Abstract
Depending on their physiological expression level, microRNAs (miRNA) address different target genes, thus have different biological functions. To identify these, the physiological expression has to be blocked. Here, we describe the use of inhibitory cholesterol-modified oligonucleotides (Antagomirs) to inhibit miRNAs selectively in primary human and murine T and B lymphocytes. Due to their lipophilic cholesterol tag Antagomirs enter primary lymphocytes efficiently and quantitatively. We show here that at concentrations of 0.125 to 1 μM, Antagomirs selectively inhibit expression of their target miRNA up to 99.5% without affecting cell viability.
Keywords: miRNA knockdown, Antagomir, Murine and human primary lymphocytes
1. Introduction
Sequence-specific, genetic inhibition of miRNA is the only way to determine its role in physiology, at the level expressed in a given cell (Stenvang et al., 2012). So far, introduction of specific inhibitory oligonucleotides into the cell had required viral and non-viral transfection protocols, the latter dependent on cationic lipids, polymer reagents or electroporation (Mantei et al., 2008). This poses a fundamental problem for the analysis of miRNA in primary T and B lymphocytes. First, they are highly refractory to non-viral transfection methods, and second, they are activated by viral infection, introducing an artifactual bias on the analysis of miRNA (Ziegler et al., 2013). Here, we describe a detailed protocol for the direct introduction of Antagomirs into murine and human primary T (Stittrich et al., 2010, Haftmann et al., 2015, Warth et al., 2015) and B lymphocytes (Knoll et al., 2013, Porstner et al., 2015) solving this fundamental problem. Antagomirs are 20–25 nucleotide long, single stranded RNA molecules, with a sequence complementary to an entire mature miRNA (Krutzfeldt et al., 2005). Their backbone consists of 2′-O-methyl (2′-O-Me) single stranded oligoribonucleotides and partially modified phosphorothioate (PS) linkers. Antagomirs have a cholesterol-tag at their 3′ end, which enables their efficient direct uptake via the cell membrane into viable cells. At concentrations of 0.125 to 1 μM, Antagomirs selectively inhibit their target miRNA up to 99.5%, without affecting viability of the T and B lymphocytes, or activating them.
2. Materials and methods
2.1. Mice
C57BL/6 mice were housed and bred under specific pathogen-free conditions. Mice were handled in accordance with good animal practice as defined by German animal welfare bodies and sacrificed by cervical dislocation.
2.2. Cell culture
Naïve CD4+ T cells and CD19+ B cells were isolated from spleens of 6–10 week old mice (magnetic activated cell sorting (MACS) microbead technology, Miltenyi Biotech). For T helper 1 lymphocyte (Th1) polarization, 3 × 106 naïve CD4+ T cells were cultured and stimulated with plate-bound αCD3/αCD28 (3 μg/ml, eBioscience), in the presence of irradiated (30 Gy) CD90-depleted splenocytes from C57BL/6 mice as antigen-presenting cells and recombinant IL-12 (5 ng/ml; R&D Systems) and anti-IL-4 (11B11) antibody. Under this condition, viable Th1 cells were restimulated every 6 days for three rounds to induce repeatedly activated Th1 cells (Haftmann et al., 2015). Ex vivo isolated murine CD19+ B cells were activated with 1 μg/ml lipopolysaccharide (LPS). Human peripheral blood mononuclear cells were isolated from whole blood by density gradient centrifugation and fluorescently labeled for CD4 and CD19.
2.3. Reagents
Lyophilized Antagomirs were custom synthesized according to Krutzfeldt et al. (2005) (Dharmacon, GE Healthcare). Antagomir sequences are as follows: Antagomir-scr 5-mU(*)mC(*)mAmCmGmCmAmGmAmUmUmCmAmUmAmA(*)mC(*)mG(*)mU(*)-3-Chol; and Antagomir-148a 5-mA(*)mC(*)mAmAmAmGmUmUmCmUmGmUmAmGmUmGmCmAmC(*)mU(*)mG(*)mA(*)-3-Chol. All ribonucleotides are 2′-O-methyl modified (mN) and (*) represents a phosphorothioate modification of the backbone. At the 3′-end of the oligonucleotides, a cholesterol molecule was added. Lyophilized Antagomirs were resolved in RNase-free water at the desired concentration at room temperature for 30 min with slight shaking.
2.4. Antagomir treatment
Naïve or resting primary murine or human T and B lymphocytes as well as murine repeatedly activated Th1 cells were washed with cold PBS at 300 g for 8 min. Lymphocytes (up to 1 × 107 cells/ml) were resuspended in 0.25 volumes of final culture volume of serum-free medium (ACCELL, Dharmacon) supplemented with Antagomir-148a or Antagomir-scr in different concentrations. Lymphocytes were incubated for 2 h at 5% CO2, 95% humidity and 37 °C. After incubation, repeatedly activated murine Th1 cells were stimulated with plate-bound αCD3/αCD28 (3 μg/ml, eBioscience). CD19+ murine B cells were activated with 1 μg/ml LPS (see also the Cell culture section). Both cell types were further cultured in RPMI 1640 (Gibco®, Life Technologies) cell culture medium with 10% FCS, 100 U/ml Penicillin/Streptomycin, 50 μM β-mercaptoethanol in vitro (RPMI was added in fourfold excess to the lymphocytes in the ACCELL/Antagomir medium).
A detailed protocol for Antagomir treatment is provided in Table 1.
Table 1.
Protocol — procedure and timing.
| Protocol | Comments | |
|---|---|---|
| Pre-incubation of lymphocytes with Antagomirs, timing 2.5 h | ||
| 1 | Wash lymphocytes (naïve or resting primary T or B cells) with cold PBS and centrifuge at 300 g for 8 min. | |
| 2 | Resuspend lymphocytes (up to 1 ∗ 107 cells/ml) in 0.25 volumes of final culture volume of serum-free medium, e.g., ACCELL (Dharmacon) (i.e., if cell culture is carried out in 5 ml, resuspend lymphocytes in 1.25 ml serum-free medium). | Critical: serum-free medium increases the efficiency of Antagomir uptake from the medium. The cholesterol-coupled Antagomirs will attach to the cell surface and are subsequently incorporated. |
| 3 | Dilute Antagomirs at the desired and titrated concentration. | Critical: even at only moderate miRNA knockdown efficiency there may already be visible phenotypical changes and impact on potential targets. Too high concentrations of Antagomirs may lead to off-target effects. Therefore, a titration of the Antagomirs and determination of suitable analysis time points in different culture settings is highly recommended prior to experimental procedures. |
| 4 | Gently mix samples and incubate for 2 h at 5% CO2, 95% humidity and 37 °C. Meanwhile prepare stimulating and differentiating medium for further cell culture. | |
| Stimulation and culture of Antagomir-treated lymphocytes in vitro | ||
| 5 | Add 0.75 volumes of serum-containing culture medium supplemented with all necessary cytokines and stimuli to culture wells/plates (e.g., in this study, repeatedly activated Th1 cells were restimulated with plate-bound αCD3/αCD28 and recombinant IL-12 and anti-IL-4 antibody). | Antagomir uptake does not depend on reactivation of lymphocytes. Resting lymphocytes treated with Antagomirs can also be monitored (e.g., Fig. 1d) and analyzed. |
| 6 | Add 0.25 volumes of Antagomir-treated cells to the medium and incubate at 5% CO2, 95% humidity and 37 °C for the desired time. | Critical: Determine the most suitable analysis time point prior to experimental procedures, e.g., by expression kinetics of miRNA, target genes and usage of reporter gene assays. |
| Control knockdown efficiency by quantitative real-time PCR analysis | ||
| 7 | At the end of the experimental procedure, perform the desired analysis and take an aliquot of the cells to determine knockdown efficiency of Antagomirs (and their potential impact on targets) by qRT-PCR analysis or Northern Blots (and/or protein detection). | |
2.5. Detection of Antagomir incorporation
Fluorescein-conjugated Antagomirs were used for quantification of Antagomir incorporation and detected by flow cytometry (MACSQuant, Miltenyi Biotec) or image stream cytometry (Amnis ImageStreamX MKII, Merck Millipore).
2.6. RNA extraction and quantitative PCR analysis
Total RNA was isolated with Direct-zol RNA kit (Zymo Research). Mature miR-148a, miR-148b, miR-152, miR-182, miR-320 and U6 small nuclear RNA (snRNA) were detected by quantitative PCR with a Taqman MicroRNA Reverse Transcription kit in combination with TaqMan MicroRNA Assays (Life Technologies) according to the manufacturer's recommendations. For normalization, the expression values were compared to values of snU6 by the change-in-threshold method (2 − ΔCT) (Haftmann et al., 2015).
Reverse transcription of mRNA was performed using the Reverse Transcription kit (Life Technologies, Germany) and cDNA was quantified by SYBR Green based real-time PCR (Roche) using the following primer pairs: murine hypoxanthine guanine phosphoribosyltransferase (HPRT) forward 5′-TCCTCCTCAGACCGCTTTT-3′, HPRT reverse 5′-CATAACCTGGTTCATCATCGC-3′, murine BIM forward 5′-CCCGGAGATACGGATTGCA-3′ and BIM reverse 5′-AACACCCTCCTTGTGTAAGTTTCGT-3′ (Haftmann et al., 2015). For normalization, the expression values were compared to values of HPRT by the change-in-threshold method (2 − ΔCT) (Livak and Schmittgen, 2001).
3. Results and discussion
Here, we show a simple and reliable method to inhibit the physiological function of miRNA in T and B lymphocytes, which are difficult to transfect by conventional methods. The introduction of Antagomirs does not require the use of transfection vehicles or electroporation. Antagomirs act in the cytoplasm by inhibiting the activity of their target miRNA.
3.1. Efficiency of Antagomir uptake in primary murine and human T and B cells
To introduce Antagomirs into ex vivo isolated murine B and T lymphocytes, the cells were incubated with fluorescein-conjugated Antagomir for 2 h in serum-free medium. Uptake of cholesterol-containing oligonucleotides is highly efficient in serum-free medium (Petrova et al., 2012). The Antagomir (Antagomir-scr) was designed not to target any known miRNA (Stittrich et al., 2010, Knoll et al., 2013, Haftmann et al., 2015, Porstner et al., 2015, Warth et al., 2015).
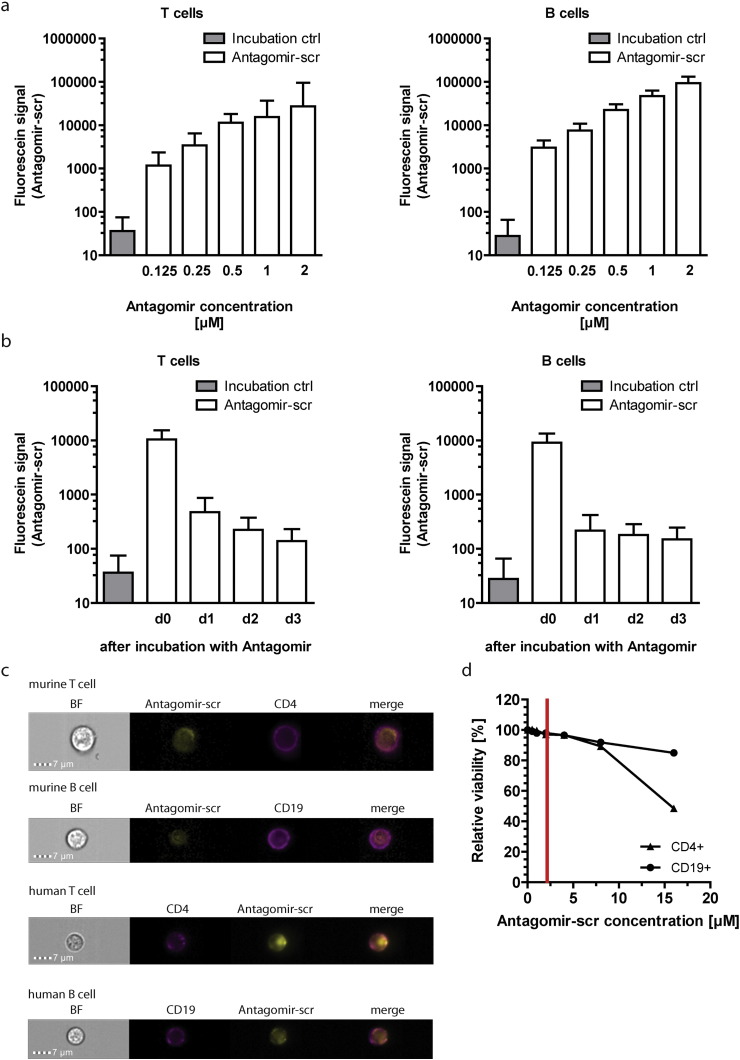
Fluorescein-conjugated Antagomir-scr was incorporated in a concentration dependent manner after 2 h incubation in primary B and T cells (Fig. 1a). Ex vivo isolated T and B cells had taken up the Antagomir quantitatively and a fluorescein signal was detectable even 3 days after activation of the cells despite proliferation (Fig. 1b). Strong fluorescence was detected at d0 and was most likely due to surface-bound Antagomir-scr which was lost by d1. The Antagomirs taken up by the cells were detectable within T as well as B lymphocytes and do not co-localize with CD4 or CD19 surface staining (Fig. 1c). Localization of Antagomirs in the cytoplasm after in vivo application has been demonstrated by others (Krutzfeldt et al., 2007), and suggests why nuclear miRNA precursors are not affected by Antagomirs (Krutzfeldt et al., 2005).
Fig. 1.
Antagomirs efficiently enter primary lymphocytes, are localized in the cytoplasm and show low cytotoxicity. Ex vivo isolated (MACS Technology, Miltenyi Biotech, Germany) unstimulated CD4+ T cells and unstimulated CD19+ B cells were treated with fluorescein-coupled non-targeting control Antagomir (Antagomir-scr) or incubated in serum-free medium without addition of Antagomir (Incubation ctrl). a) Cells were treated with concentrations of 0.125 to 2 μM of fluorescein-coupled Antagomir-scr for 2 h in serum-free medium and fluorescence was analyzed by flow cytometry; b) cells were treated with a concentration of 1 μM of fluorescein-coupled Antagomir-scr, washed after 2 h of incubation with PBS, cultured in FCS-containing RPMI medium for 3 days and fluorescence was analyzed by flow cytometry, d0 = cells after 2 h of incubation; c) cytoplasmic localization of fluorescein-coupled Antagomir-scr in live murine CD4+ T cells, murine CD19+ B cells, human CD4+ T cells and human CD19+ B cells was determined by single cell immunofluorescence analysis (Amnis ImageStreamX MKII, Merck Millipore) 24 h after incubation with Antagomirs, BF — bright field, CD4/CD19 — surface staining; d) viability of murine CD4+ Th1 cells and CD19+ B cells after treatment with different concentrations of Antagomir-scr in serum-free medium for 2 h. Viability was determined by propidium iodide staining, analyzed by flow cytometry and normalized to cells that were incubated in serum-free medium without addition of Antagomir.
When Antagomirs were applied at concentrations of 1 μM and for 3 h of incubation in serum-free medium, more than 95% of the T and B lymphocytes were viable, as determined by propidium iodide staining (Fig. 1d). An increase of the Antagomir concentration to 16 μM resulted in reduced viability of T cells to below 50%, while still more than 85% of the B cells were viable. After incubation with Antagomir concentrations of 1 μM cell viability is maintained over several days allowing for functional assays in vitro or adoptive cell transfer in murine models (Stittrich et al., 2010, Knoll et al., 2013, Haftmann et al., 2015, Porstner et al., 2015, Warth et al., 2015). Contrastingly, the introduction of antisense oligonucleotides by nucleofection or lipofection results in high variability in terms of efficiency and negatively affects cell viability and are therefore less reliable methods for miRNA knockdown in experimental settings.
3.2. Efficiency and specificity of miRNA-knockdown by Antagomir treatment
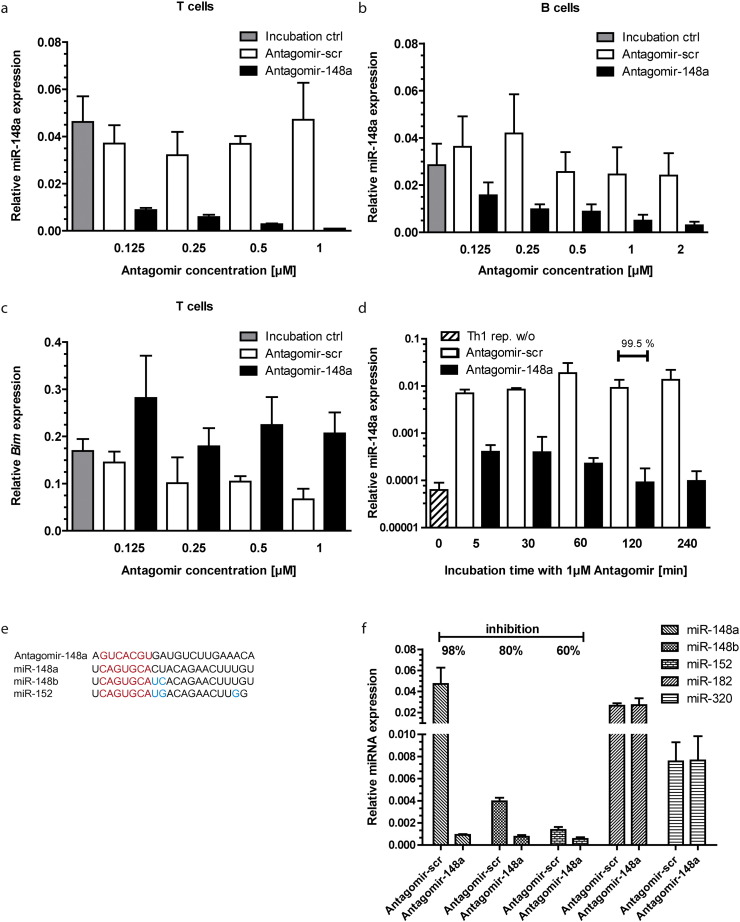
To assess the specific on-target activity of Antagomirs taken up by T and B lymphocytes, we used a non-fluorescently labeled Antagomir targeting miR-148a (Antagomir-148a). miR-148a is highly expressed in repeatedly activated murine Th1 lymphocytes following restimulation with anti-CD3 and anti-CD28 antibodies (reactivated 4 times in weekly intervals) (Haftmann et al., 2015) and in LPS-activated CD19+ B lymphocytes (Porstner et al., 2015). Repeatedly activated Th1 cells were incubated for 2 h in serum-free medium with Antagomir-148a, a non-targeting non-fluorescently labeled control Antagomir (Antagomir-scr) or without Antagomir (Incubation ctrl). Expression of miR-148a was quantitated by real-time PCR analysis. Expression of miR-148a was inhibited selectively and in a dose-dependent fashion by the Antagomir-148a at concentrations between 0.125 and 1 μM in both T and B lymphocytes. Expression of miR-148a was completely blocked in T lymphocytes at 1 μM, and inhibited by 87% in B lymphocytes at 2 μM (Fig. 2a, b). Inhibition of miR-148a results in an about 2-fold increase in the mRNA level of its target gene Bim, already at a concentration of 0.125 μM Antagomir-148a (Fig. 2c), demonstrating the specific Antagomir effect on miRNA function primarily via RNAi-independent degradation of miRNA (Krutzfeldt et al., 2007). For miRNA with unknown target genes, we suggest to use reporter gene assays for testing the functionality of Antagomir treatment (Davis et al., 2006). Such reporter constructs have specific miRNA binding sites in their 3′ untranslated region and are subsequently subjected to miRNA-mediated downregulation, which should be restored by Antagomir treatment (Davis et al., 2006).
Fig. 2.
Antagomirs silence miR-148a with a concentration-dependent efficacy in T and B cells and with high miRNA specificity. miR-148a expression was analyzed in a) repeatedly activated Th1 cells (Niesner et al., 2008, Haftmann et al., 2015) on day 3 after reactivation with 3 μg/ml of plate-bound αCD3/αCD28 and b) ex vivo isolated CD19+ B cells on day 4 after activation with 1 μg/ml LPS. The cells were treated with indicated concentrations of Antagomir-148a, a non-targeting control Antagomir (Antagomir-scr) or incubated in serum-free medium without addition of Antagomir (Incubation ctrl). miRNA expression was determined by qRT-PCR analysis and normalized to small nuclear RNA U6 (snU6); c) analysis of Bim expression on day 3 after restimulation of repeatedly activated Th1 cells treated with various concentrations of Antagomir-148a, Antagomir-scr or without Antagomir (Incubation ctrl), determined by qRT-PCR analysis and normalized to hypoxanthine-guanine phosphoribosyltransferase (HPRT) expression; d) miR-148a expression in repeatedly activated Th1 cells (Niesner et al., 2008, Haftmann et al., 2015) on day 3 after reactivation either treated with 1 μM Antagomir-148a or Antagomir-scr during different incubation times in serum-free medium in comparison to repeatedly activated Th1 cells before restimulation (Th1 rep. w/o). Repeatedly activated Th1 cells were restimulated with 3 μg/ml of plate-bound αCD3/αCD28; e) sequence homology of miR-148a, miR-148b and miR-152 and complementarity to Antagomir-148a (red indicates the miRNA seed region and blue the mismatches between miR-148b and miR-152 compared to miR-148a); f) expression levels of miR-148a, miR-148b, miR-152, miR-182 and miR-320 in repeatedly activated Th1 cells treated with 1 μM Antagomir-148a or Antagomir-scr on day 3 after restimulation with 3 μg/ml of plate-bound αCD3/αCD28, determined by qRT-PCR analysis and normalized to snU6.
To determine the optimal incubation time for miR-148a knockdown, repeatedly activated Th1 lymphocytes were incubated in serum-free medium with either 1 μM Antagomir-148a or Antagomir-scr from 5 min up to 4 h. After the various incubation periods, cells were further stimulated with αCD3 and αCD28 antibodies for 3 days. Expression of miR-148a was inhibited by 99.5% after an incubation time of 2 h and resembled miR-148a levels observed in repeatedly reactivated Th1 lymphocytes before restimulation (Fig. 2d). Five minutes of incubation with Antagomirs is sufficient to achieve a ~ 94% knockdown of miR-148a three days after activation. This may be sufficient for experiments in which prolonged serum starvation leads to massive cell death.
The specificity of Antagomir action was assessed by comparing the impact of Antagomir-148a treatment on the levels of miR-148a, miR-148b, miR-152, miR-182 and miR-320 in repeatedly reactivated T lymphocytes. Mir-148a, miR-148b and miR-152 share the same seed sequence (Fig. 2e). Antagomir-148a inhibited expression of miR-148a by 98%, miR-148b by 80% and miR-152 by 60%. Expression of miR-182 and miR-320 was not inhibited (Fig. 2f). Knockdown of the target miRNA is highly sequence-specific and miRNAs with unrelated sequence remain unaffected. Even two (miR-148b) or three mismatches (miR-152) (Fig. 2e) in closely related miRNA sequences reduce the knockdown efficiencies markedly. Off-target effects on miRNA with related sequences must be tested for each individual Antagomir. Of note, the efficiency of miRNA knockdown can be alternatively tested by Northern Blots (Krutzfeldt et al., 2005, Davis et al., 2006) instead of quantitative real time-PCR (qRT-PCR).
Antagomirs are suitable for selective miRNA inhibition in resting as well as activated lymphocytes. This method is efficient, highly reproducible and Antagomirs are incorporated to the same extent in all cells of a particular subset already after a short incubation period. In contrast, by conventional transfection- or electroporation-based methods, oligonucleotides are taken up by only by a fraction of the cells. Thus, we suggest the use of Antagomirs for miRNA inhibition in primary T and B cells as the method of choice in functional studies. We provide a typical work-flow in Table 1 and suggestions for managing experimental difficulties and troubleshooting are listed in Appendix A.
Author contribution
CH and RR established the protocol, designed and performed the experiments, analyzed the data and wrote the manuscript. JW and MP performed experiments. HDC and AR supervised the research and wrote the manuscript and MFM supervised the research, discussed the results, analyzed the data and wrote the manuscript.
Conflict of interest
The authors declare no conflicting interest in this work.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 650 and SFB 633), the IMI JU funded project BTCure, no 115241, the FORSYS (Forschungseinheiten zur Systembiologie) program and the “e:Bio — Innovationswettbewerb Systembiologie” program of the Federal Ministry of Education and Research, and the European Research Council (ERC) Advanced Grant (ERC-2010-AdG_20100317 Grant 268987). RR is a member of the Berlin-Brandenburg School of Regenerative Therapies (BSRT, DFG-GSC203). CH is a member of the International Max Planck Research School for Infectious Diseases and Immunology (IMPRS-IDI).
Appendix A. Troubleshooting
| Problem | Possible reason | Solution |
|---|---|---|
| No phenotypical change | No efficient silencing of specific miRNA | Check for knockdown efficiency, if no knockdown can be observed, see “No change in miRNA levels” section |
| No impact on target | Divergent target expression profile | Compare miRNA and target expression (mRNA and protein) kinetics and determine suitable analysis time point (impact on target mRNA and protein may vary) |
| No efficient silencing of specific miRNA | Check for knockdown efficiency, if no knockdown can be observed, see “No change in miRNA levels” section | |
| No change in miRNA levels | Antagomir does not have the correct specificity for the miRNA | Control Antagomir sequence |
| No functional Antagomir | Measure concentration of Antagomir in stock solution to ensure it was not degraded during reconstitution and/or shipment | |
| Always resuspend Antagomirs in RNase-free water, Antagomirs are extremely susceptible to RNase-mediated degradation | ||
| No efficient silencing by Antagomir | Increase Antagomir concentration (some miRNAs have a high abundance and may therefore need higher concentrations of Antagomirs or a later analysis time point for optimal knockdown effect) | |
| Increase incubation time in FCS-free medium | ||
| Decrease cell number | ||
| Test different analysis time points (Antagomirs should enter cells already shortly after treatment. If analysis time point is very late, miRNA levels could be already restored. Alternatively, highly abundant miRNAs may need a longer time period to be sufficiently knocked down) | ||
| Divergent miRNA expression profile | Define the expression kinetics of miRNA to determine suitable analysis time point | |
| Only mild change in miRNA levels | Inefficient knockdown | Increase Antagomir concentration (some miRNAs have a high abundance in the cells and may therefore need higher concentrations of Antagomirs or a later analysis time point for optimal knockdown effect. In addition, strong stimuli lead to an exacerbated proliferation and dilution of the Antagomir) |
| Increase incubation time | ||
| Decrease cell number | ||
| Test different analysis time points (Antagomirs should enter cells already shortly after treatment. If analysis time point is very late, miRNA levels could be already restored. Alternatively, highly abundant miRNAs may need a longer time period to be sufficiently knocked down) | ||
| Antagomirs are competitively taken up by non-target cells in the culture | Increase Antagomir concentration | |
| Change the ratio of target cells to non-target cells | ||
| Remove non-target cells during Antagomir treatment | ||
| No miRNA detection | Loss of miRNA during RNA purification | Use miRNA-specific RNA prep system |
| Cells died during culture | Cells are very sensitive to serum starvation | Decrease incubation time |
| Treat with rich medium, e.g., medium supplemented with ITS liquid media (Sigma Life Science, Germany) supplement in the final cell culture medium | ||
| Titrate low amounts of serum into pulse medium to find optimal balance of miRNA knockdown and cell viability | ||
| Exchange medium to provide fresh nutrients | ||
| Decrease cell culture duration |
References
- Davis S., Lollo B., Freier S., Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haftmann C., Stittrich A.B., Zimmermann J., Fang Z., Hradilkova K., Bardua M., Westendorf K., Heinz G.A., Riedel R., Siede J., Lehmann K., Weinberger E.E., Zimmel D., Lauer U., Haupl T., Sieper J., Backhaus M., Neumann C., Hoffmann U., Porstner M., Chen W., Grun J.R., Baumgrass R., Matz M., Lohning M., Scheffold A., Wittmann J., Chang H.D., Rajewsky N., Jack H.M., Radbruch A., Mashreghi M.F. miR-148a is upregulated by Twist1 and T-bet and promotes Th1-cell survival by regulating the proapoptotic gene Bim. Eur. J. Immunol. 2015;45:1192. doi: 10.1002/eji.201444633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll M., Simmons S., Bouquet C., Grun J.R., Melchers F. miR-221 redirects precursor B cells to the BM and regulates their residence. Eur. J. Immunol. 2013;43:2497. doi: 10.1002/eji.201343367. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J., Kuwajima S., Braich R., Rajeev K.G., Pena J., Tuschl T., Manoharan M., Stoffel M. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(− Delta Delta C(T)) Method. Methods. 2001;25:402. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mantei A., Rutz S., Janke M., Kirchhoff D., Jung U., Patzel V., Vogel U., Rudel T., Andreou I., Weber M., Scheffold A. siRNA stabilization prolongs gene knockdown in primary T lymphocytes. Eur. J. Immunol. 2008;38:2616. doi: 10.1002/eji.200738075. [DOI] [PubMed] [Google Scholar]
- Niesner U., Albrecht I., Janke M., Doebis C., Loddenkemper C., Lexberg M.H., Eulenburg K., Kreher S., Koeck J., Baumgrass R., Bonhagen K., Kamradt T., Enghard P., Humrich J.Y., Rutz S., Schulze-Topphoff U., Aktas O., Bartfeld S., Radbruch H., Hegazy A.N., Lohning M., Baumgart D.C., Duchmann R., Rudwaleit M., Haupl T., Gitelman I., Krenn V., Gruen J., Sieper J., Zeitz M., Wiedenmann B., Zipp F., Hamann A., Janitz M., Scheffold A., Burmester G.R., Chang H.D., Radbruch A. Autoregulation of Th1-mediated inflammation by twist1. J. Exp. Med. 2008;205:1889. doi: 10.1084/jem.20072468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova N.S., Chernikov I.V., Meschaninova M.I., Dovydenko I.S., Venyaminova A.G., Zenkova M.A., Vlassov V.V., Chernolovskaya E.L. Carrier-free cellular uptake and the gene-silencing activity of the lipophilic siRNAs is strongly affected by the length of the linker between siRNA and lipophilic group. Nucleic Acids Res. 2012;40:2330. doi: 10.1093/nar/gkr1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstner M., Winkelmann R., Daum P., Schmid J., Pracht K., Corte-Real J., Schreiber S., Haftmann C., Brandl A., Mashreghi M.F., Gelse K., Hauke M., Wirries I., Zwick M., Roth E., Radbruch A., Wittmann J., Jack H.M. miR-148a promotes plasma cell differentiation and targets the germinal center transcription factors Mitf and Bach2. Eur. J. Immunol. 2015;45:1206. doi: 10.1002/eji.201444637. [DOI] [PubMed] [Google Scholar]
- Stenvang J., Petri A., Lindow M., Obad S., Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 2012;3:1. doi: 10.1186/1758-907X-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stittrich A.B., Haftmann C., Sgouroudis E., Kuhl A.A., Hegazy A.N., Panse I., Riedel R., Flossdorf M., Dong J., Fuhrmann F., Heinz G.A., Fang Z., Li N., Bissels U., Hatam F., Jahn A., Hammoud B., Matz M., Schulze F.M., Baumgrass R., Bosio A., Mollenkopf H.J., Grun J., Thiel A., Chen W., Hofer T., Loddenkemper C., Lohning M., Chang H.D., Rajewsky N., Radbruch A., Mashreghi M.F. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat. Immunol. 2010;11:1057. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- Warth S.C., Hoefig K.P., Hiekel A., Schallenberg S., Jovanovic K., Klein L., Kretschmer K., Ansel K.M., Heissmeyer V. Induced miR-99a expression represses Mtor cooperatively with miR-150 to promote regulatory T-cell differentiation. EMBO J. 2015;34(9):1195–1213. doi: 10.15252/embj.201489589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler S., Eberle M.E., Wolfle S.J., Heeg K., Bekeredjian-Ding I. Bifunctional oligodeoxynucleotide/antagomiR constructs: evaluation of a new tool for microRNA silencing. Nucleic Acid Ther. 2013;23:427. doi: 10.1089/nat.2013.0447. [DOI] [PubMed] [Google Scholar]