Abstract
Background
The pH sensitive Helicobacter pylori ArsRS two-component system (TCS) aids survival of this neutralophile in the gastric environment by directly sensing and responding to environmental acidity. ArsS is required for acid-induced trafficking of urease and its accessory proteins to the inner membrane, allowing rapid, urea-dependent cytoplasmic and periplasmic buffering. Expression of ArsR, but not its phosphorylation, is essential for bacterial viability. The aim of this study is to characterize the roles of ArsS and ArsR in the response of H. pylori to acid.
Materials and Methods
Wildtype H. pylori and an arsR(D52N) phosphorylation deficient strain were incubated at acidic or neutral pH. Gene and protein expression, survival, membrane trafficking of urease proteins, urease activity, and internal pH were studied.
Results
Phosphorylation of ArsR is not required for acid survival. ArsS-driven trafficking of urease proteins to the membrane in acid, required for recovery of internal pH, is independent of ArsR phosphorylation. ArsR phosphorylation increases expression of the urease gene cluster, and the loss of negative feedback in a phosphorylation deficient mutant leads to an increase in total urease activity.
Conclusions
ArsRS has a dual function in acid acclimation: regulation of urease trafficking to UreI at the cytoplasmic membrane, driven by ArsS, and regulation of urease gene cluster expression, driven by phosphorylation of ArsR. ArsS and ArsR work through phosphorylation dependent and independent regulatory mechanisms to impact acid acclimation and allow gastric colonization. Furthering understanding of the intricacies of acid acclimation will impact the future development of targeted, non-antibiotic treatment regimens.
Introduction
Helicobacter pylori is a gram negative bacterium that infects the gastric mucosa of about 50% of the world’s population, leading to gastritis, gastric and duodenal ulcer disease, and gastric cancer (1). Treatment has become more difficult due mainly to emerging antibiotic resistance (2), creating a need for identification and characterization of potential non-antibiotic treatment targets. H. pylori is able to survive and grow in the acidic gastric environment, despite being bioenergetically a neutralophile, through its unique mechanism of acid acclimation (3). Acid acclimation results in periplasmic alkalization, thus mimicking a neutral pH environment at the inner membrane to allow survival and growth in the highly acidic gastric milieu (4). The main components of this system are a neutral pH optimum, cytoplasmic urease and an inner membrane localized, proton-gated urea channel, UreI (4, 5). Urease associates with the inner membrane via UreI at acidic pH, allowing direct access of urea to urease (6–8). The products of urea hydrolysis, NH3 and CO2, move back through the channel into the periplasm, facilitating rapid periplasmic alkalization (9). H. pylori expresses two acid-sensing histidine kinase proteins (ArsS and FlgS), and in the absence of these proteins, gastric infection does not occur and membrane trafficking of urease is diminished or abolished (7, 9, 10). These histidine kinase proteins are part of bacterial two component systems (TCSs) that play an important role in environmental adaptation.
Bacterial TCSs are minimally composed of a histidine kinase sensor protein usually, but not always, located in the inner membrane and a response regulator located in the cytoplasm. Responding to an environmental stimulus, the sensor component auto-phosphorylates a histidine in a conserved H box (11). Phosphate is then transferred from the H box to an aspartyl residue of the response regulator, which binds to different promoters to regulate gene expression. Although there is classically specificity of response to particular input signals, there is mounting evidence for cross talk between a sensor kinase and non-cognate response regulators (12, 13). For example, among the 29 kinase sensors expressed by E. coli, UhpB is able to phosphorylate 9 different response regulators (12). It follows that this cross-talk may be designed for optimal response to a specific environmental change.
H. pylori expresses only four kinase sensor proteins and seven response regulators (14), a relatively small number that reflects its fairly constant environmental niche in the stomach. Besides the chemotactic TCS, CheA (HP0392)/CheY (HP1067), H. pylori contains only three histidine kinases and six response regulators involved in transcriptional regulation (14–16). One of the three complete TCSs, CrdRS (HP1365/HP1364), positively regulates the expression of the copper resistance determinant CrdAB-CzcAB in response to increasing concentrations of copper ions (17).
H. pylori expresses two TCSs that respond to acidity, namely, ArsRS (HP0166/HP0165), which is inner membrane localized, and a cytoplasmic system, FlgRS (HP0703/HP0244), that regulates intermediate flagellar genes as well as pH dependent genes (18, 19). The sensor kinase of FlgRS, FlgS, has no membrane domain, therefore it responds to cytoplasmic, not periplasmic signals. Recently, it was shown that expression of FlgS is necessary for viability at pH 2.5 in the presence of 10 mM urea, and this response was independent of FlgR, its cognate response regulator (9). FlgS expression is also required for urease assembly and activation, and plays a role in cytoplasmic and periplasmic pH homeostasis (9).
Transcription of the acid acclimation operons is under control of the housekeeping σ80 factor and regulated mainly by ArsR (20). ArsR is encoded by an operon that includes the cognate transmembrane ArsS histidine kinase (21). The signal sensed by ArsS is acidification of the periplasm, transduced via protonation of at least His94 (pKa ~6.0) in the periplasmic sensory domain (10). This stimulus triggers auto-phosphorylation of ArsS and trans-phosphorylation of ArsR, increasing its DNA-binding affinity toward a specific set of promoters (22, 23). However, although arsR is an essential gene, strains carrying a point mutation that disrupts the phosphoacceptor site in the ArsR receiver domain, as well as arsS deletion mutants, are viable (23, 24). Emerging evidence indicates the existence of distinct regulatory targets controlled by phosphorylation of ArsR. The phosphorylated ArsR-dependent targets (e.g. amiE, amiF, omp11, carbonic anhydrase, hypA, ureAB, and ureI) are activated by P~ArsR upon acidification of the periplasm (25, 26). Comparison of the H. pylori acid responsive transcriptomes with the ArsRS regulon has identified genes that are acid-responsive but independent of ArsRS regulation (25–29). Among these are genes that encode proton exporters (e. g. NADH ubiquinone oxidoreductase, ubiquinol cytochrome c oxidoreductase subunits, pyruvate ferredoxin oxidoreductases, cytochrome c oxidase subunits, F1F0 ATPases) that for E. coli have increased enzyme activity in response to mild acidity (30). Therefore, the regulation of expression of proton export and cytoplasmic and periplasmic buffering are transcriptionally regulated by distinct signaling systems.
In the absence of ArsS, there is a decrease in the amount of urease and its accessory proteins found in association with the membrane and a resultant decrease in urease activity of the purified membrane fraction (7). H. pylori mutant strains containing deletions of ArsS are defective in colonization of the mouse stomach (31). Proton gating of UreI in the intact bacteria is still present, but the level of urease activity is decreased (7). Since there is evidence for phosphorylation dependent and independent regulons of ArsR, the aim of the current study was to use a phosphorylation deficient mutant of arsR, arsR(D52N), and a ΔarsS strain to identify the distinct roles of ArsS, ArsR and ArsRS in acid acclimation. Acid survival, urease activity, membrane trafficking, cytoplasmic pH, and gene and protein expression were evaluated in the arsR(D52N) strain as compared to wildtype H. pylori. ArsS and ArsR co-regulate a group of genes that require both ArsS and ArsR~P, ArsS alone regulates another group of genes or proteins either directly by phosphorylation or indirectly through an unidentified RR, and ArsR alone is required for the expression of essential genes.
Materials and Methods
Bacterial strains and culture conditions
H. pylori strain ATCC 43504 was obtained from the American Type Culture Collection (ATCC). A non-polar ATCC 43504 ΔarsS deletion mutant was constructed by allelic exchange using a kanamycin resistance gene as described previously, and downstream gene expression in the knockout was confirmed by RT-PCR amplification of the downstream HP0163 gene (7). An arsR(D52N) mutant was constructed as described below. Bacteria were grown under microaerobic conditions (5% O2, 10% CO2, 85% N2) either on Trypticase Soy Agar (TSA) plates supplemented with 5% sheep blood (Gibco BRL-Life Technologies) or in brain heart infusion (BHI) medium (Difco Laboratories) supplemented with 7% horse serum (Gibco BRL-Life Technologies) and 0.25% yeast extract (Difco Laboratories). All bacteria grown in media were in the presence of Dent selective supplement (Oxoid Limited), and the mutant strains were grown in the presence of 20 μg/mL kanamycin (Sigma).
Construction of mutant H. pylori strain 43504/arsR(D52N) by allelic exchange mutagenesis
To construct the H. pylori strain with the arsR-D52N mutation (43504/arsR(D52N)), a pBluescript II vector (Stratagene) carrying a kanamycin resistance cassette from the pUC4K vector (Amersham Biosciences) flanked by a 681bp fragment comprising the entire arsR ORF with D52N mutation, and a 1121bp fragment comprising the 5′-region arsS ORF, was introduced into H. pylori strain 43504 by natural transformation. The DNA fragment for 5′-arsS ORF was obtained by PCR amplified from chromosomal DNA of H. pylori 43504 with primer pairs arsS_5′P(925–948)-BamHI/arsS3’P(2026–2045)-SacI (Table 1). The DNA fragment for arsR ORF with D52N mutation was obtained by PCR from expression construct pET100/D-TOPO-arsR-D52N with primer pairs arsR_5′P(174431)-KpnI/arsR_3′P(173775)-EcoRI. The mutated expression construct pET100/D-TOPO-arsR-D52N was generated on the construct pET100/D-TOPO-arsR (26) using the QuikChange site-directed mutagenesis kit (Stratagene) with the sense and antisense oligonucleotides designed to contain the mutation that replaces the single nucleotide T with A in the codon 52 (GAT) of the wildtype arsR gene sequence coding for aspartic acid, which results in a mutated codon (GAA) coding for asparagine (table 1). The resulting strain (43504/arsR(D52N)) had the entire arsR gene replaced by arsR ORF with D52N mutation and a kanamycin resistance cassette. The kanamycin-selected mutant strains were confirmed by PCR.
Table 1.
Oligonucleotide primers used for construction of arsR(D52N)
| Name | Sequence (5′ to 3′)a | Siteb | Strand | Positionc |
|---|---|---|---|---|
| arsR_5′P(174431)-KpnI | attgggtaccATGATAGAAGTTTTAATGATAGAAG | Kpn I | − | 174431–174455 |
| arsR_3′P(173775)-EcoRI | gcaggaattcATATCAGTATTCTAATTTATAACCAATCC | EcoRI | + | 173775–173803 |
| arsS 5′P(925–948)-BamHI | tagtggatccGGAGTTAAGGGGTTTTGCGTTTCT | BamHI | − | 173743–173766 |
| arsS_3′P(2026–2045)-SacI | tgttgagctcATGGGCTTGGCTGGGGTTAG | Sac I | + | 172647–172666 |
Sequences in uppercase letters are derived from the genome sequences of H. pylori 26695 (14). Sequences introduced for cloning purposes are given in lowercase letters, and restriction recognition sites are underlined.
Restriction recognition sites.
Nucleotide positions refer to the genome sequence of H. pylori 26695 (14).
Sequencing of the arsR(D52N) mutant
Genomic DNA was isolated from the wildtype (strain 43504) and arsR(D52N) bacteria using the CTAB method (32). The arsR gene was amplified by PCR using the following primers: sense 5′-CTAGAATTAGTCCCCTAAAACATTAAC-3′, antisense 5′-GAGCGTTATCATAAACAAAGCGACAAC-3′. The PCR products were confirmed using a bioanalyzer (Agilent). PCR products were then TA cloned into the pCR®4-TOPO vector (Life Technologies) according to the manufacturer’s instructions. The vector was introduced into Top10 electrocompetent E. coli (Life Technologies). Plasmids were isolated (Wizard Miniprep, Promega) and digested using EcoRI restriction enzyme (Promega) to confirm presence of insert. The arsR gene was then sequenced in both wildtype and arsR(D52N) using the sense primer above (Laragen facility, Culver City, CA). Sequences were aligned using VectorNTI software and presence of mutation, as well as absence of other amino acid changes, was confirmed (data not shown).
Confirmation of expression of downstream genes in arsR(D52N)
RNA was isolated from wildtype and arsR(D52N) bacteria as described below (see “RNA isolation”). Immediately after RNA isolation was complete, cDNA was created from 6 μg RNA using the Omniscript® enzyme (Qiagen) and random primers (Life Technologies). PCR was then completed using primers to detect HP0163 (sense 5′-GCCATGCGTTAAACAAGGAT-3′, antisense 5′-ATATCCACACCGCTTTCAGC-3′), a gene downstream from arsR in the same operon. PCR products were visualized using an Agilent bioanalyzer (data not shown).
Construction of ArsS-His6 expression plasmid
The DNA fragment coding for the cytoplasmic domain of the histidine kinase, ArsS (aa 158–427, M.W. 35.3 kD) was amplified by PCR using genomic DNA from H. pylori strain 26695 as the template and primers arsS-5′P, 5′-CACCAGAGAGTTAAGATCTCAAGTGAAAC-3′ and arsS-3′P, 5′-GCTCACTTCTCTCAAATTTTCG-3′. The 5′ primer arsS-5′P contained a 4-nucleotide sequence, CACC, immediately 5′ to the codon AGA to facilitate the directional cloning into pET100/D-TOPO vector (Invitrogen), which allows expression of recombinant ArsS with an N-terminal tag containing the Xpress epitope and a 6×His tag. The nucleotide sequence of the cloned PCR products was verified by sequencing of both strands.
Overproduction and purification of ArsS-His6
ArsS-His6 recombinant protein was expressed in E.coli BL21 Star (DE3) containing plasmids pET100/D-TOPO-HP0165. Bacteria were grown in 50 mL of LB broth at 37°C to an OD600 of 0.5. Subsequently, isopropyl-1-thio-β-D-galactopyranoside was added to a final concentration of 1 mM, and incubation was continued for 2 hours. Cells were harvested and suspended in 8 mL of Native Binding Buffer (50 mM NaH2PO4, 500 mM NaCl, 10 mM Tris-HCl, pH 8.0) with 10 mM imidazole and 1 mg/mL lysozyme. After incubation on ice for 30 min, the bacteria were further disrupted by sonication. Cell debris was pelleted by centrifugation and 8 mL of the supernatant was added to the purification column containing 1.5 mL of 50% Ni-NTA Resin (Life Technologies) and incubated for 3 hours at room temperature on an orbital shaker. The column was washed with 8 mL of Native Wash Buffer (50 mM NaH2PO4, 500 mM NaCl, 10 mM Tris-HCl, pH 8.0, and 20 mM imidazole) 4 times. His-tagged proteins were eluted with 8 mL of Native Elution Buffer (50 mM NaH2PO4, 500 mM NaCl, 10 mM Tris-HCl, pH 8.0, and 250 mM imidazole). An aliquot (10 μL) from each eluted fraction was analyzed by SDS-PAGE, and the fractions with purified proteins were concentrated and desalted using Amicon Ultra-4 Centrifugal Filter Devices (Millipore). Overexpression of ArsR-His6 and ArsR(D52N)-His6 was described previously (23).
In vitro phosphorylation assay
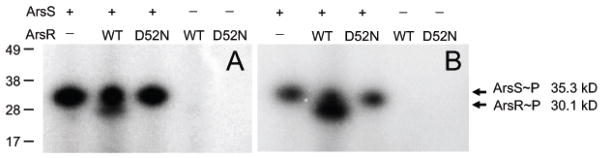
In vitro phosphotransfer from ArsS-His6 to ArsR-His6 was done using purified His6-tagged proteins in the presence of [γ-32P] ATP. Briefly, 6 pmol of ArsS-His6 was added to a buffer containing 50 mM Tris-HCl (pH 8.0), 75 mM KCl, 2 mM MgCl2, and 1 mM dithiothreitol. 10 μCi of [γ-32P] ATP was then added. 6 pmol of ArsR-His6 or ArsR(D52N)-His6 was added to 6 pmol ArsS-His6 that had been allowed to autophosphorylate for 30 minutes. Reactions were stopped after 15 and 30 minutes at 30°C by addition of 4× sample buffer, and the samples were analyzed by 4–10% Nu-PAGE. Phosphotransfer from ArsS to ArsR is time dependent. Phosphotransfer to ArsR peaked at 30 minutes with a very strong 30.1 kd band. However, due to the relatively small difference in size between the cytoplasmic domain of ArsS~P (35.3 kd) and ArsR~P, the two bands tends to fuse at 30 minutes. Therefore, both the 15 and 30 minute phosphotransfer results are shown. After drying, polyacrylamide gels were analyzed with a phosphorimager (Typhoon FLA 7000, GE Healthcare).
Survival studies
Bacteria were incubated in dialysis cassettes (Slide-A-lyzer, MWCO 10 kD, 3 mL capacity, Thermo) suspended in 1500 mL BHI adjusted to the proper pH using HCl and containing 5 mM urea as described previously (33). This system allows for maintenance of physiologic urea concentrations and constant media pH without the use of buffers, so changes can be observed over prolonged periods. Wildtype or arsR(D52N) bacteria grown on TSA plates were scraped into 7 mL BHI, then 3 mL was added to each dialysis cassette. Bacteria were incubated for 4 hours in 1500 mL media at pH 3.0 or 7.4 with 5 mM urea. After 4-hour incubation, samples were removed from dialysis cassettes, serially diluted by adding 1 mL to 9 mL media in succession, then plated in duplicate. Colonies were counted after 3 days. Data was calculated as percent of pH 7.4 control for each strain. Experiments were repeated in triplicate.
Measurement of urease activity
Wildtype and arsR(D52N) bacteria were harvested from plates, and total lysates were prepared by 3 passages through a French press at 20,000 p.s.i. Lysates were added to 100 mM sodium phosphate buffer containing 5 mM KCl, 138 mM NaCl, 0.5 mM MgCl2, 1 mM CaCl2, 10 mM glucose, 1 mM glutamine, and 5 mM [14C]urea with a specific activity of 10 mCi/mmol. The pH of the buffer used was 7.4, made by mixing 100 mM sodium phosphate monobasic and 100 mM sodium phosphate dibasic to the desired pH. The pH of the buffer during the course of the experiment did not change by more than 0.1 pH units. Urease activity was measured radiometrically (34, 35). Urease activity was measured for 30 minutes at 37°C with constant agitation. The reaction was terminated by the addition of 5N H2SO4 and incubated for 30 minutes at 37°C. The wells were placed in scintillation cocktail (HiIonicFluor; Packard Instruments), and the radioactivity measured by scintillation counting (1216 RackBeta; LKB Instruments). Protein concentration was determined by the BCA method (Pierce/Thermo). Results were expressed in μg/min/mg protein.
RNA isolation
Bacteria from six plates were resuspended in 7 mL BHI, and 3 mL was added to each of two dialysis cassettes. Dialysis cassettes were suspended in 1500 mL media at pH 3 or 7.4 with 5 mM urea as described above. Following incubation at pH 3.0 or 7.4 for 4 hours, RNA was isolated from H. pylori using a combination of the TRIzol® method and the RNeasy kit (Qiagen) as described previously (29). RNA quality was determined using an Agilent 2100 bioanalyzer. Only RNA with an RNA integrity number greater than 8 was used for analysis. Biologic replicates were completed for all experiments.
Quantitative PCR
RNA was converted to cDNA using Omniscript (Qiagen). qPCR was then completed in triplicate for each sample and each primer pair, using a Bio-Rad iCycler. Primer design was aided by the Primer3 software available at http://www-genome.wi.mit.edu/genomesoftware/other/primer3.html (36, 37). Unique primers were designed for 100–300 base pair regions of each gene in the urease gene cluster and α-carbonic anhydrase (Table 2), which is regulated by the ArsRS TCS (26).
Table 2.
qPCR primers
| Sense (5′-3′) | Anti-sense (5′-3′) | |
|---|---|---|
| ureA (HP0073) | AACCGGATGATGTGATGGAT | GGTCTGTCGCCAACATTTTT |
| ureB (HP0072) | TCTTTGGCGTAAAACCCAAC | CATAAGCCGCTTGAGACACA |
| ureI (HP0071) | TTCCTGCTGCGATTTTATCC | CATCTCACACCCAGTGTTGG |
| ureE (HP0070) | CAAGTTGGGGCTCTCTCAAG | GTGGGCTTTTCAAATGGTGT |
| ureF (HP0069) | TGCGCAAACTTCTAACATGG | GCATCGCCTTAATGTCGTTT |
| ureG (HP0068) | GGTAAAACCGCCTTGATTGA | CGGACAGCCTCCTGTTTCTA |
| ureH (HP0068) | TGCAATTAAACATCGGTCCA | CGTGGTGTTGCCCTTAAAAT |
| α-CA (HP1186) | GCAAGATAAAGCCGATTTGC | CGCCCTCTAGCGTCTTTATG |
Gene numbers refer to H. pylori strain 26695 (14)
Standard PCR performed with genomic DNA from H. pylori strain ATCC 43504 as the template was used to assure that all primer pairs resulted in amplification of a single product (data not shown). Genomic DNA was diluted serially by 10 fold to produce the standard curve for the qPCR reaction. qPCR was completed using 20 μL reactions in a 96 well plate with SYBR Green (SsoFast EvaGreen® Supermix, Bio-Rad), conditions were 92° for three minutes followed by 40 cycles of 92° for 30 seconds, 55° for 30 seconds, 72° for 40 seconds. Data were collected during the extension step and mean starting quantity was calculated using the Bio-Rad software. A melting curve was used at the end of the run to confirm that there was only one peak and only one product for each primer.
Preparation of bacterial lysates
Two plates of wildtype H. pylori or arsR(D52N) were suspended in 7 mL BHI and 3 mL of each suspension was added to a dialysis cassette as described above. Bacteria were incubated in the dialysis cassettes suspended in 1500 mL of media at pH 3.0 or 7.4 with 5 mM urea for 4 hours. Bacteria were then pelleted at 10,000 rcf, 4°C for 5 minutes. Pellets were resuspended in 3 mL ice cold 25 mM phosphate buffer with 5 mM EDTA and 150 μL bacterial protease inhibitor (Sigma). Bacteria were lysed by 3 passes through a French Press at 20,000 p.s.i. Unbroken bacteria were harvested by a 10 minute spin at 4°C, 1500 rcf. The supernate fraction was collected as total bacterial lysate. Protein concentration was determined by the BCA method (Pierce/Thermo).
Preparation of purified membranes
Wildtype H. pylori, ΔarsS, or arsR(D52N) were incubated in 10 mL BHI pH 4.5 (pH set with HCl) or pH 7.4. Incubation conditions and pH values for these experiments differed because they were designed to be comparable to previously published work (7). Inhibition of membrane trafficking in the absence of ArsS is overcome by other mechanisms (likely by contribution of cytoplasmic histidine kinase FlgS) by 90 minutes (7), therefore, 4 hour incubation was not feasible. Higher pH was required due to absence of urea. Six plates of each strain of bacteria were resuspended in 1 mL BHI, and 450 μL was added to each flask. Bacteria were incubated for 30 minutes with shaking under microaerobic conditions, then harvested by centrifugation (1500 x g, 10 minutes, 4°C). Pellets were resuspended in 3 mL ice cold 25 mM phosphate buffer with 5 mM EDTA and 150 μL bacterial protease inhibitor (Sigma). Bacteria were lysed by 3 passes through a French Press at 20,000 p.s.i. Unbroken bacteria were removed by centrifugation (1500 x g, 10 minutes, 4°C). An aliquot of the supernate was saved as total lysate, and the remainder was transferred to an ultracentrifuge tube, then centrifuged (10,000 x g, 10 minutes, 4°C). The supernate was transferred to a new tube, and membranes were pelleted (100,000 x g, 1 hour, 4°C). To further purify the membranes and remove all unattached cytoplasmic proteins, the membrane pellet was resuspended and layered on 5 mL of 25 mM phosphate buffer with 5 mM EDTA and 20% sucrose, then centrifuged (100,000 x g, 1 hour, 4°C). Centrifugation through sucrose was repeated x 2, then the membrane pellet was resuspended in 200 μL 25 mM phosphate buffer with 5mM EDTA. 0.1% SDS was added to the membranes to help resuspend the proteins. The protein concentration of the total lysate and membrane fractions was determined by the BCA method (Pierce/Thermo).
SDS-PAGE and Western blotting
Total bacterial lysates and purified membrane proteins prepared as described above were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 4–12% NuPAGE bis-tris gradient gels (Life Technologies) and transferred onto nitrocellulose (Bio-Rad). Western blot analysis was performed using multiple different antibodies. Monoclonal UreA and UreB antibodies were obtained from Austral Biologics and used at dilutions of 1:1000 and 1:20,000, respectively. Polyclonal UreI antibody, raised in rabbit and affinity purified, was used at a dilution of 1:500 (38). UreE antibody was raised in rabbit against the sequence CEWFETRKKIARFKTRQGKD (Alpha Diagnostic International) and used at a dilution of 1:250 (4). Immunolabeling was detected using the Supersignal West Pico chemiluminesce system (Pierce/Thermo).
Measurement of internal pH (pHi)
Internal (cytoplasmic) pH was measured using a fluorimeter as described previously (9). H. pylori wildtype, ΔarsS, and arsR(D52N) strains were grown overnight on TSA plates supplemented with 5% sheep blood (BD). The bacteria were removed from the plates and resuspended in 5 ml BHI medium (Difco). The membrane-permeant, pH-sensitive fluorescent probe 2,7-bis-(2-carboxyethyl)-5-carboxyfluorescein acetoxymethyl ester (BCECF-AM) was added to the bacterial suspension to a final concentration of 10 μM. The bacterial suspension was then incubated in a microaerobic (5% O2, 10% CO2, 85% N2) environment at 37°C for 30 min with shaking. The bacteria were pelleted by gentle centrifugation (2,000 x g, 5 minutes) and resuspended in 300 μL of HP medium (140 mM NaCl, 5 mM KCl, 1.3 mM CaCl2, 0.5 mM MgSO4, 10 mM glucose, 1 mM glutamine) buffered with 1 mM phosphate buffer at pH 7.4. To measure pHin, 20μL of BCECF-loaded bacteria was added to 3 mL 100 mM phosphate buffered HP medium at pH 4.5, and fluorescence was monitored using a dual-excitation (Ex1; 502 nm, Ex2; 436 nm), single-emission (Em; 530 nm) fluorimeter (Jobin Yvon Horiba Fluorolog). pH 4.5 was used due to the fluorescence range of the dye. 5 mM urea was added either at the beginning of the experiment or during the course of measurement.
Each experiment was calibrated independently. Once the fluorescence of the internal BCECF had been measured, 150 nM of the protonophore 3,3,4,5-tetrachlorosalicyanilide (TCS) was added to equilibrate the internal pH with that of the medium. HCl was then added to obtain minimum fluorescence of the dye, followed by the addition of NaOH to obtain maximum fluorescence of the dye. The internal pH was calculated using the equation:
where pKa = pKa of BCECF = 6.98, R = value at 502 nm/value at 436 nm for each data point, RA = ratio at minimum fluorescence, RB = ratio at maximum fluorescence, FA(λ2) = minimum fluorescence at 436 nm, and FB(λ2) = maximum fluorescence at 436 nm.
Results
Substitution of asparagine for aspartic acid at position 52 in ArsR abolishes phosphorylation by ArsS
Sequence analysis of arsR(D52N) confirmed the D to N mutation (data not shown). Expression of the downstream gene HP0163 at wildtype levels was confirmed by RT-PCR (data not shown), showing that the mutation was non-polar. The inability of the ArsR(D52N) strain to accept phosphotransfer from ArsS was confirmed using an in vitro phosphorylation assay (Figure 1). Although these studies do not investigate the promoter binding ability of ArsR(D52N), previous studies have shown that unphosphorylated ArsR still binds to promoters of acid activated genes (23) and the viability of the mutant shows that the substitution does not suppress promoter binding to essential genes.
Figure 1.
Phosphotransfer from His6-tagged ArsS to His6-tagged ArsR(D52N) protein is abolished. ArsS-His6 protein was incubated with [γ-32P] ATP before addition of ArsR-His6 wildtype (WT) or ArsR(D52N) mutant (D52N). Phosphotransfer from ArsS to ArsR was only seen with WT ArsR. No phosphotransfer to ArsR(D52N) was detected. Representative gels analyzed by autoradiography from a phosphotransfer assay with incubation times of 15 (A) and 30 (B) minutes are shown. The presence (+) or absence (−) of ArsS and ArsR protein used in each reaction are indicated above the lanes.
ArsR phosphorylation is not required for survival in acid
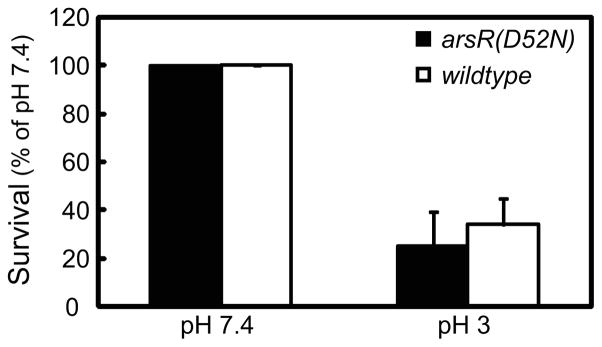
Wildtype or arsR(D52N) bacteria were incubated in dialysis cassettes at pH 3.0 or 7.4, then serially diluted and plated. The wildtype bacteria, as expected, survived at pH 3.0 in the presence of urea, with improved survival at pH 7.4. This experimental setup is unique in that it allows for incubation over a prolonged period while maintaining constant pH and urea in the environment, so overall survival at pH 3.0 with urea is lower for both strains than would be expected in a standard survival assay using a closed system (33). There was no significant difference in the pattern of survival at the two pHs in the absence of ArsR phosphorylation when compared to wildtype control (Figure 2), suggesting that phosphorylation of ArsR is not required for survival at acidic pH in the presence of urea.
Figure 2.
Phosphorylation of ArsR is not required for survival of H. pylori in acid. Wildtype and arsR(D52N) bacteria were incubated in dialysis chambers suspended in 1500 mL media for 4 hours at pH 3.0 or 7.4 with 5 mM urea. Medium pH did not change after 4 hours. Bacteria were then serially diluted and plated in duplicate. Colony counts are expressed as percent of pH 7.4 control for each strain. There was no significant difference in acid survival. N=3 complete experiments, error bars represent SEM, p=0.66.
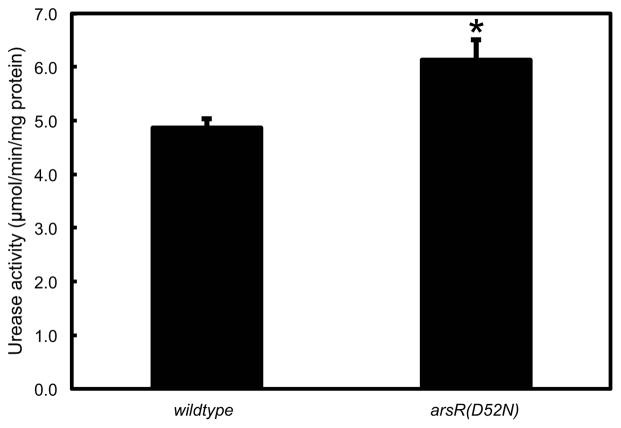
Total urease activity is greater in the arsR(D52N) strain compared to wildtype
Total cell lysates were prepared from plated bacteria. Total urease activity was measured at pH 7.4. Total urease activity was 1.2x higher in the arsR(D52N) strain as compared with the wildtype bacteria, and this difference was statistically significant (Figure 3). This is likely due to loss of the negative feedback provided by ArsR phosphorylation (21).
Figure 3.
Total urease activity is increased in the absence of ArsR phosphorylation. Wildtype and arsR(D52N) bacteria were harvested from plates (neutral pH) and lysed. Urease activity was measured. Total urease activity was increased in the arsR(D52N) strain, likely reflecting the absence of negative feedback on the urease gene cluster provided by phosphorylation of ArsR at neutral pH. N=6, error bars represent SEM, p=0.02.
ArsR phosphorylation regulates expression of the urease gene cluster
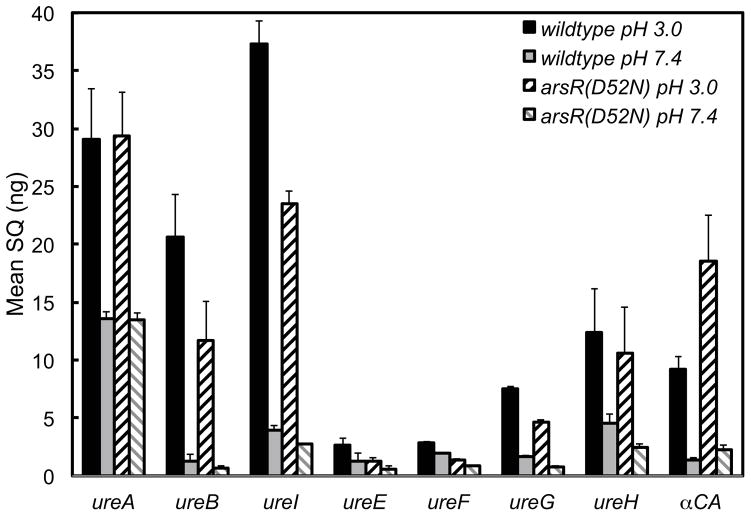
Quantitative PCR was carried out to determine the effect of ArsR phosphorylation on the expression of the genes of the 2 operons that constitute the urease gene cluster and of α-carbonic anhydrase. Wildtype and arsR(D52N) strains were incubated at pH 3.0 and 7.4 for 4 hours in dialysis cassettes, followed by RNA isolation and subsequent qPCR. The level of the ureA transcript of the arsR(D52N) strain was identical to the wildtype at both pHs, there was a 2 fold increase of transcript at pH 3.0 as compared to pH 7.4 (Figure 4). The level of ureB, ureI, ureE, ureF, ureG, and ureH transcripts of the arsR(D52N) strain was decreased at both pHs as compared with the wildtype strain, indicating that ArsR phosphorylation is needed for the constitutive expression of these genes (Figure 4). In contrast, the difference between the level of transcript at pH 3.0 and pH 7.4 was the same for both the wildtype and arsR(D52N) strains, suggesting that ArsR phosphorylation is not required for the increased transcription of these genes in response to an acid challenge, and that this response is instead driven by ArsS. Phosphorylated ArsR controls transcription of these genes beyond their baseline levels, while ArsS regulates activation of existing urease in response to acid. The level of the α-carbonic anhydrase transcript of the arsR(D52N) strain was twice that of the wildtype strain at pH 3.0 and pH 7.4 and there was an increase in transcription of both strains when challenged with acid. These data suggest that ArsR phosphorylation negatively regulates α-carbonic anhydrase but is not required for the increase in transcription seen with an acid challenge.
Figure 4.
ArsR phosphorylation affects expression of the urease gene cluster and α-carbonic anhydrase. Wildtype and arsR(D52N) strains were incubated at pH 3.0 and 7.4 for 4 hours, followed by RNA isolation and subsequent qPCR. The level of ureA transcript of the arsR(D52N) strain was identical to the wildtype at both pHs and there was a 2 fold increase at pH 3.0 vs pH 7.4. The level of ureB, ureI, ureE, ureF, ureG, and ureH transcripts of the arsR(D52N) strain were decreased at both pHs. In contrast, the relative difference between the level of transcript at pH 3.0 and pH 7.4 were the same for both the wildtype and arsR(D52N) strains. The level of α-carbonic anhydrase transcript of the arsR(D52N) strain was twice that of the wildtype strain at pH 3.0 and pH 7.4 and there was an increase in transcription of both strains when challenged with acid. n=3, error bars represent standard deviation. SQ refers to the starting quantity of RNA.
Membrane trafficking of UreA and UreB is not affected by loss of ArsR phosphorylation
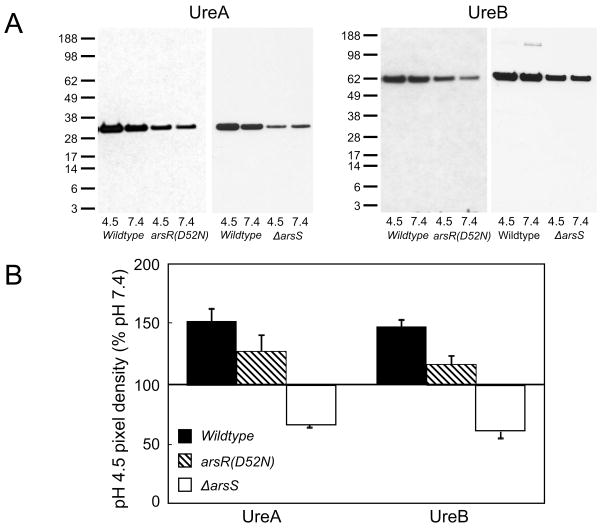
Previously it was shown that the cytoplasmic urease structural subunits, UreA and UreB, and the accessory proteins required for nickel insertion traffic to the membrane following a 30 minute incubation in acid (7). The presence of ArsS is initially required for trafficking, but with longer incubation times (>90 minutes), other compensatory mechanisms are activated to bring the urease proteins to the membrane in the absence of ArsS (7). Similar experiments were carried out to determine whether phosphorylation of ArsR is required for the movement of the urease proteins to the membrane. Membranes were prepared following a 30-minute incubation at pH 4.5 or 7.4 without urea. Overall, protein levels for UreA and UreB in the membrane fraction were decreased in the arsR(D52N) strain, but the trafficking response was not affected by the absence of phosphorylation. There was increased association of the cytoplasmic urease structural proteins, UreA and UreB, with the membrane at pH 4.5 compared to pH 7.4 in both the wildtype and the arsR(D52N) strain (Figure 5). This demonstrates that, while ArsS is required for the movement of urease proteins to the membrane in acid, this effect is not dependent on phosphorylation of ArsR.
Figure 5.
Trafficking of urease proteins to the membrane in acid is not affected by absence of phosphorylation of ArsR, but overall protein levels are decreased. Wildtype, ΔarsS and arsR(D52N) bacteria were incubated at pH 4.5 or 7.4 for 30 minutes without urea. Membranes were then purified and fractionated by SDS-PAGE. UreA and UreB were detected by Western blot. Representative blots for each antibody are shown in (A). Densitometry at pH 4.5 expressed as percent of pH 7.4 for each strain is shown in (B). N=3 for each condition, error bars represent the s.e.m.
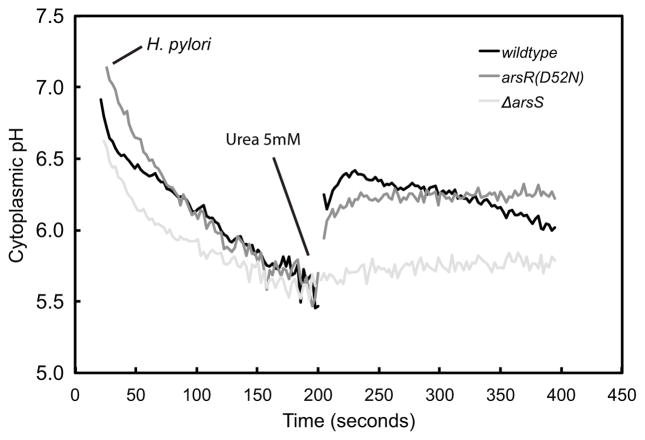
Increased cytoplasmic pH following urea addition is not affected by loss of ArsR phosphorylation but is abolished in the absence of ArsS
Cytoplasmic pH in the wildtype, ΔarsS, and arsR(D52N) strains was measured using BCECF-AM. As expected, internal pH (pHi) decreased in all 3 strains in response to external acidity (pH 4.5), to ~pH 5.5. With the addition of urea, as previously shown (4), the pHi rises to around 6.3. This increase in pHi is due to the opening of UreI, the proton-gated urea channel, and the subsequent influx and hydrolysis of urea by the preassembled UreI/urease membrane complex. The addition of urea also elicited an increase in pHi of the arsR(D52N) strain, similar to the wildtype strain. However, the rise in pHi with urea addition was absent in the ΔarsS strain (Figure 6). These results demonstrate that ArsS is required for UreI/urease complex assembly and activation and this is independent of ArsR phosphorylation. The absence of ArsS may result in either the loss of UreI/urease complex formation or urease activation or both.
Figure 6.
Urea addition increases cytoplasmic pH in acid. Cytoplasmic pH was monitored using the pH sensitive fluorophore, BCECF-AM. Cytoplasmic pH decreased in the wildtype (black), arsR(D52N) (grey) and ΔarsS (light grey) strains when presented with an acid challenge (pH 4.5). The addition of 5 mM urea resulted in a rapid increase of cytoplasmic pH in the wildtype and arsR(D52N) strains, but not the ΔarsS strain. These results demonstrate that ArsS is required for UreI/urease complex function independent of ArsR phosphorylation
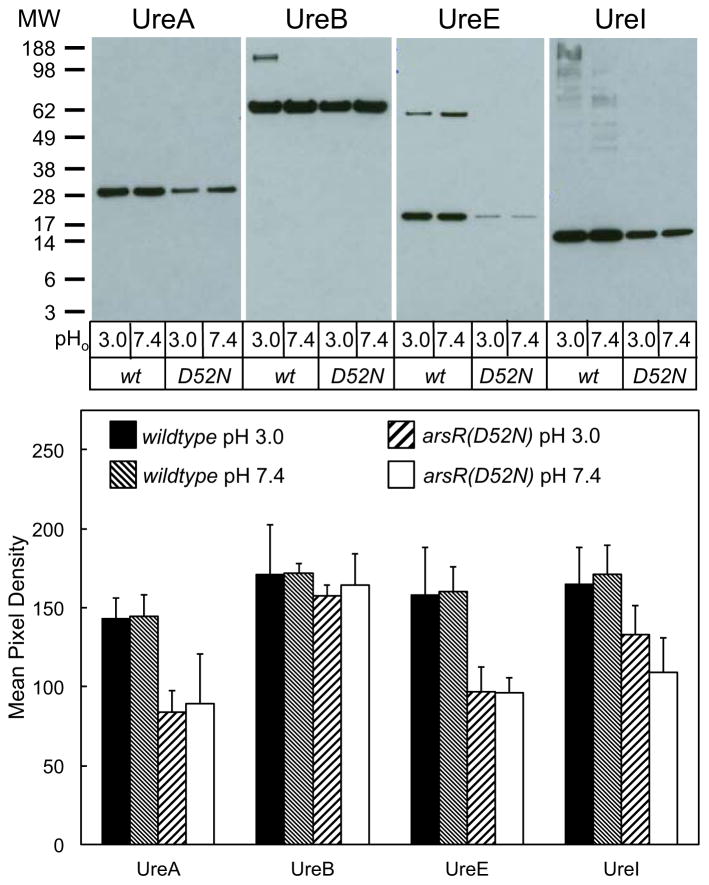
UreA, UreI, and UreE proteins are decreased in the arsR(D52N) strain in a pH independent manner
Wildtype and arsR(D52N) bacteria were incubated in dialysis chambers for 4 hours at pH 3.0 or 7.4 with urea. Bacteria were then lysed and protein was fractionated by SDS-PAGE, then analyzed by Western blot, using antibodies against UreA, UreB, UreI, and UreE. For UreA, UreE, and UreI, there was a significant decrease in the level of protein detected at both pH 3.0 and pH 7.4 in the arsR(D52N) strain as compared to the wildtype. These data show that ArsR phosphorylation increases expression of most of the urease gene cluster. However, the change in UreB was not significant at either pH as compared to wildtype (Figure 7), perhaps due to modification of expression of UreB induced by the cis-encoded, antisense ureB siRNA (39).
Figure 7.
UreA, UreE, and UreI proteins are decreased in the arsR(D52N) strain in a pH independent manner. Wildtype and arsR(D52N) bacteria were incubated in dialysis cassettes suspended in media for 4 hours at pH 3.0 or 7.4 with 5 mM urea. Medium pH did not change after 4 hours. Bacteria were then lysed, fractionated by SDS-PAGE, and proteins detected by Western blot. Representative blots for each antibody are shown in the top portion of the figure, and averaged densitometry results are shown in the graph on the bottom. N=3 for each condition, pHo = medium pH, error bars represent standard deviation.
Discussion
H. pylori has few TCSs compared to other bacteria, consistent with its exposure to the limited environment of the human stomach (40). These TCSs are critical for colonization and pathogenesis as they are involved in regulation of important virulence traits, such as chemotaxis and acid acclimation (40). The ArsRS TCS is a major contributor to acid acclimation in H. pylori. ArsS is a membrane protein shown to sense acidity (10). The response regulator ArsR is required for bacterial viability, but its growth-essential functions are independent of phosphorylation (41), as shown by the viability of the arsR(D52N) mutant, which is unable to be phosphorylated. Use of this mutant allows analysis of TCS functions in acid acclimation that are specific to each protein or to the fully functioning pair.
Phosphorylation of ArsR leads to binding to its own promoter (PArsR) and repression of gene expression (21). This self-regulatory mechanism prevents overexpression of target genes regulated by phosphorylation of ArsR (25). In the case of the urease operon, acidification of the periplasm results in the protonation of His94 in the periplasmic domain of ArsS and auto-phosphorylation (42). The phosphate is then transferred to ArsR, which initiates increased expression of the urease genes. UreI, the proton gated urea channel, allows access of urea to an augmented pool of cytoplasmic urease, which is driven to and activated at the inner membrane. Urease and accessory proteins UreE, F, G, and H are trafficked to the membrane via signaling from ArsS through an unidentified, ArsR independent pathway. The use of an alternative response regulator is not unique. There are examples in other bacteria of histidine kinase sensors functioning independently of their cognate response regulators as well as response regulators interacting with multiple histidine kinases. For example, in E. coli, the OmpR response regulator receives phosphate in vivo not only from its cognate histidine kinase, EnvZ, but also from the ArcB histidine kinase (43). In H. pylori, this response leads to periplasmic and cytoplasmic alkalization, and if allowed to continue unregulated, may lead to over alkalization and loss of survival at neutral pH (44). Repression of ArsR by its phosphorylated form would prevent this response. Accordingly, total urease activity is increased in the arsR(D52N) strain, suggesting the absence of repression and down regulation seen with phosphorylation.
The genes of the urease gene cluster are part of the ArsRS regulon. The gene cluster is comprised of two operons, one regulated by PureA that controls the expression of ureA and ureB and one regulated by PureI that controls the expression of ureI, E, F, G, and H. In acid, there is increased transcription of the urease genes and the genes that encode the urease accessory proteins. In the absence of ArsR phosphorylation, expression of the urease gene cluster is down-regulated at neutral and acidic pH. Although the expression of the urease gene cluster is reduced, the transcriptomal response to acid is maintained, with increased expression at low pH when compared to neutral pH. This finding implies that, although ArsR phosphorylation is required for increased levels of transcription of the urease genes, ArsR phosphorylation is not required for baseline constitutive production of these genes. Although there is an overall decrease in expression of the urease gene cluster in the absence of ArsR phosphorylation, the acid-induced differential in transcription of the urease genes is maintained. Other compensatory or redundant mechanisms, such as the FlgS system, are likely in place to assure the response to acid is maintained even in the setting of overall decreased gene expression in the absence of ArsR phosphorylation.
The level of UreA, UreI, and UreE proteins is decreased in the arsR(D52N) strain in a pH independent manner. However the loss of ArsR phosphorylation does not alter the amount of UreB. Despite a decrease of both RNA and protein levels of the members of the urease gene cluster, there is a small but significant increase in total lysate urease activity in the arsR(D52N) strain. This finding suggests that ArsR~P differentially regulates the activity of RNAses and proteases to alter the degradation rates of RNA and protein. The amount of urease expressed by the arsR(D52N) strain reflects the constitutive production of the enzyme. More significantly, the increased urease activity of the total lysate of the arsR(D52N) strain suggests that more of the constitutively produced apourease is active. Therefore, in the absence of ArsR phosphorylation, the decreased production of both urease RNA and protein is partially offset by an increase in activated urease. Regulation of activation of apourease allows for the rapid response of H. pylori to an acid challenge that cannot be achieved in a timely fashion by only increased transcription and translation.
Trafficking of the urease proteins to the inner membrane at acidic pH is driven by ArsS and FlgS (7, 9). In the absence of arsS, the trafficking response is diminished for the first 30 minutes, but normalizes after 90 minutes, likely driven by decreased cytoplasmic pH and subsequent activation of the cytoplasmic acid sensing histidine kinase FlgS (7). In the absence of flgS, trafficking of the urease proteins to the membrane is diminished (9). Although the total amount of urease proteins is decreased in the absence of ArsR phosphorylation, the trafficking response is unchanged. This suggests that ArsS drives this component of acid acclimation, independent of phosphorylation state of its response regulator, ArsR.
The intrabacterial trafficking of UreA and UreB to the cytoplasmic membrane of H. pylori depends on the expression of UreI (6–8). This acid-dependent relocation appears to be essential for survival of the organism in gastric acidity since it results in a 3 to 4 fold activation of the cytoplasmic urease of the organism (45). Conversely, the lower urease activity at neutral medium pH may prevent lethal alkalization of the organism (44). This is a set of responses unique to H. pylori acid acclimation.
In the absence of urea, the cytoplasmic pH of H. pylori decreases with decreasing external pH. In acid, urea addition results in an immediate and rapid increase of cytoplasmic pH, as urea is hydrolyzed at the inner-membrane, producing ammonia and carbon dioxide. The urea-induced increase in cytoplasmic pH requires the urea channel, UreI, to permit urea access to urease. In the absence of UreI, passive diffusion of 5 mM urea through the membrane is inadequate to raise internal pH. As shown here, ArsS is also required for the urea-induced increase in cytoplasmic pH, and is independent of ArsR phosphorylation. This suggests that one role of ArsS in cytoplasmic pH homeostasis is trafficking and subsequent activation of urease.
The data presented here support and confirm the role of ArsR~P in the transcriptional regulation of the urease gene cluster. Phosphorylation of ArsR increases transcription of the urease genes equally at both acidic and neutral pH, showing that the phosphorylation driven increased transcription is pH independent. Therefore the acid-induced up-regulation of the urease gene cluster is mediated through a different transcription factor and requires ArsS. ArsR phosphorylation increases the level of several proteins encoded by the urease gene cluster and is pH-independent, paralleling the transcriptomal data. ArsR~P is not needed for the acid induced trafficking and activation of urease and its accessory proteins in association with UreI, but ArsS is required. Despite decreased production of urease and the urease accessory proteins in the arsR(D52N) strain, there was a small but significant increase in urease activity of total bacterial lysate, due to the loss of negative feedback by ArsR~P. This negative feedback downregulates the expression of urease to prevent over alkalization of the cytoplasm, allowing survival even at neutral pH. The constitutive regulation of essential genes by ArsR in a phosphorylation independent manner and negative feedback of phosphorylated ArsR on itself may provide an explanation of how phosphorylation-independent activation might work. For example, in the absence of phosphorylation, ArsR may be produced in large quantities and bind to and up-regulate essential genes, overcoming the need for phosphotransfer from ArsS. This is similar to the regulation by the DesK/DesR TCS in Bacillus subtilis where, in the absence of phosphorylation, the response regulator DesR is over-produced, allowing dimerization and promoter binding of genes in its regulon (46).
Understanding the mechanisms that allow H. pylori, a neutralophile, to colonize an acidic environment provides insight into novel treatment options and bacterial physiology. ArsS senses acidity and is required for initiation of protein trafficking independent of ArsR. ArsS is important for activation of existing urease at the membrane when the pH drops. ArsR phosphorylation is required for the up-regulation of the urease gene cluster and is essential for expression of the urease gene cluster independent of pH. ArsR phosphorylation, via negative feedback on its promoter (21), prevents over-alkalization in the absence of external acidity. This may be relevant to transmission from person to person, when the bacteria must survive passage out of the stomach. From the perspective of drug development, compounds targeting phosphorylation of ArsR would need to be combined with strong acid suppression while those targeting ArsS would likely be effective in the presence of physiologic acidity.
Acknowledgments
Supported by NIH and USVA grants NIH/NCATS UL1TR000124 (UCLA CTSI, EAM), K08DK100661-01 (EAM), 2I01BX001006-05 (GS), and the UCLA CDI (EAM).
References
- 1.Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, et al. Helicobacter pylori infection and the risk of gastric carcinoma. The New England journal of medicine. 1991 Oct 17;325(16):1127–31. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 2.Malfertheiner P, Megraud F, O’Morain C, Bazzoli F, El-Omar E, Graham D, et al. Current concepts in the management of Helicobacter pylori infection: the Maastricht III Consensus Report. Gut. 2007 Jun;56(6):772–81. doi: 10.1136/gut.2006.101634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcus EA, Moshfegh AP, Sachs G, Scott DR. The periplasmic alpha-carbonic anhydrase activity of Helicobacter pylori is essential for acid acclimation. Journal of bacteriology. 2005 Jan;187(2):729–38. doi: 10.1128/JB.187.2.729-738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott DR, Marcus EA, Weeks DL, Sachs G. Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology. 2002 Jul;123(1):187–95. doi: 10.1053/gast.2002.34218. [DOI] [PubMed] [Google Scholar]
- 5.Weeks DL, Eskandari S, Scott DR, Sachs G. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science (New York, NY. 2000 Jan 21;287(5452):482–5. doi: 10.1126/science.287.5452.482. [DOI] [PubMed] [Google Scholar]
- 6.Hong W, Sano K, Morimatsu S, Scott DR, Weeks DL, Sachs G, et al. Medium pH-dependent redistribution of the urease of Helicobacter pylori. Journal of medical microbiology. 2003 Mar;52(Pt 3):211–6. doi: 10.1099/jmm.0.05072-0. [DOI] [PubMed] [Google Scholar]
- 7.Marcus EA, Sachs G, Wen Y, Feng J, Scott DR. Role of the Helicobacter pylori sensor kinase ArsS in protein trafficking and acid acclimation. Journal of bacteriology. 2012 Oct;194(20):5545–51. doi: 10.1128/JB.01263-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voland P, Weeks DL, Marcus EA, Prinz C, Sachs G, Scott D. Interactions among the seven Helicobacter pylori proteins encoded by the urease gene cluster. American journal of physiology. 2003 Jan;284(1):G96–G106. doi: 10.1152/ajpgi.00160.2002. [DOI] [PubMed] [Google Scholar]
- 9.Scott DR, Marcus EA, Wen Y, Singh S, Feng J, Sachs G. Cytoplasmic histidine kinase (HP0244)-regulated assembly of urease with UreI, a channel for urea and its metabolites, CO2, NH3, and NH4(+), is necessary for acid survival of Helicobacter pylori. Journal of bacteriology. 2010 Jan;192(1):94–103. doi: 10.1128/JB.00848-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pflock M, Dietz P, Schar J, Beier D. Genetic evidence for histidine kinase HP165 being an acid sensor of Helicobacter pylori. FEMS microbiology letters. 2004 May 1;234(1):51–61. doi: 10.1016/j.femsle.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 11.Hoch JA. Two-component and phosphorelay signal transduction. Current opinion in microbiology. 2000 Apr;3(2):165–70. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto K, Hirao K, Oshima T, Aiba H, Utsumi R, Ishihama A. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. The Journal of biological chemistry. 2005 Jan 14;280(2):1448–56. doi: 10.1074/jbc.M410104200. [DOI] [PubMed] [Google Scholar]
- 13.Rowland MA, Deeds EJ. Crosstalk and the evolution of specificity in two-component signaling. Proceedings of the National Academy of Sciences of the United States of America. 2014 Apr 15;111(15):5550–5. doi: 10.1073/pnas.1317178111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997 Aug 7;388(6642):539–47. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 15.Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999 Jan 14;397(6715):176–80. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 16.Howitt MR, Lee JY, Lertsethtakarn P, Vogelmann R, Joubert LM, Ottemann KM, et al. ChePep controls Helicobacter pylori Infection of the gastric glands and chemotaxis in the Epsilonproteobacteria. mBio. 2011;2(4) doi: 10.1128/mBio.00098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waidner B, Melchers K, Stahler FN, Kist M, Bereswill S. The Helicobacter pylori CrdRS two-component regulation system (HP1364/HP1365) is required for copper-mediated induction of the copper resistance determinant CrdA. Journal of bacteriology. 2005 Jul;187(13):4683–8. doi: 10.1128/JB.187.13.4683-4688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niehus E, Ye F, Suerbaum S, Josenhans C. Growth phase-dependent and differential transcriptional control of flagellar genes in Helicobacter pylori. Microbiology (Reading, England) 2002 Dec;148(Pt 12):3827–37. doi: 10.1099/00221287-148-12-3827. [DOI] [PubMed] [Google Scholar]
- 19.Wen Y, Feng J, Scott DR, Marcus EA, Sachs G. The pH-responsive regulon of HP0244 (flgS), the cytoplasmic histidine kinase of Helicobacter pylori. Journal of bacteriology. 2009 Jan;191(2):449–60. doi: 10.1128/JB.01219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pflock M, Kennard S, Finsterer N, Beier D. Acid-responsive gene regulation in the human pathogen Helicobacter pylori. Journal of biotechnology. 2006 Oct 20;126(1):52–60. doi: 10.1016/j.jbiotec.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 21.Dietz P, Gerlach G, Beier D. Identification of target genes regulated by the two-component system HP166-HP165 of Helicobacter pylori. Journal of bacteriology. 2002 Jan;184(2):350–62. doi: 10.1128/JB.184.2.350-362.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pflock M, Kennard S, Delany I, Scarlato V, Beier D. Acid-induced activation of the urease promoters is mediated directly by the ArsRS two-component system of Helicobacter pylori. Infection and immunity. 2005 Oct;73(10):6437–45. doi: 10.1128/IAI.73.10.6437-6445.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen Y, Feng J, Scott DR, Marcus EA, Sachs G. Involvement of the HP0165-HP0166 two-component system in expression of some acidic-pH-upregulated genes of Helicobacter pylori. Journal of bacteriology. 2006 Mar;188(5):1750–61. doi: 10.1128/JB.188.5.1750-1761.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forsyth MH, Cao P, Garcia PP, Hall JD, Cover TL. Genome-wide transcriptional profiling in a histidine kinase mutant of Helicobacter pylori identifies members of a regulon. Journal of bacteriology. 2002 Aug;184(16):4630–5. doi: 10.1128/JB.184.16.4630-4635.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pflock M, Finsterer N, Joseph B, Mollenkopf H, Meyer TF, Beier D. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. Journal of bacteriology. 2006 May;188(10):3449–62. doi: 10.1128/JB.188.10.3449-3462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wen Y, Feng J, Scott DR, Marcus EA, Sachs G. The HP0165-HP0166 two-component system (ArsRS) regulates acid-induced expression of HP1186 alpha-carbonic anhydrase in Helicobacter pylori by activating the pH-dependent promoter. Journal of bacteriology. 2007 Mar;189(6):2426–34. doi: 10.1128/JB.01492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allan E, Clayton CL, McLaren A, Wallace DM, Wren BW. Characterization of the low-pH responses of Helicobacter pylori using genomic DNA arrays. Microbiology (Reading, England) 2001 Aug;147(Pt 8):2285–92. doi: 10.1099/00221287-147-8-2285. [DOI] [PubMed] [Google Scholar]
- 28.Merrell DS, Goodrich ML, Otto G, Tompkins LS, Falkow S. pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infection and immunity. 2003 Jun;71(6):3529–39. doi: 10.1128/IAI.71.6.3529-3539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen Y, Marcus EA, Matrubutham U, Gleeson MA, Scott DR, Sachs G. Acid-adaptive genes of Helicobacter pylori. Infection and immunity. 2003 Oct;71(10):5921–39. doi: 10.1128/IAI.71.10.5921-5939.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slonczewski JL, Kirkpatrick C. Proteomic analysis of pH-dependent stress responses in Escherichia coli and Helicobacter pylori using two-dimensional gel electrophoresis. Methods in enzymology. 2002;358:228–42. doi: 10.1016/s0076-6879(02)58092-7. [DOI] [PubMed] [Google Scholar]
- 31.Loh JT, Cover TL. Requirement of histidine kinases HP0165 and HP1364 for acid resistance in Helicobacter pylori. Infection and immunity. 2006 May;74(5):3052–9. doi: 10.1128/IAI.74.5.3052-3059.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel FM, Brent K, Kingston RE, Moore DD, Smith JA, Seidman JG, et al., editors. Current Protocols in Molecular Biology. New York: Wiley; 1987. pp. 2.4.1–2.4.2. [DOI] [PubMed] [Google Scholar]
- 33.Marcus EA, Inatomi N, Nagami GT, Sachs G, Scott DR. The effects of varying acidity on Helicobacter pylori growth and the bactericidal efficacy of ampicillin. Alimentary pharmacology & therapeutics. 2012 Nov;36(10):972–9. doi: 10.1111/apt.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonald JA, Speeg KV, Jr, Campbell JW. Urease: a sensitive and specific radiometric assay. Enzymologia. 1972 Jan 31;42(1):1–9. [PubMed] [Google Scholar]
- 35.Scott DR, Weeks D, Hong C, Postius S, Melchers K, Sachs G. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology. 1998 Jan;114(1):58–70. doi: 10.1016/s0016-5085(98)70633-x. [DOI] [PubMed] [Google Scholar]
- 36.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007 May 15;23(10):1289–91. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 37.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3--new capabilities and interfaces. Nucleic acids research. 2012 Aug;40(15):e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott DR, Marcus EA, Weeks DL, Lee A, Melchers K, Sachs G. Expression of the Helicobacter pylori ureI gene is required for acidic pH activation of cytoplasmic urease. Infection and immunity. 2000 Feb;68(2):470–7. doi: 10.1128/iai.68.2.470-477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wen Y, Feng J, Scott DR, Marcus EA, Sachs G. A cis-encoded antisense small RNA regulated by the HP0165-HP0166 two-component system controls expression of ureB in Helicobacter pylori. Journal of bacteriology. 2011 Jan;193(1):40–51. doi: 10.1128/JB.00800-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joseph B, Beier D. Global analysis of two-component gene regulation in H. pylori by mutation analysis and transcriptional profiling. Methods in enzymology. 2007;423:514–30. doi: 10.1016/S0076-6879(07)23025-3. [DOI] [PubMed] [Google Scholar]
- 41.Schar J, Sickmann A, Beier D. Phosphorylation-independent activity of atypical response regulators of Helicobacter pylori. Journal of bacteriology. 2005 May;187(9):3100–9. doi: 10.1128/JB.187.9.3100-3109.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muller S, Gotz M, Beier D. Histidine residue 94 is involved in pH sensing by histidine kinase ArsS of Helicobacter pylori. PLoS One. 2009;4(9):e6930. doi: 10.1371/journal.pone.0006930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsubara M, Kitaoka SI, Takeda SI, Mizuno T. Tuning of the porin expression under anaerobic growth conditions by his-to-Asp cross-phosphorelay through both the EnvZ-osmosensor and ArcB-anaerosensor in Escherichia coli. Genes to cells: devoted to molecular & cellular mechanisms. 2000 Jul;5(7):555–69. doi: 10.1046/j.1365-2443.2000.00347.x. [DOI] [PubMed] [Google Scholar]
- 44.Clyne M, Labigne A, Drumm B. Helicobacter pylori requires an acidic environment to survive in the presence of urea. Infection and immunity. 1995 May;63(5):1669–73. doi: 10.1128/iai.63.5.1669-1673.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott DR, Marcus EA, Wen Y, Singh S, Feng J, Sachs G. Cytoplasmic histidine kinase (HP0244)-regulated assembly of urease with UreI, a channel for urea and its metabolites, CO2, NH3, and NH4+, is necessary for acid survival of Helicobacter pylori. Journal of bacteriology. 2010 Jan;192(1):94–103. doi: 10.1128/JB.00848-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trajtenberg F, Albanesi D, Ruetalo N, Botti H, Mechaly AE, Nieves M, et al. Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation. mBio. 2014;5(6):e02105. doi: 10.1128/mBio.02105-14. [DOI] [PMC free article] [PubMed] [Google Scholar]