Abstract
Background
With the increasing use of cell therapies involving immune modulatory cells, there is a need for a simple standardized method to evaluate and compare the suppressive potency of different cell products. We used the Karpas 299 (K299) cell line as the reference suppressor cell to develop a standardized suppression assay to quantitate immune-modulatory capacity of bone marrow derived mesenchymal stromal cells (BM-MSC).
Methods
Healthy donor CD4 T cells were co-cultured with the K299 cell line or with third party BM-MSC. After stimulating with anti-CD3/CD28 beads, CD154 activation and proliferation of CD4 T cells were measured to calculate suppression.
Results
The K299 cell line reproducibly suppressed both the activation and proliferation of healthy donor CD4 T cells in a dose-dependent manner. A rapid (16h) assay based on activation-suppression was selected for development. In replicate testing, there was an inherent variability of suppression of 11% coefficient of variation (CV) between different responder T cells. Suppression by BM-MSC on different responders correlated with suppression by K299. We therefore used the K299 suppression as the reference to define suppression potency of BM-MSC in K299 Suppression Units (KSU). We found that inter-donor variability, passage number, method of manufacture, and exposure of BM-MSC to steroids or interferon gamma all affected BM-MSC potency of suppression.
Conclusion
This method provides a platform for standardizing suppressor function to facilitate comparison between laboratories and for use as a cell product release assay.
Keywords: Immunosuppression, suppression potency assay, mesenchymal stromal cells, K299
INTRODUCTION
Immunologic tolerance is a critical homeostatic function to protect self from auto-immunity1–3. Various immune-regulatory cells, including regulatory T cells (Tregs)4, myeloid derived suppressor cells (MDSC)5, regulatory B cells (Bregs)6, tolerogenic dendritic cells, and mesenchymal stromal cells (MSC)7 are responsible for orchestrating this tolerance8. Immune-regulatory cells play a central role in the pathophysiology of different diseases. For example, recent evidence suggests cancer cells can hijack immune tolerance and impair cancer immunity through recruitment of MDSC and Tregs, creating an immunosuppressive cancer microenvironment9. Conversely, autoimmune diseases and graft versus host disease (GVHD) can originate from the qualitative and quantitative deficiencies of immune-regulatory cells such as Tregs10 and Bregs11. Most recently, adoptive cellular therapies with MSC12,13 and Tregs14,15 have been used in clinical trials to prevent or treat GVHD with promising outcomes.
Immune-regulatory cells have classically been characterized by their in vitro ability to suppress lymphocyte proliferation in a mixed lymphocyte reaction (MLR). However, because of inherent variability in the MLR, this approach does not allow quantitative comparisons from one experiment to the next or between laboratories. Also, peripheral blood mononuclear cells (PBMCs) instead of purified T-cells are often used as responder cells in MLR which can produce additional unpredictable variability. Furthermore, the conventional proliferation suppression assay16 requires up to 96 hours allowing for significant proliferation of the responder cells.17 In the setting of adoptive cellular therapy, a rapid potency assay is needed to release functionally validated cell products in a timely fashion.18 In addition, a standardized quantitative assay to permit objective comparisons of suppression among cell products and laboratories is essential. Rapid suppression assays (7–16 hours) measuring T cell activation markers CD15419 and CD69 on CD4 T cells have been developed for assessing Treg function.20,21 We sought to develop a rapid and easily applicable assay using a widely available suppressor cell line to serve as a standard suppressor cell for inter-laboratory comparisons. Karpas 299 (K299) is a human cell line derived from high grade non-Hodgkin lymphoma, expresses CD4, CD25, and FoxP3. This unique cell line is both phenotypically and functionally similar to Tregs, as it has been shown to suppresses T cell proliferation similar to Tregs.22,23 For these reasons, we selected this cell line as a reference standard for a quantitative suppression assay. Here, we describe the development and characteristics of an assay to evaluate the suppression potency of bone marrow derived mesenchymal stromal cells (BM-MSC) using the K299 cell line as a reference standard.
MATERIALS AND METHODS
CD4 T cell isolation and storage
Peripheral blood mononuclear cells (PBMC) from healthy volunteers were prepared by Ficoll-Hypaque density gradient centrifugation (Organon Teknika, Durham, NC, USA). CD4 T cells were isolated from healthy donor PBMC using an automated cell separator (RoboSep®: Stem Cell Technologies, Vancouver, BC, Canada). Healthy donor PBMC and isolated CD4 T cells were cryopreserved in RPMI-1640 (Life Technologies, Gaithersburg, MD, USA) with 20% fetal bovine serum (FBS) and a final concentration of 10% DMSO according to standard protocol; vials were stored in liquid nitrogen until further use. Written informed consent from all subjects was obtained in accordance with Declaration of Helsinki for the use of samples for research protocol approved by the Institutional Review Board of the National Heart, Lung, and Blood Institute.
Bone marrow derived mesenchymal stromal cell isolation and expansion
Bone marrow derived mesenchymal stromal cells (BM-MSC) were expanded from bone marrow (BM) aspirates collected from healthy volunteers at Hematology Branch, National Heart, Lung, and Blood Institute or in the Department of Transfusion Medicine, National Institutes of Health. The BM aspirates were plated in 75cm2 flask in MSC medium consisting of MEM alpha (Life Technologies, Carlsbad, CA) supplemented with 20% FBS (Sigma-Aldrich, St. Louis, MO). Non-adherent cells were removed after 24 hours, and the adherent cells were cultured for approximately 14 days with twice weekly MSC medium changes. The cells were harvested using 0.05% trypsin-EDTA (Life Technologies, Carlsbad, CA) when 70% confluence was achieved and used for further expansion. The cells were plated at a density of 4×103/cm2 in four-layer cell factory flasks (Thermo Scientific Nunc™ Cell Factory™ Systems, Waltham, MA) in MSC medium. Serial passages were obtained once the cells reached 70% confluence in successive MSC expansions. At the third or fourth passage, cells were cryopreserved in freezing medium according to standard protocol, and stored in liquid nitrogen until further use. Written informed consent from all subjects was obtained in accordance with the requirement of the Institutional Review Board of the National Heart, Lung, and Blood Institute and Department of Transfusion Medicine (DTM), National Institutes of Health. We also evaluated the clinical grade BM-MSC provided by DTM, Clinical Research Center, National Institutes of Health and Children’s National Medical Center. The clinical grade BM-MSC at Children’s National Medical Center was manufactured using the Quantum Cell Expansion System (Terumo BCT, Lakewood, CO, USA) according to the protocol as previously reported.24,25 Briefly, 4–18 hours before the initiation of BM-MSC expansion, a cell expansion set was loaded onto the device and the system was primed with PBS and 5 mg of fibronectin was loaded into the system. Either 25 mL of filtered bone marrow or 2.0– 3.5×107 expanded cells were then loaded into the bioreactor. Cells were allowed to adhere and were then perfused 5% dextrose medium at 0.1 mL/minute with a gas supply consisting of 5% CO2, 5% O2, and 90% N2. Glucose and lactate measurements were used as a surrogate of growth and the feed rate was increased according to changing levels of glucose and lactate. Cells were harvested once the lactate levels reached pre-established thresholds (0.8 mM for passage 1, 8 mM >passage 1). These products met all the minimal release criteria defined as plastic adherent cells with tri-lineage differentiation and positivity for the immune-phenotype (CD73, CD90 and CD105).
K299 cell line preparation
The human lymphoma cell line K299 was purchased from Sigma Aldrich (St. Louis, MO) and cultured at 37°C, 5% CO2 in RPMI 1640 supplemented with 10% FBS, 2 mM L-glutamine, gentamicin (50 mg/mL) and split every 2–3 days to maintain the concentration between 0.5– 2 ×106 cells/ml. All butches of cell lines are tested for Mycoplasma contamination using Mycoalert Mycoplasma Detection Kit (Lonza, Basel, Switzerland). On the day of assay, cells were harvested and irradiated 100 Gy for proliferation suppression assay.
T cell activation and proliferation assay for assessment of suppression
On the day of assay, healthy donor CD4 T cells were thawed to confirm the viability greater than 75% and rested for 2–3 hours at 37°C, 5% CO2 in T cell medium (TCM: RPMI 1640 (Life Technologies, Carlsbad, CA, USA) supplemented with 10% human AB serum (Gemini Bio-Products, West Sacramento, CA, USA), 2 mM Lglutamine, 50mg/ml gentamicin). MSC were rested at 4°C in TCM. CD4 T cells were stained with CellTrace™ Violet (CTV; Invitrogen, Grand Island, NY, USA) according to manufacturer instructions and were then stimulated with anti-CD3/CD28-coated beads (Dynabeads®, Invitrogen, Carlsbad, CA, USA) at a T cell/bead ratio of 5:1 in 96 well U bottom plate. Responder CD4 T cells were then incubated with the K299 or third-party MSC. For the activation assay, the monoclonal antibody CD154-APC (BD bioscience, Franklin Lakes, NJ) was also added to the culture. Co-cultured cells were incubated at 37°C in 5%CO2 in TCM overnight (16 hours) for activation or 72 hours for proliferation. After the corresponding incubation, the cells were washed with AnnexinV Binding Buffer (BD Biosciences, Franklin Lakes, NJ) and stained with AnnexinV-FITC or APC (BD Biosciences, Franklin Lakes, NJ) for 15 minutes in the dark at room temperature. Following this incubation, the cells were stained with Propidium Iodide (PI) (Molecular Probe, Eugene, OR), then immediately acquired on a Becton Dickinson LSRII Fortessa (BD Biosciences, Franklin Lakes, NJ). Collected data was analyzed using FlowJo software (version 8.8.6; Treestar, Oregon, USA). The viability was measured by the population of AnnexinV-negative and PI-negative T cells. This proportion of viable cells was analyzed for either CD154+ (% activation) or CTV dim (% proliferation), respectively. Suppression of T cell activation and proliferation was calculated using the equation: %Suppression = 100 − (a/b * 100), where a is the %Activation or %Proliferation in the presence of suppressor cells, and b is the %Activation or %Proliferation in the absence of suppressor cells.
Flow cytometric analysis of CD4 T cell subset
After thawing, CD4 T cells were stained with Live/Dead Fixable Violet stain (ViViD: Invitrogen, Grand Island, NY, USA) and then a monoclonal antibody panel designed to evaluate memory T cells, regulatory T cells, and Th1-Th2-Th17 cells subsets. Anti-human flow cytometry antibodies used in the panel are summarized in Supplementary Table 1. T cell memory subsets were determined within the CD4 T cell population to identify naïve cells (CCR7+CD45RO−CD4+), stem cell memory cells26,27(CCR7+CD45RO−CD95+ CD4+) central memory cells (CCR7+CD45RO+CD4+), effector memory cells (CCR7−CD45RO−CD4+), and effector memory RA (TEMRA; CCR7−CD45RO−CD27−CD45RA+CD4+). Helper T cell subsets were determined within the memory CD4 cell population by surface chemokine receptors28,29: Th1 cells (CD45RO+CCR4−CCR6−CXCR3+CD4+), Th2 cells (CD45RO+CCR4+CCR6−CXCR3−CD4+), Th1–Th17 (CD45RO+CCR4−CCR6+CXCR3+CD4+), and Th17 cells (CD45RO+CCR4+CCR6+CXCR3−CD4+). Data acquisition was performed using a Becton Dickinson LSRII Fortessa and data was analyzed using FlowJo software (Tree Star Inc. Ashland OR). At least 50,000 events per CD4 T cell population were acquired to ensure a sufficient number of cells for statistical analysis.
Manipulation of BM-MSC potency with steroids and interferon gamma (IFNγ)
Passage 3 BM-MSC were incubated overnight at 37°C with or without priming of recombinant human IFNγ (catalog# PHC4031, Life Technologies, Carlsbad, CA, USA) at a concentration of 10 ng/mL. IFN-γ primed and not-primed BM-MSC were harvested the next day using 0.05% Trypsin-EDTA and used for the activation suppression assay.
The impact of corticosteroids on the immune-suppressive effect of BM-MSC was assessed using clinical-grade methylprednisolone sodium succinate (NDC code 0009-0039-30, Pfizer, New York, NY). Dose titration was performed at the concentrations of 1000 µg/mL, 100 µg/mL, 10 µg/mL, and 1 µg/mL. CD4 T cells were co-incubated with steroids for 16 hours with and without BM-MSC (passage 3) for activation suppression assay. In both assays, suppression potency of BM-MSC was measured using K299 as a reference cell line.
Statistics and suppression standardization
All data were analyzed with PRISM 5 (GraphPad Software, Inc., California, USA). P values were calculated using one-way ANOVA, followed by a Newman-Keuls multiple comparison test. The capacity of a suppressor cell to decrease T cell activation and proliferation was calculated using K299 suppressor units (KSU). This was done by setting the % suppression in the presence of K299 for each responder within each individual test to a value of 1 by the equation a/a, where a is the % suppression in the presence of K299. Then, the KSU for other suppressors was determined using the equation b/a, where b is the % suppression in the presence of a given suppressor, and a is the % suppression in the presence of K299. The KSU value for a particular suppressor cell would be <1.0 for less suppression than K299 and > 1.0 for suppression greater than K299.
RESULTS
K299 suppresses the activation and proliferation of healthy donor CD4 T cells
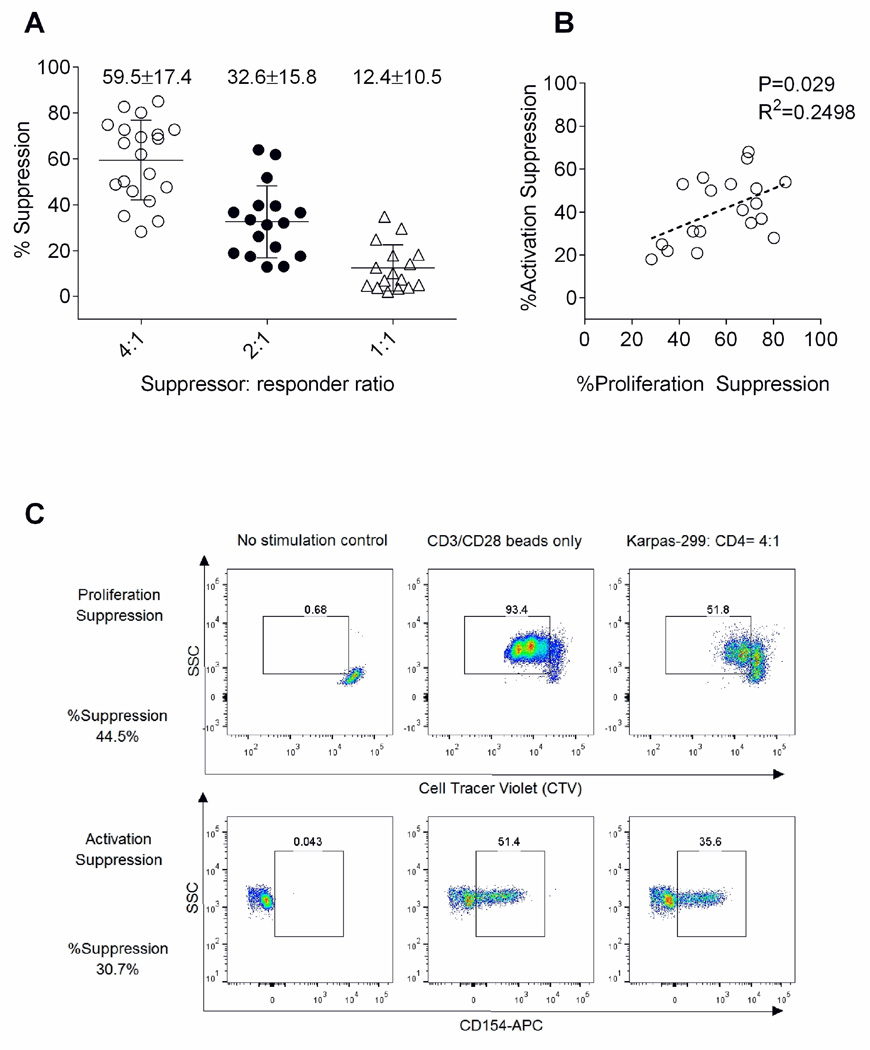
First we evaluated the suppressive potency of K299 cell lines on the CD4 T cells derived from healthy donors (n=20). The proliferation of CD4 T cells measured by CTV dilution were universally suppressed by K299 in a dose dependent manner: % suppression was 59.5±17.4% at 4:1 suppressor: responder (S: R) ratio, 32.6±15.8% at 2:1 ratio, and 12.4±10.5% at 1:1 ratio (Figure 1A). K299 also suppressed CD154 activation of CD4 T cells: % suppression 41.2±15.1% at a 4:1 ratio. The suppression of proliferation and activation were correlated (p=0.029, R2=0.2498: Figure 1B). However, the calculated % suppression values were higher in proliferation assays (Figure 1C). The robust suppression of CD4 T cell activation and proliferation by K299 supports its use as a reference cell line to standardize a T cell suppression assay. Since activation and proliferation were correlated, we choose the shorter activation assay as the most practical basis for a potency assay since the entire procedure could be performed within 24 hours.
Figure 1.
Universal suppression of healthy donor CD4+ cells by K299: (A) K299 suppressed proliferation of CD4 T cells at dose dependent manner (n=20). (B) Proliferation suppression is significantly correlated with activation suppression against the same CD4 T cells (P=0.029, R2=0.2498). (C) %Suppression values tend to be higher in proliferation assay in comparison to activation assay as shown in representative flow cytometry data.
Optimization of a suppression assay using K299
Although K299 universally suppresses healthy donor CD4 T cells, significant variances in % suppression were observed between different responders and different data collection points. We next explored variables affecting suppression by K299 cells in order to optimize the conditions of the assay.
Suppressor: responder ratio (S:R)
To define the optimum suppressor (K299) to responder (CD4 T cell) ratio, we measured activation suppression at S:R ratios from 16:1 to 1:1. The higher S:R ratio of 16:1, 8:1, 4:1 consistently showed >50% suppression, while lower suppressor: responder ratios showed significant variability (Supplementary figure 1A). Based on these findings we selected S:R ratios of 8:1 and 4:1 as a reliable and practical range.
Responder cell dose
We performed activation suppression assays using either 2×104 or 5×104 CD4 T cells per well. More consistent results were obtained when 5×104 CD4 T cells per well were used (Supplementary figure 1B). On this basis, a target dose of 5×104 CD4 T cells per well was selected.
Viability of the K299 cell line
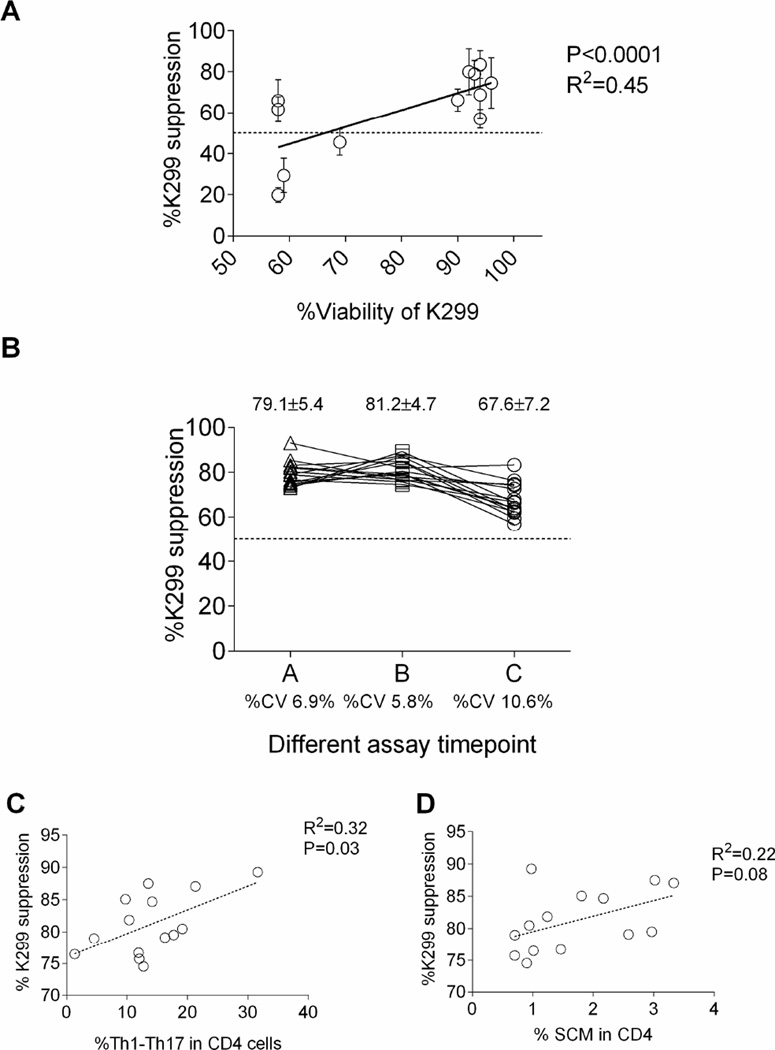
We tested K299 cell cultures with differing viabilities. There was a linear correlation of viability with potency of suppression (p< 0.0001, R2 = 0.45; Figure 2A). On this basis we selected 90% viability as a minimal threshold required for K299 cells in the suppression assay.
Figure 2.
Factors associated with K299 based suppression assay: (A) The viability of K299 cells is significantly correlated with the suppression capacity of CD4 T cells (P<0.0001, R2=0.45). (B) K299 activation suppression assay was tested using CD4 T cells from 15 different healthy donors. The %CV varied from 6.9% to 10.6% at three different time points. (C) %Th1–Th17 subset in CD4 T cells significantly correlated to the suppression susceptibility to K299 (P=0.03, R2=0.32). (D) %SCM subset in CD4 T cells showed modest correlation to the suppression susceptibility to K299 (P=0.08, R2=0.22).
Reproducibility of the suppression assay
We tested K299 activation suppression on 15 different CD4 T cells with S:R ratio of 8:1 at three time points (Figure 2B). At each time point, the mean± standard deviation of % suppression varies from 67.6±5.4% to 81.2%±4.7% while coefficient of variation (%CV) was 6.9%, 5.8%, and 10.6% respectively. Between the assays, %CV using the same responders also varies from 5.5% to 17.4%. Overall, %CV across three independent assays is calculated as 11%.
Immune-phenotype of responder CD4 T cells
To further explore the basis of variability between responders, we studied the phenotype of the CD4 selected cells used in the assays. Since CD4 T cells are composed of various memory subsets (naïve, stem cell memory26,27, central memory, effector memory cells), helper T cell subsets (Th1, Th2, Th17, Th1–17 cells28,29), and regulatory T cell subset, the composition of each subset may affect the susceptibility to K299 suppression. We analyzed the immunephenotype of CD4 T cells (n=14, representative flow data shown in Supplementary figure 2) and sought to identify a correlation with K299 suppression. The frequency of Th1-Th17 (CD45RO+CCR4−CCR6+CXCR3+) in CD4 T cells correlated significantly with suppression; R2= 0.32, P=0.03, (Figure 2C). The frequency of the stem cell memory (SCM) subset (CCR7+CD45RO− CD95+) in CD4 T cells has a trend of correlation with suppression; R2= 0.22, P=0.08 (Figure 2D). The regulatory T cell subset (FoxP3, Helios, and CD39) and T cell exhaustion markers (PD1, Lag3, PDL-1, and TIM-3) did not correlate with suppression (data not shown).
K299 can be used as a reference suppressor cell for the measurement of BM-MSC potency
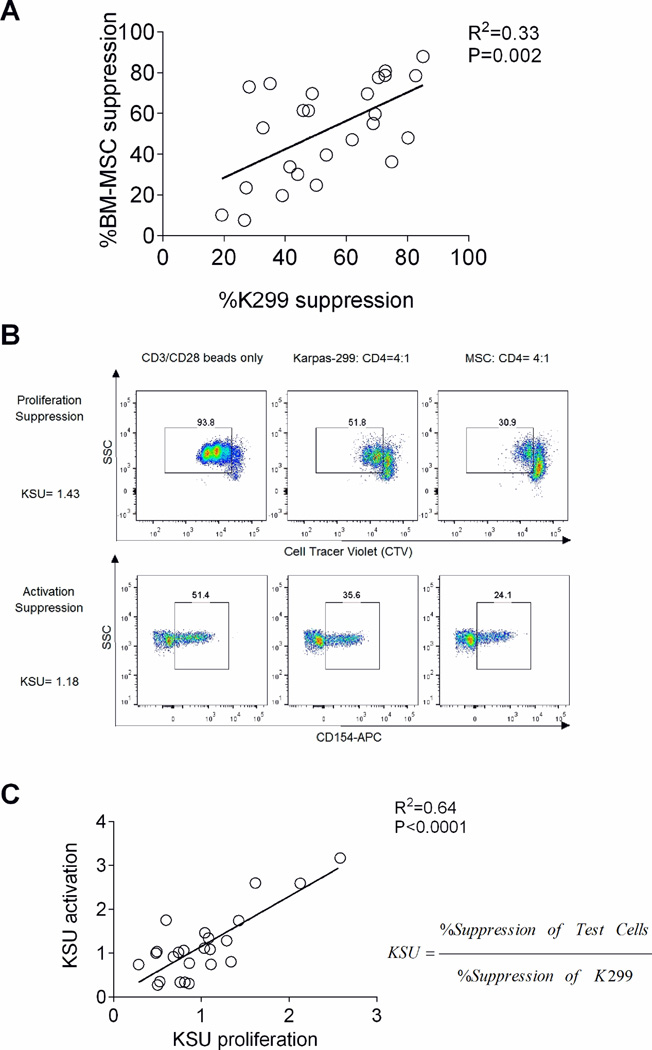
Next, we evaluated the feasibility of using K299 cell line as a reference suppressor cell line to compare suppressive potency of BM-MSC. CD4 T cells (n=24) were co-cultured with either K299 cell lines or various BM-MSC. Suppression was measured after stimulation with anti-CD3/CD28 beads. At a S:R ratio of 4:1, both K299 and BM-MSC consistently suppress CD4 T cell proliferation (Figure 3A). We showed that the degree of suppression achieved was a property of the specific sensitivity of the responder cell. Thus suppression by K299 and BM-MSC correlated linearly for any given responder cells (R=0.33, P=0.002). These results validate the use of a standardized suppression unit using the K299 cell line. We calculated suppression potency as K299 suppression units (KSU) using the formula below.
Both proliferation and activation suppression assays are able to measure potency of BM-MSC (Figure 3B). When standardized as KSU, the results between assays correlated in a linear fashion (R2=0.64, P<0.0001; Figure 3C).
Figure 3.
K299 as a reference suppressor cell line for measuring the potency of BM-MSC: (A) Proliferation suppression of K299 and BM-MSC against the same CD4 T cell responders significantly correlated with each other. (P=0.002, R2=0.33) (B) The proliferation and activation of CD4 T cells were both suppressed by K299 and BM-MSC as shown in a representative flow cytometry study of cells from one subject. (C) Both proliferation and activation suppression assays measuring BM-MSC potency are significantly correlated when calculated as KSU. (P<0.0001, R2=0.64)
The K299 suppression assay can be used to compare potency of different BM-MSC
We evaluated the potency of clinical grade BM-MSC (n=5) using K299 as a reference suppressor cells and calculated KSU for each product. Using 15 different CD4 T cell targets at S:R ratio of 8:1, the standard error of mean (SEM) of KSU was ≤0.05 (+/− 5%) for each product. We observed significant differences of suppression potency between BM-MSC from different donors (Figure 4A). Next, we tested the BM-MSC potency of different passages from the same donor and observed slightly higher KSU in early passage (passage 3) compared to late passage (passage 4) without statistical significance (Figure 4B). Finally we compared the BM-MSC potency from the same donor using different manufacturing methods. The BM-MSC (passages 3 or 4) manufactured in flasks showed higher suppression potency compared to the BM-MSC manufactured by Quantum method (Figure 4C).
Figure 4.
K299 as a reference suppressor cell line for comparing the potency of BM-MSC: (A) The K299 suppression assay demonstrated significantly different potencies between five clinical-grade BM-MSCs. (B) Early or late passage of BM-MSC from the same donor showed different potency quantified by KSU. (C) The BM-MSCs generated by different manufacturing methods from same donor showed different potency quantified by KSU.
Using the suppression assay to evaluate modulators of BM-MSC potency
Effect of interferon gamma
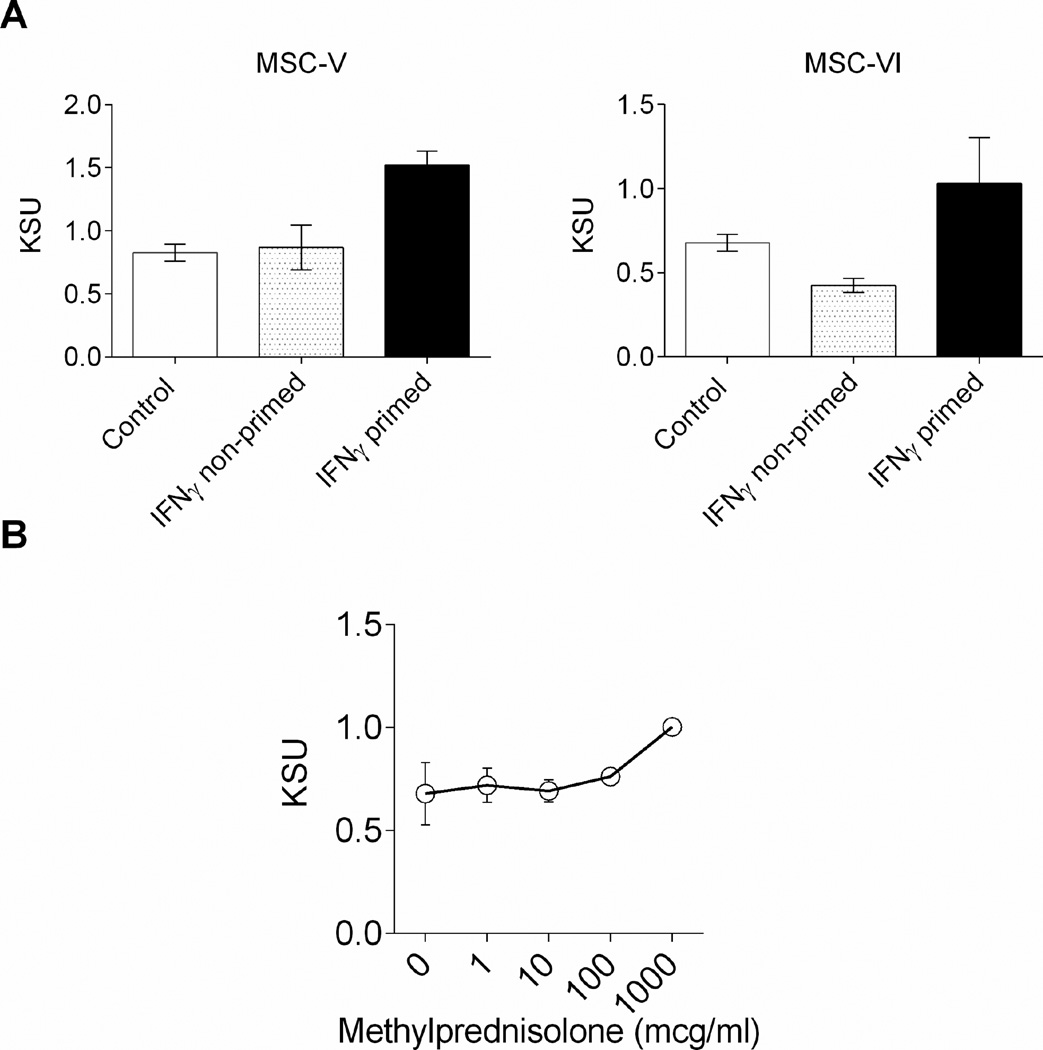
Priming with IFN-γ can modulate the suppression of T cells by BM-MSC30,31. To quantify the “licensing” effect of IFN-γ on BM-MSC, we compared suppressive potency of BM-MSC with or without overnight IFN-γ priming. IFN-γ primed BM-MSC consistently showed enhanced suppressive potency of BM-MSC compared with non-primed or freshly thawed control BM-MSC (Figure 5A).
Figure 5.
Use of the K299-based activation suppression assay to measure the impact of biological agents on BM-MSC potency: (A) IFNγ priming enhanced the suppressive potency of BM-MSC measured in KSU. The IFNγ primed BM-MSCs were compared to freshly thawed, control, or IFNγ non-primed BM-MSC (B) Methylprednisolone did not affect the potency of BM-MSC at lower dose and had additive effects at higher dose.
Effect of corticosteroids
Corticosteroids suppress the activation of CD4 T cells; however, the impact of steroids on BM-MSC is not well characterized by quantitative measurement. We tested BM-MSC suppression potency in the presence of different concentration of methylprednisolone from 1 to 1000 µg/mL. Baseline BM-MSC potency (KSU 0.67±0.15) was not impaired at low doses of corticosteroid (KSU 0.71±0.08, 0.69±0.05 at 1, and 10 µg/mL respectively) and slightly enhanced by higher doses (KSU 0.76±0.02, 1.00±0.003 at 100, and 1000 µg/mL respectively: Figure 5B).
DISCUSSION
The ability of the K299 cell line to suppress both the activation and proliferation of CD4 T cells as well as its widespread availability make it a useful standard for T cell suppression assays. At S:R ratios of 8:1 and 4:1, K299 showed >50% inhibition of activation of responder cells. We showed that if the cell viability is greater than 90%, the cell line can suppress activation with a reproducibility of +/− 5%. The main variable in the assay was the source of the responder CD4 T cells. Thus the assay can be made more reproducible if the same target is consistently used. In selecting an assay for widespread application we sought to develop a rapid activation-based suppression assay rather than the more time-consuming proliferation-based approach. The positive correlation that we found between activation and proliferation suppression justified the adoption of the simpler more rapid technique for development of the assay. From these experiments, we determined the optimal assay conditions: 5×104 responder cells per well; 4:1 or 8:1 S: R ratio and K299 cell viability of >90%. By expressing suppression as a function of the K299 suppression we could reduce the variability due to the variation in responder cell susceptibility to 10% CV and may facilitate comparisons of potency between experiments and potentially between laboratories. Finding linearity between K299 and BM-MSC at different S:R ratios and comparability of suppression between K299 and BM-MSC on more or less susceptible CD4 T cell targets, we were able to use the K299 as a standard to measure potency of BM-MSC. In this way, the activation suppression assay quantitated differences in suppressor function between different BM-MSC and different manufacturing methods. The potency of BM-MSC can be affected by the cytokine milieu such as IFNγ which is reported to enhance BM-MSC immunosuppressive function.30 We confirmed that the effect of IFNγ is powerful with a doubling of suppressive potency of BM-MSC. Since patients treated for GVHD with BM-MSC frequently receive steroids, we also explored the possible impact of steroid treatment on BMMSC function. We showed that pharmacological concentrations of methylprednisolone did not inhibit BM-MSC function and higher doses had an additive effect on T cell activation. Thus patients with steroid-refractory acute GVHD may still benefit from BM-MSC infusions with detectable plasma levels of exogenous corticosteroids.
To serve as a useful assay for general application as a measure of suppressor cell function reproducibility between laboratories, validation is needed by testing samples in several independent laboratories using a centrally distributed K299 cell line as the reference standard. In addition, in order to reduce variability, each laboratory could screen a series of donors to select a single individual with a standard susceptibility to K299 suppression of around 75% at a ratio of 8:1 (as indicated in Figure 2A). The selected sample could be shared with individual labs and tested as a control to maintain consistency between the assays.
In conclusion, while attempts to optimize and develop reproducible in vitro suppressor assays in an effort to determine release criteria for BM-MSC based on their function, it should also be noted that they still remain an imperfect means of judging potency where a relationship between an in vitro assay and a clinical outcome is implied. The assay used here while serving as a surrogate for immune modulation of GVHD, for example, may be inappropriate for determining efficacy of BM-MSC in wound repair. Indeed, even within the context of measuring responses in acute GVHD, it has yet to be shown that measuring T cell suppression in the BM-MSC product will have any predictive value for response. Furthermore, the selection of an optimal reference suppressor line remains controversial since the mechanism of action in T cell suppression can differ between various immune-regulatory cells.8 The comparison of K299 to other proposed reference cell lines such as an MSC cell line16 would be required to resolve the issue of whether lineage specific suppression assays are needed in the relevant culture setting with xeno-free standardized medium.
Only consistent repetitive testing of BM-MSC function in a single assay such as the one we describe, in a large series of patients with GVHD, consistently treated in the same schedule would determine the ultimate utility of this type of assay.
Supplementary Material
Supplementary Figure 1. Optimization of the K299-based suppression assay : (A) The higher suppression: responder (S:R) ratio (16:1, 8:1, 4:1) demonstrated consistent CD4 T cell suppression above 50%; in contrast, lower S:R showed lower levels of suppression with significant variances. (B) The higher number of responder cells per well showed more consistent %suppression with less variance.
Supplementary Figure 2. Representative gating strategy for assessing T cell immune-phenotype. (A) Memory cell subset were determined within the CD4 T cell population to identify naïve cells (CCR7+CD45RO−CD4+), stem cell memory cell (CCR7+CD45RO−CD95+CD4+) central memory cells (CCR7+CD45RO+CD4+), effector memory cells (CCR7−CD45RO−CD4+), and effector memory RA cells (TEMRA; CCR7−CD45RO−CD27−CD45RA+CD4+). (B) Helper T cell subsets were determined within the memory CD4 cell population by surface chemokine receptors: Th1 cells (CD45RO+CCR4−CCR6− CXCR3+CD4+), Th2 cells (CD45RO+CCR4+CCR6−CXCR3−CD4+), Th1–Th17 (CD45RO+CCR4−CCR6+CXCR3+CD4+), and Th17 cells (CD45RO+CCR4+CCR6+CXCR3−CD4+).
ACKNOWLEDGEMENT
We thank F Chinian (Hematology Branch, NHLBI, NIH, USA), Dr. Hanh Khuu, Naoza Collins-Johnson Minh Tran (Department of Transfusion Medicine, Clinical Research Center, NIH, USA) for their technical support. We also thank all volunteers who participated in this study.
FUNDING: This manuscript preparation was supported by the Intramural Research Program of the National Institutes of Health, at the National Heart, Lung, and Blood Institute. Board of Visitors Grant awarded to CMB and PJH at Children’s National Health System.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS:
Study concept and design: S.M., B.S., N.F.H, S.I, A.J.B.
In vitro experiment and data collection: S.M., B.S., N.F.H, S.I., N.D., A.P.G., D.S., K.K., H.P., C.B., D.S.
Analysis and interpretation of data: S.M., B.S., N.F.H., H.P., C.B., S.I., A.J.B
Drafting of manuscript: S.M., B.S., N.F.H., M.B., C.B., S.I., A.J.B. Obtained funding and study supervision: A.J.B.
DISCLOSURE: All authors declare no conflict of interest.
Reference
- 1.Owen RD. Immunogenetic Consequences of Vascular Anastomoses between Bovine Twins. Science. 1945;102(2651):400–401. doi: 10.1126/science.102.2651.400. [DOI] [PubMed] [Google Scholar]
- 2.Billingham RE, Brent L, Medawar PB. Actively acquired tolerance of foreign cells. Nature. 1953;172(4379):603–606. doi: 10.1038/172603a0. [DOI] [PubMed] [Google Scholar]
- 3.Silverstein AM. Ontogeny of the Immune Response. Science. 1964;144(3625):1423–1428. doi: 10.1126/science.144.3625.1423. [DOI] [PubMed] [Google Scholar]
- 4.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133(5):775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Talmadge JE, Gabrilovich DI. History of myeloid-derived suppressor cells. Nat Rev Cancer. 2013;13(10):739–752. doi: 10.1038/nrc3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizoguchi A, Mizoguchi E, Takedatsu H, Blumberg RS, Bhan AK. Chronic intestinal inflammatory condition generates IL-10-producing regulatory B cell subset characterized by CD1d upregulation. Immunity. 2002;16(2):219–230. doi: 10.1016/s1074-7613(02)00274-1. [DOI] [PubMed] [Google Scholar]
- 7.Keating A. Mesenchymal stromal cells: new directions. Cell Stem Cell. 2012;10(6):709–716. doi: 10.1016/j.stem.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Wood KJ, Bushell A, Hester J. Regulatory immune cells in transplantation. Nat Rev Immunol. 2012;12(6):417–430. doi: 10.1038/nri3227. [DOI] [PubMed] [Google Scholar]
- 9.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331(6024):1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 10.Sakaguchi S, Miyara M, Costantino CM, Hafler DA. FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol. 2010;10(7):490–500. doi: 10.1038/nri2785. [DOI] [PubMed] [Google Scholar]
- 11.Lund FE, Randall TD. Effector and regulatory B cells: modulators of CD4+ T cell immunity. Nat Rev Immunol. 2010;10(4):236–247. doi: 10.1038/nri2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Blanc K, Rasmusson I, Sundberg B, et al. Treatment of severe acute graft-versus-host disease with third party haploidentical mesenchymal stem cells. Lancet. 2004;363(9419):1439–1441. doi: 10.1016/S0140-6736(04)16104-7. [DOI] [PubMed] [Google Scholar]
- 13.Le Blanc K, Frassoni F, Ball L, et al. Mesenchymal stem cells for treatment of steroid-resistant, severe, acute graft-versus-host disease: a phase II study. Lancet. 2008;371(9624):1579–1586. doi: 10.1016/S0140-6736(08)60690-X. [DOI] [PubMed] [Google Scholar]
- 14.Di Ianni M, Falzetti F, Carotti A, et al. Tregs prevent GVHD and promote immune reconstitution in HLA-haploidentical transplantation. Blood. 2011;117(14):3921–3928. doi: 10.1182/blood-2010-10-311894. [DOI] [PubMed] [Google Scholar]
- 15.Martelli MF, Di Ianni M, Ruggeri L, et al. HLA-haploidentical transplantation with regulatory and conventional T-cell adoptive immunotherapy prevents acute leukemia relapse. Blood. 2014;124(4):638–644. doi: 10.1182/blood-2014-03-564401. [DOI] [PubMed] [Google Scholar]
- 16.Bloom DD, Centanni JM, Bhatia N, et al. A reproducible immunopotency assay to measure mesenchymal stromal cell-mediated T-cell suppression. Cytotherapy. 2015;17(2):140–151. doi: 10.1016/j.jcyt.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wells AD, Gudmundsdottir H, Turka LA. Following the fate of individual T cells throughout activation and clonal expansion. Signals from T cell receptor and CD28 differentially regulate the induction and duration of a proliferative response. J Clin Invest. 1997;100(12):3173–3183. doi: 10.1172/JCI119873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galipeau J, Krampera M. The challenge of defining mesenchymal stromal cell potency assays and their potential use as release criteria. Cytotherapy. 2015;17(2):125–127. doi: 10.1016/j.jcyt.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Chattopadhyay PK, Yu J, Roederer M. A live-cell assay to detect antigen-specific CD4+ T cells with diverse cytokine profiles. Nat Med. 2005;11(10):1113–1117. doi: 10.1038/nm1293. [DOI] [PubMed] [Google Scholar]
- 20.Canavan JB, Afzali B, Scotta C, et al. A rapid diagnostic test for human regulatory T-cell function to enable regulatory T-cell therapy. Blood. 2012;119(8):e57–e66. doi: 10.1182/blood-2011-09-380048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruitenberg JJ, Boyce C, Hingorani R, Putnam A, Ghanekar SA. Rapid assessment of in vitro expanded human regulatory T cell function. J Immunol Methods. 2011;372(1–2):95–106. doi: 10.1016/j.jim.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Fischer P, Nacheva E, Mason DY, et al. A Ki-1 (CD30)-positive human cell line (Karpas 299) established from a high-grade non-Hodgkin's lymphoma, showing a 2;5 translocation and rearrangement of the T-cell receptor beta-chain gene. Blood. 1988;72(1):234–240. [PubMed] [Google Scholar]
- 23.Wolke C, Tadje J, Bukowska A, et al. Assigning the phenotype of a natural regulatory T-cell to the human T-cell line, KARPAS-299. Int J Mol Med. 2006;17(2):275–278. [PubMed] [Google Scholar]
- 24.Hanley PJ, Mei Z, Durett AG, et al. Efficient manufacturing of therapeutic mesenchymal stromal cells with the use of the Quantum Cell Expansion System. Cytotherapy. 2014;16(8):1048–1058. doi: 10.1016/j.jcyt.2014.01.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin-Manso G, Hanley PJ. Using the quantum cell expansion system for the automated expansion of clinical-grade bone marrow-derived human mesenchymal stromal cells. Methods Mol Biol. 2015;1283:53–63. doi: 10.1007/7651_2014_164. [DOI] [PubMed] [Google Scholar]
- 26.Gattinoni L, Lugli E, Ji Y, et al. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17(10):1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lugli E, Gattinoni L, Roberto A, et al. Identification, isolation and in vitro expansion of human and nonhuman primate T stem cell memory cells. Nat Protoc. 2013;8(1):33–42. doi: 10.1038/nprot.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Acosta-Rodriguez EV, Rivino L, Geginat J, et al. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. 2007;8(6):639–646. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- 29.Gosselin A, Monteiro P, Chomont N, et al. Peripheral blood CCR4+CCR6+ and CXCR3+CCR6+CD4+ T cells are highly permissive to HIV-1 infection. J Immunol. 2010;184(3):1604–1616. doi: 10.4049/jimmunol.0903058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chinnadurai R, Copland IB, Patel SR, Galipeau J. IDO-independent suppression of T cell effector function by IFN-gamma-licensed human mesenchymal stromal cells. J Immunol. 2014;192(4):1491–1501. doi: 10.4049/jimmunol.1301828. [DOI] [PubMed] [Google Scholar]
- 31.Krampera M, Cosmi L, Angeli R, et al. Role for interferon-gamma in the immunomodulatory activity of human bone marrow mesenchymal stem cells. Stem Cells. 2006;24(2):386–398. doi: 10.1634/stemcells.2005-0008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Optimization of the K299-based suppression assay : (A) The higher suppression: responder (S:R) ratio (16:1, 8:1, 4:1) demonstrated consistent CD4 T cell suppression above 50%; in contrast, lower S:R showed lower levels of suppression with significant variances. (B) The higher number of responder cells per well showed more consistent %suppression with less variance.
Supplementary Figure 2. Representative gating strategy for assessing T cell immune-phenotype. (A) Memory cell subset were determined within the CD4 T cell population to identify naïve cells (CCR7+CD45RO−CD4+), stem cell memory cell (CCR7+CD45RO−CD95+CD4+) central memory cells (CCR7+CD45RO+CD4+), effector memory cells (CCR7−CD45RO−CD4+), and effector memory RA cells (TEMRA; CCR7−CD45RO−CD27−CD45RA+CD4+). (B) Helper T cell subsets were determined within the memory CD4 cell population by surface chemokine receptors: Th1 cells (CD45RO+CCR4−CCR6− CXCR3+CD4+), Th2 cells (CD45RO+CCR4+CCR6−CXCR3−CD4+), Th1–Th17 (CD45RO+CCR4−CCR6+CXCR3+CD4+), and Th17 cells (CD45RO+CCR4+CCR6+CXCR3−CD4+).