Abstract
Human carbamoyl phosphate synthetase (CPS1), a 1500-residue multidomain enzyme, catalyzes the first step of ammonia detoxification to urea requiring N-acetyl-L-glutamate (NAG) as essential activator to prevent ammonia/amino acids depletion. Here we present the crystal structures of CPS1 in the absence and in the presence of NAG, clarifying the on/off-switching of the urea cycle by NAG. By binding at the C-terminal domain of CPS1, NAG triggers long-range conformational changes affecting the two distant phosphorylation domains. These changes, concerted with the binding of nucleotides, result in a dramatic remodeling that stabilizes the catalytically competent conformation and the building of the ~35 Å-long tunnel that allows migration of the carbamate intermediate from its site of formation to the second phosphorylation site, where carbamoyl phosphate is produced. These structures allow rationalizing the effects of mutations found in patients with CPS1 deficiency (presenting hyperammonemia, mental retardation and even death), as exemplified here for some mutations.
CPS1 activation by NAG1,2 has paramount importance, as shown by human NAG synthase deficiency3,4, which is indistinguishable clinically from CPS1 deficiency but which is successfully treated by giving to the patient the NAG analog and orphan drug N-carbamoyl-L-glutamate3,4. CPS1 is a true switch operated by NAG levels, which is on when the NAG level is high, with highly active ammonia detoxification, whereas when the NAG level is low (off situation) CPS1 is less active, decreasing urea cycle operation5. Given the high Km for glutamate of NAG synthase6 and the short half-life of NAG in vivo7, NAG levels reflect the nitrogenous burden manifested in the glutamate level. In this way, ammonia detoxification stops before too low ammonia levels cause depletion of ammonia-derived amino acids such as glutamate and glutamine and the turning on of undue protein catabolism8.
CPS1 is a complex multidomain enzyme9,10,11,12,13 having two separate active centers13,14,15 that catalyze a three-step reaction involving two highly unstable intraenzymatic intermediates16,17 and intermediate migration between both active centers13 (Fig. 1a). Bicarbonate is phosphorylated by one ATP molecule (ATPA) to give carboxyphosphate, which is then attacked by ammonia, yielding carbamate, which is phosphorylated by another ATP molecule (ATPB) to produce carbamoyl phosphate (Fig. 1a)13,14,15,16,17. Both phosphorylation steps occur at different sites13,14,15 and depend on NAG2,16,18,19. This effector exemplifies extreme allosteric activation, since in its absence CPS1 exhibits ≤2% of the activity at NAG saturation2,20, largely because NAG increases the Vmax and decreases the KmATP of CPS120. Conversely, ATP strongly increases the affinity of CPS1 for NAG21. Thus, there is cross-talk between the NAG and nucleotide sites of CPS1, with NAG and ATP bindings being mutually favoured. In addition, CPS1 affinities for its essential ionic activators potassium and magnesium are increased importantly by NAG20.
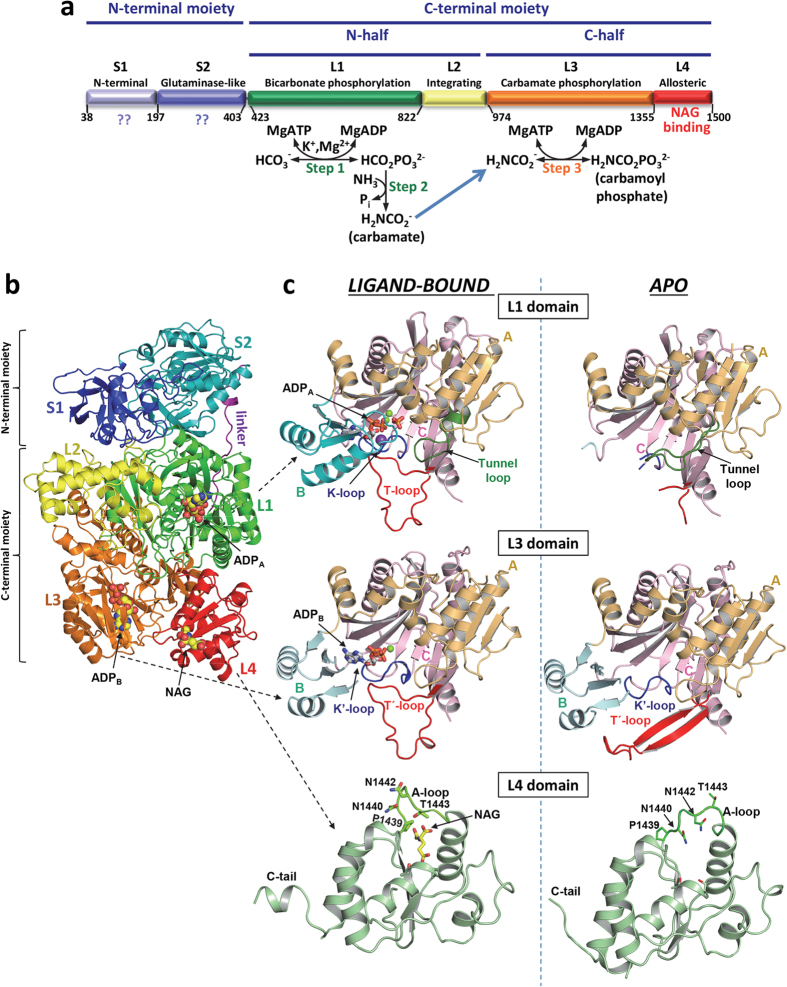
Figure 1. Structure of human CPS1.
(a) Scheme of the mature CPS1 polypeptide, indicating its two moieties (top), which are homologous to the small and large subunits of CPS from E. coli. Also indicated are the two sequence-related halves of the large moiety (middle). Domain composition with names and boundaries, given as residue numbers, is indicated in the lower bar. The three steps of the CPS1 reaction are shown below the corresponding domains where they occur. The blue arrow indicates carbamate migration between the phosphorylation active centers, a process contributed mainly by residues from both phosphorylation domains. (b) Cartoon representation of the CPS1 monomer, with domains shown in different colors, and labeled. The structure depicted corresponds to the ligand-bound conformation. The two ADPs and NAG molecules found bound to CPS1 are shown in space-filling representation. In (c), domains L1, L3 and L4 are shown expanded, in the ligand-bound (left) and apo (right) forms. In L1 and L3 the three subdomains A, B and C, and important loops mentioned in the text are labeled and represented with different colors. In the L4 domain some residues from the A-loop are shown (as sticks) to highlight the large structural changes of these side chains when NAG is bound. The ADP molecules, a phosphate and NAG are shown in rods representation, magnesium ions are shown as green spheres and the potassium ion bound to the K-loop is shown as a violet sphere.
We clarify here the structural mechanism of CPS1 activation by NAG, by determining crystal structures of human CPS1 in NAG-free form (apo form) at 2.4 Å resolution and in NAG and ADP complexed form (ligand-bound form) at 2.6 Å resolution. These two forms represent or approximate the inactive and the active forms of the enzyme, respectively. Their comparison has revealed important conformational changes that involve the NAG binding site in the C-terminal region of the enzyme and the two nucleotide binding sites that are more than ~35 Å away, providing structural clues on CPS1 regulation and functioning. Furthermore, as exemplified here with a few CPS1 mutations, these structures set the frame for rationalizing the effects of the >130 clinical single amino acid substitutions reported in patients with CPS1 deficiency (CPS1D) that are spread over the entire CPS1 molecule22. Our structures might also help understand how the reported acylations of some CPS1 lysines and their deacylation by Sirt5 could control urea cycle activity, linking CPS1 activity control with aging processes and with the hyperammonemia of glutaric aciduria23,24,25,26.
Results and Discussion
Overall human CPS1 architecture
The crystals of the apo and the ligand-bound forms of human recombinant CPS127 (lacking the N-terminal mitochondrial targeting peptide, residues 1–37; replaced by a His-tag to facilitate purification) contained in their asymmetric units (Table 1), one or two similar dimers (Supplementary Fig. 1), respectively. These dimers resulted from contacts involving <3% of the molecular surface (1300 or 1100 Å2 per subunit in the apo and ligand-bound forms, respectively), indicating poor dimer stability (PISA server, http://www.ebi.ac.uk/pdbe/pisa)28, consistent with the behavior of human CPS1 in solution as a monomer-dimer system in rapid equilibrium in which the monomer predominates2,27,29. Dimerization, mediated by exposed parts of the C-terminal domain and of the end of the preceding domain, is not analyzed here any further, since CPS1 monomers and dimers are active when NAG is present30.
Table 1. Data collection and refinement statistics.
| apo CPS1 | Ligand-bound CPS1 | |
|---|---|---|
| Data collection | ||
| Space group | P21 | P1 |
| Cell dimensions | ||
| a, b, c (Å) | 99.3, 133.5, 142.9 | 78.9, 98.6, 214.9 |
| α, β, γ (º) | 90.0, 102.5, 90.0 | 90.7, 98.7, 90.1 |
| Matthews Coefficient | 2.81 | 2.51 |
| Resolution (Å) | 44.5-2.40 (2.53-2.40)* | 40.0-2.60 (2.74-2.60)* |
| Total Reflections | 475,967 (59,848)* | 329,157 (40,514)* |
| Unique Reflections | 136,595 (17,237)* | 178,654 (24,542)* |
| Rsym (%) | 11.4 (34.2)* | 8.1 (32.1)* |
| I/σI | 4.8 (2.0)* | 7.9 (2.1)* |
| Completeness (%) | 96.3 (83.4)* | 90.7 (84.9)* |
| Redundancy | 3.5 (3.5)* | 1.8 (1.7)* |
| CC1/2 (%)66 | 98.5 (87.4)* | 98.5(51.6)* |
| Refinement | ||
| Resolution (Å) | 43.95-2.40 | 40.00-2.60 |
| No. reflections | 129,708 | 169,655 |
| Rwork/Rfree (%) | 16.5/19.6 | 19.5/22.9 |
| No. atoms or molecules: | ||
| Protein (monomers) | 2 | 4 |
| Protein (atoms) | 21,001 | 44,040 |
| Ligands (molecules or ions) | ||
| ADP | 0 | 8 |
| NAG | 0 | 4 |
| Phosphate | 0 | 4 |
| Ni/Mg/K/Cl | 2/0/0/0 | 4/12/18/3 |
| Water | 819 | 640 |
| Average B-factors | ||
| Protein atoms | 40.5 | 46.1 |
| Ligands | ||
| ADP | – | 26.2 |
| NAG | – | 21.5 |
| Phosphate | – | 21.9 |
| Ni/Mg/K/Cl | 34.5/–/–/– | 50.4/23.3/41.5/33.7 |
| Water | 35.5 | 23.4 |
| Ramachandran plot62 | ||
| Favored (%) | 96.6 | 96.9 |
| Outliers (%) | 0.41 | 0.18 |
| R. m. s. deviations | ||
| Bond lengths (Å) | 0.009 | 0.011 |
| Bond angles (º) | 1.29 | 1.40 |
*Values in parenthesis correspond to the highest resolution shell.
The CPS1 monomer presents the previously inferred9,10,11,12,13 6-globular domain architecture (Fig. 1a,b and Supplementary Fig. 2), with 2-domain and 4-domain respective N- and C-terminal moieties that correspond, respectively, to the small and large subunits of bacterial CPSs (these are heterodimeric, intrinsically active and NAG-independent CPSs)31,32, with an extensive interface of ~2400 Å2 between these moieties. An extended 20-amino acid ~50 Å linker, well conserved among the CPS1 family, which connects these moieties (Fig. 1b), appears quite mobile (up to 7 Å displacements in its central region observed in different CPS1 subunits).
Of the S1 and S2 domains that compose the N-terminal moiety, S2 is an ancestral but inactive2,9 glutamine amidotransferase/glutaminase-like domain. The four domains that compose the C-terminal moiety include two homologous catalytic domains9,13, L1 and L3, which phosphorylate respectively bicarbonate and carbamate13. L1 and L3 are linked by an intervening domain (L2) recently defined as an integrating domain33, which exhibits a unique fold of seven α-helices and two β-strands (Fig. 1b). The C-terminal or allosteric domain L4 presents a methylglyoxal synthase-like fold34 and was shown to bind NAG35. Helices, strands and loops will be named here as shown in Supplementary Fig. 2, giving the domain (S1, S2, etc.) and then the sequential numbering of the secondary structure element within the domain (S2α1, L4β2, etc.).
Although exhibiting the same overall architecture (averaged r.m.s.d., 1.3 Å for the superposition of 1285 Cα atoms), the apo and ligand-bound forms present large differences (Supplementary Figs 2 and 3) that arise from structural rearrangements in domain L4 and in both phosphorylation domains L1 and L3 (Fig. 1c and Supplementary Fig. 3). The differences also reflect reorientations of domains (Supplementary Video 1), particularly of domain L4 with respect to domains L1 and L3, with respective rotations of 13° and 9.3° (Supplementary Table 1) and with changes in the buried surface areas between these domains (Supplementary Table 2).
NAG binding and signal transmission to the phosphorylation domains
One NAG molecule was found sitting with full occupancy in each subunit of ligand-bound CPS1 (Supplementary Fig. 1), in a pocket of domain L4 that is close to the interface with domain L3 (Fig. 2a). The NAG molecule utilizes its three Cα substituents for stereospecific anchoring on the enzyme (Fig. 2b). These findings agree with the results of solution studies revealing the binding of one NAG molecule per enzyme subunit21, of structure-reactivity investigations on the requirements of the ligand for binding at the NAG site of CPS136, and with the exclusive photoaffinity labeling of domain L4 with the NAG analogue N-chloroacetyl-L-glutamate35. Thus, NAG binding to CPS1 as observed in the crystal structure appears to genuinely reflect the binding of this effector to the enzyme in vivo.
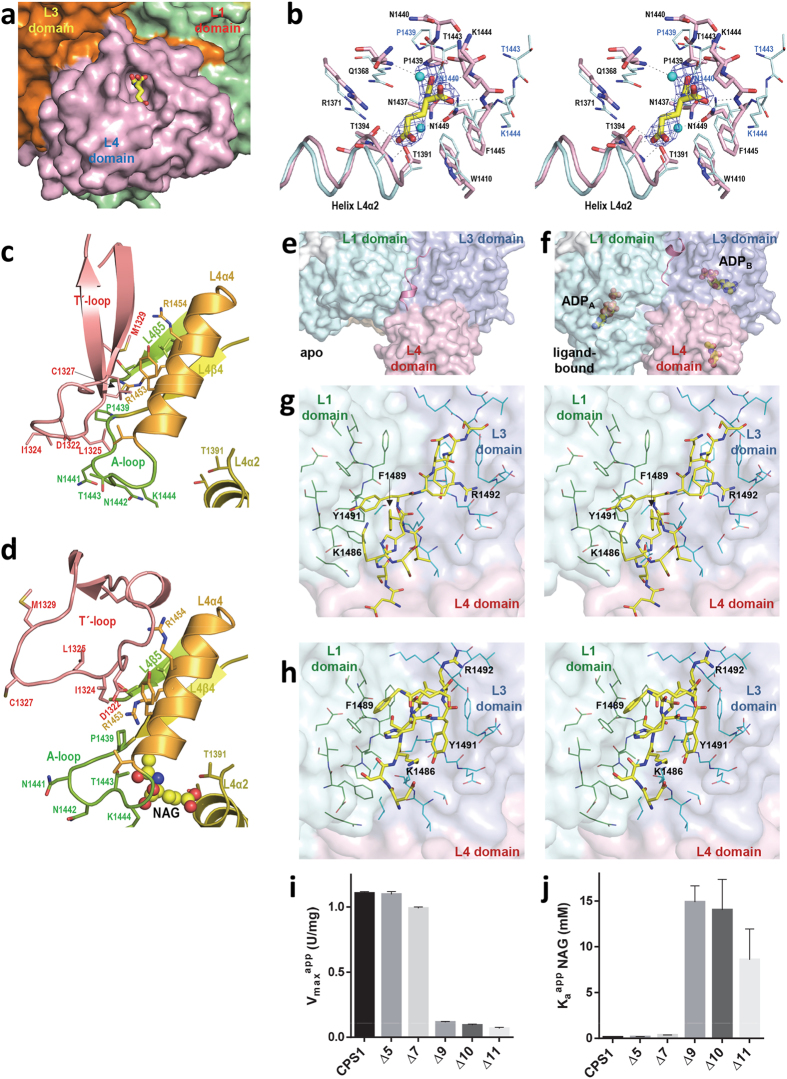
Figure 2. The allosteric L4 domain.
(a) View of NAG bound in its binding pocket in the L4 domain, not far from the interface with the L3 domain. (b) Stereo view of the superposition of the NAG site of the apo (carbon atoms colored cyan) and ligand-bound (protein and NAG carbon atoms colored pink and yellow, respectively) forms, identifying the residues by labeling them. When the position of one residue changes much, the residue is labeled in blue for the apo form. The unbiased (Fo-Fc) omit electron density map obtained prior to building the ligand is shown as a blue grid contoured at 2σ. (c,d) Helix L4α4 and some adjacent regions of the L4 domain and the T′-loop from domain L3 are shown, in the (c) apo and (d) ligand-bound forms. NAG is shown in spheres representation. The side-chains of some residues are shown in sticks representation and some secondary structure elements and residues are identified by labeling in the same color as the structural element to which they belong. (e,f) C-tail from domain L4 interacting with neighbor domains L1 and L3, in the apo and ligand-bound forms, and (g,h) respective close-up stereo-views of (e,f) with some residues labeled to highlight the radical changes of both the C-tail structure and their interactions with neighbor domains. (i,j) Influence of C-terminal truncations of the indicated length (Δn denotes the number of deleted residues starting from the C-terminus) on (g) the activity of the enzyme at saturation of NAG (Vmaxapp), and on (h) the concentration of NAG (KaappNAG) needed for attaining an activity of 0.5 × Vmaxapp. CPS1 denotes the wild-type enzyme.
Of the three non-hydrogen substituents of the Cα of NAG, the α-COO− interacts extensively with the encircling L4β4-L4α4 loop (A-loop; residues 1439–1445; Fig. 1c, bottom), forming hydrogen bonds with backbone nitrogen atoms (Gln1368, Lys1444, Phe1445) and with the Asn1449 side chain. The acetamide moiety of NAG is hydrogen-bonded via its O atom with the -OH of Thr1443 and inserts its methyl group in a sub-pocket formed by Asn1437, Pro1439 and Asn1449. Only minor adjustments in this sub-pocket are needed for accepting the amido group of the NAG analog N-carbamyl-L-glutamate used therapeutically in NAG synthase deficiency3,4. The third substituent of the NAG Cα makes hydrophobic contacts with Gln1368 and Trp1410 and faces with its δ-COO− the N-end of helix L4α2 (residues 1391–1400), forming hydrogen bonds with this helix via backbone atoms (Ala1393 and Thr1394) and side chains atoms (Thr1391 and Thr1394) (Fig. 2b). These interactions account for the observations36 that all alterations tested in the three substituents of the NAG Cα reduced the affinity for CPS1, excepting the addition of a phenyl group to the Cβ of NAG, which was without substantial effect. This lack of effect is explained by the present structure, given the exposure of the face of the Cβ of NAG, where the phenyl substituent would be found, and the nearness of the hydrophobic chains of Trp1410 and Phe1445 (Fig. 2b), which could interact with the phenyl group of the modified ligand.
Despite the triggering by NAG of extreme allosteric activation, domain L4 does not exhibit large structural differences when the apo and the ligand-bound crystals are compared (r.m.s.d., 1.1 Å for 124 superposed Cα atoms). Only three sites of this domain exhibit significant differences (Supplementary Fig. 3). One such site is the hinge (Gly1354-Pro1358) for reorientation of the L4 domain relative to domain L3, at the connection between these domains (Supplementary Video 1). In the NAG-interacting A-loop, the seven residues forming this loop alter their conformation (Fig. 1c, bottom panel and Fig. 2c,d), with movements of up to 9 Å of Asn1440 and Thr1443. The C-tail (residues 1481–1500) (Supplementary Fig. 3) that projects away from domain L4 and sits along a groove between both phosphorylation domains, closely interacting with them (Fig. 2e-h), forms a short helix in the ligand-bound form that is absent in the apo form. This change is associated with drastically different C-tail interactions with neighbour domains in both crystal forms (Fig. 2g,h and Supplementary Video 1). Given the remoteness from the bound NAG (Fig. 1c, bottom panels), these C-tail differences do not result from direct interactions of the tail with the activator. However, limited proteolysis studies had shown that these last 20 residues of rat CPS1 are extremely important for activation, with their deletion causing a >30 fold increase in the KaNAG (the NAG concentration needed for half-maximal activation) and in the KmATP 11. Human CPS1 variants with progressively shorter C-ends show that the KaNAG is abruptly increased (thus, the apparent affinity for NAG is decreased) and the apparent Vmax is importantly decreased when the last nine residues are eliminated (Fig. 2i,j), confirming the importance of the C-tail.
L4 domain residues contributing to CPS1 activation by NAG should change due to NAG binding and should be involved in interdomain interactions that transmit the NAG signal to the phosphorylation domains. Excluding the C-tail, only three residues from the L4 domain, Thr1443 and the pair Pro1439/Arg1453, fulfill these requirements (Fig. 2c,d). In the ligand-bound form, Thr1443 interacts with NAG, while in the apo form it interacts with Leu1325 from the L3 domain (Fig. 2b–d). Changes in Pro1439 associated with NAG binding (Figs 1c and 2b–d) affect Arg1453, which makes a salt bridge with Asp1322 and interacts weakly with Ile1324 (both from the L3 domain) in the ligand-bound form, while in the apo form Arg1453 interacts with Cys1327 and Met1329 (Fig. 2c,d). All the residues of the L3 domain mediating these interactions belong to the L3β15-L3β16 loop (residues 1311–1333), named here the T′-loop (from signal transmission loop), which transitions from a β-hairpin in the apo form to a 20 Å-wide expanded loop in the ligand-bound form (Fig. 1c, middle panels, Fig. 2c,d and Supplementary Figs 2 and 3). The changing interactions of Thr1443 and Arg1453 with the T′-loop appear to stabilize one or the other form of this loop in parallel with whether NAG is or is not bound, possibly being essential for signal transmission in the NAG-operated switch. Indeed, the finding37 in patients with severe NAGS deficiency of the mutations Pro1439Leu, Thr1443Ala, Arg1453Gln and Arg1453Trp, emphasize the key role played by these three L4 domain residues. Furthermore, the introduction in CPS1 of the Asp1322Leu, Arg1453Gln and Arg1453Trp mutations caused enzyme inactivation without apparent impairment of enzyme folding37,38, as expected if NAG activation were abolished by preventing the transmission of the activating signal from L4 to the remainder of the enzyme.
Superposing the apo conformation of domain L4 (excluding the C-tail) onto the ligand-bound conformation of CPS1 results in Arg1453 clashing with Ile1324, which should destabilize the apo conformation of Pro1439. This agrees with the enhancement of NAG binding via the stabilization of the active conformation of CPS1, as exemplified by using cryoprotectants, which are NAG-independent activators of CPS12,39. Superposing the ligand-bound conformation of domain L4 (excluding the C-tail) onto the apo conformation of CPS1 results in only a minor contact of Asn1441 with Phe1143 and in some contacts of Arg1453 mainly with Cys1327. Therefore, NAG binding to domain L4 contributes to trigger structural changes towards the active conformation in neighbor domain L3, though the reorganization required affects very few residues.
Changes in the phosphorylation domains and in nucleotide binding
As expected from the mutual cross-talk between NAG binding and nucleotide binding in CPS120,21, the active form of the enzyme represented by the ligand-bound form not only has NAG bound in each monomer, but also binds two molecules of the ADP added in the crystallization mixture (Fig. 1b and Supplementary Fig. 1). Each ADP sits in one phosphorylation domain (L1 and L3 domains, Fig. 1c), with the adenine ring sandwiched between the central β sheets of the B and C subdomains (for the boundaries between the A, B and C subdomains, see Supplementary Fig. 2), as is characteristic for the ATP-grasp fold40. Also as it might have been expected for domains having arisen from an ancestral duplication9, the two phosphorylation domains form a pseudohomodimer (Fig. 1b) with nearly exact twofold pseudosymmetry (174° and 178.5° rotations and r.m.s.d. values of 2.1 and 1.8 Å for superposition of 269 and 330 Cα atoms in the apo and ligand-bound forms, respectively).
Although being homologous and carrying out phosphorylations of the analogous compounds bicarbonate and carbamate (Fig. 1a), the catalytic properties of both phosphorylation domains L1 and L3 are not identical41. These differences manifest themselves in the present structures. The structures of the L3 domain are highly similar in the apo and ligand-bound forms (r.m.s.d., 0.83 Å for 340 residues). Virtually all elements of this domain are observed in both crystal forms (Fig. 1c, middle panels). In contrast, in the case of the L1 domain there are ~90 residues that are disordered (and thus that are not seen) in the apo form and that become ordered when the enzyme has the ligands bound to it (Fig. 1c, top panels). These structural findings parallel the observations that ATPB binds to domain L3 irrespective of whether NAG is or is not bound20,42, whereas the affinity of the enzyme for ATPA, the ATP molecule that binds in domain L1, is extremely low in the absence of NAG20. Indeed, the crystal structures account for this decreased affinity, since in the apo form, (Fig. 1c, top right panel) the entire B subdomain of domain L1, and loop L1β11-L1β12 (residues 654–662; called here the K-loop) could not be traced, indicating that they are mobile or disordered. In contrast, in the ligand-bound form (Fig. 1c, top left panel) the entire B subdomain is well traced in this domain, closing on the bound ADP, with loop L1β11-L1β12 being fully structured, having at its center a coordinated potassium ion (thus the name K-loop) and being involved in intimate interactions with the bound ADP (Fig. 3a,c). Interestingly, the L1β17-L1β18 loop (residues 777–793; named here the T-loop because of its equivalence in the L1 domain to the T'-loop of the L3 domain) could not be traced in the apo form. This loop became fixed and structured in the ligand-bound form (Fig. 1c, top panels), presenting the same wide open loop shape as the T'-loop (compare with Fig. 1c, middle left panel). This structural fixation appears largely due to the interactions of this loop with the widened T′-loop (Fig. 3c). Indeed, the T'-loop was the only part of the L3 domain that exhibited important conformational differences between the apo and the ligand-bound forms (Fig. 1c, middle panels and Supplementary Figs 2 and 3).
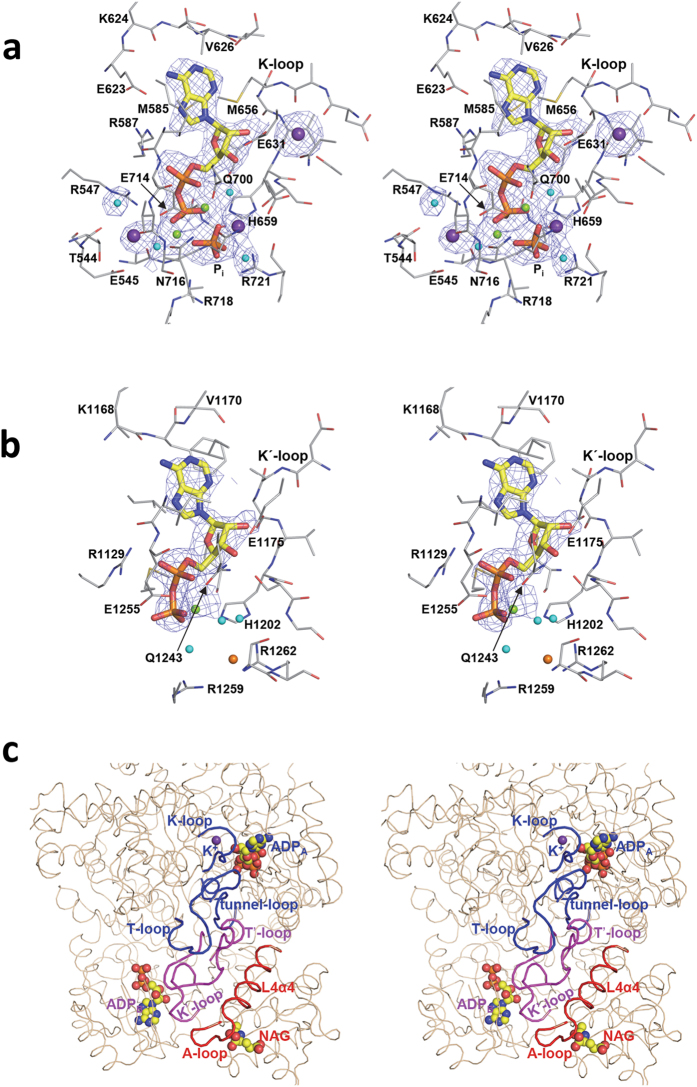
Figure 3. Stereo views of the binding sites for the two molecules of the nucleotide that are used in the reaction, and for the loop system used in signal transmission.
ADP binding in domains L1 (a) and L3 (b). Protein and ADP and phosphate are shown in sticks representation (carbon atoms in grey for the protein and in yellow for the ligands). The lone phosphate is labeled Pi. Potassium, magnesium, chloride ions and water molecules are spheres colored purple, green, orange and cyan, respectively. The (Fo-Fc) electron density maps, computed without the ligands, are shown as a blue grid contoured at 2σ. (c) Close up of the structure of the majority of the C-terminal moiety, to show the binding of both ADP molecules and of NAG and to highlight the relations of helix L4α4 and of the loops more centrally involved in signal transmission. Whereas the overall structure is in brown thin string, the highlighted elements are in thicker string and are labeled, with red, magenta and blue coloring depending on whether they belong to L4, L3 or L1, respectively.
The two phosphorylation steps of the CPS1 reaction also differ in that the step of bicarbonate phosphorylation requires potassium and excess magnesium over that for complexing the nucleotide, whereas these needs are not patent for the carbamate phosphorylation step (Fig. 1a, reactions)19,41. In agreement with these differences, in the ligand-bound form the L3 domain had no potassium complexed to its K'-loop and it contained only the magnesium ion that is complexed to the ADP, whereas the L1 domain besides having also two bound magnesium ions, hosted a potassium ion coordinated to its K-loop (Figs 1c and 3a,b). These two magnesium ions are coordinated to both the ADP molecule and to one nearby phosphate molecule (Fig. 3a) that, given its location relative to the ADP, may represent the phosphate moiety of the carboxyphosphate intermediate made at this phosphorylation site by the attack of bicarbonate on the ATP. Indeed, although no bicarbonate was observed in the structures, the fact that the mutation to Met of the nearby Thr544 (Fig. 3a) found in a patient with CPS1 deficiency27 selectively increased ~60-fold the Km for bicarbonate, points to the binding of bicarbonate in the vicinity of the phosphate. In any case, this phosphate interacts with the highly conserved Arg721 (Fig. 3a), possibly reflecting the corresponding binding of this arginine to ATPA, increasing the susceptibility of the γ-phosphate to the attack by bicarbonate. This arginine, which in the absence of ligands interacts with Glu440, interacts in the ligand-bound form not only with the phosphate but also with Gly436, a residue that is at the tip of the L1β1-L1α1 loop (the tunnel-loop, residues 430–443) of subdomain A. This last interaction should help stabilize this loop in the conformation that delineates the carbamate tunnel that characterizes the activated form of the enzyme (Fig. 4a, center; discussed below). It is interesting that Arg1262, which is the residue corresponding in domain L3 to Arg721 of domain L1, interacts with a chloride ion that sits approximately at an equivalent position to that of the phosphate in the L1 domain (Fig. 3b).
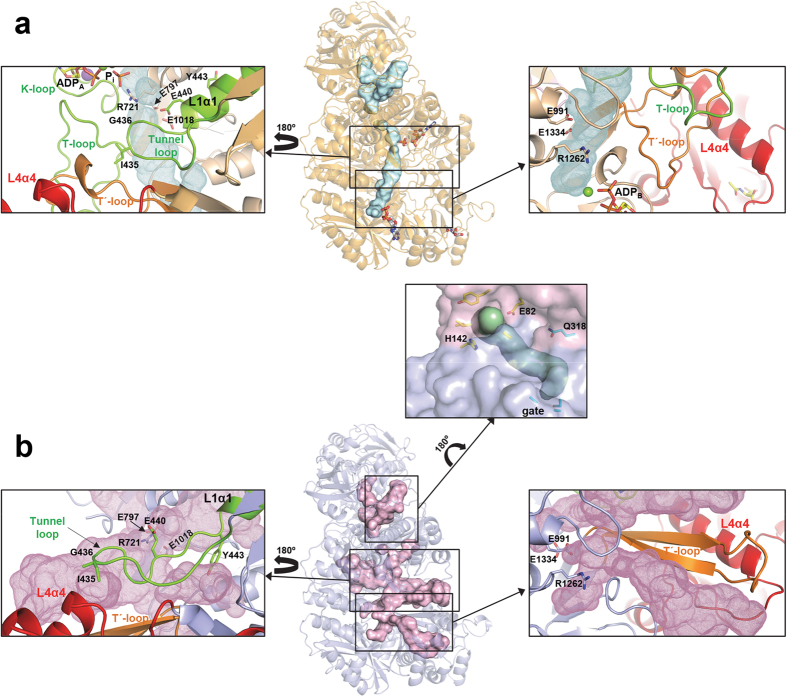
Figure 4. Tunnels and cavities in CPS1.
Overall views (center) of tunnels and cavities (surface representation) in (a) the ligand-bound and (b) the apo forms of CPS1. The carbamate tunnel linking the two phosphorylation active sites (see text) is well defined only in the ligand-bound conformation. Boxes at both sides show detailed views of the structural elements forming the carbamate tunnel. The tunnel loop (colored green) and the T′-loop (colored orange) occlude in the ligand-bound form many of the cavities found in the apo form. Some side chains are explicitly shown to highlight the structural changes. A possible ammonia tunnel, which remains invariant between the apo and ligand bound forms, is depicted in the central box. Residues at the entrance to this tunnel and at the conserved gate are shown with carbon atoms colored yellow and cyan, respectively.
As already discussed, the L3 domain, and in particular the T′-loop, can play an important role in allosteric activation due to both its extensive interdomain interactions (Fig. 3c) and the already commented important changes between the apo and ligand-bound forms (Fig. 1c, middle panels, Figs 2c,d and 4, right boxes, and Fig. 5). Thus, the ~20 Å-wide loop observed in the ligand-bound form interacts not only with helix L4α4 (to which Arg1453 belongs), but also with the T-loop, the tunnel-loop and subdomain B from domain L1, and with the K′-loop from domain L3, indicating that this T′-loop can be a hub for NAG signal transmission to the catalytic machinery (Fig. 3c).
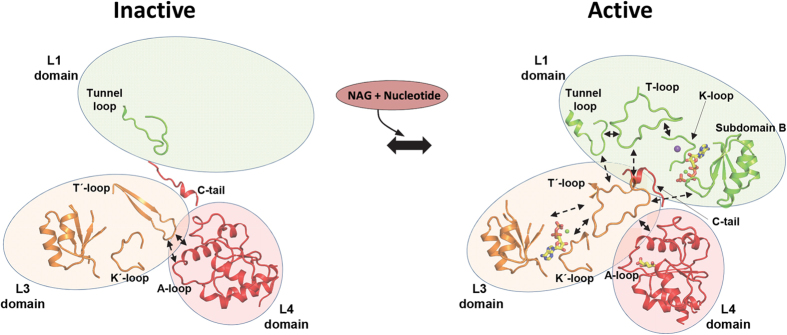
Figure 5. Scheme of CPS1 activation.
Schematic representation of the structural changes and interactions (indicated as double dashed arrows) between the different elements involved in the activation of CPS1 upon binding of NAG and the nucleotide. Ligands are shown in sticks representation. Binding of NAG alters the A-loop, which in the apo form interacts with the T′-loop from the L3 domain. This loop experiences a radical reorganization between the apo and ligand-bound forms. The T′-loop interacts also with the T-loop (from the L1-domain) influencing both ATP binding sites mainly through their interaction with the K′- and K-loops, respectively. These changes together with the remodeling of the tunnel-loop and reorientations between domains result in the building of a functional carbamate tunnel. Stabilization of the modified architecture is achieved through the C-tail fixing the relative orientations of domains L1, L3 and L4.
The carbamate tunnel is aberrant in the apo form and becomes functional in the ligand-bound form
Carbamate, a highly labile compound produced in domain L1 by the reaction between carboxyphosphate and ammonia, has to be transported intramolecularly ~35 Å, shielded from water, to the active center of domain L3, where it is phosphorylated to give carbamoyl phosphate (Fig. 1a)13. This transport can take place through a tunnel connecting the active centers of domains L1 and L3, similarly to what was proposed for Escherichia coli CPS32,34,43 (EcCPS). In CPS1 the possible carbamate tunnel, roughly linear and with a diameter of 2.6–5 Å, is well defined only in the ligand-bound form (Fig. 4a), where it is formed by residues evenly belonging to domains L1 and L3, according to the quasi-two fold symmetry that relates these two domains. At each end of the tunnel Arg721 and Arg1262 can participate, as indicated before, in the binding to the nucleotide γ-phosphate in domains L1 and L3, respectively. These arginine residues, together with five conserved glutamic acid residues at the beginning (glutamates 440, 797 and 1018) and at the end (glutamates 991 and 1334) of the tunnel, can play a role in the entry and exit of carbamate to and from the tunnel43,44,45.
In the apo form of CPS1 cavities and tunnels increase and change dramatically with respect to the ones seen in the ligand-bound form (Fig. 4a,b), with several expansions, in particular in the central part of the tunnel, and three large lateral branches that open at the molecular surface (Fig. 4b). Two of these branches are almost perpendicular to the direction joining the phosphorylation centers, with one branch parallel to the tunnel-loop helix L1α1 (Fig. 4b, left box). In the ligand-bound form these two branches are occluded mainly by the lengthening by 1.5 turns of helix L1α1 and by the related remodeling of the tunnel-loop. This loop, placed between domains L1, L3 and L4, shrinks and establishes new hydrogen bonds mainly with loop L3β2-L3β3 (residues 1014–1027; only Glu1018 is seen in Fig. 4, left boxes) and with the T′-loop (Figs 3c and 4a left box and Fig. 5). Similarly, a third lateral branch is occluded in the ligand-bound form by, in particular, the remodeling of the T′-loop (Fig. 4, right boxes). Formation of a functional carbamate tunnel in the active-like ligand-bound form appears to indicate that the main restraint for the large cavities and tunnels found in the apo form is to allow the structural rearrangements required for CPS1 activation. In summary, the structural changes that lead to definition of the appropriate carbamate tunnel in the ligand-bound form appear to be key elements in the process of CPS1 activation.
An incomplete catalytic triad for glutamine use, and a potential ammonia tunnel.
Glutamine is utilized as the endogenous source of ammonia by all types of CPS31,46,47,48 except CPS12. The inability of human CPS1 to use glutamine is explained by the replacement by serine (Ser294) in this enzyme of the key catalytic cysteine of the catalytic triad of histidine, glutamate and cysteine34,49 used for glutamine hydrolysis by other CPSs (Supplementary Fig. 4). However, other factors might contribute also to the lack of glutaminase activity of human CPS1, since Rana catesbeiana CPS1 conserves the catalytic triad but is unable to utilize glutamine, recovering the ancestral glutaminase activity by mutating to leucine a lysine that corresponds to Thr295 of human CPS150. In any case, CPS1 must take ammonia from the environment and should deliver it to the carboxyphosphate production site at the beginning of the carbamate tunnel. Cavities and tunnels observed in the CPS1 N-terminal moiety in both the apo and ligand-bound forms (Fig. 4), are likely related with this ammonia intake and delivery. Although CPS1 maintains many elements of the EcCPS ammonia tunnel going from the ancestral glutaminase site to the conserved gate (CPS1 residues Cys648, Ala667 and Ala729) at the end of this tunnel51 in domain L1, Gln318 replaces Gly293 of EcCPS and blocks this tunnel before the gate. However, there is an alternative potential path for intake of external ammonia. This path could correspond to the tunnel that skips the ancestral glutaminase active site of CPS1, starting at the molecular surface, between residues Glu82 and His142, and continuing parallel to the interface between the N- and C-terminal moieties, reaching the gate of the carbamate tunnel (Fig. 4b, box at the center). Charged residues at the entrance of this alternative tunnel might facilitate the entry of uncharged ammonia, the true CPS1 substrate14, rather than of NH4+.
The CPS1 structure allows rationalizing the effects of clinical mutations
The large number of "private" missense mutations with little recurrence found in individual patients with CPS1 deficiency22 (Supplementary Figs 2 and 5) and the lack in most cases of appropriate pedigree information, render crucial to clarify whether a given mutation is disease-causing or is merely a rare unreported trivial polymorphism. The present structures are valuable to rationalize the consequences of individual mutations, as exemplified here with the reported clinical mutations that affect the L4 domain22 (Supplementary Fig. 5b). These mutations appear a good sampling set since their effects were examined experimentally using recombinant CPS127,37,38. In the crystal structure, the Arg1371Leu, Thr1391Met and Thr1443Met mutations eliminate key polar interactions with NAG (Fig. 2b and Supplementary Fig. 5b), agreeing with the observations that these mutations increase two orders of magnitude the KaNAG 27,37. As indicated before, the key role in signal transmission of the Arg1453/Asp1322 ion pair (Fig. 2d) predicts abolition of NAG activation by the Arg1453Trp and Arg1453Gln clinical mutations, again agreeing with the experimental observation that these mutants were inactive38. By altering the A-loop of the NAG site that changes its conformation in the ligand-bound form of CPS1 (Fig. 1c, bottom panels; and Fig. 2c,d), the Pro1439Leu mutation may be expected to impair NAG binding, as observed (~15-fold increase in Ka for NAG)37. The Pro1462Arg mutation, by drastically affecting a structurally very important residue at the beginning of L4β5, should impair CPS1 activation, given the interaction of L4β5 with the wide form of the T′-loop that is observed in the ligand-bound crystal (Supplementary Fig. 5b). Correspondingly, this mutation decreased Vmax ~30-fold37. In contrast, Pro1411 is in an external loop that is not involved in NAG binding neither in the interactions with other domains or in dimer formation (Supplementary Fig. 5b). Thus, the Pro1411Leu substitution should have little effect, agreeing with its finding in a patient without a drastic disease phenotype37, and with the observation that this substitution did not cause important changes in the properties of the in vitro expressed enzyme38. The mutation Tyr1491His38, affecting a C-tail residue, may be predicted to drastically affect the NAG activation process, and, indeed, the affinity for NAG was decreased ~60-fold by this mutation38. Even misfolding or destabilizing mutations were amenable to rationalization of their effects. The mutation Leu1381Ser, causing misfolding and degradation, and the Ala1378Thr and Leu1398Val mutations, which decreased the thermal stability of CPS127,37, affect the crowded, highly hydrophobic patch that glues the central β sheet and the α3 layer in the α3β5α2 sandwich that constitutes the L4 domain fold (Supplementary Fig. 5b). Differences between wild-type and mutant forms in side-chain polarity and/or in size can account for the misfolding or the thermal destabilization observed with these mutations. Overall, it was possible to rationalize the effects of most of the reported clinical mutations that map in this domain, suggesting that similarly accurate rationalizations, and even predictions, could be made concerning the effects of the mutations affecting other domains.
Final remarks
The present structures clarify the activation mechanism of this intrinsically inactive enzyme. NAG, by binding to domain L4, triggers changes in the A-loop and in Arg1453 (Fig. 1c, bottom panels and Fig. 2c,d) that result in changing interactions with the T′-loop of domain L3, which reorganizes completely from a β-hairpin in the apo form to a widened loop in the ligand-bound form (Figs 1c and 5). In this last form, the T′-loop interacts also with the tunnel-loop and the T-loop of the L1 domain, thus transferring the activating information to the bicarbonate-phosphorylating domain, whereas inside domain L3 the T′-loop interacts with the K′-loop (Figs 3c and 5). Binding of a nucleotide to domain L3 should cause only limited stabilization of the active conformation, given the small remodeling required, mainly of the K′-loop. In fact, nucleotide binding at this site was reported to be independent of NAG20,42. The sole binding of either NAG alone or of a nucleotide to domain L3 most likely could not trigger efficient activation. In domain L1, the T-loop interacts with the K-loop, which changes, from disordered in the apo form, to establishing strong interactions with the bound nucleotide and with a potassium ion in the ligand-bound form (Figs 3c and 5). Therefore, a bound nucleotide in the bicarbonate phosphorylation domain L1 should have a major stabilizing effect on the active conformation, in line with the observations made in NAG binding studies21. However, by the same reason, binding of the nucleotide to a not yet active conformation is not favored, again in line with studies of the reaction in the absence of NAG20.
The activation process also involves changes in the relative orientations of the L4 domain and the phosphorylation domains. These changes are reflected in ~10° rotation of L1 and L3 (Supplementary Table 1), and the extensive loop remodeling resulting in the delineation of a well-defined carbamate tunnel (Fig. 4) that should enable shielded passage of carbamate from its formation site in domain L1 to its phosphorylation site in domain L3. However, ADP release from domain L1 at the end of each catalytic cycle should destabilize the active conformation. The C-tail may lock the active conformation by providing a stability barrier, preventing the return of the different domains and of the carbamate tunnel to the orientations and shapes observed in inactive CPS1. This locking may explain the lag effects observed both during activation when the enzyme is first exposed to a mixture of NAG and the CPS1 substrates, or during deactivation (even if NAG is lost, CPS1 remains active for some time)15,20,52. C-tail control also agrees with the observation that the last 20 C-terminal residues are important for the cross-talk between activator and phosphorylation sites that characterizes the allosteric activation of CPS1 (Fig. 2i,j)11.
The present structures also shed light on why CPS1 cannot use glutamine, given the substitution of the catalytic Cys by a Ser. However, glutamine binding might be impeded by other glutamine binding site alterations, and, in fact, the passage of glutamine-derived ammonia might be prevented by changes in the ancestral ammonia tunnel that connects the remnant of the glutaminase site with the large subunit32,34. In a mutant of frog CPS1 with artificially gained glutaminase activity, ammonia was excreted, later re-entering through another path, agreeing with the blockage of the ancestral ammonia tunnel50, and with the existence of an alternative path for external ammonia intake, that could correspond to the one proposed here (Fig. 4b, central box). A less clearly defined similar alternative tunnel might also be present in EcCPS (PDB 1JDB). Intake of free ammonia through this alternative tunnel would be in agreement with the lack of activity with glutamine as substrate of EcCPS variants in which the tunnel from glutamine was blocked or perforated, since these variants were normally active with external ammonia44,53,54. In contrast, variants altering the gate affected importantly the activity with both glutamine and free ammonia51.
Despite the important functional differences with CPS1, the glutamine-dependent, NAG insensitive, CPS from E. coli, a heterodimer regulated by ornithine whose active-like structure was determined nearly 20 years ago32, has been used for homology modeling of CPS1 in numerous structure-function studies of missense mutations of human CPS122,27,33,37,38,55. Due to the intricate network of interactions and remodeling found in the structures of CPS1, it is now clear that variants can interfere at the level of substrate binding and catalysis, but also by altering the tunneling of intermediate products or by interfering with the rearrangements required by regulation and activation. Results from this work provide a unique structural framework to interpret the molecular pathologies, as exemplified above, and might also help assess the consequences of the post-translational modifications that are presently being identified in CPS123,24,25,26 some of which appear to be endowed with activity-regulating roles and to be susceptible to removal by the anti-aging protein Sirt5. Therefore, our structural studies may open the gate to further structure-functional studies that might connect the present knowledge to superposed levels of regulation and of biological significance.
Materials and Methods
Wild type and mutant enzyme production, purification, site-directed mutagenesis and enzyme activity assay
With the aim to generate enzyme forms lacking 11, 10, 9, 7 and 5 C-terminal residues, stop codons were introduced to replace the CPS1 coding codons at positions (as codon numbers) 1490, 1491, 1492, 1494 and 1496, using site-directed mutagenesis by the overlapping extension method27 (Quickchange kit from Stratagene), utilizing appropriate forward and reverse oligonucleotide primers (Supplementary Table 3). We corroborated by sequencing the correctness of the constructs, the presence of the desired codon change and the absence of unwanted mutations.
Either wild-type or mutant human mature CPS1 with an N-terminal His6-tag was expressed in a baculovirus/insect cell system and purified as previously described27. For crystallization, the wild-type enzyme was concentrated to 8–10 mg/ml and was placed in a solution of 50 mM glycyl-glycine pH 7.2, 50 mM KCl, 5% glycerol, by repeated rounds of centrifugal ultrafiltration at 4 °C.
The effect of NAG concentration on CPS1 activity was determined in an ornithine transcarbamylase-coupled assay. In this assay carbamoyl phosphate production was monitored colorimetrically as citrulline27 after 10-min incubation of the enzyme (wild-type or mutant form) at 37 °C. The assay mixture containing 50 mM glycyl-glycine pH 7.4, 70 mM KCl, 1 mM dithiothreitol, 20 mM MgSO4, 5 mM ATP, 35 mM NH4Cl, 50 mM KHCO3, 10 mM NAG, 5 mM L-ornithine, 4 U/ml of ornithine transcarbamoylase and variable concentrations (0–50 mM) of NAG (neutralized with KOH).
Crystallization
Initial crystallization screenings at 21 °C used a HoneyBee X8 robot (Genomics Solutions), the sitting-drop vapor-diffusion method in 96-well MRC plates, different protein concentrations, and several commercial crystallization screen kits from Qiagen, Jena Bioscience and Hampton Research. The apo crystals were obtained using a 1/1 mixture of protein solution (8 mg/ml, containing 1 mM AMPPNP, 5 mM MgCl2, 1 mM NAG and 10 mM KCl) and of crystallization solution (20% w/v PEG 3350 and 0.2 M trisodium citrate). The cryobuffer used for crystal harvesting into liquid nitrogen was the crystallization solution enriched with 20% (w/v) PEG 400. The ligand-bound crystal was obtained at 4 °C by supplementing the protein with 20 mM ADP, 60 mM MgCl2, 10 mM NAG and 40 mM KCl, using as crystallization solution 0.4 M MgCl2 and 0.2 M ammonium tartrate, utilizing for harvesting into liquid nitrogen a cryoprotectant solution containing the mother liquor plus 10% (w/v) PEG 3350 and 30% glycerol.
Data collection and processing
X-ray diffraction data of apo and ligand-bound crystals were collected at 100 K (Oxford Cryosystems) at 0.961 Å and 0.979 Å wavelengths, respectively. Apo crystals were analyzed in the ID14-4 beamline of the ESRF (Grenoble, France) using an ADSC Quantum Q325r detector. Ligand-bound CPS1 was analyzed at beamline XALOC (ALBA, Barcelona, Spain) using a Dectris PILATUS3 6 M detector. Datasets processed with XDS56, converted with POINTLESS57, were scaled with SCALA57 at resolutions of up to 2.4 Å and 2.6 Å for apo and ligand-bound crystals, respectively. The apo crystal belonged to space group P21 (unit cell parameters: a = 99.3 Å, b = 133.5 Å, c = 142.9 Å and β = 102.5º) and contained two CPS1 subunits in the asymmetric unit. The crystal of ligand-bound CPS1 belonged to space group P1 (a = 78.92 Å, b = 98.56 Å, c = 214.89 Å, and α = 90.66°, β = 98.65°, γ = 90.08°) and contained four CPS1 subunits in the unit cell. Table 1 gives details and statistics of crystallographic data.
Phasing, model building, and refinement
Molecular replacement was successful with data for the apo crystal reprocessed in space group P1, using PHASER58 and, as search models, the heterodimer of EcCPS (PDB 1JDB)34 and the C-terminal domain of human CPS1 (PDB 2YVQ). This initial solution consisted of four CPS subunits in the P1 unit cell presenting clear deviations from a P21 packing. Model improvement was achieved by an iterative process of rigid body refinement of domains forming each CPS subunit, followed by restrained refinement, including non-crystallographic symmetry restraints, with REFMAC559 and graphic model-rebuilding with Coot60. A perfect pseudo-merohedral twinning (-h, -k, -l) was detected and taken into account by REFMAC5. The four subunits contained in the P1 cell experienced relatively large reorganizations approaching, as refinement progressed, a packing corresponding to a P21 space group. Therefore, when the Rfree was below 26% the structure of one of the CPS subunits was used to obtain a solution by molecular replacement with the original P21 data. Refinement was continued in this P21 space group. Isotropic B factors and TLS were used in the last steps of refinement, with TLS groups chosen according to the previously determined domains27. All diffraction data were used throughout the refinement process except the 5% randomly selected data used to calculate the Rfree. Stereochemistry of the model was checked and improved by analysis with PDB_REDO61 and MolProbity62 (final score was 1.16 (100th percentile)).
Ligand-bound crystal data were initially integrated and scaled in space group P21. Molecular replacement with PHASER58 using a monomer of the previously solved apo model provided a solution with acceptable electron density map. Positioning of domains was improved by rigid body refinement. Several fragments absent in the apo model, as well as some regions showing important structural differences, were manually rebuilt with Coot60, in accordance to the density map and partially using the EcCPS model from PDB 1JDB as reference. This model was subjected to restrained refinement using REFMAC559. However, refinement became stuck at high R/Rfree values. Detailed inspection of the data and analysis with SFCHECK and Xtriage from the PHENIX package63 showed that the crystal presented pseudosymmetry and the correct space group was P1. Therefore, diffraction data were reprocessed and scaled in P1, resulting in a solution with four CPS1 subunits in the unit cell. Restrained refinement, with twinning-correction only in the first steps, and automatically generated non-crystallographic symmetry restraints, was performed with REFMAC559. Ligands were incorporated to the model with Coot60. B factors and TLS (1 group per monomer) were used in the last refinement steps. The nature of the ions was decided during refinement taking into account the size of the electron density, the geometry of the coordination sphere with, in particular, the coordination distances and the type of ligands, and the presence of the ion in the crystallization drop. In the case of Ni, it was not present but could have leached out from the Ni-affinity column used for enzyme purification. Thermal factors of most ions were similar to the ones of the neighbouring protein atoms, further supporting the nature of the ions assigned. Stereochemistry of the model was improved and checked as described for the apo model. The MolProbity score was 1.45 (100th percentile).
The spaces of the cavities corresponding to the ammonia and carbamate transport tunnels, were analyzed with MOLE64. HOLLOW65 was used to fill the channels with dummy atoms for representation. Graphical representations of the structures were generated with PyMOL (http://www.pymol.org/).
Additional Information
Accession codes: Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 5DOT and 5DOU.
How to cite this article: Cima, S. et al. Structure of human carbamoyl phosphate synthetase: deciphering the on/off switch of human ureagenesis. Sci. Rep. 5, 16950; doi: 10.1038/srep16950 (2015).
Supplementary Material
Acknowledgments
This work was supported by grants from The Fundación Alicia Koplowitz, the Valencian (PrometeoII/2014/029 to V.R) and Spanish Governments (BFU2011-30407 and BFU2014-58229-P to V.R; SAF2010-17933 to J.C; BFU2012-36827 to I.F.). The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under BioStruct-X (grant agreement Nº283570), within proposal 7687. We thank the Servicio de Cristalogénesis of the Instituto de Biomedicina de Valencia for robotized crystal growth and Alberto Marina for X-ray diffraction of the crystals produced in the initial crystal screen. We thank the ESRF (Grenoble, France) and ALBA (Barcelona, Spain) synchrotrons for access to respective beamlines ID14-4 and Xaloc and for the collaboration of their staffs that contributed to the results presented here.
Footnotes
Author Contributions S.d.C. and L.M.P. performed the crystallographic experiments. C.D. and A.I.M. prepared the protein and carried out the biochemical experiments. S.d.C., L.M.P. and I.F. processed the crystallographic data and built the models. J.C and V.R. envisaged and planned the work and supervised its development. S.d.C., L.M.P., I.F. and V.R. analyzed and discussed the results and wrote the paper. J.C. and V.R. provided core funding. All the authors read the paper, commented on the contents and agreed with the publication of the results.
References
- Cohen P. P., Hall L. M. & Metzenberg R. L. Isolation and characterization of a naturally occurring stimulator of citrulline biosynthesis. Nature 178, 1468–1469 (1956). [DOI] [PubMed] [Google Scholar]
- Rubio V., Ramponi G. & Grisolia S. Carbamoyl phosphate synthetase I of human liver. Purification, some properties and immunological cross-reactivity with the rat liver enzyme. Biochim. Biophys. Acta, 659, 150–160 (1981). [DOI] [PubMed] [Google Scholar]
- Bachmann C., Colombo J. P. & Jaggi K. N-acetylglutamate synthetase (NAGS) deficiency: diagnosis, clinical observations and treatment. Adv. Exp. Med. Biol. 153, 39–45 (1982). [DOI] [PubMed] [Google Scholar]
- Brusilow S. W. & Horwich A. L. Urea cycle enzymes. In Scriver C. R., Beaudet A. L., Sly W. S. & Valle D., editors; Child, B., Kinzler, K. W. & Vogelstein, B., associated editors. The Metabolic and Molecular Bases of Inherited Disease. 8th edition. New York, McGraw-Hill. Vol 2, pp 1909–1963 (2001). [Google Scholar]
- Stewart P. M. & Walser M. Short term regulation of ureagenesis. J. Biol. Chem. 255, 5270–5280 (1980). [PubMed] [Google Scholar]
- Bachmann C., Krähenbühl S. & Colombo J. P. Purification and properties of acetyl-CoA:L-glutamate N-acetyltransferase from human liver. Biochem. J. 205, 123–127 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T., Mori M. & Tatibana M. Regulation of N-acetyl-L-glutamate degradation in mammalian liver. J. Biochem. 91, 563–569 (1982). [DOI] [PubMed] [Google Scholar]
- Kadowaki M. & Kanazawa T. Amino acids as regulators of proteolysis. J. Nutr. 133, 2052S–2056S (2003). [DOI] [PubMed] [Google Scholar]
- Nyunoya H., Broglie K. E., Widgren E. E. & Lusty C. J. Characterization and derivation of the gene coding for mitochondrial carbamyl phosphate synthetase I of rat. J. Biol. Chem. 260, 9346–9356 (1985). [PubMed] [Google Scholar]
- Powers-Lee S. G. & Corina K. Domain structure of rat liver carbamoyl phosphate synthetase I. J. Biol. Chem. 261, 15349–15352 (1986). [PubMed] [Google Scholar]
- Marshall M. & Fahien L. A. Proteolysis as a probe of ligand-associated conformational changes in rat carbamyl phosphate synthetase I. Arch. Biochem. Biophys. 262, 455–470 (1988). [DOI] [PubMed] [Google Scholar]
- Evans D. R. & Balon M. A. Controlled proteolysis of ammonia-dependent carbamoyl- phosphate synthetase I from Syrian hamster liver. Biochim. Biophys. Acta 953, 185–196 (1988). [DOI] [PubMed] [Google Scholar]
- Rubio V. Structure-function studies in carbamoyl phosphate synthetases. Biochem. Soc. Trans. 21, 198–202 (1993). [DOI] [PubMed] [Google Scholar]
- Rubio V. & Grisolia S. Mechanism of mitochondrial carbamoyl-phosphate synthetase: synthesis and properties of active CO2, precursor of carbamoyl phosphate. Biochemistry 16, 321–329 (1977). [DOI] [PubMed] [Google Scholar]
- Britton H. G., Rubio V. & Grisolia S. Mechanism of carbamoyl-phosphate synthetase. Properties of the two binding sites for ATP. Eur. J. Biochem. 102, 521–530 (1979). [DOI] [PubMed] [Google Scholar]
- Metzenberg R. L., Marshall M. & Cohen P. P. Carbamyl phosphate synthetase: studies on the mechanism of action. J. Biol. Chem. 233, 1560–1564 (1958). [PubMed] [Google Scholar]
- Rubio V., Britton H. G., Grisolia S., Sproat B. S. & Lowe G. Mechanism of activation of bicarbonate ion by mitochondrial carbamoyl-phosphate synthetase: formation of enzyme-bound adenosine diphosphate from the adenosine triphosphate that yields inorganic phosphate. Biochemistry 20, 1969–1974 (1981). [DOI] [PubMed] [Google Scholar]
- Guthöhrlein G. & Knappe J. Structure and function of carbamoylphosphate synthetase. On the mechanism of bicarbonate activation. Eur. J. Biochem. 8, 207–214 (1969). [DOI] [PubMed] [Google Scholar]
- Lusty C. J. Carbamyl phosphate synthetase. Bicarbonate-dependent hydrolysis of ATP and potassium activation. J. Biol. Chem. 253, 4270–4278 (1978). [PubMed] [Google Scholar]
- Rubio V., Britton H. G. & Grisolia S. Mitochondrial carbamoyl phosphate synthetase activity in the absence of N-acetyl-L-glutamate. Mechanism of activation by this cofactor. Eur. J. Biochem. 134, 337–343 (1983). [DOI] [PubMed] [Google Scholar]
- Alonso E. & Rubio V. Binding of N-acetyl-L-glutamate to rat liver carbamoyl phosphate synthetase (ammonia). Eur. J. Biochem. 135, 331–337 (1983). [DOI] [PubMed] [Google Scholar]
- Häberle J. et al. Molecular defects in human carbamoyl phosphate synthetase I: Mutational spectrum, diagnostic and protein structure considerations. Hum Mutat 32, 579–589 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Lomb D. J., Haigis M. C. & Guarente L. SIRT 5 deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell 137, 560–570 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M. et al. Overexpression of SIRT5 confirms the involvement in deacetylation and activation of carbamoyl phosphate synthetase 1. Biochem. Biophys. Res. Commun. 393, 73–78 (2010). [DOI] [PubMed] [Google Scholar]
- Du J. et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M. et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 19, 605–617 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Fernandez C. et al. Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as a key tool. Hum. Mutat. 34, 1149–1159 (2013). [DOI] [PubMed] [Google Scholar]
- Krissinel E. & Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- Pierson D. L. & Brien J. M. Human carbamylphosphate synthetase I. Stabilization, purification, and partial characterization of the enzyme from human liver. J. Biol. Chem. 255, 7891–7895 (1980). [PubMed] [Google Scholar]
- Lusty C. J. Catalytically active monomer and dimer forms of rat liver carbamoyl-phosphate synthetase. Biochemistry 20, 3665–3674 (1981). [DOI] [PubMed] [Google Scholar]
- Meister A. Mechanism and regulation of the glutamine-dependent carbamyl phosphate synthetase of Escherichia coli. Adv. Enzymol. Relat. Areas Mol. Biol. 62, 315–374 (1989). [DOI] [PubMed] [Google Scholar]
- Thoden J. B., Holden H. M., Wesenberg G., Raushel F. M. & Rayment I. Structure of carbamoyl phosphate synthetase: a journey of 96 Å from substrate to product. Biochemistry 36, 6305–6316 (1997). [DOI] [PubMed] [Google Scholar]
- Díez-Fernández C., Hu L., Cervera J., Häberle J. & Rubio V. Understanding carbamoyl phosphate synthetase (CPS1) deficiency by using the recombinantly purified human enzyme: effects of CPS1 mutations that concentrate in a central domain of unknown function. Mol. Genet. Metab. 112, 123–132 (2014). [DOI] [PubMed] [Google Scholar]
- Thoden J. B., Raushel F. M., Benning M. M., Rayment I. & Holden H. M. The structure of carbamoyl phosphate synthetase determined to 2.1 Å resolution. Acta Crystallogr. D. Biol. Crystallogr. 55, 8–24 (1999). [DOI] [PubMed] [Google Scholar]
- Rodriguez-Aparicio L. B., Guadalajara A. M. & Rubio V. Physical location of the site for N-acetyl-L-glutamate, the allosteric activator of carbamoyl phosphate synthetase, in the 20-kilodalton COOH-terminal domain. Biochemistry 28, 3070–3074 (1989). [DOI] [PubMed] [Google Scholar]
- Britton H. G., García-España A., Goya P., Rozas I. & Rubio V. A structure-reactivity study of the binding of acetylglutamate to carbamoyl phosphate synthetase I. Eur. J. Biochem. 188, 47–53 (1990). [DOI] [PubMed] [Google Scholar]
- Díez-Fernández C., Gallego J., Häberle J., Cervera J. & Rubio V. The study of carbamoyl phosphate synthetase 1 deficiency sheds light on the mechanism for switching on/off the urea cycle. J. Genet. Genomics 42, 249–260 (2015). [DOI] [PubMed] [Google Scholar]
- Pekkala S. et al. Understanding carbamoyl-phosphate synthetase 1 (CPS1) deficiency by using expression studies and structure-based analysis. Hum. Mutat. 31, 801–808 (2010). [DOI] [PubMed] [Google Scholar]
- Rubio V., Britton H. G. & Grisolía S. Activation of carbamoyl phosphate synthetase by cryoprotectants. Mol. Cell. Biochem. 53–54, 279–298 (1983). [DOI] [PubMed] [Google Scholar]
- Fawaz M. V., Topper M. E. & Firestine S. M. The ATP-grasp enzymes. Bioorg. Chem. 39, 185–191 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusty C. J. The molecular structure and function of carbamyl phosphate synthetase I. Trans. N.Y. Acad Sci. 41, 103–115 (1983). [DOI] [PubMed] [Google Scholar]
- Alonso E. & Rubio V. Affinity cleavage of carbamoyl-phosphate synthetase I localizes regions of the enzyme interacting with the molecule of ATP that phosphorylates carbamate. Eur. J. Biochem. 229, 377–384 (1995). [DOI] [PubMed] [Google Scholar]
- Kim J., Howell S., Huang X. & Raushel F. M. Structural defects within the carbamate tunnel of carbamoyl phosphate synthetase. Biochemistry 41, 12575–12581 (2002). [DOI] [PubMed] [Google Scholar]
- Kim J. & Raushel F. M. Access to the carbamate tunnel of carbamoyl phosphate synthetase. Arch. Biochem. Biophys. 425, 33–41 (2004). [DOI] [PubMed] [Google Scholar]
- Lund L., Fan Y., Shao Q., Gao Y. Q. & Raushel F. M. Carbamate transport in carbamoyl phosphate synthetase: a theoretical and experimental investigation. J. Am. Chem. Soc. 132, 3870–3878 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makoff A. J. & Radford A. Genetics and biochemistry of carbamoyl phosphate biosynthesis and its utilization in the pyrimidine biosynthetic pathway. Microbiol. Rev. 42, 307–328 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. E. Pyrimidine nucleotide biosynthesis in animals: genes, enzymes and regulation of UMP biosynthesis. Annu. Rev. Biochem. 49, 253–279 (1980). [DOI] [PubMed] [Google Scholar]
- Anderson P. M. Glutamine- and N-acetylglutamate-dependent carbamoyl phosphate synthetase in elasmobranchs. Science 208, 291–293 (1980). [DOI] [PubMed] [Google Scholar]
- Simmer J. P., Kelly R. E., Rinker A. G. Jr, Scully J. L. & Evans D. R. Mammalian carbamyl phosphate synthetase (CPS). DNA sequence and evolution of the CPS domain of the Syrian hamster multifunctional protein CAD. J. Biol. Chem. 265, 10395–10402 (1990). [PubMed] [Google Scholar]
- Saeed-Kothe A. & Powers-Lee S. G. Gain of glutaminase function in mutants of the ammonia-specific frog carbamoyl phosphate synthetase. J. Biol. Chem. 278, 26722–26726 (2003). [DOI] [PubMed] [Google Scholar]
- Fan Y., Lund L., Shao Q., Gao Y. Q. & Raushel F. M. A combined theoretical and experimental study of the ammonia tunnel in carbamoyl phosphate synthetase. J. Am. Chem. Soc. 131, 10211–10219 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton H. G. & Rubio V. Carbamoyl-phosphate synthetase I. Kinetics of binding and dissociation of acetylglutamate and of activation and deactivation. Eur. J. Biochem. 171, 615–622 (1988). [DOI] [PubMed] [Google Scholar]
- Huang X. & Raushel F. M. An engineered blockage within the ammonia tunnel of carbamoyl phosphate synthetase prevents the use of glutamine as a substrate but not ammonia. Biochemistry 39, 3240–3247 (2000). [DOI] [PubMed] [Google Scholar]
- Huang X. & Raushel F. M. Restricted passage of reaction intermediates through the ammonia tunnel of carbamoyl phosphate synthetase. J. Biol. Chem. 275, 26233–26240 (2000). [DOI] [PubMed] [Google Scholar]
- Yefimenko I., Fresquet V., Marco-Marín C., Rubio V. & Cervera J. Understanding carbamoyl phosphate synthetase deficiency: impact of clinical mutations on enzyme functionality. J. Mol. Biol. 349, 127–141 (2005). [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. R. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006). [DOI] [PubMed] [Google Scholar]
- McCoy A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–67 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W. G. & Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten R. P. et al. PDB_REDO: automated re-refinement of X-ray structure models in the PDB. J. Appl. Crystallogr. 42, 376–384 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnal D. et al. MOLE 2.0: advanced approach for analysis of biomacromolecular channels. J. Cheminform. 5, 39 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho B. K. & Gruswitz F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8, 49 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs K. & Karplus P. A. Better models by discarding data? Acta Crystallogr. D Biol. Crystallogr. 69, 1215–1222 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.