IL-17 proteins, the IL-17 sequences identified in the three lamprey species all possess the four characteristic conserved cysteines (Supplemental Fig. 2). Based on phylogeny and conservation in amino acid residues, relative to the well-defined IL-17 family members in mammals, the lamprey IL-17D.1 and IL-17D.2 sequences are close to mammalian IL-17D, whereas IL-17B, IL-17E and IL-17C sequences are similar to mammalian IL-17B, IL-17E, and IL-17C sequences, respectively (Fig. 1, Table I, Supplemental Fig. 2). Notably, lamprey orthologues of mammalian IL-17A and IL-17F were not found.
FIGURE 1.
Phylogenetic comparison of lamprey IL-17s with mammalian IL-17s. The C-terminal conserved domain was used in the analysis. The phylogenetic tree is constructed by the NJ method. The numbers indicate bootstrap confidence values obtained for each node after 1000 replications. The accession numbers for sequences used in this analysis were as follows: Human IL-17A, NP 002181; Dog IL-17A, NP 001159350; Mouse IL-17A, NP 034682; Rat IL-17A, NP 001100367; Horse IL-17A, NP 001137264; Monkey IL-17F, XP 002746687; Rat IL-17F, NP 001015011; Mouse IL-17F, NP 665855; Opossum IL-17F, XP 001370182; Human IL-17F, NP 443104; Human IL-17B, NP 055258; Dog IL-17B, XP 546311; Mouse IL-17B, NP 062381; Cow IL-17B, NP 001178974; Sheep IL-17B, XP 004009006; Human IL-17C, NP 037410; Mouse IL-17C, NP 665833; Guinea pig IL-17C, XP 003460965; Dog IL-17C, XP 851256; Opossum IL-17C, XP 007477360; Human IL-17D, NP 612141; Mouse IL-17D, NP 665836; Cow IL-17D, XP 871741; Pig IL-17D, XP 005653873; Opossum IL-17D, XP 001375129; Human IL-17E, NP 073626; Cow IL-17E, XP 605190; Guinea pig IL-17E, XP 003474349; Sheep IL-17E, NP 001182148; Mouse IL-17E, NP 542767; Sea Lamprey IL-17D.1, KR059941; Sea Lamprey IL-17C, KR059956; European lamprey IL-17D.2, KR059945; European lamprey IL-17B, KR059946; European lamprey IL-17E, KR059947; European lamprey IL-17D.1, KR059948; and European lamprey IL-17C, KR059949. The same phylogenetic relationship was demonstrated by the ML method (data not shown).
Expression patterns of sea lamprey IL-17 genes
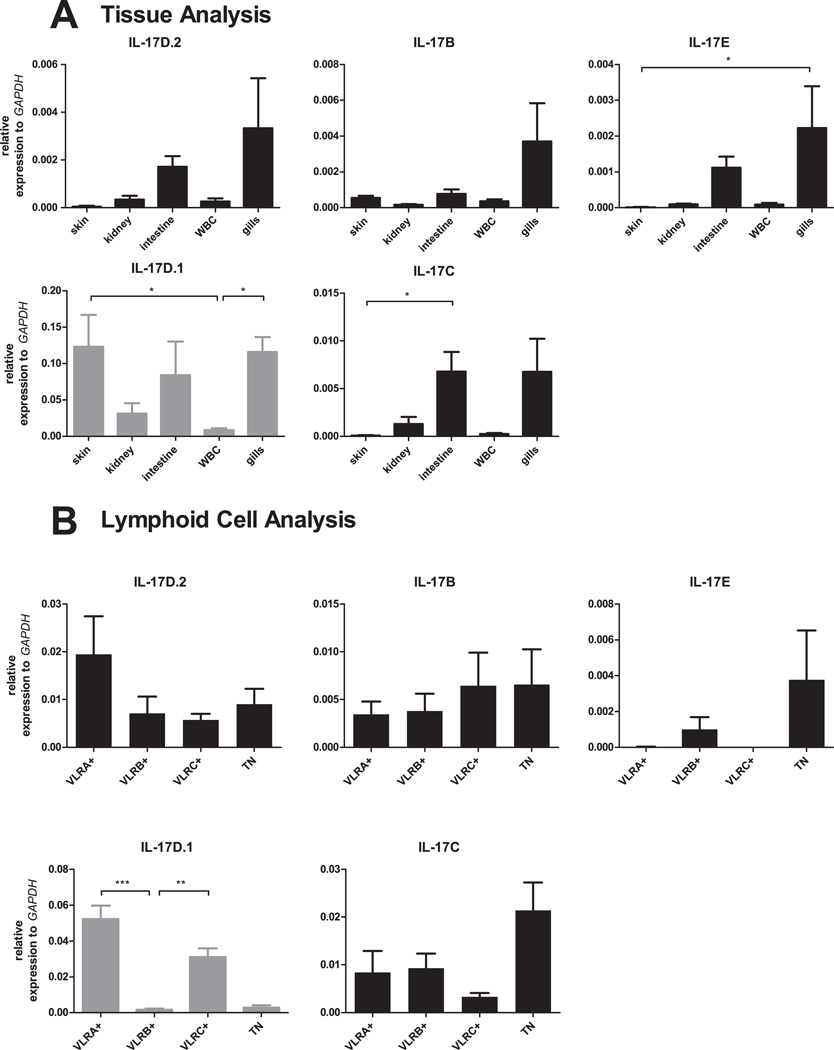
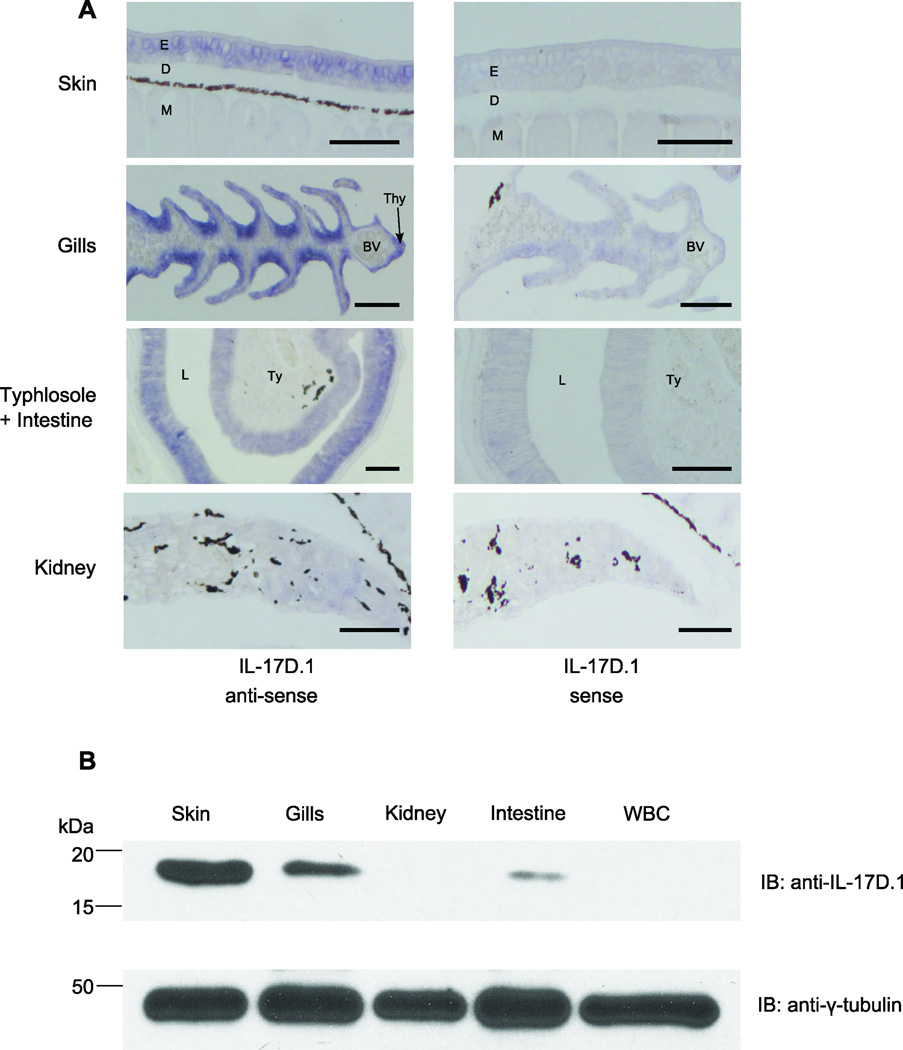
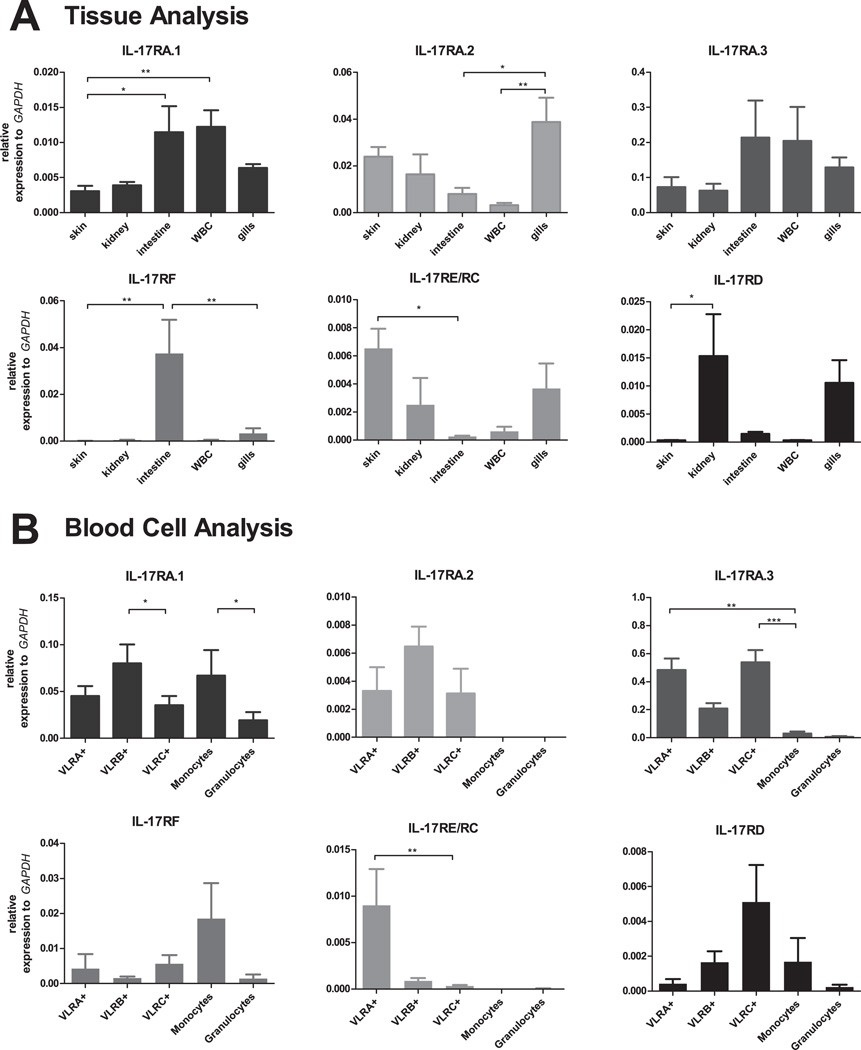
The expression patterns of IL-17 genes were determined by real-time PCR analysis after validating the specificities of primers by sequencing amplicons generated by RT-PCR (data not shown). We found that IL-17D.1 was expressed at highest levels in skin and gills, with lower levels being observed in the kidneys and white blood cells (WBC) (Fig. 2A). The other IL-17s in lampreys, IL-17D.2, IL-17B, IL-17E and IL-17C, were expressed primarily in the intestine and gills. RNA in situ hybridization analysis indicated that the epithelial cells in the larval skin, gill filaments and intestine expressed IL-17D.1 transcripts (Fig. 3A). In a prior study, VLRA+ lymphocytes were found to preferentially express an IL-17 gene that corresponds to the IL-17D.1 member of the gene family defined here (41). In the present studies, we sorted the VLRA+, VLRB+, VLRC+ and VLR triple-negative cells within the lymphocyte gate and examined their expression of IL-17D.1. The VLRA+ and VLRC+ lymphocytes were both found to express IL-17D.1 preferentially. IL-17D.2, IL-17B, and IL-17C expression were detected in all lymphocyte populations, whereas the expression of IL-17E was detected in the VLRB+ lymphocytes and triple-negative cells (Fig. 2B). The relatively high level of expression that we observed for IL-17D.1 facilitated the derivation of a full-length IL-17D.1 clone by the use of nested PCR. In order to confirm the results of our RNA expression studies for IL-17D.1 at the protein level, we produced mAbs that recognize the recombinant IL-17D.1 protein (Supplemental Fig. 1B). Using one of these mAbs, expression of IL-17D.1 was demonstrable in skin, gills and intestine of lamprey larvae by Western blot analysis (Fig. 3B); notably, IL-17D.1 protein levels correlated well with the RNA expression levels (Fig. 2, Fig. 3). These findings confirmed that IL-17D.1 is expressed by cells at the barrier surfaces in the gills, skin and intestine and by circulating VLRA+ and VLRC+ T cell-like lymphocytes.
FIGURE 2.
Sea lamprey expression of IL-17 transcripts. (A) Tissue distribution. Relative expression levels of IL-17D.2, IL-17B, IL-17E, IL-17D.1 and IL-17C transcripts were determined by real-time PCR analysis of skin, kidneys, intestine, white blood cells (WBC) and gills of sea lamprey larvae (n=6 larvae). (B) IL-17D.1 expression by sorted VLRA+, VLRB+, VLRC+ and triple negative (TN) lymphocyte populations (n=6 larvae). Relative expression level of IL-17D.1 was determined by real-time PCR. mRNA abundance relative to that of GAPDH: 2−ΔCt, ΔCt = Cttarget gene −CtGAPDH. Error bars indicate SEM. The scales of the y-axis are different on each graph.
FIGURE 3.
Sea lamprey expression of IL-17D.1. (A) Cellular expression of IL-17D.1 transcripts in different tissues. Tissue sections of skin, gills, kidney, typhlosole and intestine of lamprey larvae were hybridized with sense (control) and anti-sense riboprobes. Positive hybridization signals with IL-17D.1 riboprobes are indicated by blue color. E, epidermis; D, dermis; M, muscle; Thy, thymoid; BV, blood vessel; L, lumen; Ty, typhlosole. Scale bars, 100 µm. (B) Tissue distribution of IL-17D.1 protein expression. Whole tissue lysates from skin, gills, kidneys, intestine, and white blood cells (WBC) were resolved by reducing SDS-PAGE and blotted with a mouse monoclonal anti-IL-17D.1 antibody (clone 114–178). γ-Tubulin served as the loading control.
Identification of IL-17R genes
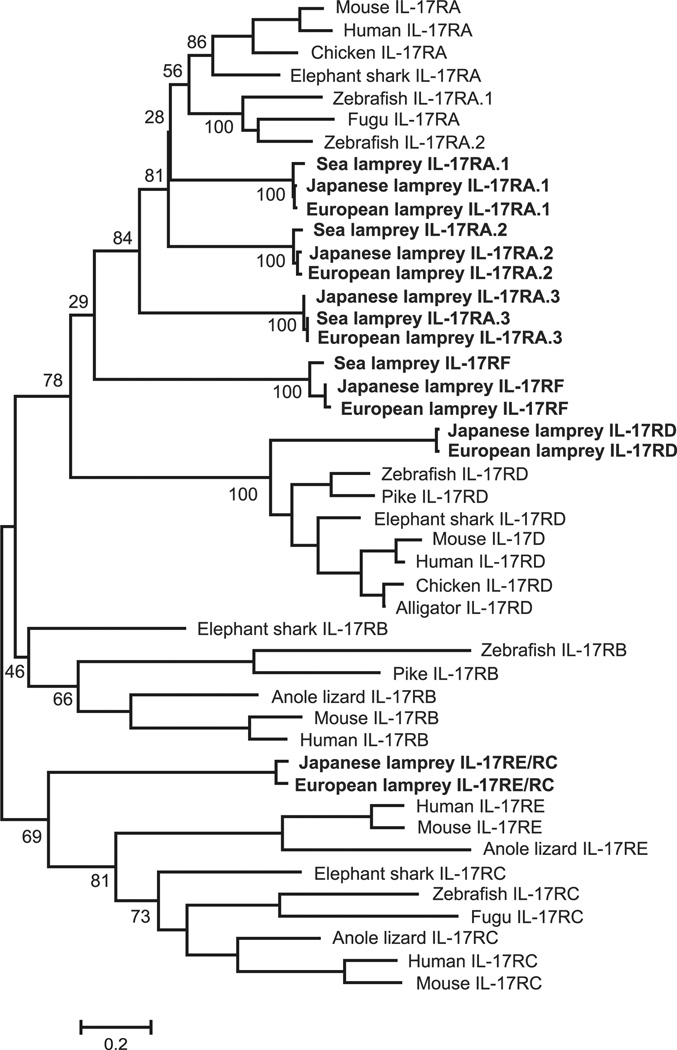
To identify IL-17R family genes in lampreys, we performed tBLASTn searches of available sea lamprey and Japanese lamprey genome sequences using mammalian IL-17R family genes as query sequences. For both lamprey species, we identified six distinct IL-17R sequences. Six different IL-17R sequences were also identified in the transcriptomes of European brook lamprey (Fig. 4, Supplemental Table 1). For all IL-17R family genes, we could confirm the presence of the coding region for the characteristic conserved SEFIR domain (Supplemental Fig. 3) with the exception of two sea lamprey IL-17R sequences (IL-17RE/RC and IL-17RD). We attribute this failure to the incompleteness of the sea lamprey genome sequence. Phylogenetic analysis of the deduced IL-17Rs C-terminal amino acid sequences indicated that they clustered into six groups and exhibited unambiguous one to one orthologous relationships between the three lamprey species (Fig. 4). Lamprey IL-17RA.1, IL-17RA.2 and IL-17RA.3 are phylogenetically close to the mammalian IL-17RA sequences, whereas IL-17RE/RC and IL-17RD cluster with mammalian IL-17RE and IL-17RD respectively (Fig. 4, Supplemental Fig. 3). The top blast BLAST hits in the similarity search against the NCBI protein database also supported these conclusions (not shown).
FIGURE 4.
Phylogenetic comparison of lamprey IL-17Rs with mammalian IL-17Rs. The C-terminal conserved SEFIR domain was used in the analysis. The phylogenetic tree is constructed by the NJ method. Due to incompleteness of sea lamprey genome sequence, the two partial IL-17R sequences (IL-17RE/RC and IL-17RD) were excluded. The numbers indicate bootstrap confidence values obtained for each node after 1000 replications. The accession numbers for sequences used in this analysis were as follows: Mouse IL-17RA, NP 032385; Rat IL-17RA, NP 001101353; Horse IL-17RA, XP 005610881; Opossum IL-17RA, XP 007503896; Human IL-17RA, EAW57738; Mouse IL-17RB, NP 062529; Human IL-17RB, AAF86051; Horse IL-17RB, XP 005600603; Dog IL-17RB, XP 005632474; Opossum IL-17RB, XP 007500672; Mouse IL-17RD, NP 602319; Human IL-17RD, NP 060033; Rabbit IL-17RD, XP 002713305; Cow IL-17RD, NP 001192356; Guinea pig IL-17RD, XP 003480139; Human IL-17RC, NP 703191; Cow IL-17RC, XP 005222441; Dog IL-17RC, XP 005632241; Mouse IL-17RC, NP 849273; Platypus IL-17RC, XP 007657284; Human IL-17RE, NP 705616; Pig IL-17RE, XP 005669819; Mouse IL-17RE, NP 665825; Sheep IL-17RE, XP 004018654; Cow IL-17RE, XP 005907154 ; Sea Lamprey IL-17RA.1, KR059942; Sea Lamprey IL-17RA.2, KR059943; Sea Lamprey IL-17RA.3, KR059944; European lamprey IL-17RA.1, KR059950; European lamprey IL-17RA.2, KR059951; European lamprey IL-17RA.3, KR059952; European lamprey IL-17RF, KR059953; European lamprey IL-17RE/RC, KR059954; and European lamprey IL-17RD, KR059955. The same phylogenetic relationship was demonstrated by the ML method (data not shown).
IL-17R expression
When expression of the IL-17R family members was examined in different tissues by real-time PCR, IL-17RA.3 was found to be expressed in the intestine, blood cells, skin, kidneys and gills (Fig. 5A). IL-17RA.1 was expressed at highest levels in the intestine and blood cells, with weaker expression being detectable in skin, kidneys and gills, whereas IL-17RA.2 expression was most easily detectable in the gills. IL-17RF expression was expressed preferentially in the intestine, IL-17RE/RC in skin and IL-17RD in the kidney. When different leukocyte populations were isolated from blood samples and examined for expression of the different IL-17Rs, IL-17RA.1 was found to be expressed preferentially by VLRB+ lymphocytes, as noted previously (41), and also by monocytes (Fig. 5B). IL-17RA.3 expression was detected principally in the VLRA+ and VLRC+ lymphocytes, but not in the monocytes and granulocytes. IL-17RA.2 expression was detectable at low levels in all three VLR+ lymphocyte populations; IL-17RF and IL-17RD expression was detectable in all blood cell populations examined, albeit at very low levels. Interestingly, IL-17RE/RC was preferentially expressed by the VLRA+ lymphocytes, whereas the level of IL-17RD expression was highest in the VLRC+ lymphocytes (Fig. 5B).
FIGURE 5.
Sea lamprey expression of IL-17R transcripts. (A) Tissue distribution. The relative expression levels of IL-17Rs were determined by real-time PCR in skin, kidneys, intestine, white blood cells (WBC) and gills (n=6 larvae). (B) Expression of sea lamprey IL-17Rs by different WBC types. Relative IL-17R expression levels were determined by real-time PCR analysis of sorted lymphocyte, monocyte and granulocyte populations (n=3–6 larvae). mRNA abundance relative to that of GAPDH: 2−ΔCt, ΔCt = Cttarget gene −CtGAPDH. Error bars indicate SEM. The scales of the y-axis are different on each graph.
Cellular and molecular interactions of IL-17D.1
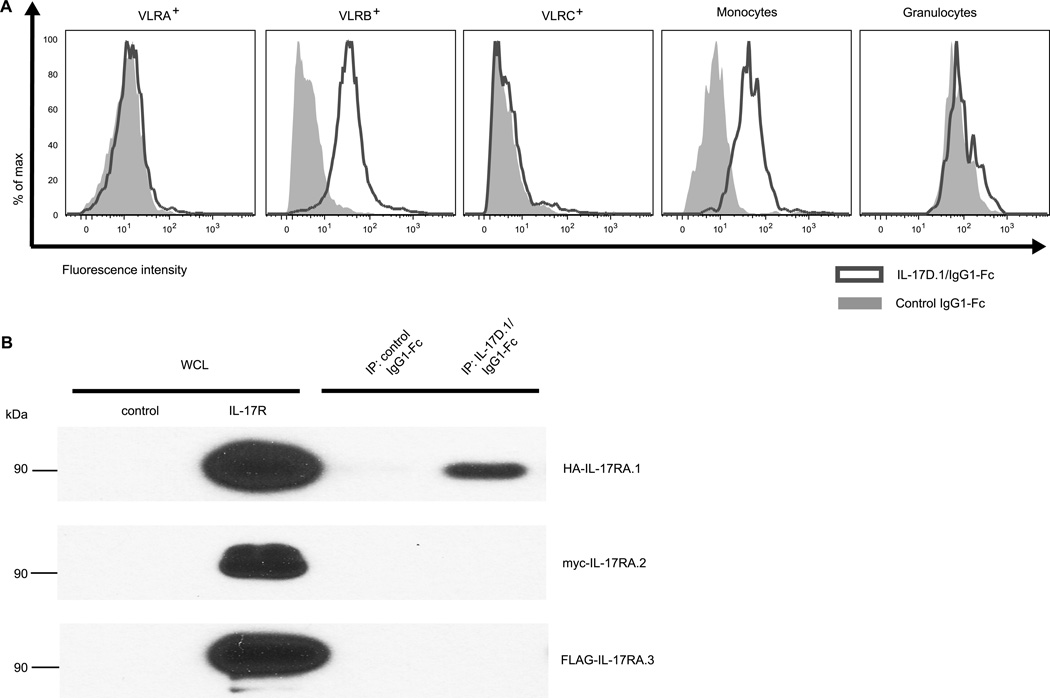
Since we were able to obtain the full-length cDNA of IL-17D.1, we focused on this cytokine and its potential receptor(s) in the following studies. The chimeric IL-17D.1/IgG1-Fc protein that was used to generate anti-IL-17D.1 mAbs was also used to assess IL-17D.1 binding by different types of blood cells (Supplemental Fig. 1A). In these experiments, an unrelated recombinant hagfish protein (VLRA) fused with the human IgG1-Fc was used as a control and goat anti-human IgG antibodies as the secondary detection reagent. In flow cytometric analyses, binding of the chimeric IL-17D.1/IgG1-Fc protein by both VLRB+ lymphocytes and monocytes was observed, while the control IgG1-Fc fusion protein did not bind to any of the white blood cell types (Fig. 6A). Notably, the IL-17D.1/IgG1-Fc protein failed to bind to VLRA+ or VLRC+ lymphocytes and granulocytes, indicating cell-type specificity of expression of the IL-17D.1 receptor.
FIGURE 6.
Binding of IL-17D.1 to recombinant IL-17RA.1 and to the surface of B-like cells and monocytes. (A) Analysis of IL-17D.1/IgG1-Fc binding by different types of white blood cells. Blood cells were incubated with either control IgG1-Fc (grey area) or IL-17D.1/IgG1-Fc (solid line) and stained with a PE-conjugated secondary antibody and co-stained with mAbs specific for VLRA (R110), VLRB (4C4), VLRC (3A5), monocytes (8A1) and granulocytes (2D4). The lymphocyte gate excluding dead cells and doublets was further gated based on the surface staining by these mAbs. From the gated population, IL-17D.1/IgG1-Fc staining was then determined. The fluorescence intensity is shown on a log scale. The selective binding by VLRB+ lymphocytes and monocytes was observed in four independent experiments. (B) Association of recombinant IL-17D.1 and IL-17RA.1 proteins. HEK-293T cells were transfected with control vector, HA-IL-17RA.1, myc-IL-17RA.2 or FLAG-IL-17RA.3 vector. After 60 hours, whole cell lysates (WCL) were immunoprecipitated (IP) with IL-17D.1/IgG1-Fc or control IgG1-Fc proteins followed by protein G beads. Cell lysates and immunoprecipitated proteins were resolved on reducing SDS-PAGE before Western blotting with anti-HA, anti-myc or anti-FLAG antibodies.
The full-length cDNAs of three IL-17R family members in sea lamprey (IL-17RA.1, IL-17RA.2, and IL-17RA.3) were obtained by using RACE and nested PCR. HEK-293T cells were transfected with plasmids expressing IL-17RA.1 (HA-tagged) or IL-17RA.2 (myc-tagged) or IL-17RA.3 (FLAG-tagged) respectively. Flow cytometric analysis of the transfected cells using anti-tag antibodies confirmed that all three IL-17Rs were expressed by the transfectants (data not shown). In order to examine which of these receptors are capable of interacting with IL-17D.1, immunoprecipitation analysis of these recombinant IL-17Rs was carried out. The results indicated that IL-17D.1 associated with IL-17RA.1, but not with either IL-17RA.2 or IL-17RA.3 (Fig. 6B). These results, together with the findings described above suggest the potential for the VLRA+ and VLRC+ lymphocytes to interact with the VLRB+ lymphocytes and monocytes via IL-17D.1 production by the T-like cells and IL-17RA.1 by the B-like cells and monocytes.
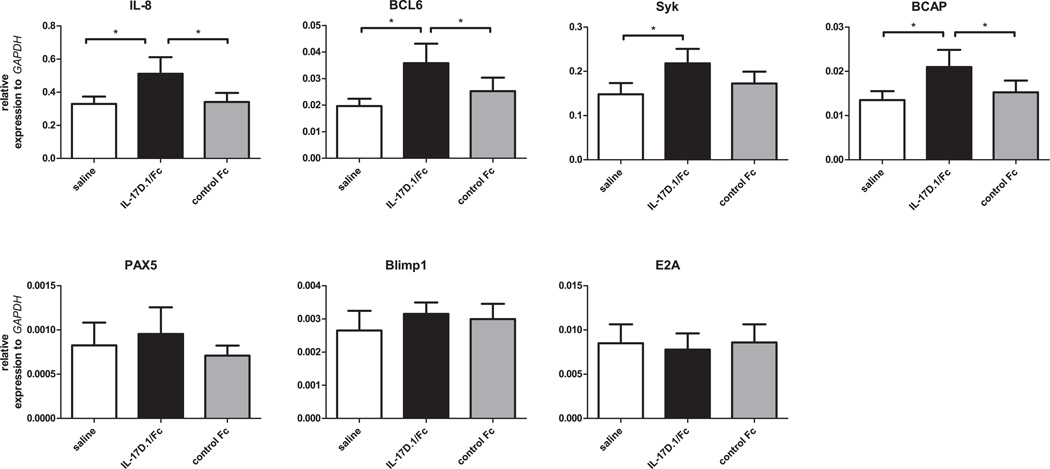
IL-17D.1 treatment effects on blood leukocytes
Since IL-17D.1 was shown to bind to VLRB+ lymphocytes and monocytes by flow cytometry, we examined the ex vivo effect of IL-17D.1 on the transcriptional status of blood leukocytes. In these experiments, blood leukocytes were incubated alone with either IL-17D.1/IgG1-Fc or the control protein (extracellular region of IL-17RA.1/IgG1-Fc). After 16 hours incubation, we examined the expression levels of various candidate genes. We focused in particular on genes that are preferentially expressed by VLRB+ cells, a presumptive target of IL-17D.1’s action: the chemokine IL-8, the transcription factors E2A, PAX5, Blimp1 and BCL6, and Syk and BCAP that are known components of the BCR-mediated signaling pathways in mammals (41, 43, 50). The results of our real-time PCR analysis indicate that IL-17D.1/IgG1-Fc significantly induced the upregulation of IL-8, BCL6 and BCAP gene expression, but had no detectable effect on the expression levels of E2A, PAX5 and Blimp1 (Fig. 7). These studies suggested that recombinant IL-17D.1 is biologically active and can induce gene expression changes in B-like cells that express the cognate receptor.
FIGURE 7.
Induction of gene expression in lamprey blood leukocytes by IL-17D.1. Blood leukocytes were incubated with saline, 2µg/ml IL-17D.1/IgG1-Fc, or 2µg/ml control Fc for 16 hours. The expression levels of various genes were examined by real-time PCR after incubation (n=5 larvae). mRNA abundance relative to that of GAPDH: 2−ΔCt, ΔCt = Cttarget gene −CtGAPDH. Error bars indicate SEM. The scales of the y-axis are different on each graph.
Discussion
In our analysis of the sea lamprey IL-17 ligand and receptor gene families, we have identified five members of the IL-17 family of ligands (IL-17D.2 through IL-17C) and six members of the IL-17 receptor family (IL-17RA.1 through IL-17RD). All five lamprey IL-17s encode the conserved C-terminal region with four cysteines that likely form the characteristic cystine-knot noted in human IL-17F; other cysteine residues in lamprey IL-17s may participate in homodimer or heterodimer formation via interchain disulfide bonds. It is noteworthy that, by examining genomic and transcriptome datasets of three lamprey species, we were unable to identify lamprey orthologues of mammalian IL-17A and IL-17F.
IL-17D.1 (mammalian IL-17D like) was found to be highly expressed at both mRNA and protein levels in skin, intestine and gills of sea lamprey larvae, thereby resembling the tissue distribution observed for IL-17D expression in teleost fish. IL-17D.1 expression was localized to epithelial cells in these barrier tissues, an expression pattern also noted for some of the IL-17 family members in mammals. Stimulation with heat killed E. coli and agonists to TLR2 or TLR5 induced the production of IL-17C from human colon epithelial cells (34). Similar to IL-17C, IL-17E is expressed in epithelial cells in response to parasites, viruses, fungi, and protease allergens (51).
Furthermore, colonic epithelial cells express IL-17E in conventionally-reared mice, but not in germ-free mice (52). The expression of IL-17 cytokines by epithelial cells is thought to be regulated through pattern recognition receptor (PRR) sensing of pathogens or commensal microbiota (53). TLR expression in gills, intestine, kidney and other lamprey tissues (54) potentially could affect the basal production of IL-17D.1 to control infections and promote the VLR-mediated adaptive immune responses. The sensitivity of RNA in situ hybridization proved to be insufficient to detect the low levels of IL-17D.1 expression in the lamprey blood cells. However, by real-time PCR, the T-like VLRA+ and VLRC+ cells were shown to express IL-17D.1. In mammals, Th17 cells, γδ T cells and innate lymphoid cells have all been shown to produce proinflammatory IL-17 molecules (55, 56).
With respect to IL-17Rs, we note that all receptors exhibit the conserved intracellular domain that is an important functional domain for signal transduction from these receptors. This domain is also found in Act1, which serves an important role in IL-17R mediated signaling (57); Act1 is recruited to IL-17R through SEFIR-SEFIR interaction (58) and in turn it recruits and ubiquitinates TRAF6 to mediate downstream activation of NF-κB (33, 59). The identification of the lamprey orthologues of mammalian IL-17s, IL-17Rs, Act1, TRAF6 and components of NF-κB signaling (C-Rel; NF-κB p105; IκB-α) suggests that the IL-17 signaling pathway is evolutionarily conserved (60–63). Three of the lamprey IL-17R orthologues, IL-17RA.1, IL-17RA.2 and IL-17RA.3, are most closely related to mammalian IL-17RA. The sequences of IL-17RE/RC and IL-17RD cluster with mammalian IL-17RE and IL-17RD sequences, respectively. The existence of IL-17RA and IL-17RD orthologues in amphioxus, lampreys, cartilaginous fish and teleost supports the idea that amphioxus IL-17RA and IL-17RD are the ancestors of vertebrate IL-17RA and IL-17RD respectively (38). All of these IL-17R family members have also been identified in the shark Callorhinchus milii, a cartilaginous fish representative (38).
IL-17RA is preferentially expressed in hematopoietic tissues of mammals (29, 64). Our transcriptional analysis indicated that IL-17RA.3 is most highly expressed in skin, kidney, intestine, white blood cells and gills. Lamprey IL-17RA.1 is expressed predominantly in the intestine and white blood cells, while IL-17RA.2 expression is highest in the gills of lamprey larvae. The expression of IL-17RF, IL-17RE/RC and IL-17RD could be detected in intestine, skin and kidneys respectively, suggesting that they may have differential roles in different tissues. Among the white blood cell types, the VLRB+ B-like lymphocytes and monocytes expressed IL-17RA.1 preferentially, whereas VLRA+ and VLRC+ T-like lymphocytes expressed IL-17RA.3 preferentially.
When recombinant IL-17D.1/IgG1-Fc fusion protein was used to determine which of the IL-17Rs produced by transfected cells could bind IL-17D.1, we observed the association of IL-17D.1 with IL-17RA.1, but not with IL-17RA.2 or IL-17RA.3. In this context, it is interesting to note that lampreys possess three representatives of IL-17Rs (IL-17RA.1, IL-17RA.2 and IL-17RA.3) that are closely related to mammalian IL-17RA, which serves as a common receptor subunit and participates in the recognition of several IL-17 family cytokines. Our data thus suggest that, despite these similarities, functional differences may exist at least between IL-17RA.1 and IL-17RA.2 or IL-17RA.3.
By using the chimeric IL-17D.1/IgG1-Fc and unrelated chimeric proteins controls, we also found that IL-17D.1 binds to VLRB+ lymphocytes and monocytes, but not to VLRA+ lymphocytes, VLRC+ lymphocytes and granulocytes. Transcriptional analysis indicated that VLRB+ B-like lymphocytes and monocytes preferentially express IL-17RA.1. Collectively, these findings indicate that VLRB+ lymphocytes and monocytes express the complementary receptor for IL-17D.1 and, by analogy with findings in mammals, imply that the T-like VLRA+ and VLRC+ cells may produce IL-17D.1 to regulate B-like VLRB+ cell and monocyte function in lampreys. Regulation of B cell function by IL-17s is not without precendent in the mammalian system. In the BXD2 mouse model of autoimmunity, IL-17A upregulates the B cell expression of regulator of G-protein signaling 13 and 16 (Rgs13 and Rgs16) to reduce the chemotactic response and to promote the formation of autoreactive germinal centers (65). In the K/BxN mouse model of inflammatory arthritis, IL-17A is required for efficient germinal center formation via a direct effect on B cells (66). In addition, IL-17A promotes immunoglobulin isotype switching in mice (67), and induces monocyte adhesion and differentiation of an anti-inflammatory macrophages subset (68, 69). IL-17A also facilitates monocyte migration through its interaction with IL-17RA and IL-17RC on monocytes in humans (70) .
IL-17D.1/IgG1-Fc treatment of lamprey blood leukocytes increased the expression levels of IL-8, BCL6 and BCAP, three genes that are preferentially expressed by VLRB+ cells. Induction of IL-8 in fibroblasts, endothelial cells, epithelial cells and synoviocytes by IL-17 has been reported in numerous studies (55, 71–74). BCAP has been shown to serve as an adaptor molecule linking CD19 and BCR-associated kinases to the PI3K pathway in B cells (75, 76), and as a TIR-domain-containing adaptor in the TLR signaling pathway to regulate inflammatory responses (77). The increase of BCAP expression levels by IL-17D.1 suggests that lamprey IL-17D.1 may modulate B-like cell signaling and TLR signaling through its effect on BCAP expression. In grass carp, recombinant IL-17D promotes IL-1β, TNF-α and IL-8 expression in head kidney cells (20) and in rainbow trout, IL-17A/F2 stimulates splenocyte expression of IL-6, IL-8 and anti-microbial peptide BD-3 (16). Future studies will be required to elucidate the functional consequences of the IL-17D.1 and IL-17RA.1 interactions between innate and adaptive immune system cells in lampreys.
In conclusion, we have identified five IL-17 genes and six IL-17R genes in sea lampreys. IL-17D.1 is mainly expressed in the epithelium of skin, gills and intestine, while IL-17R genes are ubiquitously expressed. T-like VLRA+ and VLRC+ lymphocytes in lampreys preferentially produce IL-17D.1, the most prominently expressed IL-17 family member. Conversely, B-like VLRB+ lymphocytes and monocytes express IL-17RA.1, which can bind IL-17D.1. Our collective findings suggest that functional interactions between the lamprey T-like cells and other cells of the adaptive and innate immune systems are facilitated by this highly conserved cytokine and cytokine receptor interaction.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Health Grants AI072435 and GM100151 and the Georgia Research Alliance, by the Max Planck Society and the European Research Council under the European Union's Seventh Framework Programme (FP7/2007–2013), ERC grant agreement n° ERC-2012-AdG – 323126.
We thank Jianxu Li, Brant Herrin and Cuiling Yu for helpful suggestions and discussion; Sommer A. Durham and Robert E. Karaffa, II for help with cell sorting; Ulrike Bönisch, Yu-Wen Chung-Davidson and Weiming Li for sequencing; Sarah Diehl, Miho Sera and Tadashi Imanishi for data processing; Xiaoyan Sun for statistical advice.
Abbreviations used in this article
- SEFIR
similar expression to fibroblast growth factor genes and IL-17R
- VLRs
variable lymphocyte receptors
- NJ
neighbor-joining
- ML
maximum likelihood
- IgG1-Fc
Fc region of human IgG1
- PEI
polyethylenimine
- FSC
forward scatter
- SSC
side scatter
- RACE
rapid amplification of cDNA ends
- ORFs
open reading frames
References
- 1.Roberts S, Gueguen Y, de Lorgeril J, Goetz F. Rapid accumulation of an interleukin 17 homolog transcript in Crassostrea gigas hemocytes following bacterial exposure. Dev. & Comp. Immunol. 2008;32:1099–1104. doi: 10.1016/j.dci.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Wu S-Z, Huang X-D, Li Q, He M-X. Interleukin-17 in pearl oyster (Pinctada fucata): molecular cloning and functional characterization. Fish Shellfish Immunol. 2013;34:1050–1056. doi: 10.1016/j.fsi.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Hibino T, Loza-Coll M, Messier C, Majeske AJ, Cohen AH, Terwilliger DP, Buckley KM, Brockton V, Nair SV, Berney K, Fugmann SD, Anderson MK, Pancer Z, Cameron RA, Smith LC, Rast JP. The immune gene repertoire encoded in the purple sea urchin genome. Dev. Biol. 2006;300:349–365. doi: 10.1016/j.ydbio.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 4.Vizzini A, Di Falco F, Parrinello D, Sanfratello MA, Mazzarella C, Parrinello N, Cammarata M. Ciona intestinalis interleukin 17-like genes expression is upregulated by LPS challenge. Dev. Comp. Immunol. 2014;48:129–137. doi: 10.1016/j.dci.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 5.Huang S, Yuan S, Guo L, Yu YY, Li J, Wu T, Liu T, Yang M, Wu K, Liu H, Ge J, Yu YY, Huang H, Dong M, Yu C, Chen S, Xu A. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 2008;18:1112–1126. doi: 10.1101/gr.069674.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu. Rev. Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 7.Li H, Chen J, Huang A, Stinson J, Heldens S, Foster J, Dowd P, Gurney AL, Wood WI. Cloning and characterization of IL-17B and IL-17C, two new members of the IL-17 cytokine family. Proc. Natl. Acad. Sci. 2000;97:773–778. doi: 10.1073/pnas.97.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Starnes T, Broxmeyer HE, Robertson MJ, Hromas R. Cutting Edge: IL-17D, a Novel Member of the IL-17 Family, Stimulates Cytokine Production and Inhibits Hemopoiesis. J. Immunol. 2002;169:642–646. doi: 10.4049/jimmunol.169.2.642. [DOI] [PubMed] [Google Scholar]
- 9.O’Sullivan T, Saddawi-Konefka R, Gross E, Tran M, Mayfield SP, Ikeda H, Bui JD. Interleukin-17D mediates tumor rejection through recruitment of natural killer cells. Cell Rep. 2014;7:989–998. doi: 10.1016/j.celrep.2014.03.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fort MM, Cheung J, Yen D, Li J, Zurawski SM, Lo S, Menon S, Clifford T, Hunte B, Lesley R, Muchamuel T, Hurst SD, Zurawski G, Leach MW, Gorman DM, Rennick DM. IL-25 Induces IL-4, IL-5, and IL-13 and Th2-Associated Pathologies In Vivo. 2001;15:985–995. doi: 10.1016/s1074-7613(01)00243-6. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y-H, Angkasekwinai P, Lu N, Voo KS, Arima K, Hanabuchi S, Hippe A, Corrigan CJ, Dong C, Homey B, Yao Z, Ying S, Huston DP, Liu Y-J. IL-25 augments type 2 immune responses by enhancing the expansion and functions of TSLP-DC-activated Th2 memory cells. J. Exp. Med. 2007;204:1837–1847. doi: 10.1084/jem.20070406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hymowitz SG, Filvaroff EH, Yin J, Lee J, Cai L, Risser P, Maruoka M, Mao W, Foster J, Kelley RF, Pan G, Gurney AL, de Vos AM, Starovasnik MA. IL-17s adopt a cystine knot fold: structure and activity of a novel cytokine, IL-17F, and implications for receptor binding. EMBO J. 2001;20:5332–5341. doi: 10.1093/emboj/20.19.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S, Song X, Chrunyk BA, Shanker S, Hoth LR, Marr ES, Griffor MC. Crystal structures of interleukin 17A and its complex with IL-17 receptor A. Nat. Commun. 2013;4:1888. doi: 10.1038/ncomms2880. [DOI] [PubMed] [Google Scholar]
- 14.McDonald NQ, Lapatto R, Murray-Rust J, Gunning J, Wlodawer A, Blundell TL. New protein fold revealed by a 2.3-A resolution crystal structure of nerve growth factor. Nature. 1991;354:411–414. doi: 10.1038/354411a0. [DOI] [PubMed] [Google Scholar]
- 15.Gunimaladevi I, Savan R, Sakai M. Identification, cloning and characterization of interleukin-17 and its family from zebrafish. Fish Shellfish Immunol. 2006;21:393–403. doi: 10.1016/j.fsi.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Monte MM, Wang T, Holland JW, Zou J, Secombes CJ. Cloning and characterization of rainbow trout interleukin-17A/F2 (IL-17A/F2) and IL-17 receptor A: expression during infection and bioactivity of recombinant IL-17A/F2. Infect. Immun. 2013;81:340–353. doi: 10.1128/IAI.00599-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korenaga H, Kono T, Sakai M. Isolation of seven IL-17 family genes from the Japanese pufferfish Takifugu rubripes. Fish Shellfish Immunol. 2010;28:809–818. doi: 10.1016/j.fsi.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 18.Wang T, Martin Sa M, Secombes CJ. Two interleukin-17C-like genes exist in rainbow trout Oncorhynchus mykiss that are differentially expressed and modulated. Dev. Comp. Immunol. 2010;34:491–500. doi: 10.1016/j.dci.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Kumari J, Larsen AN, Bogwald J, Dalmo RA. Interleukin-17D in Atlantic salmon (Salmo salar): molecular characterization, 3D modelling and promoter analysis. Fish Shellfish Immunol. 2009;27:647–659. doi: 10.1016/j.fsi.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 20.Du L, Qin L, Wang X, Zhang A, Wei H, Zhou H. Characterization of grass carp (Ctenopharyngodon idella) IL-17D: molecular cloning, functional implication and signal transduction. Dev. Comp. Immunol. 2014;42:220–228. doi: 10.1016/j.dci.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Li C, Thongda W, Luo Y, Beck B, Peatman E. Characterization and mucosal responses of interleukin 17 family ligand and receptor genes in channel catfish Ictalurus punctatus. Fish Shellfish Immunol. 2014;38:47–55. doi: 10.1016/j.fsi.2014.02.020. [DOI] [PubMed] [Google Scholar]
- 22.Tsutsui S, Nakamura O, Watanabe T. Lamprey (Lethenteron japonicum) IL-17 upregulated by LPS-stimulation in the skin cells. Immunogenetics. 2007;59:873–882. doi: 10.1007/s00251-007-0254-2. [DOI] [PubMed] [Google Scholar]
- 23.Kono T, Korenaga H, Sakai M. Genomics of fish IL-17 ligand and receptors: a review. Fish Shellfish Immunol. 2011;31:635–643. doi: 10.1016/j.fsi.2010.11.028. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Zhang Y, Zhang Y, Xiang Z, Tong Y, Qu F, Yu Z. Genomic characterization and expression analysis of five novel IL-17 genes in the Pacific oyster, Crassostrea gigas. Fish Shellfish Immunol. 2014;40:455–465. doi: 10.1016/j.fsi.2014.07.026. [DOI] [PubMed] [Google Scholar]
- 25.Valenzuela-Muñoz V, Gallardo-Escárate C. Molecular cloning and expression of IRAK-4, IL-17 and I-κB genes in Haliotis rufescens challenged with Vibrio anguillarum. Fish Shellfish Immunol. 2014;36:503–509. doi: 10.1016/j.fsi.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 26.Kuestner RE, Taft DW, Haran A, Brandt CS, Brender T, Lum K, Harder B, Okada S, Ostrander CD, Kreindler JL, Aujla SJ, Reardon B, Moore M, Shea P, Schreckhise R, Bukowski TR, Presnell S, Guerra-Lewis P, Parrish-Novak J, Ellsworth JL, Jaspers S, Lewis KE, Appleby M, Kolls JK, Rixon M, West JW, Gao Z, Levin SD. Identification of the IL-17 Receptor Related Molecule IL-17RC as the Receptor for IL-17F. J. Immunol. 2007;179:5462–5473. doi: 10.4049/jimmunol.179.8.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toy D, Kugler D, Wolfson M, Van den Bos T, Gurgel J, Derry J, Tocker J, Peschon J. Cutting Edge: Interleukin 17 Signals through a Heteromeric Receptor Complex. J. Immunol. 2006;177:36–39. doi: 10.4049/jimmunol.177.1.36. [DOI] [PubMed] [Google Scholar]
- 28.Wright JF, Bennett F, Li B, Brooks J, Luxenberg DP, Whitters MJ, Tomkinson KN, Fitz LJ, Wolfman NM, Collins M, Dunussi-Joannopoulos K, Chatterjee-Kishore M, Carreno BM. The Human IL-17F/IL-17A Heterodimeric Cytokine Signals through the IL-17RA/IL-17RC Receptor Complex. J. Immunol. 2008;181:2799–2805. doi: 10.4049/jimmunol.181.4.2799. [DOI] [PubMed] [Google Scholar]
- 29.Yao Z, Fanslow WC, Seldin MF, Rousseau A-M, Painter SL, Comeau MR, Cohen JI, Spriggs MK. Herpesvirus Saimiri encodes a new cytokine, IL-17, which binds to a novel cytokine receptor. Immunity. 1995;3:811–821. doi: 10.1016/1074-7613(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 30.Ye P, Rodriguez FH, Kanaly S, Stocking KL, Schurr J, Schwarzenberger P, Oliver P, Huang W, Zhang P, Zhang J, Shellito JE, Bagby GJ, Nelson S, Charrier K, Peschon JJ, Kolls JK. Requirement of Interleukin 17 Receptor Signaling for Lung Cxc Chemokine and Granulocyte Colony-Stimulating Factor Expression, Neutrophil Recruitment, and Host Defense. J. Exp. Med. 2001;194:519–528. doi: 10.1084/jem.194.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang W, Na L, Fidel PL, Schwarzenberger P. Requirement of Interleukin-17A for Systemic Anti-Candida albicans Host Defense in Mice. J. Infect. Dis. 2004;190:624–631. doi: 10.1086/422329. [DOI] [PubMed] [Google Scholar]
- 32.Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, Ho AW, Hai JH, Yu JJ, Jung JW, Filler SG, Masso-Welch P, Edgerton M, Gaffen SL. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J. Exp. Med. 2009;206:299–311. doi: 10.1084/jem.20081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaffen SL. Structure and signalling in the IL-17 receptor family. Nat Rev Immunol. 2009;9:556–567. doi: 10.1038/nri2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramirez-Carrozzi V, Sambandam A, Luis E, Lin Z, Jeet S, Lesch J, Hackney J, Kim J, Zhou M, Lai J, Modrusan Z, Sai T, Lee W, Xu M, Caplazi P, Diehl L, de Voss J, Balazs M, Gonzalez L, Singh H, Ouyang W, Pappu R. IL-17C regulates the innate immune function of epithelial cells in an autocrine manner. Nat. Immunol. 2011;12:1159–1166. doi: 10.1038/ni.2156. [DOI] [PubMed] [Google Scholar]
- 35.Chang SH, Reynolds JM, Pappu BP, Chen G, Martinez GJ, Dong C. Interleukin-17C Promotes Th17 Cell Responses and Autoimmune Disease via Interleukin-17 Receptor E. Immunity. 2011;35:611–621. doi: 10.1016/j.immuni.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song X, Zhu S, Shi P, Liu Y, Shi Y, Levin SD, Qian Y. IL-17RE is the functional receptor for IL-17C and mediates mucosal immunity to infection with intestinal pathogens. Nat. Immunol. 2011;12:1151–1158. doi: 10.1038/ni.2155. [DOI] [PubMed] [Google Scholar]
- 37.Terajima D, Yamada S, Uchino R, Ikawa S, Ikeda M, Shida K, Arai Y, Wang H-G, Satoh N, Satake M. Identification and Sequence of Seventy-nine New Transcripts Expressed in Hemocytes of Ciona intestinalis, Three of Which May Be Involved in Characteristic Cell-cell Communication. DNA Res. 2003;10:203–212. doi: 10.1093/dnares/10.5.203. [DOI] [PubMed] [Google Scholar]
- 38.Wu B, Jin M, Zhang Y, Wei T, Bai Z. Evolution of the IL17 receptor family in chordates: a new subfamily IL17REL. Immunogenetics. 2011;63:835–845. doi: 10.1007/s00251-011-0554-4. [DOI] [PubMed] [Google Scholar]
- 39.Tsang M, Friesel R, Kudoh T, Dawid IB. Identification of Sef, a novel modulator of FGF signalling. Nat. Cell Biol. 2002;4:165–169. doi: 10.1038/ncb749. [DOI] [PubMed] [Google Scholar]
- 40.Alder MN, Herrin BR, Sadlonova A, Stockard CR, Grizzle WE, Gartland LA, Gartland GL, Boydston JA, Turnbough CL, Cooper MD. Antibody responses of variable lymphocyte receptors in the lamprey. Nat. Immunol. 2008;9:319–327. doi: 10.1038/ni1562. [DOI] [PubMed] [Google Scholar]
- 41.Guo P, Hirano M, Herrin BR, Li J, Yu C, Sadlonova A, Cooper MD. Dual nature of the adaptive immune system in lampreys. Nature. 2009;459:796–801. doi: 10.1038/nature08068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kasamatsu J, Sutoh Y, Fugo K, Otsuka N, Iwabuchi K, Kasahara M. Identification of a third variable lymphocyte receptor in the lamprey. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14304–14308. doi: 10.1073/pnas.1001910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirano M, Guo P, McCurley N, Schorpp M, Das S, Boehm T, Cooper MD. Evolutionary implications of a third lymphocyte lineage in lampreys. Nature. 2013;501:435–438. doi: 10.1038/nature12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herrin BR, Hirano M, Li J, Das S, Sutoh Y, Cooper MD. Molecular Biology of B Cells. Elsevier; 2015. [Google Scholar]
- 45.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larkin Ma, Blackshields G, Brown NP, Chenna R, McGettigan Pa, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 47.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 49.Bajoghli B, Guo P, Aghaallaei N, Hirano M, Strohmeier C, McCurley N, Bockman DE, Schorpp M, Cooper MD, Boehm T. A thymus candidate in lampreys. Nature. 2011;470:90–94. doi: 10.1038/nature09655. [DOI] [PubMed] [Google Scholar]
- 50.Najakshin AM, Mechetina LV, Alabyev BY, Taranin AV. Identification of an IL-8 homolog in lamprey (Lampetra fluviatilis): early evolutionary divergence of chemokines. Eur. J. Immunol. 1999;29:375–382. doi: 10.1002/(SICI)1521-4141(199902)29:02<375::AID-IMMU375>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 51.Pappu R, Rutz S, Ouyang W. Regulation of epithelial immunity by IL-17 family cytokines. Trends Immunol. 2012;33:343–349. doi: 10.1016/j.it.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 52.Zaph C, Du Y, Saenz SA, Nair MG, Perrigoue JG, Taylor BC, Troy AE, Kobuley DE, Kastelein RA, Cua DJ, Yu Y, Artis D. Commensal-dependent expression of IL-25 regulates the IL-23-IL-17 axis in the intestine. J. Exp. Med. 2008;205:2191–2198. doi: 10.1084/jem.20080720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wells JM, Rossi O, Meijerink M, van Baarlen P. Epithelial crosstalk at the microbiota-mucosal interface. Proc. Natl. Acad. Sci. U. S. A. 2011;(108 Suppl):4607–4614. doi: 10.1073/pnas.1000092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kasamatsu J, Oshiumi H, Matsumoto M, Kasahara M, Seya T. Phylogenetic and expression analysis of lamprey toll-like receptors. Dev. Comp. Immunol. 2010;34:855–865. doi: 10.1016/j.dci.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 55.Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu. Rev. Immunol. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 56.Cua DJ, Tato CM. Innate IL-17-producing cells: the sentinels of the immune system. Nat. Rev. Immunol. 2010;10:479–489. doi: 10.1038/nri2800. [DOI] [PubMed] [Google Scholar]
- 57.Novatchkova M, Leibbrandt A, Werzowa J, Neubüser A, Eisenhaber F. The STIR-domain superfamily in signal transduction, development and immunity. Trends Biochem. Sci. 2003;28:226–229. doi: 10.1016/S0968-0004(03)00067-7. [DOI] [PubMed] [Google Scholar]
- 58.Qian Y, Liu C, Hartupee J, Altuntas CZ, Gulen MF, Jane-Wit D, Xiao J, Lu Y, Giltiay N, Liu J, Kordula T, Zhang Q-W, Vallance B, Swaidani S, Aronica M, Tuohy VK, Hamilton T, Li X. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat. Immunol. 2007;8:247–256. doi: 10.1038/ni1439. [DOI] [PubMed] [Google Scholar]
- 59.Liu C, Qian W, Qian Y, Giltiay NV, Lu Y, Swaidani S, Misra S, Deng L, Chen ZJ, Li X. Act1, a U-box E3 ubiquitin ligase for IL-17 signaling. Sci. Signal. 2009;2 doi: 10.1126/scisignal.2000382. ra63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu B, Jin M, Gong J, Du X, Bai Z. Dynamic evolution of CIKS (TRAF3IP2/Act1) in metazoans. Dev. Comp. Immunol. 2011;35:1186–1192. doi: 10.1016/j.dci.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 61.Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, Campbell MS, Yandell MD, Manousaki T, Meyer A, Bloom OE, Morgan JR, Buxbaum JD, Sachidanandam R, Sims C, Garruss AS, Cook M, Krumlauf R, Wiedemann LM, Sower Sa, Decatur WA, Hall JA, Amemiya CT, Saha NR, Buckley KM, Rast JP, Das S, Hirano M, McCurley N, Guo P, Rohner N, Tabin CJ, Piccinelli P, Elgar G, Ruffier M, Aken BL, Searle SMJ, Muffato M, Pignatelli M, Herrero J, Jones M, Brown CT, Chung-Davidson Y-W, Nanlohy KG, Libants SV, Yeh C-Y, McCauley DW, Langeland JA, Pancer Z, Fritzsch B, de Jong PJ, Zhu B, Fulton LL, Theising B, Flicek P, Bronner ME, Warren WC, Clifton SW, Wilson RK, Li W. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat. Genet. 2013;45:415–421. doi: 10.1038/ng.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pancer Z, Mayer WE, Klein J, Cooper MD. Prototypic T cell receptor and CD4-like coreceptor are expressed by lymphocytes in the agnathan sea lamprey. Proc. Natl. Acad. Sci. 2004;101:13273–13278. doi: 10.1073/pnas.0405529101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yermolenko ZI. Transcriptome Analysis of Sea Lamprey Embryogenesis Transcriptome Analysis of Sea Lamprey Embryogenesis. Set. Hall Univ. Diss. Theses. 2014 [Google Scholar]
- 64.Ishigame H, Kakuta S, Nagai T, Kadoki M, Nambu A, Komiyama Y, Fujikado N, Tanahashi Y, Akitsu A, Kotaki H, Sudo K, Nakae S, Sasakawa C, Iwakura Y. Differential roles of interleukin-17A and −17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity. 2009;30:108–119. doi: 10.1016/j.immuni.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 65.Hsu H-C, Yang P, Wang J, Wu Q, Myers R, Chen J, Yi J, Guentert T, Tousson A, Stanus AL, Le TL, Lorenz RG, Xu H, Kolls JK, Carter RH, Chaplin DD, Williams RW, Mountz JD. Interleukin 17-producing T helper cells and interleukin 17 orchestrate autoreactive germinal center development in autoimmune BXD2 mice. Nat. Immunol. 2008;9:166–175. doi: 10.1038/ni1552. [DOI] [PubMed] [Google Scholar]
- 66.Wu H-J, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mitsdoerffer M, Lee Y, Jäger A, Kim H-J, Korn T, Kolls JK, Cantor H, Bettelli E, Kuchroo VK. Proinflammatory T helper type 17 cells are effective B-cell helpers. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14292–14297. doi: 10.1073/pnas.1009234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Erbel C, Akhavanpoor M, Okuyucu D, Wangler S, Dietz A, Zhao L, Stellos K, Little KM, Lasitschka F, Doesch A, Hakimi M, Dengler TJ, Giese T, Blessing E, Katus HA, Gleissner CA. IL-17A influences essential functions of the monocyte/macrophage lineage and is involved in advanced murine and human atherosclerosis. J. Immunol. 2014;193:4344–4355. doi: 10.4049/jimmunol.1400181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zizzo G, Cohen PL. IL-17 stimulates differentiation of human anti-inflammatory macrophages and phagocytosis of apoptotic neutrophils in response to IL-10 and glucocorticoids. J. Immunol. 2013;190:5237–5246. doi: 10.4049/jimmunol.1203017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shahrara S, Pickens SR, Dorfleutner A, Pope RM. IL-17 induces monocyte migration in rheumatoid arthritis. J. Immunol. 2009;182:3884–3891. doi: 10.4049/jimmunol.0802246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gaffen SL. Recent advances in the IL-17 cytokine family. Curr. Opin. Immunol. 2011;23:613–619. doi: 10.1016/j.coi.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hirata T, Osuga Y, Hamasaki K, Yoshino O, Ito M, Hasegawa A, Takemura Y, Hirota Y, Nose E, Morimoto C, Harada M, Koga K, Tajima T, Saito S, Yano T, Taketani Y. Interleukin (IL)-17A stimulates IL-8 secretion, cyclooxygensase-2 expression, and cell proliferation of endometriotic stromal cells. Endocrinology. 2008;149:1260–1267. doi: 10.1210/en.2007-0749. [DOI] [PubMed] [Google Scholar]
- 73.Hwang S-Y, Kim J-Y, Kim K-W, Park M-K, Moon Y, Kim W-U, Kim H-Y. IL-17 induces production of IL-6 and IL-8 in rheumatoid arthritis synovial fibroblasts via NF-kappaB- and PI3-kinase/Akt-dependent pathways. Arthritis Res. Ther. 2004;6:R120–R128. doi: 10.1186/ar1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iwakura Y, Ishigame H, Saijo S, Nakae S. Functional specialization of interleukin-17 family members. Immunity. 2011;34:149–162. doi: 10.1016/j.immuni.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 75.Okada T, Maeda A, Iwamatsu A, Gotoh K, Kurosaki T. BCAP: the tyrosine kinase substrate that connects B cell receptor to phosphoinositide 3-kinase activation. Immunity. 2000;13:817–827. doi: 10.1016/s1074-7613(00)00079-0. [DOI] [PubMed] [Google Scholar]
- 76.Inabe K. Tyrosine phosphorylation of B-cell adaptor for phosphoinositide 3-kinase is required for Akt activation in response to CD19 engagement. Blood. 2002;99:584–589. doi: 10.1182/blood.v99.2.584. [DOI] [PubMed] [Google Scholar]
- 77.Troutman TD, Hu W, Fulenchek S, Yamazaki T, Kurosaki T, Bazan JF, Pasare C. Role for B-cell adapter for PI3K (BCAP) as a signaling adapter linking Toll-like receptors (TLRs) to serine/threonine kinases PI3K/Akt. Proc. Natl. Acad. Sci. U. S. A. 2012;109:273–278. doi: 10.1073/pnas.1118579109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.