Abstract
Background
Multi-drug resistant (MDR) Enterobacteriaceae are on the increase worldwide and their spread has become a global challenge. Escalating the challenge is the possibility that many of these are Carbapenemase-producing Enterobacteriaceae (CPE). This further complicates patient management. The magnitude of MDR-CPE in many developed settings has been reported, however, there is paucity of data from resource limited settings. We evaluated the epidemiology of MDR-CPE of clinical origin in South Western Uganda.
Methods
From September 2013 to June 2014, all Enterobacteriaceae isolated from diverse specimens obtained from patients attending Mbarara Regional Referral Hospital, South-western Uganda, were screened for MDR in a laboratory-based cross sectional study. Isolates found to be MDR were screened for carbapenem susceptibility/resistance phenotypically by Kirby Bauer disc diffusion method following CLSI guidelines and genetically using the multiplex real-time Polymerase Chain Reaction (RT-PCR).
Results
Of the 658 strains isolated, 183 (27.8%) were MDR and 68 (37.15%) of those MDR exhibited at least one form of carbapenem resistance with 23 (12.57%) and 56 (30.60%) isolates expressing phenotypic and genetic resistance, respectively. Eleven MDR-CPE (6.01%) isolates exhibited both phenotypic and genotypic resistance to carbapenems. Only blaVIM and blaOXA-48 genes were detected among the genetically resistant isolates.
Conclusion
The high prevalence of MDR-CPE calls for aggressive infection control and prevention strategies, including reinforcement of hand hygiene, using contact precautions and early detection of CPE through use of targeted surveillance and molecular techniques in resource limited settings.
Keywords: Enterobacteriaceae, carbapenemase, multi-drug resistant, CPE, ESBL
1. INTRODUCTION
Carbapenems are crucial for the management of life-threatening infections [1] and are often the antimicrobials of last resort to treat infections due to extended-spectrum β-lactamase (ESBL)-or plasmid-mediated AmpC (pAmpC)-producing organisms of the Enterobacteriaceae family [1]. These pathogens are frequently also resistant to other antibiotic classes and are described as multi-drug resistant (MDR) [2–4].
Carbapenem-resistant Enterobacteriaceae (CRE), are a family of germs that are difficult to treat because they have high levels of resistance to antibiotics [5]. Resistance in these organisms is largely due to production of beta-lactamase enzymes [6] like Klebsiella pneumoniae carbapenemase (KPC), New Delhi Metallo-betalactamase (NDM) and Verona Integron-Mediated Metallo-β-lactamase (VIM), that break down carbapenems and make them ineffective [5].
Acquired class A (KPC), class B (IMP, VIM, NDM), or class D (OXA-48, OXA-181) carbapenemases are the most important determinants sustaining resistance to carbapenems [6–7]. The corresponding genes are mostly plasmid-located and associated with various mobile genetic structures (insertion sequences, integrons, transposons), further enhancing their spread.
Unfortunately, the prevalence of CPE has increased worldwide during the past 10 years, seriously compromising the therapeutic armamentarium [7–10]. There is paucity of data regarding their prevalence in Resource Limited Settings and as such, not much has been/ is being done to contain them. Although it is a requirement in hospital pharmacies to have a prescription note, in private pharmacies, patients access antibiotics freely over the counter without necessarily presenting a prescription note.
To ensure their containment, wide dissemination of information that will enable development of evidence-based strategies involving microbiologists, clinicians and other stakeholders is essential. Herein, we report the epidemiology of MDR Enterobacteriaceae isolates of clinical origin in a low income setting.
2. MATERIALS AND METHODS
We conducted a cross-sectional study at Mbarara Regional Referral Hospital (MRRH), Mbarara, Uganda, from September 2013 to June 2014. MRRH is the regional referral hospital in south Western Uganda. It provides public healthcare with general and teaching hospital facilities and has a capacity of more than 600 beds. The study was approved by the Faculty of Medicine Research and Ethics Committee (FREC), the Institutional Review Committee (IRC) of Mbarara University of Science and Technology and the Uganda National Council for Science and Technology.
Viable isolates of the Enterobacteriaceae family obtained from various clinical specimens of all patients attending MRRH during the study period were identified following standard microbiological procedures and then screened for phenotypic multi-drug resistance using Kirby-Bauer disc diffusion method following CLSI guidelines [11].
Isolates that screened positive for MDR were screened for carbapenem susceptibility/resistance phenotypically by Kirby Bauer disc diffusion method following CLSI guidelines [11]. Briefly, a 10 µg imipenem disc was placed on lawn culture of the isolate on Mueller Hinton agar and Phenotypic expression of a Carbapenemase was taken to be detected if the diameter of zone of inhibition was ≤19mm and, genetically using the multiplex real-time Polymerase Chain Reaction (RT-PCR) at Epicentre Mbarara Research Centre Laboratory. We used the QIAamp® DNA Min kit (QIAGEN, GmbH Ebensburg, German) for extraction and the Qiagen Multiplex PCR kit (QIAGEN, GmbH Ebensburg, German) for the amplification. PCR for the following carbapenemase genes blaIMP; blaVIM; blaOXA-48; blaKPC; was done as described previously [12–15].
Standardized controls were incorporated at all levels of analysis/ testing to rule out contamination and ensure good performance of the kits and processes. The control bacterial strains (Klebsiella pneumoniae 211 (T), Klebsiella pneumoniae 714, DSMZ Escherichia coli 9377 and Escherichia coli ATCC 25922) and DNA products were obtained from Institute of Microbiology, Gissen, Germany.
All the data were summarized as proportions. The primary outcome of interest was resistance to carbapenems. Prevalence ratios for the phenotypic and genetic characterization were obtained. Kappa statistics for the comparison between phenotypic and genotypic characterization were obtained. STATA, version 13 (StataCorp, College Station, Texas, USA) was used for all the analyses. A p-value ≤ 0.05 was considered to be statistically significant. The graphs and pie-charts were drawn using Microsoft Excel 2010.
3. RESULTS AND DISCUSSION
Of the 658 Enterobacteriaceae strains isolated, 183(27.8%), representing 22 different species of Enterobacteriaceae from a total of 11 types of clinical samples (Fig. 2), were found to be MDR and were screened for carbapenem resistance. Escherichia coli and Klebsiella pneumoniae were the most common isolated strains (54.1% & 18.6%, respectively) (Fig. 2). Of the 183 MDR isolates (Table 3), 68 (37.15%) exhibited at least one form of carbapenem resistance with 23 (12.5%, 95% CI: 7.7% – 17.4%) and 56 (30.6%, 95% CI: 23.8% – 37.3%) isolates expressing phenotypic and genetic resistance, respectively (Table 1, Table 2). Eleven (6.01%) isolates exhibited both phenotypic and genotypic resistance to carbapenems (Table 2). The isolates were recovered from both in-patients and out- patients.
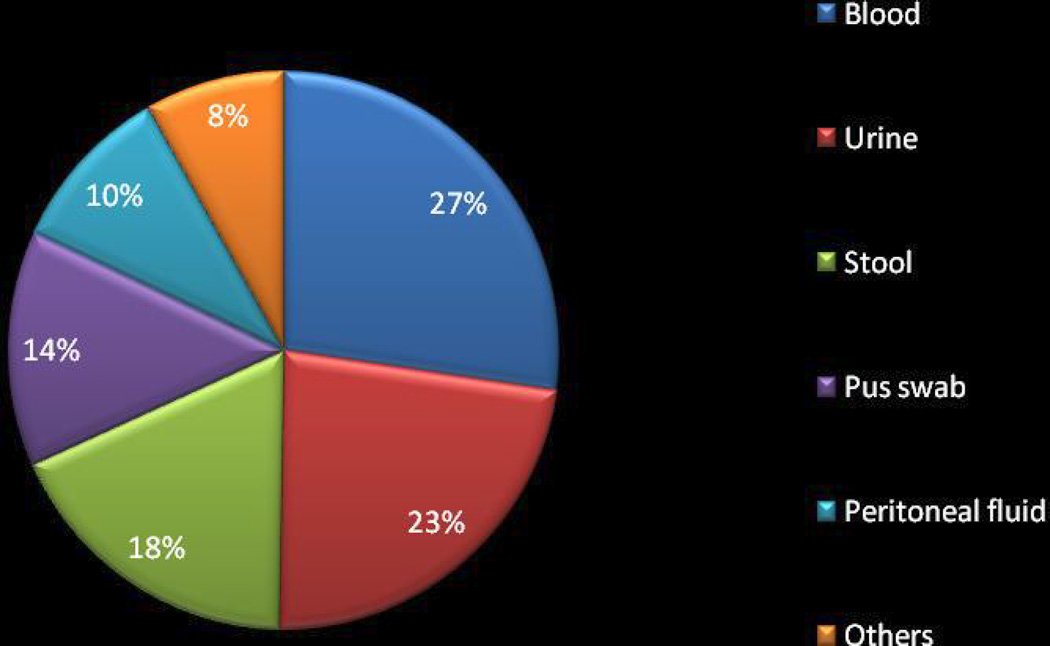
Fig. 2. Distribution of clinical specimens used in the study (N=183).
Others include; HVS, CSF, Pleural fluid, Sputum, Nasal swab
Table 3.
Distribution of different CRDG among the multi-drug resistant Gram negative bacteria
| Isolate/organism | Carbapenem-resistance determining genes, No. (%) |
Total detection |
|
|---|---|---|---|
| VIM | OXA-48 | ||
| Escherichia coli (n=99) | 20(46.5) | 6(46.1) | 26 |
| Klebsiella pneumoniae (n=35) | 17(39.5) | 5(38.5) | 22 |
| Proteus mirabilis (n=10) | 1(2.3) | 1(7.7) | 2 |
| Salmonella spp (n=10) | 3(7.1) | 3 | |
| Morganella morganii (n=3) | 1(2.3) | 1 | |
| Enterobacter sakazaki (n=3) | 1(7.7) | 1 | |
| Stenotrophomonas spp (n=2) | 1(2.3) | 1 | |
| Total | 43 (100) | 13(100) | 56 |
Table 1.
Phenotypic and genetic characterization
| Variable | n (%) |
|---|---|
| Phenotypic | |
| Ceftriaxone | 181 (98.91) |
| Ceftazidime | 181 (98.91) |
| Cefot/cefuroxime | 181 (98.91) |
| Ciprofloxacin | 155 (84.70) |
| Imipenem | 9 (4.92) |
| Total phenotypic | 23 (12.57) |
| Genotypic resistance | |
| Genotypic markers of resistance | |
| VIM | 43 (23.50) |
| OXA-48 | 13 (7.10) |
| Total genotypic resistance | 56 (30.60) |
Table 2.
Phenotypic and genetic resistance
| Genetic characterization | Total | |||
|---|---|---|---|---|
|
Phenotypic characterization |
Result | Resistant | Susceptible | |
| Resistant | 11 | 12 | 23 | |
| Susceptible | 45 | 115 | 160 | |
| Total | 56 | 127 | 183 | |
K= 0.12, p-value =0.0276/ 0.02/ <0.05
The study obtained a prevalence of 30.6% of Carbapenem Resistance Determining Genes (CRDG) among multidrug resistant gram negative bacilli. This high prevalence is supported by previous reports in other parts of the world, 42.0% in Tanzania, between 31% and 55% in India and 43% in Tunisia [3,7–10,16], that necessitates immediate public concern and action.
Eleven isolates exhibited both genetic and phenotypic resistance. However, 12/23 phenotypically resistant isolates did not exhibit any of the tested genetic markers of resistance. This could be due to possession of other markers that were not tested in this study or new variants. Of the 56 isolates that expressed genetic markers of resistance, 45 isolates did not express phenotypic resistance. This is possibly due to possession, by some isolates, of silent genes that only exhibit phenotypic characteristics under conducive conditions.
The proportion of resistance by genetic characterization was significantly higher than that by phenotypic characterization (p= 0.0000, K= 0.12) (Table 2). Owing to this, molecular techniques enable early detection of resistance, better patient management and are a basis for early re-enforcement of infection control.
Of the most common carbapenem resistance determining genes (blaVIM, blaKPC, blaOXA-48, blaIMP-A, blaIMP-B and blaIMP-C) tested, only blaVIM and blaOXA-48 genes were detected (Fig. 1) among the genetically resistant isolates (Table 3). Genetic resistance in these isolates was therefore due to production of carbapenemases as previously reported [6,16]. VIM, which has been reported in several parts of the world [7,16] was the more frequent (43/56, 76.8%) carbapenemase identified in this population.
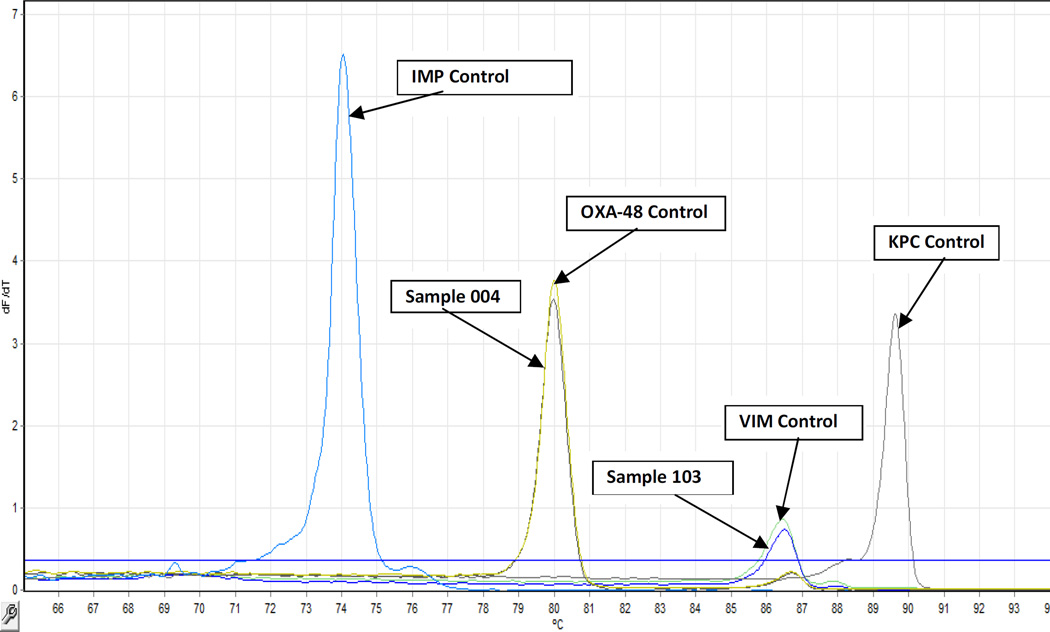
Fig. 1. RT-PCR results.
An example of Results from Real –time multiplex PCR melting curves of amplicons generated by primers targeting four Carbapenemase types. The gene targets, from left to right, are as follows: bla IMP type (Tm 74.1°C), bla OXA-48 (Tm 80°C), blaVIM (86.6°C), blaKPC (89.7°C). Sample 004 and Sample 103 are clinical isolates that carried a carbapenem resistance determining gene
4. CONCLUSION AND RECOMMENDATIONS
The high prevalence of MDR-CPE calls for aggressive infection control and prevention strategies, including reinforcement of hand hygiene, using contact precautions and early detection of CPE through use of targeted surveillance and molecular techniques in resource limited settings. Also there is need to establish local antibiotic resistance surveillance teams so as to monitor and steward antibiotic use. There is need to study the clinical significance of PCR results in resource limited settings where phenotypic testing remains the most feasible method of resistance detection/prediction.
Further sequencing of these isolates could avail more information about genetic markers not considered in this study or new variants.
ACKNOWLEDGEMENTS
We are grateful to the MRRH Microbiology department (Mr. Lwanga Nkangi and Mr. James Mwesigye) for availing the isolates and their technical support. We thank Epicentre Mbarara Research Centre administration for authorizing us to use their laboratory and the laboratory staff, especially Daniel Omoding and Ronald Sekaayi for their technical support.
FUNDING
This study was funded by MEPI-MESAU Grant Number 5R24TW008886 supported by OGAC, NIH and HRSA.
Footnotes
Authors’ contributions
This work was carried out in collaboration between all authors. Author LMA participated in study conception, design, data collection, data analysis, manuscript writing and revision. Author VK participated in data analysis, manuscript writing and revision. Authors DN and YB participated in data collection and manuscript revision. Author JB participated in study design and manuscript revision. All the authors read and approved the final manuscript.
COMPETING INTERESTS
Authors have declared that no competing interests exist.
REFERENCES
- 1.Levy HG, Gould I, Endimiani A, Ramón PP, Daikos G, Po-Ren H, Mehtar S, Petrikkos G, Casellas JM, Daciuk L, Paciel D, Novelli A, Saginur R, Pryluka D, Medina J, Savio E. Detection, Treatment and Prevention of Carbapenemase-Producing Enterobacteriaceae: Recommendations from an International Working Group. Journal of chemotherapy. 2013 doi: 10.1179/1973947812Y.0000000062. [DOI] [PubMed] [Google Scholar]
- 2.Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. Carbapenems: past, present, and future. Antimicrob Agents Chemother. 2011;55:4943–4960. doi: 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pitout JD, Laupland KB. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging publichealth concern. Lancet Infect Dis. 2008;8:159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 4.Perez F, Endimiani A, Hujer KM, Bonomo RA. The continuing challenge of ESBLs. Curr Opin Pharmacol. 2007;7:459–469. doi: 10.1016/j.coph.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Centres for Disease Control and Prevention (CDC) Detection of Verona Integron-Encoded Metallo-Beta-Lactamase in Klebsiella pneumoniae…Unites States. MMWR. 2010;59(37):1212. [PubMed] [Google Scholar]
- 6.Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The World Health Organization (WHO) Geneva, Switzerland: 2012. The evolving threat of antimicrobial resistance - options for action. [Google Scholar]
- 9.Cantón R, Akóva M, Carmeli Y, Giske CG, Glupczynski Y, Gniadkowski M, Livermore DM, Miriagou V, Naas T, Rossolini GM, Samuelsen Ø, Seifert H, Woodford N, Nordmann P, et al. the European Network on Carbapenemases: Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect. 2012;18:413–431. doi: 10.1111/j.1469-0691.2012.03821.x. [DOI] [PubMed] [Google Scholar]
- 10.Casellas JM. Antibacterial drug resistance in Latin America: consequences for infectious disease control. Rev Panam Salud Pública. 2012;30:519–528. [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute/NCCLS. Nineteenth informational supplement. Wayne, PA: CLSI; 2012. Performance Standards for Antimicrobial Susceptibility Testing. M100-S22. [Google Scholar]
- 12.Bradford PA, Bratu C, Urban C, Visalli M, Mariano N, Landman D, Rahal JJ, Brooks S, Cebular S, Quale J. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbanem hydrolyzing KPC-2 and Inhibithor-resitant TEM30 β-lactamases in New York. Clin Infect Dis. 2004;39(1):55–60. doi: 10.1086/421495. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Zhou Z, Zang Y, Yu Y. Emergence of NDM-1 producing Acinetobacter baumannii in China. J Antimicrob Chemother. 2011;66(6):1255–1259. doi: 10.1093/jac/dkr082. [DOI] [PubMed] [Google Scholar]
- 14.Mendes ER, Kiyota KA, Monteiro J, Castanheira M, Andrade SS, Gales AC, Pignatari AC, Tufik S. Rapid detection and identification of Metallo-β-lactamase-encoding genes by multiplex real time PCR assay and melt curve analysis. J Clin Microbiol. 2007;45(2):544–547. doi: 10.1128/JCM.01728-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SY, Park YJ, Yu JK, Kim HS, Park YS, Yoon JB, Yoo JY, Lee K. Prevalence and mechanisms of decreased susceptibility to carbapenems in Klebsiella pneumonia isolates. Diagn Microbiol Infect Dis. 2007;57(1):85–91. doi: 10.1016/j.diagmicrobio.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 16.Mushi Martha F, Mshana Stephen E, Imirzalioglu Can, Bwanga Freddie. Carbapenemase Genes among Multidrug Resistant Gram Negative Clinical Isolates from a Tertiary Hospital in Mwanza, Tanzania. Bio Med Research International Volume. 2014:6. doi: 10.1155/2014/303104. Article ID 303104. [DOI] [PMC free article] [PubMed] [Google Scholar]