Cancer cells hijack and remodel existing metabolic pathways for their benefit. Argininosuccinate synthase (ASS1) is a urea cycle enzyme that is essential in the conversion of nitrogen from ammonia and aspartate to urea. A decrease in nitrogen flux through ASS1 in the liver causes the urea cycle disorder citrullinemia1. In contrast to the well-studied consequences of loss of ASS1 activity on ureagenesis, the purpose of its somatic silencing in multiple cancers is largely unknown2. Here, we show that decreased activity of ASS1 in cancers supports proliferation by activating CAD (carbamoyl-phosphate synthase 2, aspartate transcarbamylase, dihydroorotase complex) and facilitating pyrimidines synthesis.
Our studies were initiated by delineating the consequences of loss of ASS1 activation in human, in the two types of citrullinemia. We found that in citrullinemia type I (CTLN I) that is caused by enzymatic deficiency of ASS1, there is increased pyrimidine synthesis and proliferation as compared to citrullinemia type II (CTLN II), in which there is decreased substrate availability for ASS1 due to deficiency of the aspartate transporter, citrin. Building on these results, we demonstrate that ASS1 deficiency in cancer increases cytosolic aspartate levels which increases CAD activation by upregulating its substrate availability and also by increasing its phosphorylation by S6K1 through the mammalian target of Rapamycin (mTOR) pathway. Decreasing CAD activity by blocking citrin, the mTOR signaling or pyrimidine synthesis, decreases proliferation and thus may serve as a therapeutic strategy in multiple cancers where ASS1 is downregulated.
Our results demonstrate that ASS1 downregulation is a novel mechanism to support cancerous proliferation and provide a metabolic link between the urea cycle enzymes and pyrimidine synthesis.
In contrast to the well-delineated biochemical and clinical consequences of loss-of-function germline mutations in ASS1 which have not been reported to include cancer, studies have shown a correlation between somatic deficiency of ASS1 in cancer and poor prognosis, for which the mechanism remain obscure3,2. Outside the liver, ASS1 is expressed in most tissues where it catalyzes the penultimate step in the synthesis of arginine. Argininosuccinate lyase (ASL), the enzyme downstream of ASS1 is directly responsible for arginine synthesis4 (Figure 1A). A well-established sequel of ASS1 and or ASL deficiency, is arginine auxotrophy5 and thus, arginine catabolizing enzymes have been used as therapy in ASS1 depleted tumors with limited benefit, especially in melanoma, wherein the cancer cells develop resistance by re-expressing ASS1 within days3. Since there are cancers in which both these genes are epigenetically silenced6, ASS1 deficiency in cancers might have an arginine-independent effect which might be related to its substrate, aspartate (Figure 1A).
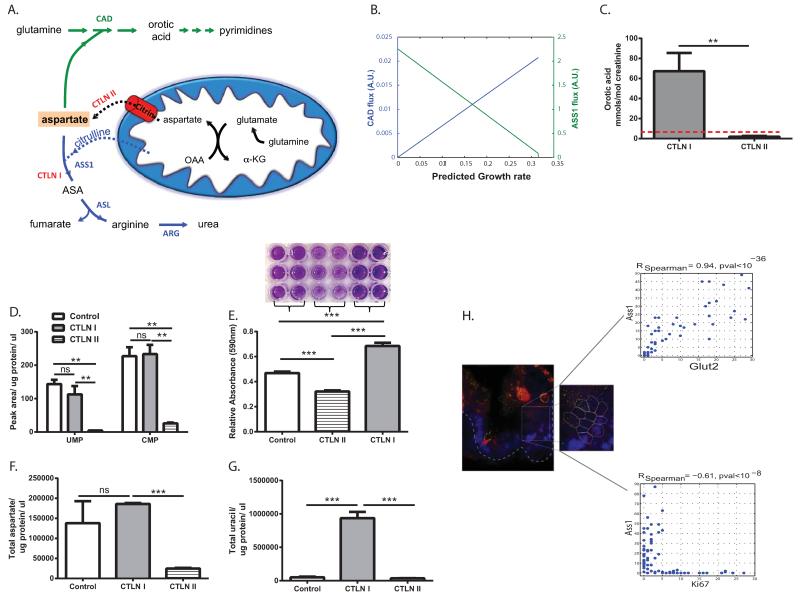
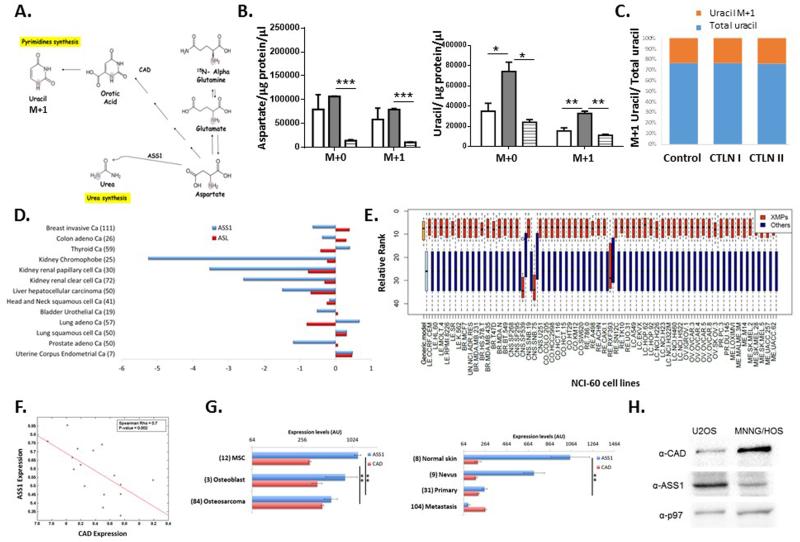
Figure 1. ASS1 inactivation correlates with noncancerous proliferation.
(A) Illustration of the metabolic flux involved in nitrogen contributions to nucleic acid synthesis. The aspartate nitrogen can be used for synthesis of pyrimidines (green path) or urea (blue path). In ASS1 deficiency (CTLN I) there is a potential diversion of the aspartate towards pyrimidine whereas in citrin deficiency (CTLN II) aspartate is not transported across the mitochondria. ASS1- argininosuccinate synthase; ASL – argininosuccinate lyase; ASA- argininosuccinate; ARG1- arginase; CAD- carbamoyl-phosphate synthase 2, aspartate transcarbamylase and dihydroorotase. (B) Prediction by the generic human model; decreasing ASS1 activation (green line) results in an increase in the cellular growth rate and in the flux through the CAD reaction (blue line), A.U.- arbitrary units. (C) Urinary orotic acid levels are elevated significantly in patients with CTLN I as compared to normative values in control subjects (0.3 up to 2.8 mmol/ mol creatinine, depicted by the red dashed line), and to those with CTLN II, **p-Value<0.005 using log transformed data for T-Test analysis. (n= 5 with CTLN I and 4 with CTLN II). (D-G) These experiments were repeated twice with pooled cells from two patients with CTLN II, from one subject with CTLN I and from three control subjects. Statistical analysis was performed using one-way ANOVA, Error bars represent SER. (D) Significantly lower levels of pyrimidines in CTLN II fibroblasts as compared to CTLN I and to normal fibroblasts. TMP was undetectable. (E) Upper panel- Crystal violet staining on day 5 shows increased proliferation in CTLN I as compared to CTLN II and to normal control. Lower panel- A quantification graph for the staining. Anova; ***p-Value< 0.0005. (F-G) Primary fibroblasts from CTLN I subjects have an increased levels of aspartate (F) and uracil (G) as compared to fibroblasts from patients with CTLN II and to normal controls (ANOVA, Tukey HSD, *p<0.0005). This is a representative graph of three independent experiments. (H) Left panel: A small-intestine tissue section of a mouse was hybridized with FISH probes libraries for single Ass1 (green dots) and for Ki67 mRNA molecules (red dots). Nuclei were stained with DAPI (blue). Dashed line outlines the crypt bottom. White lines mark cell borders according to co-immunofluorescence staining with FITC-E-cadherin. A magnifying window highlights a clear demarcation of Ass1 mRNA localizing to the differentiated cells of the crypt, (Magnification 100×). Goblet cells exhibit some non-specific fluorescence appearing in multiple channels. Right Panel: Quantified correlations between Ass1 and Glut2, an enterocyte marker (Upper panel), and between Ass1 and Ki67 (Lower panel).
In the cytosol, aspartate serves as a substrate for both ASS1 and the enzymatic complex CAD. We thus hypothesized that decreased ASS1 activity might enhance aspartate availability for CAD for the synthesis of pyrimidine nucleotides to promote proliferation (Figure 1A). If correct, deficiency in the mitochondrial aspartate transporter, citrin, would be expected to decrease aspartate availability for both ASS1 and CAD and hence restrict proliferation (Figure 1A).
We first assessed the correlation between ASS1 levels and proliferation in non-cancerous states. A generic stoichiometric model of human metabolism7, 17 predicted that inactivation of ASS1 is significantly associated with an increase in growth rate, and is additionally predicted to increase flux through the reaction catalyzed by CAD (Figure 1B). Thus, we expected subjects with ASS1 deficiency (CTLN I), to have increased synthesis of pyrimidines due to increased utilization of aspartate by CAD, as compared to those with CTLN II in whom aspartate availability to CAD is decreased (Figure 1A). Indeed, urinary levels of orotic acid, a product reflecting the synthetic activity of CAD, were significantly elevated in human subjects with CTLN I as compared to the normative values from control population and to subjects with CTLN II (Figure 1A and 1C). Moreover, we found that CTLN I fibroblasts have increased synthesis of pyrimidines and proliferation as compared to CTLN II cells (Figure 1D-E). Using 15N5-α- glutamine we further show that CTLN I cells generate more total as well as labeled M+1 aspartate and M+1 uracil, compared to control and CTLN II fibroblasts (Figure 1F-G and Extended data Figure 1A-C). Hence, there is a specific decrease in aspartate transport from the mitochondria in CTLN II, leading to reduced aspartate availability for pyrimidine synthesis and restricting proliferation. Interestingly, growth restriction has been reported in humans with CTLN II8 but no growth aberrancies have been reported in CTLN I, further providing a clinical human context to the findings and suggesting that in physiological proliferation, aspartate deficiency has more severe clinical consequences than its enrichment. To corroborate our results in another model system, we analyzed Ass1 mRNA levels in wild-type newborn mouse intestines which express high levels of ASS1 and contain both proliferating and differentiating cells in the crypts and villi, respectively9. We found a significant correlation between the levels of Ass1 and Glut2, a mature enterocyte marker in the differentiated enterocytes in the villi, whereas, a significant inverse correlation was observed between Ass1 and Ki-67, a marker of proliferation, in the proliferating cells in the crypts (Figure 1H). Thus, ASS1 inactivation has an important role in proliferation of non-cancerous cells, in increasing aspartate availability for pyrimidine synthesis by CAD.
We next evaluated whether this mechanism could be the reason for the downregulation of ASS1 in cancer. According to the well-established “Warburg effect”, different metabolites are diverted from their “routine pathways” for the synthesis of biological molecules that are essential for cell division and growth. We hence conducted an analysis of ASS1 expression data in cancer cell lines from the NCI-60 collection and found a significant inverse correlation between ASS1 expression levels and the reported doubling time of the cancerous cells (Figure 2A). To further test whether this correlation is explicable by diversion of aspartate flux, we utilized our modeling program and predicted that with ASS1 inactivation, there is an accompanying significant increase in aspartate flux through the relevant metabolic reactions for nucleic acid synthesis (Extended data Table 1). In contrast, modeling the inactivation of ASL predicted an endogenous arginine depletion that does not directly affect the flux towards nucleic acid synthesis (Data not shown). Furthermore, analysis of The Cancer Genome Atlas (TCGA) database for ASL and ASS1 expression shows that these genes can both be downregulated in the same cancers, suggesting that they are not mutually exclusive (Extended data Figure 1D). Thus, ASS1 silencing in cancerous proliferation might have an arginine independent effect that is related to nucleotide synthesis.
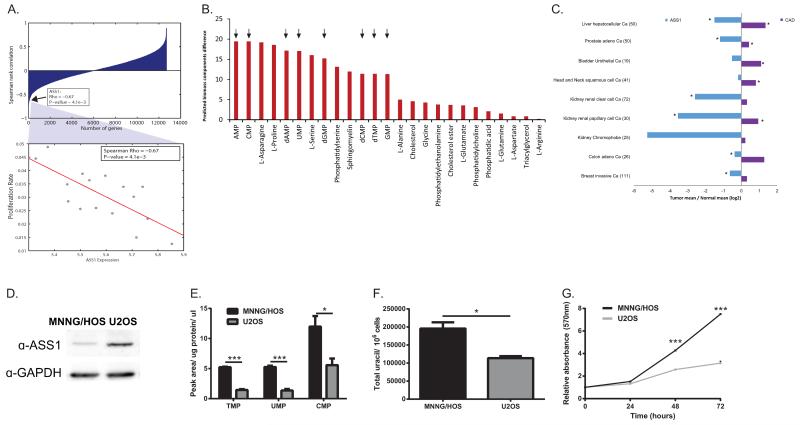
Figure 2. ASS1-deficient tumors have increased proliferation rate and an increase in pyrimidine levels.
(A) Upper panel- ASS1 is ranked within the top 24 genes out of 14,000, that show a significant inverse correlation with proliferation rate among the 16 NCI-60 cancer cell lines in which ASS1 is downregulated; Spearman rank correlation between the expression of each gene and its associated proliferation rates. Lower panel- A magnifying view of the correlation between proliferation rate and ASS1 expression levels. (B) Predicted differences in the production rate of biomass components following the inactivation of ASS1. The production of nucleic acids marked in arrows, is predicted to have a large increase following ASS1 inactivation in the majority of the NCI-60 cell-lines. The figure represents the results obtained using the LOX IMVI cell-line model, however the same results were seen for all NCI- 60 cell lines as well as in using the human generic model. The models used are based on a series of simplifying assumptions as described by us previously in details10 (C) Analysis of the TCGA database of matched tumor-normal tissue pairs showing that CAD expression is elevated significantly in tumors with ASS1 downregulation as compared to normal tissue, (p-Value<0.05). (D) An immunoblot of osteosarcoma cell lines showing decreased expression of ASS1 compared to the loading control GAPDH in MNNG/HOS as compared to U2OS. (E) Osteosarcoma cells with ASS1 downregulation have a significant increase in pyrimidine levels as measured by LC/MS, n≥3 (*p-Value<0.05; ***p-Value<0.0005). Error bars represent SER (F) Osteosarcoma cells with ASS1 downregulation have a significant increase in uracil, n=3 (*p-Value<0.05). Error bars represent SER (G) Osteosarcoma cells with ASS1 downregulation have a significant increase in proliferation as measured by MTT assay, (***p-Value<0.0005).
Using specific metabolic models tailored for each of the NCI-60 cell-lines10, we further predicted that 8 out of the 13 metabolites computationally shown to be increased with ASS1 inactivation, are nucleic acids (Figure 2B and Extended data Figure 1E). Additionally, specific analysis of the TCGA database of tumors where ASS1 expression is downregulated shows a significant upregulation in the expression of CAD, as compared to the paired normal tissue (Figure 2C). We further confirmed the inverse upregulation in the expression of CAD versus ASS1 at the mRNA level in the NCI-60 cancer cell lines database as well as in independent databases for patients with osteosarcoma11 and melanoma12 and found that downregulation of ASS1 and upregulation of CAD are in concordance with cancerous phenotype (Extended data Figure 1F-G). In addition, we demonstrate the inverse expression levels between ASS1 and CAD at the protein level using osteosarcoma and melanoma cell lines that differ in their expression pattern of ASS1 (Extended data Figure 1H and Extended data Figure 2A). To validate these modelling and global informatics analyses with experimental evidence, we studied osteosarcoma cell lines in which ASS1 was either deficient (MNNG/HOS) or present (U2OS) (Figure 2D and Supplementary Figure 1). Metabolic analysis confirmed that cells deficient in ASS1 had an increase in pyrimidine levels, increase in the level of uracil as well as a significantly increased proliferation rate (Figure 2E-G) as compared to osteosarcoma cells that have higher levels of ASS1. We additionally verified these results in melanoma cell lines that differ in their level of ASS1 expression (Extended data Figure 2B-D).
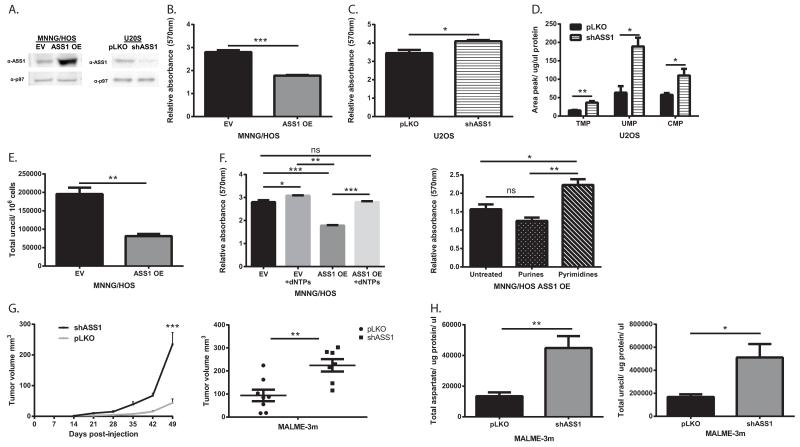
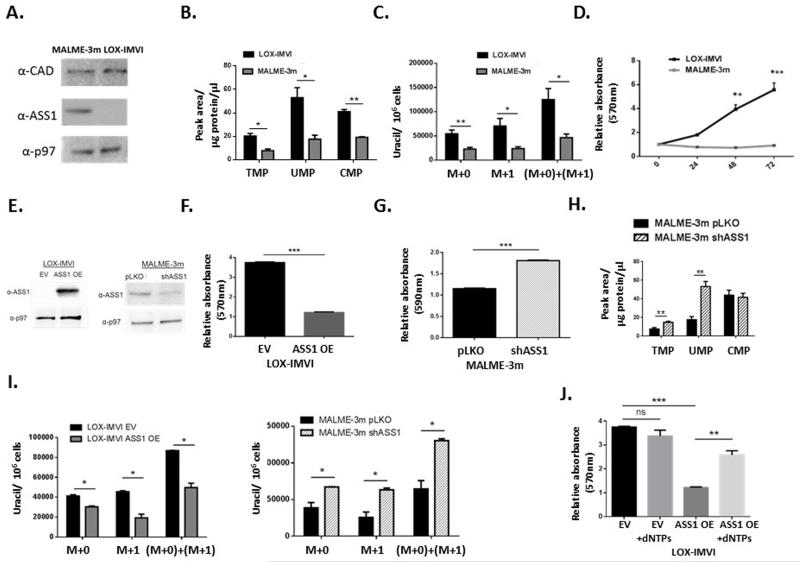
To more definitively dissect the connection between ASS1 expression and proliferation from other metabolic changes that occur in cancer cells, we overexpressed ASS1 in MNNG/HOS, and knocked it down in U2OS cells (Figure 3A). Our results clearly show that changes in ASS1 levels inversely alter the proliferation rate and pyrimidine synthesis in these cells (Figure 3B-E and Extended data Figure 3A-F). If the major determinant by which ASS1 overexpression decreases proliferation is through diverting aspartate metabolism away from pyrimidine synthesis, supplementation with nucleic acids should restore proliferation. Indeed, supplementing the media with nucleic acids and specifically with pyrimidines, significantly restores the proliferation of ASS1 overexpressing cells to a similar level as the parental cell-line (Figure 3F and Extended data Figure 2E-J). Thus, in two distinct forms of cancers, changes in ASS1 expression levels directly affect aspartate utilization for pyrimidine synthesis and proliferation. Importantly, similar results were obtained in-vivo where mice injected with melanoma cells knocked down for ASS1, developed tumors that grew more rapidly and had higher levels of total and M+1 labeled aspartate and uracil as compared to the parental tumor cells that expressed the empty vector (Figure 3G-H and Extended data Figure 3G).
Figure 3. ASS1 expression levels in cancer determine aspartate availability for pyrimidine synthesis.
(A) Immunoblots of osteosarcoma cells following transduction with either ASS1 overexpression construct (left) or with ASS1-shRNA (right). (B) MTT proliferation assay showing a significant decrease following ASS1 overexpression and a significant increase in proliferation following transduction with shASS1. (***p-Value<0.0005). (C) in osteosarcoma cells. The proliferation values are shown for day 3, after normalizing the data for the reading on day 1, n≥3 (*p-Value<0.05). (D) LC/MS measurements of pyrimidine levels showing a significant increase following the use of ASS1-shRNA in osteosarcoma cells, n≥3. (*p-Value=0.05, **p-Value=0.005). (E) Total Uracil is decreased significantly in osteosarcoma cells with ASS1 overexpression, n≥3 (**p-Value=0.005). (F) Left panel- dNTP supplementation rescues proliferation after ASS1 overexpression in osteosarcoma cells. Right panel- Pyrimidines significantly rescue proliferation in ASS1 overexpressing MNNG/HOS cells, n≥3, (*p-Value <0.05, **p-Value <0.005, ***p-Value<0.0005). (G-H) 10^7 MALME-3m melanoma cells transduced with either pLKO empty vector or with shASS1, were injected subcutaneously to immune deficient mice. The experiment was repeated three times. Following euthanization, the tumors were removed, measured and incubated with labeled 15Nα-GLN for 6 hours. Two weeks after injection, the group injected with melanoma cells expressing shASS1 developed tumors that grew more rapidly in size (G- Left panel) and were hence significantly larger when removed (G- Right panel) The experiment was repeated 3 times with similar results, (**p-Value<0.005, ***p-Value<0.0005). (H) Tumors with shASS1 had higher levels of total aspartate (Left panel) and total uracil (Right panel) as compared to tumors expressing the empty vector. Statistical analysis was performed using repeated measurements analysis of variance, n=15 (*p-Value<0.05; **p-Value<0.005). In all panels, error bars represent SER
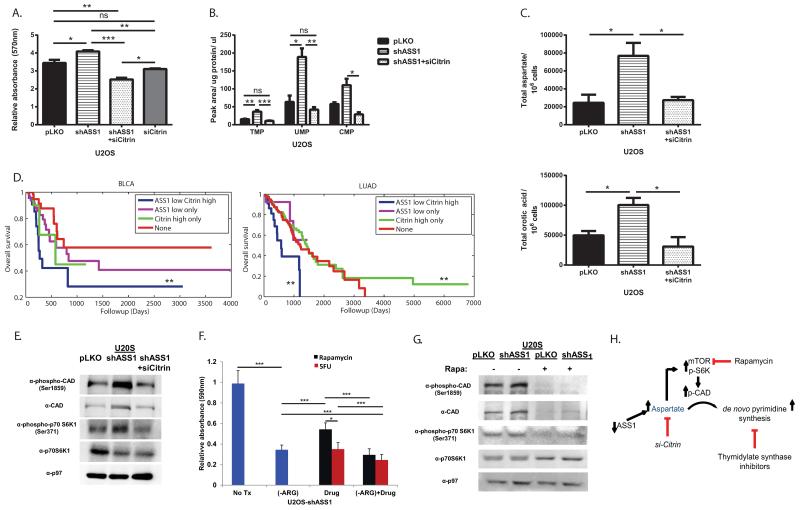
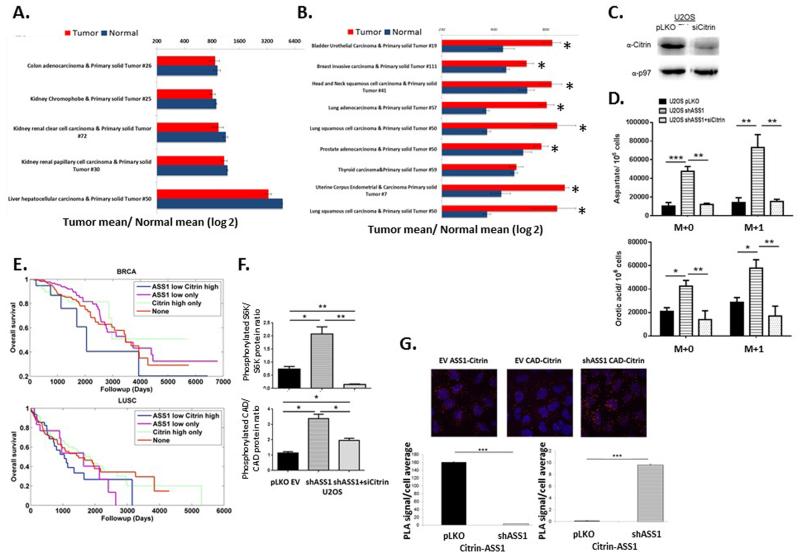
An expected synergistic way to increase aspartate delivery for pyrimidine synthesis would be by upregulation of citrin. Analysis of the TCGA database showed that in tissues that normally do not express citrin at high levels13, there is a significantly elevated expression in the cancerous state (Extended data Figure 4A -B). In addition, in the liver where citrin is strongly expressed, a recent publication of ASS1 expression in hepatocellular carcinoma showed that downregulation of ASS1 is associated with a more malignant cancerous phenotype14. These results, together with our study of primary human fibroblast cells (Figure 1C-G), imply that proliferation induced by loss of ASS1 in tumors might be counteracted by inhibiting citrin. Indeed, si-citrin in U2OS decreases proliferation significantly when ASS1 levels are reduced (Figure 4A). Use of si-citrin decrease also pyrimidine, total and labeled M+1 aspartate and M+1 orotic acid levels (Figure 4B-C and Extended data Figure 4C-D). As citrin is part of the malate-aspartate shuttle, its deficiency is expected to affect several aspects in cell survival and growth. Our results indicate that citrin function in transferring mitochondrial-derived aspartate is important for supplying substrate for pyrimidine synthesis, especially in cancers with ASS1 downregulation. These findings are therapeutically relevant as survival analysis of several cancers in the TCGA database reveal that cancers with both decreased ASS1 expression and high citrin levels have a trend for significantly worse prognosis (Figure 4D, Extended data Figure 4E and Extended data Table 2).
Figure 4. Decreasing CAD activation decreases proliferation in ASS1-deficient cancers.
(A) MTT assay showing that decreasing citrin levels significantly decreases proliferation in U2OS osteosarcoma even after a significant proliferation increase is accomplished by ASS1 downregulation, n≥3 (*p-Value<0.05; **p-Value<0.005; ***p-Value<0.0005). (B) Decreasing citrin levels decreases pyrimidine levels in U2OS cells with ASS1 downregulation, n≥3 (*p-Value<0.05). (C) GCMS measurements showing that U2OS with shASS1 has a significant increase in total aspartate (Upper panel), as well as in total orotic acid (Lower panel), that are reversed when transfected with si-citrin, n≥3 (ANOVA, Tukey HSD, *p-Value<0.05; **p-Value<0.005). Error bars represent SER. (D) Kaplan-Meier (KM) survival analysis for 2 different cancer types; BLCA - bladder cancer and LUAD - lung adenocarcinoma, showing significant poor survival for cancers with low ASS1 and high citrin expression levels, n≥3. **Log rank p-value≤0.005. (E) Immunoblot of osteosarcoma cells for the mTOR pathway downstream effectors- S6K1 and CAD showing increase phosphorylation following shASS1 that is reversed when cells are transfected with si-citrin. (F) A quantification graph of crystal violet staining of osteosarcoma cells transduced with shASS1 following drug treatments. All treatments were significant as compared to no treatment. In addition, the results show a significant additive beneficial effect of decreased proliferation in response to treatment with medium depleted of arginine together with either mTOR or pyrimidine synthesis inhibitors; (Rapamycin or 5FU respectively). Of note, as isolated treatment, 5FU was more beneficial than Rapamycin. Cells were grown in medium depleted of arginine, in complete medium with either Rapamycin or 5FU, and in arginine depleted medium together with either Rapamycin or 5FU, n=9 ***p-value<0.0005). (G) A western blot showing decreased activation of the mTOR proteins following Rapamycin treatment (H) A Schematic presentation for potential interventions in pyrimidine synthesis in ASS1 deficient tumors.
The utilization of citrin-derived aspartate by CAD requires CAD activation. Recently, CAD was shown to be activated by ribosomal protein S6 kinase (S6K1), regulated by the mTOR pathway15. When ASS1 expression in cancer cells is decreased, we find increased phosphorylation of S6K1 and CAD that is decreased by si-citrin, implying that aspartate levels are important in regulating the mTOR pathway activation (Figure 4E and Extended data Figure 4F). In addition, we show a significant increase in the location proximity between CAD and citrin following ASS1 downregulation (Extended data Figure 4G). Thus, aspartate regulates pyrimidine levels by regulating CAD’s substrate availability, protein localization, and activity. In concordance, we see a decrease in proliferation when ASS1 deficient cells are treated with either the mTOR inhibitor Rapamycin or with thymidylate synthase inhibitor- Fluorouracil (5FU) (Figure 4F). Importantly, Rapamycin treatment is accompanied by a decrease in CAD phosphorylation (Figure 4G). Hence, targeting aspartate transport could be an additional therapeutic option in cancers with ASS1 silencing; especially in cancers that develop resistance to arginine depleting agents (Figure 4H).
In summary, our studies demonstrate that ASS1, a urea cycle enzyme, regulates pyrimidine synthesis in cancerous proliferation by regulating CAD activation, via regulating aspartate levels. There are several clinical trials in patients with ASS1 deficient-hepatocellular carcinoma and mesothelioma, which combine arginine-depleting agents with thymidylate synthase inhibitors as capecitabine and pemetrexed (clinicaltrials.gov NCT02089633, NCT02029690). We believe our study provides the rationale for such therapeutic modalities and hence has direct translational relevance.
Supplementary Methods
Methods
Measurements in human subjects
The fibroblast studies were performed on anonymized cells devoid of all identifiers. The data analysis involving urine orotic acid levels were performed under a protocol approved by the Institutional Review Board of Baylor College of Medicine. Urine samples were prepared by mixing 200 μl of with isotopic internal standard 15N2-orotic acid (Cambridge Isotope Laboratories). Orotic acid and orotidine were assayed on a Micromass Quattro mass spectrometer (Waters). HPLC was performed on a Waters ODS-AQ analytical column [150 ×2.0mm (i.d.), 5-μm bead size]. Mobile phase was isocratic 0.05 M ammonium formate (pH 4.0). The MS/MS system was set at a flow rate of 0.2 ml/min. Mass spectrometer was operated in the Electrospray ionization (ESI) negative multiple-reaction-monitoring (MRM) mode. Nitrogen was used as nebulizer gas at flow rate of 60-90 l/hr and desolvation gas 500 l/hr. Other optimized mass spectrometer parameters were cone voltage −15V, capillary −3250V and collision voltage – 10V.
Genome Scale Metabolic Modelling (GSSM)
A metabolic network consisting of m metabolites and n reactions can be represented by a stoichiometric matrix S, where the entry Sij represents the stoichiometric coefficient of metabolite i in reaction j 22. A CBM model imposes mass balance, directionality and flux capacity constraints on the space of possible fluxes in the metabolic network’s reactions through a set of linear equations:
| (1) |
| (2) |
Where ν stands for the flux vector for all of the reactions in the model (i.e. the flux distribution). The exchange of metabolites with the environment is represented as a set of exchange (transport) reactions, enabling a pre-defined set of metabolites to be either taken up or secreted from the growth media. The steady-state assumption represented in Equation (1) constrains the production rate of each metabolite to be equal to its consumption rate. Enzymatic directionality and flux capacity constraints define lower and upper bounds on the fluxes and are embedded in Equation (2). In the following, flux vectors satisfying these conditions will be referred to as feasible steady-state flux distributions. The analyses were performed under the RPMI-1640 medium. We used the biomass function introduced in Folger et al16.
Predicting growth rate, metabolite production and flux distribution through metabolic modelling
To determine the relation between ASS1 activity, CAD activity and growth rate, we utilized the generic human model and simulated the inactivation and activation of the reaction catalysed by ASS1. The inactivation was simulated by constraining the flux through the ASS1 reaction to zero, while the activation was simulated by enforcing increased positive flux through the ASS1 reaction up to the maximal possible flux, as computed via Flux Variability Analysis17. At each such point, the maximal growth rate is computed via Flux Balance Analysis (FBA) 17. Additionally, we estimated the flux through the reaction catalysed by CAD under maximal growth rate based on 1000 different feasible flux samples18.
We next utilized genome-scale metabolic models for each of the NCI-60 cancer cell lines based on their gene expression measurements10. In each cell-line model we performed the following analyses: (1) we computed the production of each biomass component under both the inactivation and maximal activation of ASS1, as described above. The difference between the predicted production rates of each biomass component in the two states was then computed based on the results of this optimization problem. (2) Similarly, we examined the flux change of each reaction under maximal biomass production in both the inactivation and activation states, as described above. In each of these states we sampled the solution space and obtained 1000 feasible flux distributions18. Focusing on the reactions associated with aspartate and glutamine, we computed the fold-change in flux rate together with its significance level. The latter was computed via a two-sided Wilcoxon rank sum test. The largest fold-change among these reactions was predicted for the reactions catalysed by the CAD enzyme.
TCGA data analysis
For each tumor, normalized gene expression levels measured using RSEM19, were obtained from the RNASeqV2 data sets at the TCGA portal (https://tcga-data.nci.nih.gov/tcga/). Only matched tumor-normal pairs were used. For each tumor type, we computed the mean expression levels in the tumor and normal samples, added a pseudo-count of 1 to each mean, and plotted the ratio between the means.
Metabolomics Analysis
Osteosarcoma or Melanoma cell line were seeded at 10^6 cells per 10cm palate and incubated with either 4mM L-GLUTAMINE, (ALPHA-15N, 98%, Cambridge Isotope Laboratories, Inc.) or 4mM L-GLUTAMINE, (AMIDE-15N, 98%+, Cambridge Isotope Laboratories, Inc.) for 24 hours. Subsequently, cells were washed with ice cold saline, lysed with 50% methanol in water and quickly scraped followed by three freeze thaw cycles in liquid nitrogen. The insoluble material was pelleted in a cooled centrifuge (4 °C) and the supernatant was collected for consequent GC-MS analysis. Samples were dried under air flow at 42°C using Techne Dry-Block Heater with sample concentrator (Bibby Scientific Limited, UK) and the dried samples were treated with 40 μl of a methoxyamine hydrochloride solution (20 mg/ml in pyridine) at 37 °C for 90 min while shaking followed by incubation with 70 μl N, O-Bis (trimethylsilyl) trifluoroacetamide (Sigma) at 37 °C for additional 30 min.
Gas chromatography/mass spectrometry
GC–MS analysis was performed using a gas chromatograph (7820AN, Agilent Technologies, USA) interfaced with a mass spectrometer (5975 Agilent Technologies, USA). A HP-5ms capillary column 30 m × 250 μm × 0.25 μm (19091S-433, Agilent Technologies, USA) was used. Helium carrier gas was maintained at a constant flow rate of 1.0 mL min–1. The GC column temperature was programmed from 70 to 150 °C via a ramp of 4 °C min–1, 250–215 °C via a ramp of 9 °C min–1, 215–300 °C via a ramp of 25 °C min–1 and maintained at 300 °C for additional 5 min. The MS was by electron impact ionization and operated in full scan mode from m/z, 30–500. The inlet and MS transfer line temperatures were maintained at 280 °C, and the ion source temperature was 250 °C. Sample injection (1 μL) was in splitless mode.
Nucleic acid analysis
Materials
Ammonium acetate (Fisher Scientific) and ammonium bicarbonate (Fluka) of LC-MS grade were used. Sodium salts of AMP, CMP, GMP, TMP and UMP were obtained from Sigma-Aldrich. Acetonitrile of LC grade was supplied from Merck. Water with resistivity 18.2 MΩ was obtained using Direct 3-Q UV system (Millipore).
Extract preparation
The obtained samples were concentrated in speedvac to eliminate methanol, and then lyophilized till dryness, re-suspended in 200 μl of water and purified on polymeric weak anion columns Strata-XL-AW 100u (30mg/1ml, Phenomenex) as following. Each column was conditioned by passing 1 ml of methanol, then 1 ml of formic acid/methanol/water (2/25/73), and equilibrated with 1 ml of water. The samples were loaded, and each column was washed with 1 ml of water and 1 ml of 50% methanol. The purified samples eluted with 1 ml of ammonia/methanol/water (2/25/73) followed by 1 ml of ammonia/methanol/water (2/50/50) were collected, concentrated in speedvac to remove methanol, and lyophilized. Before LCMC analysis the obtained residues were re-dissolved in 100 μl of water, centrifuged 5 min. at 21,000 g to rid of insoluble material.
LCMS analysis
The LC-MS/MS instrument consisted of Acuity I-class UPLC system (Waters) and Xevo TQ-S triple quadruple mass spectrometer (Waters) equipped with an electrospray ion source and operated in positive ion mode was used for analysis of nucleoside monophosphates. MassLynx and TargetLynx software (v.4.1, Waters) were applied for the acquisition and analysis of data. Chromatographic separation was done on a 100 × 2.1-mm i.d., 1.8-μm UPLC HSS T3 column equipped with 50 × 2.1-mm i.d., 1.8-μm UPLC HSS T3 pre-column (both Waters Acuity) with mobile phases A (10 mM ammonium acetate and 5 mM ammonium hydrocarbonate buffer, pH 7.0 adjusted with 10% acetic acid) and B (acetonitrile) at a flow rate of 0.3 ml/min and column temperature 35°C. A gradient was as follows: 0–6 min the column was hold at 0% B, then 6-6.5 min linear increase till 100% B, 6.5-7.0 min hold at 100% B, 7.0-7.5 min back to 0% B and equilibration at 0% B for 2.5 minutes. Samples kept at 8°C were automatically injected in a volume of 3 μl.
For mass spectrometry argon was used as the collision gas with flow 0.25 ml/min. The capillary voltage was set to 2.90 kV, source temperature - 150°C, desolvation temperature - 350°C, desolvation gas flow - 650 L/min. Analytics were detected using multiple reaction monitoring (MRM) applying the parameters listed in Supplementary Table 1.
Hybridizations and imaging
Single molecule FISH (smFISH) was performed with probe libraries for Ass1 (74 probes, sequences described below in supplementary methods) and Ki67 (96 probes20). Imaging was performed as previously described20. smFISH images were filtered with a Laplacian of Gaussian filter of size 15 pixels and standard deviation of 1.5 pixels. Image is a maximum projection of 10 stacks spaced 0.3 um apart in the Z-direction. Each dot in these figures represents a cell and the quantification dots were counted on 8 Z-stacks spaced 0.3μm apart (total tissue volume of 2.4μm).
Proximity ligation assay
The assay was performed as published21 using Sigma Aldrich kit (Cat # DUO 92004-30-RXN). Antibodies used for detection were diluted in PBS; ASS1 (1:200, #ab170952, abcam), citrin (1:100, #H00010165-M01, clone # 4F4, abnova) and anti-CAD (1:100, ab40800, abcam).
Cell cultures
All cell lines were authenticated; Melanoma cell line LOX IMVI, MALME-3m and Osteosarcoma cell lines, MNNG/HOS, U2OS were purchased from ATTC and cultured using standard procedures in a 37 °C humidified incubator with 5% CO2 in Roswell Park Memorial Institute Medium (RPMI) (Invitrogen) supplemented with 10-20% heat-inactivated fetal bovine serum, 10% pen-strep and 2 mM glutamine. All cells are tested routinely for Mycoplasma using Mycoplasma EZ-PCR test kit (#20-700-20, Biological Industries, Kibbutz Beit Ha’emek).
Proliferation Assays
MTT
Cells were seeded in 12-well plates at 4-8*104 cells/well in a triplicate. After 6 hours for adherence of the cells, 0.1 mg/ml of MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5 diphenyltetrazolium bromide) (Catalog #: CAS 298-93-1, Calbiochem) in PBS was added to each cell type, starting at 0 h, in 24 h intervals. Deoxynucleotide Set (Catalog #: DNTP100-1KT, Sigma-Aldrich) was added to the cells in the same time intervals at a final concentration of 10uM. Cells were lysed with DMSO. Absorbance was measured at 570 nm.
Crystal Violet Staining
Cells were seeded in 12-well plates at 40,000-100,000 cells/well in a triplicate. Time 0 was calculated as the time the cells became adherent, which was after about 6 hours from plating. For each time point, cells were washed with PBS ×1 and fixed in 4% PFA (in PBS). Cells were then stained with 0.1% Crystal Violet (Catalog #: C0775, Sigma-Aldrich) for 20 minutes (1ml per well) and washed with water. Cells were then incubated with 10% acetic acid for 20 minutes with shaking. Extract was then diluted 1:4 in water and absorbance was measured at 590 nm every 24 hours.
Protein and RNA analysis
Western blotting
Cells were lysed in RIPA (Sigma-Aldrich) and 0.5% protease inhibitor cocktail (Calbiochem). Following centrifugation, the supernatant was collected and protein content was evaluated by the Bradford assay. 100μg from each sample under reducing conditions were loaded into each lane and separated by electrophoresis on a 10% SDS polyacrylamide gel. Following electrophoresis, proteins were transferred to Immobilon transfer membranes (Tamar, Jerusalem, Israel). Nonspecific binding was blocked by incubation with TBST (10 mM Tris–HCl (pH 8.0), 150 mM NaCl, 0.1 % Tween 20) containing 3% Albumin from Bovine Serum for 1h at room temperature. Membranes were subsequently incubated with antibodies against ASS1 (1:500, sc-99178, Santa Cruz Biotechnology)22, p97 (1:10,000, PA5-22257, Thermo Scientific), GAPDH (1:1000, 14C10, #2118, Cell Signaling)23, CAD (1:1000, ab40800, abcam)24, phospho-CAD (Ser1859) (1:1000, #12662, Cell Signaling)15, p70 S6 Kinase (1:1000, #9202, Cell Signaling), phospho-p70 S6 Kinase (Ser371) (1:1000, #9208, Cell Signaling)25. Antibody was detected using peroxidase-conjugated AffiniPure goat anti-rabbit IgG or goat anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA) and enhanced chemiluminescence western blotting detection reagents (EZ-Gel, Biological Industries).
Gels were quantified by Gel Doc™ XR+ (BioRad) and analyzed by ImageLab 4.1 software (BioRad). The band area was calculated by the intensity of the band. The obtained value was then divided by the value obtained from the loading control.
RNA extraction and cDNA synthesis
RNA was extracted from cells by using PerfectPure RNA Cultured Cell Kit (5′-PRIME). Complementary DNA was synthesized from 1ug RNA by using qScript™ cDNA Synthesis Kit (Quanta).
Quantitative Real-Time PCR
Detection of ASS1 on cDNAs (see above) was performed using cyber green PCR master mix (Tamar, Jerusalem, Israel) and the required primers. Primers’ sequences are: human ASS1; Forward: 5′-TTATAACCTGGGATGGGCACC-3′, Reverse: 5′-TGGACATAGCGTCTGGGATTG-3′, human HPRT; Forward: 5′-ATTGACACTGGCAAAACAATGC-3′, Reverse: 5′-TCCAACACTTCGTGGGGTCC-3′. Analysis was performed using StepOne real-time PCR technology (Applied Biosystems,CA).
Transient transfection
Cells were seeded in 12-well plates at 30,000 cells/well, or in 10cm plates at 106 cells/plate, in a triplicate. At the following day, cells were transfected with either 20pmol or 600pmol siRNA siGenome SMARTpool targeted to Citrin mRNA (Catalog #: M-007472-01, Thermo scientific), respectively. Transfection was done with Lipofectamine® 2000 Reagent (Catalog #: 11668-019, Invitrogen) in the presence of Opti-MEM® I Reduced Serum Medium (Catalog #: 31985-062, Invitrogen). 4 hours after transfection, medium was replaced and experiments were performed starting 72 hours post transfection.
Infection
Over- expression
Cells were infected with pLenti3.3/TR and with pLenti6.3/TO/V5-DEST-based lent viral vector with or without the human ASS1 transcript. Transduced cells were selected with 1mg/ml Geneticin and with 7.5μg/ml Blasticidin for each plasmid, respectively. When induction of expression was needed, cells were added with 10μg/ml Tetracycline/Doxycycline.
shRNA
Cells were infected with pLKO-based lenti viral vector with or without the human ASS1 shRNA encoding one or two separate sequences combined (Catalog #: RHS4533-EG445, GE Healthcare, Dharmacon). Transduced cells were selected with 2μg/ml Puromycin.
Arginine deprivation combined with drug treatments
U2OS human osteosarcoma cell-line was seeded in 6-well plates at 80,000 cells/well. The following day, cells were treated with either 100nM Rapamycin (#R0395, Sigma-Aldrich) or with 10μM 5FU (#F6627, Sigma-Aldrich) in regular medium, with 10% dialyzed FCS-arginine-free-RPMI (#06-1104-34-1A, Biological Industries, Kibbutz beit-haemek, Israel), or with both arginine depleted medium and one of these drugs. Rapamycin and 5FU were renewed into the medium every day, whereas fresh arginine-free medium was supplemented twice a week.
Animal studies
According to the approved IACUC protocol 17270415-2, tumors did not exceed the limits of more than 10% of the animal weight and were not longer than 1.5cm in length in any dimension (Supplementary Figure 2). 10^7 MALME-3m melanoma cells suspended in 500mcl with 5% Matrigel (#4132053 Corning) were injected subcutaneously to 8-12 weeks old male SCID mice that were purchased from Harlan. There were 22 SCID from which 5-6 mice were used for each cell line at each out of the three experiments performed. No randomization was used. Mice were monitored for survival and tumor burden twice a week by a veterinarian investigator who was blinded to the expected outcome. Tumors were measured using a caliber. After euthanization, tumors were removed and incubated in medium containing 15N labeled glutamine for 6 hours followed by GCMS analysis. Tumor size was calculated as published26.
Building Cell Models
We utilized genome-scale metabolic models of NCI-60 cancer cell lines. The reconstruction method (based on the yet unpublished methods termed PRIME in Yizhak et al, personal communication) requires several key inputs: (a) the generic human model7; (b) gene expression data for each cell line from1919 ENREF 2619, and (c) growth rate measurements (available at the NCI website: http://dtp.nci.nih.gov/docs/misc/common_files/cell_list.html). The algorithm then reconstructs a specific metabolic model for each sample by modifying the upper bounds of reactions in accordance with the expression of the individual gene microarray values.
Specifically, the model reconstruction process is as follows:
-
(1)
Decompose reversible reactions into unidirectional forward and backward reactions.
-
(2)
Evaluate the correlation between the expression of each reaction in the network and the measured growth rate. The expression of a reaction is defined as the mean over the expression of the enzymes catalyzing it.
-
(3)
Modify upper bounds on reactions demonstrating significant correlation to the growth rate (after correcting for multiple hypothesis using FDR) in a manner that is linearly related to expression value.
Ass1 probes
Gcgcatccagggttataagc;gcggagcagggctgagagag;gcccaggcggcagtggcaga;ggccagatgaaccactcagt;agtccgtgtagtctgctttc;gattataggtacaggtccct;ccttgctggacatcttgtct;ctgtaggccagaaccacaga;gcaggaggtgtccaggccac;gttccttcagccacacgagg;taggcgatgacatcatagcc;cttctggccaatgttggcca;tcctggcttcctcaaagtct;gccccaagcttcagcgcctt;atcctcaatgaacacctttt;cttccacaaattccttgctc;tggacagcaggccagatgaa;gtcctcgtagagtgcactgg;gagaggtgcccaggagatag;cgagctatgcaaggcctggc;ctgggcaatctccacctgtc;acacatacttggccccttca;ccctttcccgtggcgccgtg;ctcaaagcggacctggtcat;gtgccagtgaatagcaggtg;ggagcgatgaccttaatctg;tgtaaaactcaggcatcctc;atcatttcggcccttgaacc;gttgctttgcatactccatc;gtgacagggatggggattcc;catactccaggggctcttgg;tgatgtgcatgaggttttca;tccaggatcccagcctcata;aggtgcttgattcttggggt;gagtttttgtgtagagaccc;ttgggtgctttggcagggtc;tatctcaaggacatctgggc;cagggaccccttttttgaat;tctttgatgttggtcacctt;ggatgtggtgcgggttgtgc;tcaggtacatgaagagttcc;ccgtgcttgcccgcaacttc;cacgatgtcaatgcgaccca;tcattccaatgaagcggttc;gtctcgtagatacctcggga;gtaaaggatggtccctgctg;cctctatgtctaaatgagcg;acttcccgatccatcgtgaa;caggccctgcttgatt;cgagctctgcgaatttgagg;ctgtgccagaaacctgtgta;gcgaacaaattcacattcag;cctgggacttctggatacag;tgcaccttcccttctacccg;ttggcccttgaagacagaca;actcccgaccgaggatgtac;tcattgtagagtgaaagtgg;tgcacgttcatgctcaccag;gtcgatgggctcatagtcgc;tgatattgatgaagccagtg;tactccttcagcctgagcga;gaccttgctctgaaggcgat;ttgtcagggtctatttggca;gagtggggaggcccgctcct;gctgaagcctgggagagctg;caaatttatcacaacaatta;ggtggagaacaagctacaat;gacacagcagccccagtcag;aggctgtgggggggcggggg;gctataggggaccagggaac;ccttggatgaccacttttgt;agctgcccgccaccctccct;attgtcattttatgctttct;aagactaatgtaacttcttt
Statistics
All statistical analyses were performed using Tukey HSD or independent-samples Student’s T-test of multiple or two groups, respectively. Log-transformed data were used where differences in variance were significant and variances were correlated with means. The sample size was chosen in advance based on common practice of the described experiment and is mentioned for each experiment. Each experiment was conducted with biological and technical replicates and repeated at least three times unless specified otherwise. Based on pre-established criteria, individual outlier data points that were more than 2 standard deviations away from the mean were excluded from the data analysis. Statistical tests were done using Statsoft’s STATISTICA, ver. 10. All error bars represent SER. P<0.05 was considered significant in all analyses (* denotes P<0.05, **<P,0.005, ***P<0.0005).
Kaplan Meir (KM) - For each cancer type, the KM-plot indicates the survival rates of the 4 different patients group as labeled. Analysis was performed for the cancer types for which there was sufficient survival data.
Extended Data
Extended data Figure 1: ASS1 deficiency correlates with aspartate utilization by CAD in cancerous and non-cancerous cells.
(A) Schematic flux tracing of the Alpha labeled nitrogen of glutamine (15N-α-Glutamine) to nucleic acid synthesis via aspartate. (B) The ratio between M+1 labeled/ total level of uracil in fibroblasts is similar between citrullinemia patients and control, n≥3. Error bars represent SER. (C) Labeled levels of M+1 aspartate (left) and M+1 uracil (Right) synthesized from 15N-α-labeled glutamine, are higher in fibroblasts from CTLN I as compared to fibroblasts from controls and CTLNII patients, n≥3. (D) TCGA analysis of tumor-normal paired tissues for gene expression comparison shows the expression levels of ASL and ASS1 in different cancers. (E) A graph plot generated from the modelling data for the production capacity of metabolites following ASS1 inactivation in each of the NCI-60 cell lines as well as in the generic model. The reddish bars represent the ranking of nucleic acids while the blueish bars represent the ranking of all other metabolites. (F) Correlation analysis of NCI-60 cell lines shows a significant inverse correlation between ASS1 and CAD expression levels. (G) Osteosarcoma (Upper panel) and melanoma (Lower panel) microarray data was obtained from the NCBI EO database (accessions GSE33383 and GSE46517, respectively). Raw expression levels were plotted and significance was computed using t-test on log2-transformed expression levels. The number of patients for each subtype is shown in parenthesis on the left. (H) A western blot for CAD and ASS1 shows higher expression level of CAD in MNNG/HOS human osteosarcoma cell line which has low expression level of ASS1 in comparison to U2OS which has higher expression levels of ASS1. p97 is shown as loading control.
Extended data Figure 2: ASS1 inactivation in melanoma correlates with increased proliferation.
(A) An immunoblot showing different expression levels of ASS1 and CAD in two different cancer cell lines of melanoma. (B) Melanoma cells with ASS1 downregulation have a significant increase in pyrimidine levels as measured by LC/MS, n≥3. (C) Melanoma cells with ASS1 downregulation have a significant increase in total Uracil, n=4. (D) Melanoma cells with ASS1 downregulation have a significant increase in proliferation as measured by MTT assay, n=2. (E) Immunoblots of melanoma cells for ASS1 levels following transduction with either ASS1 over expression construct or with shASS1. (F) Proliferation assays showing a significant decrease in proliferation following ASS1 overexpression in melanoma using MTT, n=3. (G) Crystal violet quantification for melanoma cells following transduction with shASS1 demonstrating increase in proliferation, n=3. (H) LC/MS measurements of pyrimidine levels showing a significant increase following the use of shASS1 in melanoma cells, n≥3. (I) Left panel- Total uracil levels are decreased significantly in melanoma cells with ASS1 over expression and increased in melanoma cells with shASS1- Right panel, n≥2. (J) Significant increase in proliferation of melanoma cells by dNTP’s after ASS1 overexpression, n=3. In all panels, error bars represent SER.
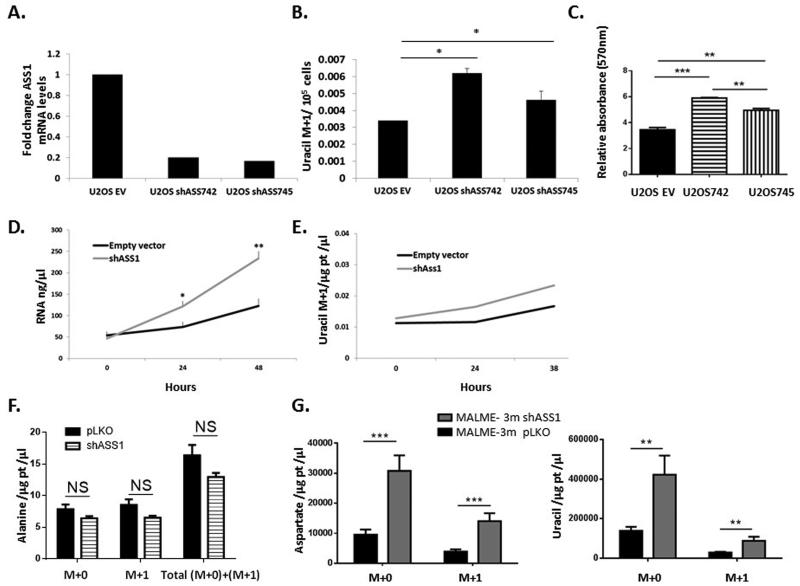
Extended data figure 3: Downregulation of ASS1 levels increases pyrimidine synthesis.
(A) Osteosarcoma cells were transduced with two different shASS1 vectors- shASS742 and shASS745. Both clones decreased ASS1 levels efficiently to approximately 20% expression (Left panel) resulting in a significant increase in uracil M+1 levels (B) and in proliferation, n≥3 (C). (D) RNA levels measured in U2OS at 24 hour intervals show increase levels of RNA in U2OS infected with shASS1 as compared to the empty vector. (E) Uracil M+1 levels increase more in U2OS infected with shASS1 as compared to the empty vector during 38 hours of measurements. (F) The levels of total and labeled M+1 alanine synthesis from 15N-a- glutamine does not change significantly following ASS1 downregulation, n=3. (G) Tumors with shASS1 had higher levels of M+1 aspartate (Left panel) and M+1 uracil (Right panel) synthesized from 15Nα-glutamine, as compared to tumors expressing the empty vector, n=15. In all panels, error bars represent SER.
Extended data figure 4: Cancers with ASS1 downregulation are addicted to aspartate.
(A-B) Analysis of the TCGA data base of matched tumor-normal pairs showing no significant difference in the expression level of citrin in tissues with a high base line expression of citrin, (A) and significant elevation in tumors in which the normal tissue has low basal expression of citrin (B) (*p<0.001). (C) Immunoblot showing the expression level of citrin in osteosarcoma cells following si-citrin.(D) Labeled and unlabeled aspartate (Upper panel) and uracil (Lower panel) are elevated significantly in cancers with ASS1 downregulation and are comparable to control in cells with both ASS1 and Citrin downregulation; n≥3. Error bars represent SER. (E) Kaplan-Meier (KM) survival analysis for 2 different cancer types; BRCA- breast cancer (Upper panel) and LUSC- lung squamous cell carcinoma (Lower panel), showing poor survival trend for cancers with low ASS1 and high citrin. For each cancer type the KM-plot indicates the survival rates of 4 patients group: ASS1 low expression - citrin high expression; ASS1 low expression; citrin high expression; none of the above groups. Analysis was performed for the cancer types for which there was sufficient survival data. (F) A quantification graph of a western blot showing decreased CAD and S6K phosphorylation following treatment of U2OS with si-citrine. Error bars represent SER. (G) Proximity ligation assay showing increased proximity between CAD and citrin following ASS1 knockdown in U2OS cells (Upper panel-red dots). The left and middle pictures show the proximity between ASS1 and CAD to citrin in U2OS infected with empty vector while the right picture shows the proximity between CAD and citrin following infection of U2OS with shASS1. The Lower panel shows quantification of proximity ligation assays performed on U2OS infected with either empty vector (EV) or with shASS1 using antibodies for citrin, ASS1 and CAD. The pictures were quantified using ImageJ.
Extended data Table 1: ASS1 inactivation is predicted to increase aspartate flux for nucleic acid synthesis.
Predicted fold-change in flux rates through pathways associated with aspartate and glutamine, when comparing ASS1 inactivation vs. activation state. The most significant change is predicted to effect the pyrimidine biosynthesis pathway followed by purine synthesis pathway, (two-sided Wilcoxon ranksum P-value < 8.4e-198, Methods).
| Aspartate | |||
|---|---|---|---|
| Metabolic Pathway | Catalyzing Enzymes | Inactive ASS1/Active ASS1 | P-value |
| Pyrimidine Biosynthesis | (790.1), CAD |
|
8.47E-198 |
| IMP Biosynthesis | (10606.1) PAICS |
|
<1e-300 |
| Nucleotides | (159.1) Adenylosuccinate synthase |
|
1.55E-265 |
Extended data Table 2: Kaplan Meier Log Rank data analysis shows significant worsening in the survival of patients with low ASS1 and high citrin expression levels in bladder cancer and lung adenocarcinoma.
The table shows the number of patients for which data was available in each group as well as the pair wise p-Value of comparison between the corresponding groups; BLCA- bladder cancer, BRCA- breast cancer, LUAD- lung adenocarcinoma, LUSC- lung squamous carcinoma.
| BLCA | Number of patients | ASS1 low only | Citrin high only | None | |
|---|---|---|---|---|---|
| ASS1 low Citrin high | 26 | 0.062029244 | 0.41802416 | 0.002920577 | |
| ASS1 low only | 437 | 0 | 0.939396939 | 0.133262071 | |
| Citrin high only | 83 | 0 | 0 | 0.397997761 | |
| None | 415 |
| BRCA | Number of patients | ASS1 low only | Citrin high only | None | |
|---|---|---|---|---|---|
| ASS1 low Citrin high | 25 | 0.091470414 | 0.348538412 | 0.215689269 | |
| ASS1 low only | 76 | 0 | 0.513641931 | 0.247876216 | |
| Citrin high only | 14 | 0 | 0 | 0.784222648 | |
| None | 90 |
| LUAD | Number of patients | ASS1 low only | Citrin high only | None | |
|---|---|---|---|---|---|
| ASS1 low Citrin high | 34 | 0.086250611 | 0.000771207 | 0.005978456 | |
| ASS1 low only | 16 | 0 | 0.989469401 | 0.778260123 | |
| Citrin high only | 181 | 0 | 0 | 0.440612674 | |
| None | 189 |
| LUSC | Number of patients | ASS1 low only | Citrin high only | None | |
|---|---|---|---|---|---|
| ASS1 low Citrin high | 56 | 0.889254024 | 0.131597923 | 0.271250284 | |
| ASS1 low only | 35 | 0 | 0.234416519 | 0.540649678 | |
| Citrin high only | 173 | 0 | 0 | 0.710240332 | |
| None | 127 |
Supplementary Material
Acknowledgments
We thank A. Gross and B. Lee for helpful discussions. We acknowledge and thank the Weizmann Institute for providing financial and infrastructural support and the Baylor College of Medicine Biochemical lab for their data sharing. We greatly appreciate the statistical analysis by Ron Rotkopf and the technical contributions of Arye Tishbee, Tom Kaufman, Dana Laufer and Ilana Rogachev. AE is incumbent of the Leah Omenn Career Development Chair and is supported by research grants from the European research program (CIG618113, ERC614204), the Israel Science Foundation (1343/13; 1952/13) and from the Minerva grant award (711730). AE received additional support from the Adelis Foundation, the Henry S. and Anne S. Reich Research Fund, the Dukler Fund for Cancer Research, the Paul Sparr Foundation, the Saul and Theresa Esman Foundation, from Joseph Piko Baruch, and from the estate of Fannie Sherr. L.A. was supported by a postdoctoral fellowship from Teva and KY was supported by the Azrieli Foundation Fellowship Award. S.N.S.C. is supported by Baylor College of Medicine IDDRC Grant (1 U54 HD083092) and by the Doris Duke Charitable Foundation (DDCF 2013095). I.U. was supported by a grant from the Rising Tide Foundation and by a research grant from The Abramson Family Center for Young Scientists.
References
- 1.Dimmock D, et al. Citrin deficiency, a perplexing global disorder. Mol Genet Metab. 2009;96:44–49. doi: 10.1016/j.ymgme.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Delage B, et al. Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer. Int J Cancer. 2010;126:2762–2772. doi: 10.1002/ijc.25202. [DOI] [PubMed] [Google Scholar]
- 3.Long Y, et al. Arginine deiminase resistance in melanoma cells is associated with metabolic reprogramming, glucose dependence, and glutamine addiction. Mol Cancer Ther. 2013;12:2581–2590. doi: 10.1158/1535-7163.MCT-13-0302. doi:10.1158/1535-7163.MCT-13-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris SM., Jr. Recent advances in arginine metabolism: roles and regulation of the arginases. Br J Pharmacol. 2009;157:922–930. doi: 10.1111/j.1476-5381.2009.00278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wheatley DN. Controlling cancer by restricting arginine availability--arginine-catabolizing enzymes as anticancer agents. Anti-cancer drugs. 2004;15:825–833. doi: 10.1097/00001813-200410000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Syed N, et al. Epigenetic status of argininosuccinate synthetase and argininosuccinate lyase modulates autophagy and cell death in glioblastoma. Cell death & disease. 2013;4:e458. doi: 10.1038/cddis.2012.197. doi:10.1038/cddis.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duarte NC, et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci U S A. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. doi:10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi K, Saheki T, Song YZ. In: GeneReviews. Pagon RA, et al., editors. 2005. [Google Scholar]
- 9.Marion V, et al. Hepatic adaptation compensates inactivation of intestinal arginine biosynthesis in suckling mice. PLoS One. 2013;8:e67021. doi: 10.1371/journal.pone.0067021. doi:10.1371/journal.pone.0067021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yizhak K, et al. Phenotype-based cell-specific metabolic modeling reveals metabolic liabilities of cancer. eLife. 2014;3 doi: 10.7554/eLife.03641. doi:10.7554/eLife.03641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuijjer ML, et al. IR/IGF1R signaling as potential target for treatment of high-grade osteosarcoma. BMC cancer. 2013;13:245. doi: 10.1186/1471-2407-13-245. doi:10.1186/1471-2407-13-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kabbarah O, et al. Integrative genome comparison of primary and metastatic melanomas. PLoS One. 2010;5:e10770. doi: 10.1371/journal.pone.0010770. doi:10.1371/journal.pone.0010770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.del Arco A, et al. Expression of the aspartate/glutamate mitochondrial carriers aralar1 and citrin during development and in adult rat tissues. Eur J Biochem. 2002;269:3313–3320. doi: 10.1046/j.1432-1033.2002.03018.x. [DOI] [PubMed] [Google Scholar]
- 14.Tan GS, et al. Novel proteomic biomarker panel for prediction of aggressive metastatic hepatocellular carcinoma relapse in surgically resectable patients. Journal of proteome research. 2014;13:4833–4846. doi: 10.1021/pr500229n. doi:10.1021/pr500229n. [DOI] [PubMed] [Google Scholar]
- 15.Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. doi:10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Folger O, et al. Predicting selective drug targets in cancer through metabolic networks. Molecular systems biology. 2011;7:501. doi: 10.1038/msb.2011.35. doi:10.1038/msb.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varma AP. Metabolic flux balancing: Basic concepts, scientific and practical use. Bio. Technology. 1994;12:994–998. B.O. [Google Scholar]
- 18.Bordel S, Agren R, Nielsen J. Sampling the solution space in genome-scale metabolic networks reveals transcriptional regulation in key enzymes. PLoS computational biology. 2010;6:e1000859. doi: 10.1371/journal.pcbi.1000859. doi:10.1371/journal.pcbi.1000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JK, et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc Natl Acad Sci U S A. 2007;104:13086–13091. doi: 10.1073/pnas.0610292104. doi:10.1073/pnas.0610292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itzkovitz S, et al. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat Cell Biol. 2012;14:106–114. doi: 10.1038/ncb2384. doi:10.1038/ncb2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu GJ, et al. Protein tag-mediated conjugation of oligonucleotides to recombinant affinity binders for proximity ligation. New biotechnology. 2013;30:144–152. doi: 10.1016/j.nbt.2012.05.005. doi:10.1016/j.nbt.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Hao G, Xie L, Gross SS. Argininosuccinate synthetase is reversibly inactivated by S-nitrosylation in vitro and in vivo. J Biol Chem. 2004;279:36192–36200. doi: 10.1074/jbc.M404866200. doi:10.1074/jbc.M404866200. [DOI] [PubMed] [Google Scholar]
- 23.Bjorklund NL, Sadagoparamanujam VM, Taglialatela G. Selective, quantitative measurement of releasable synaptic zinc in human autopsy hippocampal brain tissue from Alzheimer’s disease patients. Journal of neuroscience methods. 2012;203:146–151. doi: 10.1016/j.jneumeth.2011.09.008. doi:10.1016/j.jneumeth.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robitaille AM, et al. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339:1320–1323. doi: 10.1126/science.1228771. doi:10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, et al. Signal transduction pathways involved in phosphorylation and activation of p70S6K following exposure to UVA irradiation. J Biol Chem. 2001;276:20913–20923. doi: 10.1074/jbc.M009047200. doi:10.1074/jbc.M009047200. [DOI] [PubMed] [Google Scholar]
- 26.Pinthus JH, et al. WISH-PC2: a unique xenograft model of human prostatic small cell carcinoma. Cancer Res. 2000;60:6563–6567. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.