Significance
Centrosomes organize the microtubule cytoskeleton in interphase and mitosis. During mitosis, the centrosomes are important for the formation and positioning of the bipolar mitotic spindle on which chromosomes are segregated. The presence of more than two centrosomes can drive mitotic chromosome segregation errors and the formation of aneuploid cells. Centrosome amplification is a common feature of aneuploid cancer cells, but a long-standing question is whether this is a cause or a consequence of tumor development. To assess this question, we generated mice in which centrosome amplification can be induced widely. Despite chronic centrosome amplification, tumorigenesis was not enhanced, demonstrating that an excess of centrosomes is not sufficient to drive tumor development.
Keywords: centrosome amplification, tumorigenesis, p53, Plk4 kinase
Abstract
Centrosomes are microtubule-organizing centers that facilitate bipolar mitotic spindle assembly and chromosome segregation. Recognizing that centrosome amplification is a common feature of aneuploid cancer cells, we tested whether supernumerary centrosomes are sufficient to drive tumor development. To do this, we constructed and analyzed mice in which centrosome amplification can be induced by a Cre-recombinase–mediated increase in expression of Polo-like kinase 4 (Plk4). Elevated Plk4 in mouse fibroblasts produced supernumerary centrosomes and enhanced the expected mitotic errors, but proliferation continued only after inactivation of the p53 tumor suppressor. Increasing Plk4 levels in mice with functional p53 produced centrosome amplification in liver and skin, but this did not promote spontaneous tumor development in these tissues or enhance the growth of chemically induced skin tumors. In the absence of p53, Plk4 overexpression generated widespread centrosome amplification, but did not drive additional tumors or affect development of the fatal thymic lymphomas that arise in animals lacking p53. We conclude that, independent of p53 status, supernumerary centrosomes are not sufficient to drive tumor formation.
Since their initial description by Theodore Boveri in 1900 (1), centrosomes have been recognized as the main microtubule-organizing centers of animal cells and organize bipolar microtubule spindle assembly and function during mitosis. To ensure that chromosomes are divided faithfully into the two daughter cells, the number of centrosomes must be precisely controlled. Cells begin the cycle with a single centrosome that duplicates exactly once to give rise to two centrosomes that form the poles of the mitotic spindle (2, 3). The acquisition of more than two centrosomes, a state known as centrosome amplification, can lead to chromosome segregation errors and subsequent aneuploidy (4–7). In addition, centrosome abnormalities have been proposed to lead to alterations in microtubule nucleation and organization that promote the loss of cell and tissue architecture observed in cancers. Consistent with this, recent work has shown that supernumerary centrosomes can promote cellular invasion in an in vitro model (8).
Centrosome amplification is commonly observed in hematologic malignancies and solid tumors, and a clear link exists between centrosome amplification and aneuploidy in a wide variety of cancer cell lines (6, 7, 9, 10). Furthermore, the presence of supernumerary centrosomes correlates with increased tumor aggressiveness and poor prognosis in human patients (11). Experiments with transplanted larval brain and wing disk tissues in Drosophila have shown that the presence of extra centrosomes can initiate tumorigenesis with (12) or without (13, 14) driving appreciable increases in the level of aneuploidy.
Despite the strong link between centrosome amplification and tumorigenesis, extra centrosomes negatively impact the fitness of mammalian cells and tissues. Induction of centrosome amplification in nontransformed human telomerase-expressing (hTERT) RPE-1 cells triggers a p53-dependent cell cycle arrest (15), whereas driving centrosome amplification in mice brain leads to a developmental loss of neural stem cells by p53-dependent apoptosis (16).
More than a century ago, Boveri suggested a link between acquisition of too many centrosomes and tumorigenesis (17). Nevertheless, whether and how centrosome amplification impacts mammalian tumor development remains untested. Here we have developed a mouse model in which centrosome amplification can be induced by Cre-recombinase–mediated elevation in Plk4 expression. In the presence of the p53 tumor suppressor, widespread elevation of Plk4 drove the production and accumulation of too many centrosomes in liver and skin cells, but this did not accelerate tumorigenesis. Chronic elevation of Plk4 levels in mice without functional p53 produced widespread accumulation of cells with centrosome amplification. Even here, however, centrosome amplification did not drive new tumors or affect the development of thymic tumors driven by loss of p53. Thus, in either the presence or the absence of p53, centrosome amplification is not a universal driver of tumor development in mammals.
Results
Creation of a Mouse Model to Study the Effects of Centrosome Amplification.
Centrosome duplication is controlled by Polo-like kinase 4 (Plk4), and increased expression of Plk4 gives rise to the formation of multiple centrosomes in the same cell cycle (15, 18–21). To establish the effects of centrosome amplification in vivo, we developed a transgenic mouse line in which murine Plk4-EYFP could be conditionally increased in cells after expression of Cre recombinase (Fig. 1A). In the absence of Cre activity, a chicken β-actin/rabbit β-globin promoter (CAG) (22) drove expression of histone H2B-mRFP, followed by a transcription termination sequence flanked by lox P sites that served to silence expression of a downstream Plk4-EYFP gene. Thus, expression of H2B-mRFP served as an initial marker for transgene expression.
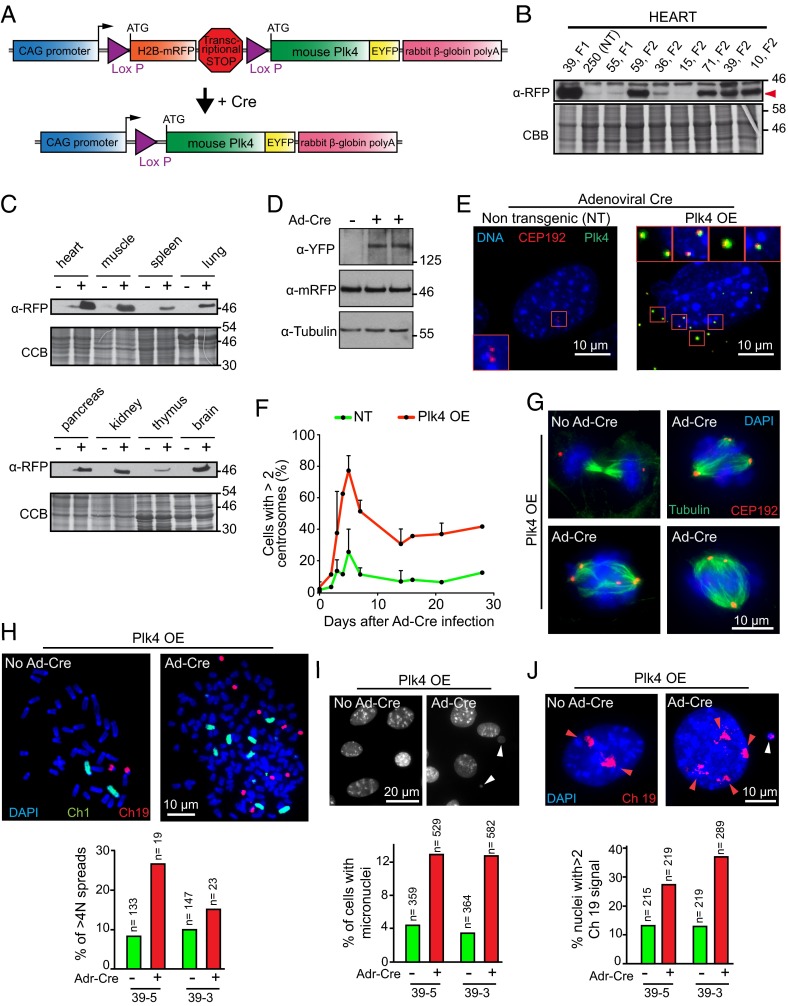
Fig. 1.
Generation of a mouse model for Cre-inducible Plk4 expression. (A) Schematic of the gene construct used to generate inducible Plk4 OE transgenic mice. A chicken β-actin (CAG) promoter initially directs production of H2B-mRFP. Action of Cre at the two Lox P sites will excise the H2B-mRFP gene and the transcriptional stop cassette, thereby activating Plk4-EYFP expression. (B) Immunoblot of H2B-mRFP in heart tissue lysates from various Plk4 OE mouse founder lines. Numbers indicate the founder line. NT, nontransgenic. The red arrowhead indicates the mRFP signal. (C) Immunoblots to determine the expression of H2B-mRFP in various tissues from the transgenic mouse line 39. (−), control (nontransgenic) mice; (+), Plk4 OE mice. (D) Immunoblot of lysates from MEFs showing the accumulation of the Plk4-EYFP protein. (E) Immunofluorescence images of MEFs at 2 d after transduction with Ad-Cre. Plk4-EYFP (green) is visualized only after Cre expression. Accumulation of supernumerary centrosomes was tracked using CEP192 (red). (Scale bar: 10 μm.) (F) Quantification of centrosome amplification (>2 centrosomes per cell) in MEFs at various times after transduction with Ad-Cre. Points represent the mean of two independent experiments. Error bars indicate SD. (G) Immunofluorescence images showing mitotic figures in Plk4 OE MEFs at 5 d after Ad-Cre transduction or without treatment. (Scale bar: 10 μm.) (H) Representative immunofluorescence image of a chromosome spread of an MEF derived from a Plk4 OE mouse at 5 d after treatment with Ad-Cre. Chromosomes 1 and 19 are shown in green and red, respectively. Forty chromosomes are visible in the “no Ad-Cre” condition, and 167 chromosomes are visible in the “Ad-Cre” condition. The graph represents the proportion of cell spreads with >4N DNA content. The quantification was made in two independent populations of MEFs (39-3, 39-5). (I) Representative immunofluorescence image of MEFs derived from a Plk4 OE animal at 5 d after treatment with Ad-Cre. White arrowheads indicate micronuclei. The graph represents the proportion of cells with micronuclei. The quantification was made in two independent populations of MEFs (39-3, 39-5). (J) Representative immunofluorescence image of MEFs, derived from a Plk4 OE animal, at 5 d after treatment with Ad-Cre. The cells were treated with a chromosome painting probe to visualize chromosome 19 (red). Red arrowheads point to chromosome 19 signals. The white arrowhead indicates a chromosome 19 signal in a micronucleus. The graph represents the proportion of cells presenting more than two signals for chromosome 19. The quantification was made in two independent populations of MEFs (39-3, 39-5).
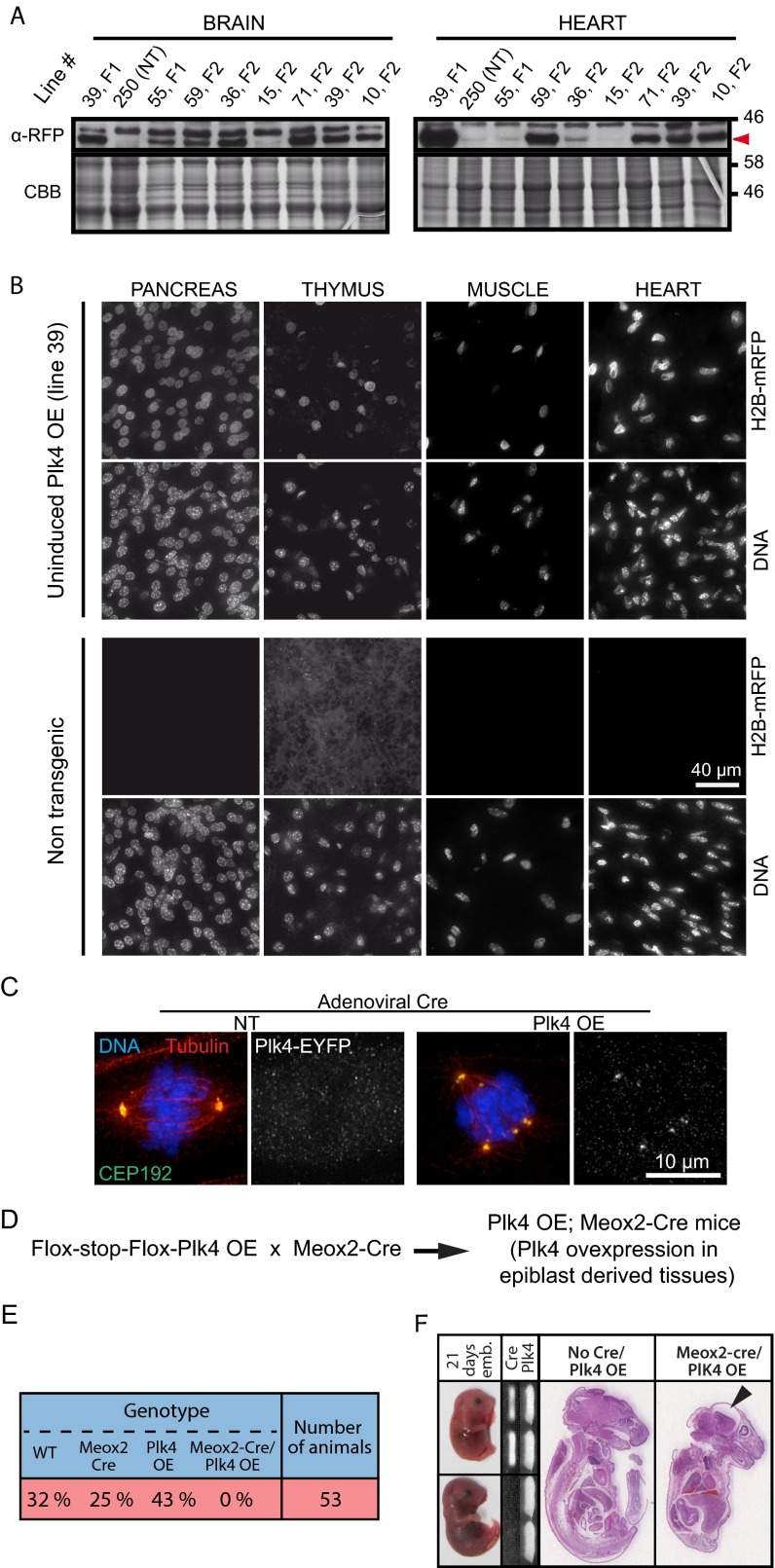
Seven mice founder lines were produced. Founder line 39 was determined to have the highest level of H2B-mRFP expression and thus was selected for further characterization (hereinafter referred to as Plk4 OE) (Fig. 1B and Fig. S1A). Immunoblotting of tissue lysates revealed H2B-mRFP expression in a wide range of tissues (Fig. 1C). Nevertheless, immunofluorescence analysis of individual cells in tissue sections revealed wide variability in the fraction of H2B-mRFP–positive nuclei across different tissues (Fig. S1B).
Fig. S1.
Chronic Plk4 overexpression beginning early in embryogenesis is lethal. (A) Immunoblots of brain and heart tissue lysates obtained from different inducible Plk4 mouse lines. Numbers indicate the founder line. Note that the Right panel presenting heart tissue lysates also appears in Fig. 1B. NT, nontransgenic. Red arrowhead indicates the mRFP signal. (B) Immunofluorescence images of tissues from nontransgenic control or the Plk4 OE mouse line (line 39). H2B-mRFP–positive nuclei are present in all tissues anlayzed. (Scale bar: 40 μm.) (C) Immunofluorescence images showing mitotic spindles in nontransgenic and Plk4 OE MEFs at 2 d after Ad-Cre transduction. (Scale bar: 10 μm.) (D) Outline of the breeding scheme used to obtain Meox2-Cre;Plk4 OE animals. (E) The Plk4 transgenic line 39 was crossed to mice expressing Cre under the control of the Meox2 promoter. The table shows the proportion of animals obtained with the indicated genotypes. No Meox2-Cre;Plk4 OE animals were obtained, indicating that early and ubiquitous Plk4 overexpression leads to embryonic lethality. (F) Meox2-Cre;Plk4 OE embryos where collected at 21 d postfertilization. (Left) Images documenting delayed development in Meox2-Cre;Plk4 OE embryos. (Middle) PCR genotyping of the embryos. (Right) H&E staining of sections of control (no Meox2-Cre; Plk4 OE) and Meox2-Cre;Plk4 OE embryos. The black arrowhead points to microcephaly in the Meox2-Cre;Plk4 OE embryo.
Supernumerary Centrosomes Promote Chromosome Segregation Errors and Aneuploidy in Primary Fibroblasts.
To test the ability of transgenic Plk4-EYFP to drive centrosome amplification, we derived mouse embryonic fibroblasts (MEFs) from Plk4 OE embryos. Infection of the MEFs with adenovirus encoding Cre recombinase (Ad-Cre) produced excision of the H2B-mRFP gene and activated expression of Plk4-EYFP (Fig. 1D). At 5 d after infection, 77% of Plk4 OE cells exhibited supernumerary centrosomes (Fig. 1 E and F), indicating that our transgenic model was capable of driving centrosome amplification in vitro. As expected, the extra centrosomes were functional, acting as microtubule-organizing centers during mitosis and contributing to the formation of abnormal mitotic figures (Fig. 1G and Fig. S1C).
We next assessed whether supernumerary centrosomes where capable of promoting mitotic errors and aneuploidy in MEFs. Using populations of MEFs prepared from independent embryos carrying the Plk4 OE transgene, at 5 d after Ad-Cre transduction we analyzed metaphase chromosome spreads (Fig. 1H), formation of micronuclei (Fig. 1I), and number of copies of chromosome 19 in interphase nuclei (Fig. 1J). In all three cases, there was a significant increase in mitotic errors and aneuploidy in the Ad-Cre–treated cells compared with untreated control cells, indicating that supernumerary centrosomes in MEFs derived from Plk4 OE mice can generate aneuploidy.
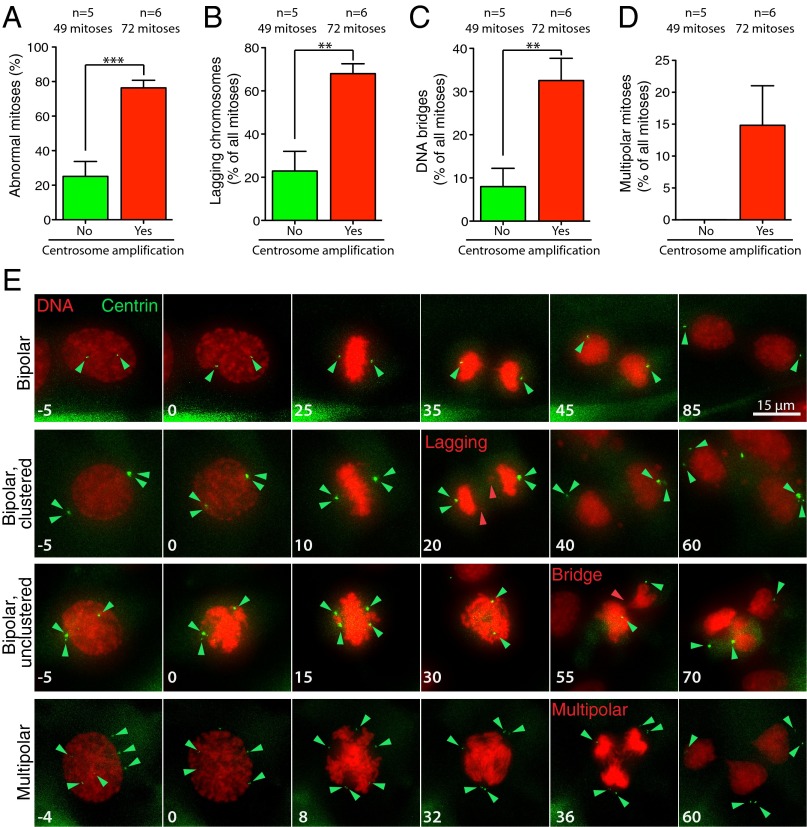
To evaluate the effect of the supernumerary centrosomes on mitotic chromosome segregation, we performed live imaging of skin fibroblasts expressing histone H2B-RFP and Centrin-GFP. This analysis revealed that extra centrosomes led to a threefold increase in the frequency of abnormal mitotic divisions in skin fibroblasts (75% abnormal divisions in cells with supernumerary centrosomes, compared with 25% in cells with two centrosomes) (Fig. 2 A and E). Cells with supernumerary centrosomes showed a greater than threefold increase in lagging chromosomes and DNA bridges in anaphase (Fig. 2 B–E). In addition, whereas multipolar divisions were absent in cells with two centrosomes, multipolar divisions were observed in 15% of mitoses in cells with supernumerary centrosomes (Fig. 2 D and E). Taken together, these data show that supernumerary centrosomes act as microtubule nucleating centers and are competent to induce chromosome segregation errors in vitro.
Fig. 2.
Supernumerary centrosomes promote chromosome mis-segregation in fibroblasts. (A) Quantification of chromosome segregation errors in the Centrin 1-GFP–positive cell population at 48 h after 4-OHT treatment in ERT-Cre;Centrin-GFP and ERT-Cre;Centrin-GFP;Plk4 OE skin fibroblasts with p53f/+, p53f/f, or p53+/+ alleles. Bars represent the mean. Error bars indicate SEM. (B) Quantification of lagging chromosomes during anaphase in cells as described in A. Bars represent the mean of at least five independent experiments. Error bars indicate SEM. ***P < 0.005; P value of unpaired t test calculated on the mean values from five and six independent measurements. (C) Quantification of DNA bridges during anaphase in cells as described in A. Bars represent the mean of at least five independent experiments. Error bars indicate SEM. **P < 0.01; P value of unpaired t test calculated on the mean values from five and six independent measurements. (D) Quantification of multipolar mitoses in cells as described in A. Bars represent the mean of at least five independent experiments. Error bars indicate SEM. **P < 0.01; P value of unpaired t test calculated on the mean values from five and six independent measurements. (E) Representative image from time-lapse filming of skin fibroblasts undergoing mitosis with or without extra centrosomes. Green arrowheads point to centrosomes. Red arrowheads point to chromosome segregation defects. Numbers indicate time in minutes.
Plk4-Mediated Centrosome Amplification in Vivo.
To drive expression of Plk4-EYFP in vivo, we crossed Plk4 OE transgenic animals to mice expressing Cre recombinase under control of the endogenous Meox-2 promoter (23) (Fig. S1D). Meox-2-Cre is expressed from embryonic day 5 in epiblast-derived tissues. Plk4 OE;Meox2-Cre animals died shortly after birth as a result of microcephalic brain development (Fig. S1 E and F). A similar defect was previously observed when Plk4 was selectively overexpressed during brain development of mice (16).
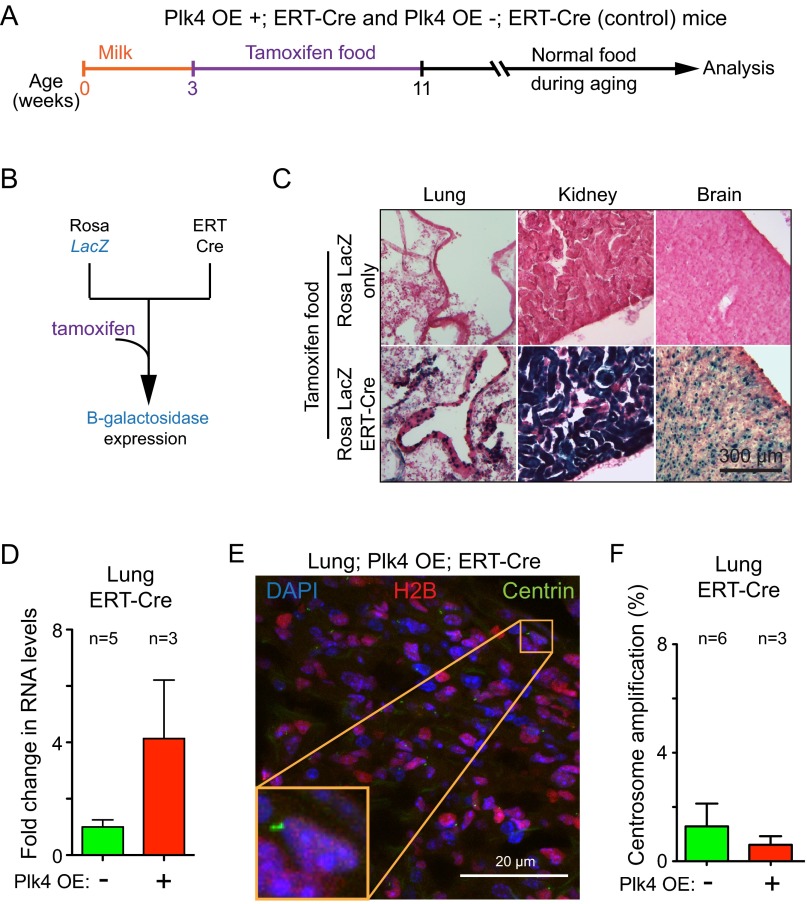
To overcome the early embryonic lethality associated with Plk4 overexpression, we took advantage of a tamoxifen inducible Cre recombinase (ERT-Cre) to induce Cre recombination in young mice (24). We first crossed ERT-Cre mice to mice expressing a Cre-inducible β-galactosidase reporter (25) and assayed Cre activity after exposing animals to tamoxifen in their diet for 2 mo (Fig. S2 A and B). Staining of tissue sections with X-gal revealed Cre activity in all organs examined: heart, muscle, thymus, spleen, pancreas, stomach, intestine, colon, skin, salivary glands, liver, thymus, kidney, lung, and brain (Fig. S2C). These data demonstrate the ability to induce widespread Cre activity using tamoxifen administered through the diet.
Fig. S2.
Centrosome amplification is not tolerated in vivo in the lung. (A) Protocol for activation of expression of Plk4-EYFP in Plk4-OE;ERT-Cre mice. Mice were fed tamoxifen for a 2-mo period beginning at weaning age. (B) Breeding scheme to generate a mouse line containing a tamoxifen-inducible Cre (ERT-Cre) together with a Cre-inducible β-galactosidase transgene. (C) Images show X-gal staining, and thus β-galactosidase activity, in a variety of tissues from a ERT-Cre;Lac Z mouse treated as in A. Control indicates mice containing only the Cre-activatable Lac Z gene without ERT-Cre transgene. (D) Fold change in Plk4 mRNA in the lungs of Plk4 OE;ERT-Cre;Centrin-GFP mice compared with control ERT-Cre;Centrin-GFP mice. Measurements were taken at 2 mo after tamoxifen treatment. Fold change is relative to nontransgenic control animals (−). Bars represent the mean, and error bars indicate SEM. n = 3 Plk4 OE (+) mice and n = 5 nontransgenic controls (−). (E) Representative immunofluorescence image of a lung section from a mouse overexpressing Plk4 after tamoxifen treatment. (F) Percentage of centrosome amplification within cells expressing Centrin 1-GFP in control lungs vs. Plk4 OE lungs. Bars represent the mean, and error bars indicate SEM. n = 3 Plk4 OE (+) mice and n = 6 nontransgenic controls (−). A minimum of 107 centrin-positive cells were counted for each data point.
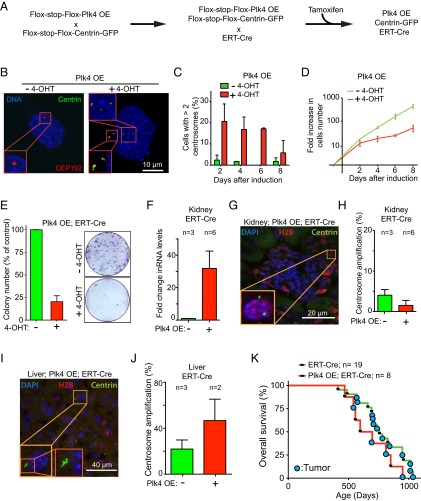
To visualize centrosomes in tissue sections, we introduced a Rosa26-targeted, lox-STOP-lox-Centrin 1-GFP construct into Plk4 OE;ERT-Cre animals. In triply transgenic animals (Plk4 OE;ERT-Cre;Centrin-GFP), the action of Cre inactivates H2B-mRFP expression and activates both Plk4 and Centrin-GFP expression, the latter providing a marker to count centrosomes in cells exposed to active Cre (Fig. 3A). We derived skin fibroblasts from Plk4 OE;ERT-Cre;Centrin-GFP mice and treated them with 4-hydroxytamoxifen (4-OHT) to assess induction of centrosome amplification. At 48 h after 4-OHT treatment, 21% of cells exhibited centrosome amplification, compared with only 2% of untreated control cells (Fig. 3 B and C). Nevertheless, at 8 d after induction of Plk4 overexpression, the frequency of centrosome amplification had decreased to 5%, indicating a fitness disadvantage in cells with elevated Plk4 and supernumerary centrosomes (Fig. 3C). Consistently, fibroblasts with extra centrosomes showed a reduced proliferation rate and clonogenic survival compared with untreated control cells (Fig. 3 D and E).
Fig. 3.
Centrosome amplification is not tolerated in vitro and in most tissues in vivo. (A) Breeding scheme to obtain mice containing the tamoxifen-inducible Cre (ERT-Cre), the Cre-inducible Plk4 transgene, and the Cre-inducible Centrin-GFP centrosomal marker. (B) Representative immunofluorescence image of a skin fibroblast derived from a Plk4 OE;ERT-Cre;Centrin-GFP animal treated for 48 h with or without 4-OHT. (C) Centrosome quantification in cells as described in B. Bars represent the mean of two independent experiments. Error bars indicate SEM. A minimum of 60 cells were analyzed for each condition. (D) Change in cell numbers over time in Plk4 OE skin fibroblasts in which Plk4 expression was induced (+ tamoxifen) or not (− tamoxifen). Lines represent the mean of two experiments. Error bars indicate SEM. (E) Clonogenic analysis of Plk4 OE; ERT-Cre;Centrin-GFP skin fibroblasts treated with (+) or without (−) 4-OHT. The graph shows the percentage of clones that grew in Plk4-overexpressing (+ tamoxifen) cells compared with uninduced controls (− tamoxifen). Bars represent the mean of two experiments. Error bars indicate SEM. Two plates were analyzed for each data point. (F) Fold change in Plk4 mRNA in the kidney of Plk4 OE; ERT-Cre;Centrin-GFP mice compared with control ERT-Cre;Centrin-GFP mice. Measurements were taken at 2 mo after tamoxifen treatment. Bars represent the mean and error bars indicate SEM. n = 3 mice with Plk4 OE (+) and n = 6 nontransgenic control mice (−). (G) Representative immunofluorescence image of a kidney section (a glomerulus is visible in the middle of the image) from Plk4 OE mice after tamoxifen treatment. (H) Percentage of centrosome amplification in cells expressing Centrin 1-GFP in the kidney of tamoxifen-treated Plk4-overexpressing mice (+) and nontransgenic control mice (−). Bars represent the mean, and error bars indicate SEM. n = 3 mice with Plk4 OE (+) and n = 6 nontransgenic animals (−). A minimum of 86 centrin-positive cells were counted for each animal. (I) Representative immunofluorescence image of a liver section from a Plk4 OE mouse after tamoxifen treatment. (J) Percentage of centrosome amplification within cells expressing Centrin 1-GFP in control livers vs. Plk4 OE livers. Measurements were obtained at 2 mo after tamoxifen treatment. Bars represent the mean, and error bars indicate SEM. n = 2 mice with Plk4 OE (+) and n = 3 nontransgenic mice (−). A minimum of 25 centrin-positive cells were counted for each animal. (K) Survival analysis of mouse cohorts treated with tamoxifen citrate food (as in Fig. S2A). Animals developing overt tumors are marked with a blue circle.
Overexpression of Plk4 in the liver produced a more than twofold elevation of centrosome amplification (to 47%) compared with the basal level observed in control animals not bearing the Plk4 transgene (22%) (Fig. 3 I and J). Similarly, an increase in Plk4 RNA drove centrosome amplification in 20% of keratinocytes within the skin epidermis (26). However, despite 4-fold and 30-fold elevations, respectively, in the level of Plk4 transcripts in lung and kidney (Fig. 3F and Fig. S2D), cells in either tissue did not accumulate supernumerary centrosomes above the proportion found in control animals (Fig. 3 G and H and Fig. S2 E and F). Similarly, no overt centrosome amplification was observed in the spleen and pancreas. Thus, in mice with active p53, Cre-mediated activation of Plk4 overexpression is sufficient to drive centrosome amplification in permissive tissues, including the liver (Fig. 3) and skin (26). Nevertheless, cohorts of Plk4-overexpressing mice showed no difference in overall survival compared with control animals (Fig. 3K).
p53 Limits Continued Cycling of Cells That Have Acquired Supernumerary Centrosomes.
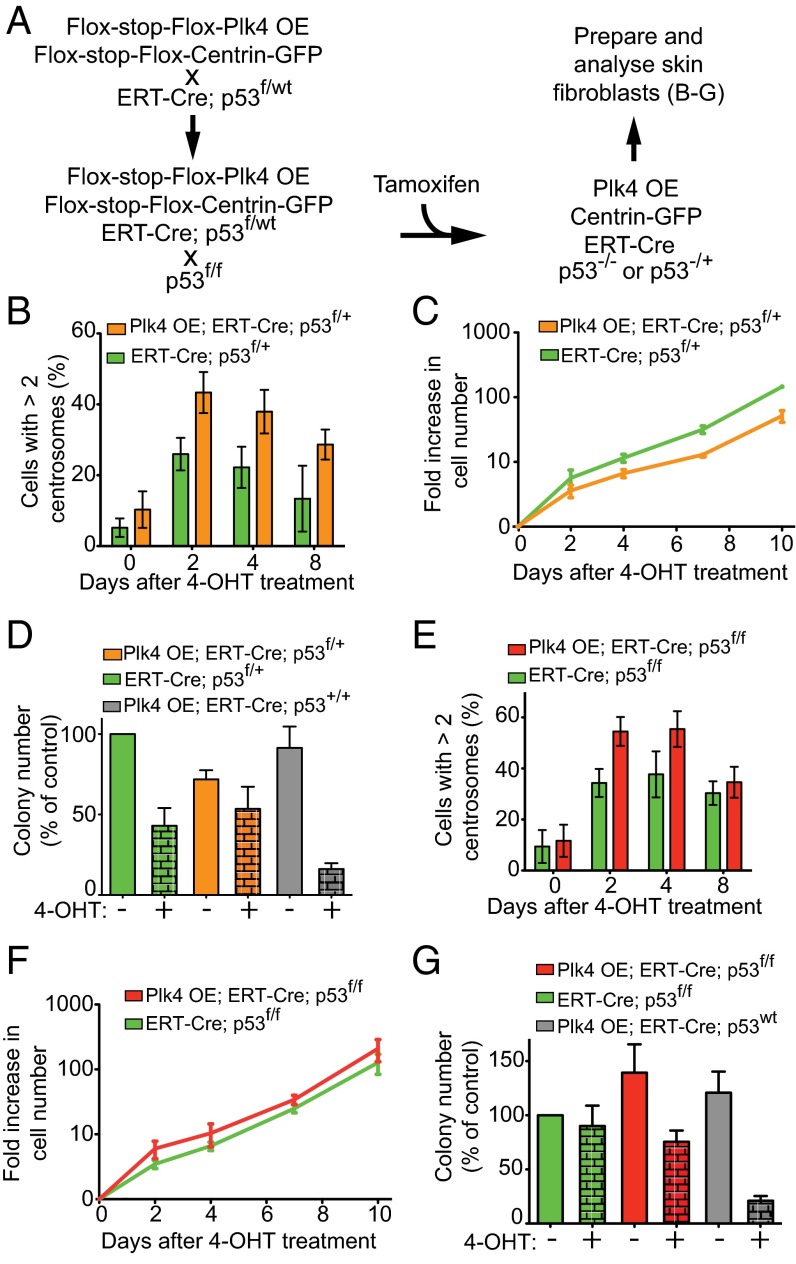
Previous studies have implicated the existence of one or more p53-dependent pathways that prevent the proliferation of cells with supernumerary centrosomes both in cell culture (15) and in the mouse brain (16). To test whether p53 is responsible for eliminating cells that acquire extra centrosomes, we introduced a conditional allele of p53 into Plk4 OE mice (27). Quadruply modified animals (Plk4 OE;ERT-Cre;Centrin-GFP;p53f/+) were created harboring Plk4 OE, ERT-Cre, conditional Centrin 1-GFP, one conditional (floxed) p53 allele, and one wild type (WT) p53 allele (Fig. 4A). Thus, Cre induction will activate the Plk4 and Centrin 1-GFP transgenes while simultaneously inactivating one allele of p53. Consequently, GFP-positive centrosomes mark cells with successful induction of Cre and in which both increased Plk4 levels and reduced p53 expression are expected.
Fig. 4.
p53 reduction or inactivation allows Plk4-induced centrosome amplification in cell culture. (A) Breeding scheme to obtain mice with a Cre-inducible Plk4 transgene, tamoxifen-inducible Cre (ERT-Cre), a Cre-inducible Centrin-GFP allele, and one or two Cre-inactivable p53 alleles. (B) Quantification of centrosome amplification within the Centrin-GFP–positive cell population after 4-OHT treatment. Measurements were done at various times after 4-OHT treatment in Plk4 OE; ERT-Cre;Centrin-GFP;p53f/+ skin fibroblasts and ERT-Cre;Centrin-GFP;p53f/+ control fibroblasts. Bars represent the mean of a minimum of three independent experiments (except for day 8 without the Plk4 OE where two independent experiments were quantified). Error bars indicate SEM. A minimum of 47 cells were analyzed for each data point. (C) Quantification of the change in cell number over time after 4-OHT treatment of Plk4 OE; ERT-Cre;Centrin-GFP;p53f/+ skin fibroblasts and ERT-Cre;Centrin-GFP;p53f/+ control fibroblasts. Lines represent the mean of two experiments. Error bars indicate SEM. (D) Clonogenic survival of Plk4 OE;ERT-Cre;Centrin-GFP;p53f/+ skin fibroblasts and ERT-Cre;Centrin-GFP;p53f/+ control fibroblasts treated with 4-OHT. Bars represent the mean of a minimum of three independent experiments. Error bars indicate SEM. Two plates were analyzed for each data point. (E) Quantification of centrosome amplification within the Centrin-GFP–positive cell population after 4-OHT treatment. Measurements were made at various times after tamoxifen treatment in Plk4 OE; ERT-Cre;Centrin-GFP;p53f/f skin fibroblasts and ERT-Cre;Centrin-GFP;p53f/f control fibroblasts. Bars represent the mean of at least three independent experiments. Error bars indicate SEM. A minimum of 48 cells were analyzed for each data point. (F) Quantification of change in cell numbers over time after 4-OHT treatment of Plk4 OE;ERT-Cre;Centrin-GFP;p53f/f skin fibroblasts and ERT-Cre;Centrin-GFP;p53f/f control fibroblasts. Lines represent the mean of two independent experiments. Error bars indicate SEM. (G) Clonogenic capabilities of Plk4 OE; ERT-Cre;Centrin-GFP;p53f/f skin fibroblasts and ERT-Cre;Centrin-GFP;p53f/f control fibroblasts treated with 4-OHT. Bars represent the mean of a minimum of three independent experiments. Error bars indicate SEM. Two plates were analyzed for each data point.
Using these Plk4 OE;ERT-Cre;Centrin-GFP;p53f/+ mice, we first tested whether centrosome amplification was tolerated in p53 heterozygous cells. Cre-dependent Plk4 overexpression and inactivation of the floxed p53 allele in Plk4 OE;ERT-Cre;Centrin-GFP;p53f/+ skin fibroblasts promoted centrosome amplification, with almost one-half (∼43%) of the cells accumulating more than two centrosomes by 2 d after tamoxifen treatment (Fig. 4B). Heterozygosity for p53 allowed the continued growth of Plk4 OE cells with extra centrosomes in vitro (Fig. 4C) and rescued their ability to form colonies in long-term growth assays (Fig. 4D). The continued cell cycling of cells with extra centrosomes following the inactivation of one p53 allele suggests that supernumerary centrosomes promote chromosome segregation errors that enable the loss of heterozygosity of the remaining WT p53 allele.
We next tested how complete p53 loss affected cells with Plk4-driven supernumerary centrosomes. For this, ERT-Cre was used to inactivate p53 in p53f/f skin fibroblasts also carrying Cre-activatable Plk4 OE and Centrin 1-GFP genes (Plk4 OE;ERT-Cre;Centrin-GFP;p53f/f). As expected based on previous work (28), increased Plk4 levels augmented centrosome amplification in fibroblasts lacking p53: 54% of cells contained more than two centrosomes within 2 d of Plk4 induction (Fig. 4E), compared with 34% in p53 null fibroblasts without Plk4 overexpression and 9% in WT fibroblasts. Importantly, p53 null fibroblasts with a Plk4-dependent elevation in extra centrosomes proliferated at a similar rate to that of control cells lacking p53 (Fig. 3F), and the clonogenic survival of cells with supernumerary centrosomes was enhanced by loss of p53 (Fig. 4G). We conclude that p53 suppresses the maintenance and proliferation of cells with supernumerary centrosomes in vitro.
Plk4-Driven Centrosome Amplification Does Not Affect Tumorigenesis in p53 Heterozygous Mice.
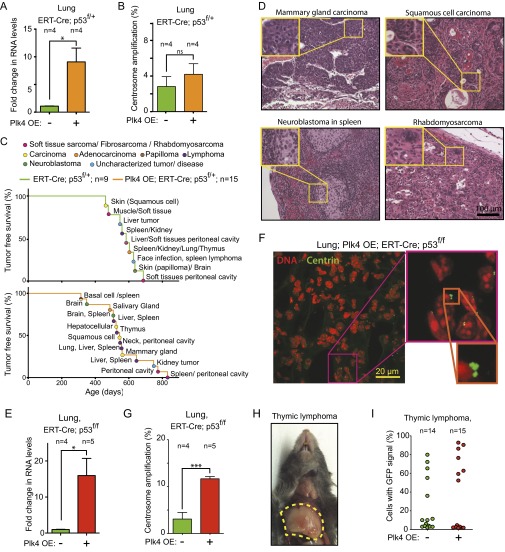
To assess whether centrosome amplification affected tumor development in mice with reduced levels of p53, we assembled cohorts of Plk4 OE mice with ERT-Cre, conditional Centrin 1-GFP, and one conditional p53 allele (p53f/+). Mice were fed for 2 mo with tamoxifen to activate the Plk4 and Centrin 1-GFP genes while inactivating p53 (Fig. S2A). Despite the fact that loss of a single allele of p53 permitted the growth of cells with supernumerary centrosomes in vitro (Fig. 4 C and D), Plk4 overexpression did not alter centrosome numbers in the cells in the lung and kidney of p53+/− mice (Fig. 5 A and B and Fig. S3 A and B), consistent with continuing p53-dependent arrest and/or death of cells that underwent aberrant mitosis with too many centrosomes.
Fig. 5.
Plk4-driven centrosome amplification does not affect the development of tumors resulting from p53 loss. (A) Fold change in Plk4 mRNA in the kidney of Plk4 OE; ERT-Cre;Centrin-GFP;p53f/+ mice compared with control ERT-Cre;Centrin-GFP;p53f/+ mice. Measurements were taken at 2 mo after tamoxifen treatment. Bars represent the mean, and error bars indicate SEM. n = 4 animals for each group. *P < 0.05; P value of unpaired t test calculated on the mean values from four independent measurements. (B) Percentage of centrosome amplification within cells expressing Centrin-GFP in the kidney of Plk4 OE; ERT-Cre;Centrin-GFP;p53f/+ mice and in control ERT-Cre;Centrin-GFP;p53f/+ mice. Measurements were taken at 2 mo after tamoxifen treatment. Bars represent the mean, and error bars indicate SEM. n = 6 mice with Plk4 OE activation and n = 6 without Plk4 OE activation. ns, P > 0.05; P value of unpaired t test calculated on the mean values from six independent measurements. A minimum of 177 centrin-positive cells were counted for each animal. (C) Tumor-free survival of Plk4 OE; ERT-Cre;Centrin-GFP;p53f/+ mice and control ERT-Cre;Centrin-GFP;p53f/+ mice after 2 mo of tamoxifen treatment. n = 15 mice with tamoxifen treatment and n = 9 without tamoxifen treatment. (D) Fold change in Plk4 mRNA in kidneys of Plk4 OE; ERT-Cre;Centrin-GFP;p53f/f mice compared with control ERT-Cre;Centrin-GFP;p53f/f mice. Measurements were taken at 2 mo after tamoxifen treatment. Bars represent the mean, and error bars indicate SEM. n = 6 mice for tamoxifen-induced and n = 4 for uninduced. *P < 0.05; P value of unpaired t test calculated on the mean values from four and six independent measurements. (E) Representative immunofluorescence image of a kidney section (a glomerulus is visible in the middle of the image) from a Plk4 OE; ERT-Cre;Centrin-GFP;p53f/f mouse after tamoxifen treatment. (F) Percentage of centrosome amplification within cells expressing Centrin-GFP in the kidney of Plk4 OE; ERT-Cre;Centrin-GFP;p53f/f mice and in control ERT-Cre;Centrin-GFP;p53f/f mice. Measurements were taken two months after tamoxifen treatment. Bars represent the mean, and error bars indicate SEM. n = 6 mice with Plk4 OE and n = 4 controls. *P < 0.05; P value of unpaired t test calculated on the mean values from four and six independent measurements. A minimum of 172 centrin-positive cells were counted for each animal. (G) Tumor-free survival of Plk4 OE; ERT-Cre;Centrin-GFP;p53f/f mice and control ERT-Cre;Centrin-GFP;p53f/f mice after 2 mo of tamoxifen treatment. n = 25 mice with Plk4 OE and n = 27 controls. (H) (Left) Representative immunofluorescence image of a thymic lymphoma section. (Right) Quantitation of centrosome amplification in individual tumors scored within cells containing a Centrin-GFP signal. Tumors from 15 mice with the Plk4 OE gene and 14 mice without the Plk4 OE gene were analyzed. A minimum of 40 centrin-positive cells were analyzed for each tumor. Bars represent the mean, and errors bars indicate mean and SEM. (I) Mitotic figures in H&E staining of thymic lymphoma sections from Plk4 OE;ERT-Cre;p53f/f mice after tamoxifen induction of Cre to activate the Plk4 OE gene and inactivate p53. Red arrowheads point to segregation errors. The graph shows the proportion of abnormal anaphases (lagging chromosomes, chromosome bridges, and multipolar anaphases). Bars show the mean and error bars show the SEM from measurements from six individual tumors for each condition. ***P < 0.001; P value of an unpaired t test calculated on the mean values of measurements from six independent tumors. A total of 107 anaphases were analyzed for the control condition, and 121 anaphases were analyzed for the Plk4 OE condition.
Fig. S3.
Plk4 overexpression does not affect the type of tumor arising from heterozygous loss of p53. (A) Fold change in Plk4 mRNA in the lungs of Plk4 OE; ERT-Cre;Centrin-GFP;p53f/+ mice compared with control ERT-Cre;Centrin-GFP;p53f/+ mice. Measurements were taken at 2 mo after tamoxifen treatment. Bars represent the mean, and error bars indicate SEM. n = 4 mice with Plk4 OE and n = 4 mice without Plk4 OE. *P < 0.05; P value of unpaired t test calculated on the mean values from four independent measurements. (B) Percentage of centrosome amplification within cells expressing Centrin 1-GFP in the lung of Plk4 OE;ERT-Cre;Centrin-GFP;p53f/+ mice and control ERT-Cre;Centrin-GFP;p53f/+ mice. Measurements were taken at 2 mo after tamoxifen treatment. Bars represent the mean, and error bars indicate SEM. n = 6 mice with Plk4 OE and n = 4 mice without Plk4 OE. A minimum of 116 centrin-positive cells were counted for each data point. ns, P value > 0.05; P value of unpaired t test calculated on the mean values from four independent measurements. (C) Tumor-free survival and tumor spectrum of Plk4 OE;ERT-Cre;Centrin-GFP;p53f/+ mice and control ERT-Cre;Centrin-GFP;p53f/+ mice after 2 mo of feeding with food containing tamoxifen. At each event, the location and tumor type are described using a color code. (D) H&E-stained sections of tumors developed in Plk4 OE;ERT-Cre;Centrin-GFP;p53f/+ mice (mammary gland carcinoma, squamous cell carcinoma, and neuroblastoma within spleen) and in ERT-Cre;Centrin1-GFP;p53f/+ control mice (rhabdomyosarcoma). (E) Fold change in Plk4 mRNA in the lungs of Plk4 OE;ERT-Cre;Centrin-GFP;p53f/f mice compared with control ERT-Cre;Centrin-GFP;p53f/f mice. Measurements were taken at 2 mo after tamoxifen treatment. Bars represent the mean, and error bars indicate SEM. n = 5 mice with Plk4 OE and n = 4 mice without Plk4 OE. *P < 0.05; P value of unpaired t test calculated on the mean values from four and five independent measurements. (F) Representative immunofluorescence image of a lung section from a Plk4 OE;ERT-Cre;Centrin-GFP;p53f/f mouse after tamoxifen treatment. (G) Percentage of centrosome amplification within cells expressing Centrin 1-GFP in lungs of Plk4 OE;ERT-Cre;Centrin1-GFP;p53f/f mice and in control ERT-Cre;Centrin1-GFP;p53f/f mice. Measurements were taken at 2 mo after tamoxifen treatment. Bars represent the mean, and error bars indicate SEM. n = 5 mice with Plk4 OE and n = 4 mice without Plk4 OE. ***P < 0.001; P value of unpaired t test calculated on the mean values from four and five independent measurements. A minimum of 101 centrin-positive cells were counted for each animal. (H) Image of a thymic lymphoma developed in a Plk4 OE;ERT-Cre;Centrin-GFP;p53f/f mouse fed with tamoxifen. The dotted line highlights the tumor. (I) Percentage of Centrin 1-GFP–positive cells in individual thymic lymphomas. Tumors were from 15 ERT-Cre;Centrin-GFP;p53f/f animals and 14 Plk4 OE;ERT-Cre;Centrin1-GFP;p53f/f animals. A minimum of 40 centrin-positive cells were analyzed for each tumor.
p53 heterozygous animals develop an increased frequency of a variety of tumor types, typically accompanied by loss of heterozygosity for p53 (29–31). To determine whether Plk4 overexpression and resultant centrosome amplification influenced tumor development in p53 heterozygous mice, we fed Plk4 OE;ERT-Cre;Centrin-GFP;p53f/+ mice tamoxifen for 2 mo starting at weaning. The resultant Plk4 overexpression, including in lung and kidney, had no effect on tumor initiation or tumor-free survival (Fig. 5C) and the spectrum of tumors that formed was similar to that found in mice heterozygous for p53 (Fig. S3 C and D).
Plk4-Driven Centrosome Amplification Does Not Affect Tumorigenesis in p53 Null Mice.
To assess whether Plk4-dependent centrosome amplification affects tumor development in the absence of p53, we assembled cohorts of mice possessing ERT-Cre, the conditional Plk4 OE, conditional Centrin 1-GFP, and two conditional p53 alleles (i.e., p53f/f). Centrosome number was analyzed in mice fed for 2 mo with tamoxifen to activate the Plk4 and Centrin-GFP genes and inactivate both p53 alleles (Fig. S2A). Plk4 mRNA levels were strongly increased (by 16- and 48-fold, respectively) in the lung and kidney of tamoxifen-fed Plk4 OE;ERT-Cre;Centrin-GFP;p53f/f animals (Fig. 5 D and Fig. S3E). Importantly, cells with centrosome amplification were increased fourfold in lung and fivefold in kidney in these mice Fig. 5 E and F and (Fig. S3 F and G).
As expected from the high penetrance of thymic lymphomas reported previously in a different p53 null mouse (31), tamoxifen-induced Cre expression in p53f/f animals led to the development of fatal thymic lymphomas (Fig. S3H) starting at 144 d of age and occurring with complete penetrance by 235 d (Fig. 5G). Induction of Cre to both activate the Plk4 OE transgene and inactivate the p53f/f alleles also produced lymphomas in 100% of the animals, with an onset and overall survival time identical to those seen in p53 null mice without Plk4 overexpression (produced by tamoxifen-induction of Cre in ERT-Cre;Centrin-GFP;p53f/f mice) (Fig. 5G).
Cre-dependent activation of the conditional Centrin 1-GFP transgene was used to mark centrosomes in cells in which p53 was inactivated. A striking heterogeneity was observed in the fraction of cells with Centrin 1-GFP–positive centrosomes in thymic tumors from both p53f/f and Plk4 OE;p53f/f mice (from ∼2% to ∼90%) (Fig. S3I). This heterogeneity was mimicked by a similar divergence in the level of centrosome amplification (between ∼1% and 30%) in the tumors from p53f/f or Plk4 OE;p53f/f mice (Fig. 5H). Plk4 OE-induced centrosome amplification promoted increased genetic instability, as demonstrated by a significant increase in the frequency of abnormal anaphases in the tumors from Plk4 OE mice (elevated from 50.4% in p53f/f to 69% in Plk4 OE;p53f/f; P < 0.0001, t test with six control tumors and six Plk4 OE tumors) (Fig. 5I). Thus, Plk4-driven centrosome amplification can drive increased genetic instability in p53 null thymic tumors, but this does not reduce (or extend) tumor-free survival from loss of p53 or enhance thymic tumor initiation and growth.
Centrosome Amplification and Induced Tumorigenesis in Skin.
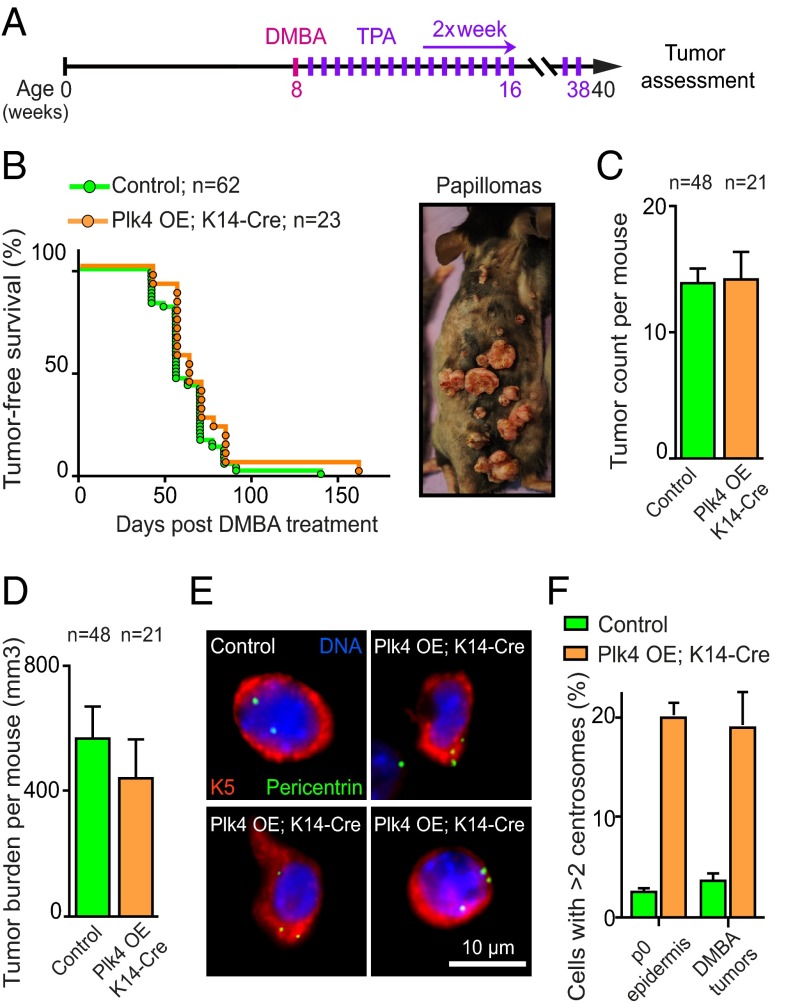
Recognizing that >10% of basal epidermal cells of the skin have extra centrosomes throughout adult life when Plk4 overexpression is induced in those cells (26), we tested whether centrosome amplification influenced tumor development in a classical skin carcinogenesis model in which topical application of the chemical mutagen 7,12-dimethylbenz(a)anthracene (DMBA) is followed by multiple applications of the tumor-promoter 12-O-tetradecanoyl-phorbol-13-acetate (TPA) (32) (Fig. 6A). Treatment with DMBA/TPA induces papillomas, a proportion of which can progress into squamous cell carcinomas.
Fig. 6.
Centrosome amplification does not enhance DMBA/TPA-induced tumorigenesis. (A) Schematic showing the DMBA and TPA treatment course. (B) Tumor-free survival curves of Plk4 OE;K14-Cre and control (Plk4 OE, K14-Cre and WT) mice treated with DMBA/TPA. (Inset) A Plk4 OE;K14-Cre mouse with lesions at various stages of tumor progression. A total of 62 control animals (22 Plk4 OE, 20 K14-Cre, and 20 WT) and 23 Plk4 OE;K14-Cre animals were monitored. (C and D) Quantification of tumor number (C) and total tumor volume (D) per mouse assessed after 32 wk of DMBA/TPA treatment. Bars show the mean, and error bars indicate the SEM from 48 control animals and 21 Plk4 OE; K14-Cre animals. (E) Representative image of cells isolated from DMBA/TPA-induced tumors from control or Plk4 OE;K14-Cre mice. (F) Quantification of centrosome number in cells isolated from P0 epidermis and DMBA/TPA-induced tumors. Bars show the mean and error bars represents the SEM from at least four individual animals and tumors for each condition.
To achieve Plk4 overexpression uniquely in the epidermis, Plk4 OE mice were crossed to animals expressing Cre under control of the keratin 14 promoter (K14-Cre) (33). A DMBA/TPA treatment protocol was applied to the right side of the back skin in a cohort of Plk4 OE;K14-Cre and control (Plk4 OE, K14-Cre, and WT) mice, and tumor development was followed over time. Initial analysis confirmed that skin epidermis was permissive for centrosome amplification in the presence of active p53 (Fig. 6 E and F), with >20% of cells containing more than two centrosomes in the Plk4 OE;K14-Cre mice. The median tumor-free survival for Plk4 OE;K14-Cre (n = 23) and control animals (n = 62) was indistinguishable, however (Fig. 6B).
Both cohorts developed similar numbers of tumors per animal, with the Plk4 OE;K14-Cre mice exhibiting a statistically insignificant trend toward lower overall tumor volume per mouse (Fig. 6 C and D). At no point did animals develop tumors on the untreated left side of the back. Analysis of cells within the tumors revealed cells with extra centrosomes at comparable percentages to those measured in newborns (Fig. 6 E and F). Given the lack of change in overall or tumor-free survival in Plk4 OE;K14-Cre and control mice (26), we conclude that centrosome amplification in the skin does not enhance chemical carcinogenesis-induced tumorigenesis or spontaneous tumorigenesis during aging.
Discussion
Centrosome amplification has been observed in a wide array of human tumors (9, 11, 34–36) and is capable of promoting chromosomal instability and aneuploidy both in vitro (6, 7) and in vivo (12, 16). These observations have promoted the idea that abnormalities in centrosome number may drive events in tumor development. Here we have tested this proposal using a mouse model in which the levels of Plk4 can be conditionally increased to drive centrosome amplification in vivo. Our data reveal that despite widespread Plk4 elevation, p53 acts to prevent accumulation of cells with extra centrosomes in most tissues of the mouse. Chronic Plk4 elevation did not increase spontaneous tumors during aging in either p53+/+ or p53+/− mice. This held true even in the few permissive tissues, including the liver and the proliferative basal layer of the skin epidermis, in which cells with amplified centrosomes did accumulate after Plk4 overexpression. In addition, chronic elevation of Plk4 and centrosome number in p53 null mice did not affect the development and timing of lethal thymic lymphomas or promote development of other tumor types within the restricted lifespan of p53−/− mice. Taken together, our evidence establishes that centrosome amplification is not sufficient to drive tumor formation in mammals, with or without active p53.
Both centrosome amplification (37, 38) and centrosome loss (39, 40) have previously been reported to lead to a p53-dependent cell cycle arrest in skin fibroblasts. Centrosome amplification is also selected against during brain development in mice (16) and in Drosophila (12). Adding to this knowledge, we now show that p53 prevents widespread centrosome amplification in many tissue types in mice (Fig. 5). It remains to be determined whether p53 directly controls centrosome duplication or rather acts to block the proliferation of cells that spontaneously accumulate extra centrosomes through errors in centrosome duplication or failure of cytokinesis.
We found that increased Plk4 levels exacerbated centrosome amplification in mice lacking p53, but failed to influence the development of thymic lymphomas that occur with complete penetrance in the absence of p53 (Fig. 5). The level of centrosome amplification in the thymic lymphomas varied widely but, surprisingly, was not increased in the Plk4 OE mice. This finding suggests that excessive levels of centrosome amplification were selected against during the evolution of these tumors. To avoid the highly aggressive thymic lymphomas that develop in the absence of p53 activity while maintaining sustained levels of centrosome amplification in multiple tissues of adult mice, an attractive future approach would be to analyze athymic Plk4 OE;p53−/− mice (e.g., after surgical thymectomy in young mice) or to combine the Plk4 OE;p53f/f mouse model with tissue-specific expression of Cre to induce Plk4 overexpression and inactivation of p53.
Plk4 overexpression drove the formation of supernumerary centrosomes in the skin and liver of mice, indicating that these tissues tolerate centrosome amplification even in the presence of active p53. Nevertheless, increased centrosome numbers did not promote spontaneous tumor development in these tissues. Furthermore, although loss of the remaining WT allele of p53 will drive the development of a variety of tumors in aging p53+/− animals (29–31), surprisingly, centrosome amplification in the liver and skin did not alter tumor development in p53+/− animals.
We note that in addition to its tolerance for centrosome amplification in p53 competent mice, the mouse livers displayed variable and evolving levels of polyploidy (41). Thus, a parallel can be drawn between hepatocytes that can naturally acquire extra centrosomes by failing one or more cytokinesis but still maintain proliferative capabilities and cancer cells with extra centrosomes that are frequently tetraploid or hypertriploid (42, 43). Despite the absence of tumor formation in the livers of p53+/− animals in our in vivo system, the idea of tetraploidization as an intermediate in cell transformation and tumor initiation has been proposed and tested in different in vitro systems (44, 45). The recent development of a mouse in which cytokinesis failure can be experimentally triggered in skin fibroblasts in vivo (46) potentially could allow testing of acute Plk4-driven centrosome amplification combined with cytokinesis failure as a means to test whether elevated centrosome amplification can favor tumor development in induced tetraploid cells.
Our results in a mammalian organism contrast with what has been reported in flies, in which chronic overexpression of SAK, the fly homolog of Plk4, generated cells with extra centrosomes that were capable of forming tumors when transplanted into WT hosts (12, 14). The mechanistic determinants for the differing outcomes between the mammalian and insect examples have not been established. One potential explanation for the difference is that to suppress tumorigenesis and support the much longer mammalian life span, cell growth control is substantially more stringent in mammals than in flies.
Beyond our analysis of how centrosome amplification affects spontaneous tumorigenesis during aging or from loss of the p53 tumor suppressor, we also tested how similar amplification affected DMBA/TPA-induced papillomas that arise from cell proliferation in the presence of an induced Ras mutation (32). We found that chronic increase in centrosome number did not affect the number of lesions or promote an increase in tumor volume. Recognizing that extra centrosomes have been reported to promote cellular invasion in vitro (8), we then examined invasion of the basement membrane of the epidermis, the initial step in metastasis to other organs. We found no increase in invasive behavior in papillomas with Plk4-driven centrosome amplification, and no change in the rate at which these papillomas progressed into aggressive squamous cell carcinomas. Given that loss of the p19Arf tumor suppressor has been shown to stimulate papilloma transition to squamous cell carcinomas and their metastatic capabilities (47), an examination of the invasion potential of papillomas with p19Arf deficiency and induced centrosome amplification is warranted.
The experimental approaches used here exploited conditional elevation of Plk4 expression to drive what is expected to be continuing centrosome amplification. It is possible that a continuing excess of centrosomes produces a level of genome instability that suppresses any initial increase in tumorigenic potential arising from increased chromosome segregation errors. Therefore, analyzing the correlation between the levels of induced centrosome amplification and the extent of the resulting aneuploidy would be a valuable test for assessing the sensitivity of different tissues to centrosome amplification-driven aneuploidy. We note, however, that even in the current tests, centrosome amplification would be expected to be transient in at least a proportion of cells in which a mitotic chromosome segregation error included loss of the chromosome carrying the Plk4 OE transgene. We suggest that a future, more comprehensive test of whether (and/or how) transiently induced centrosome amplification affects tumorigenesis would be of great interest. Nevertheless, our current evidence demonstrates that chronic centrosome amplification is not sufficient to drive tumor formation in mammals independent of p53 status.
Experimental Procedures
Animals.
Transgenic mice bearing the conditional Plk4 transgene were generated by injecting the construct shown in Fig. 1A into the pronucleus of C57BL/6 fertilized eggs. Mice were subsequently backcrossed onto a C57BL/6 background. To generate Cre-inducible Centrin 1-GFP mice, a lox-STOP-lox-Centrin 1-GFP cassette was targeted to the ROSA26 locus under control of the endogenous ROSA26 promoter in 129 × 1/SvJ ES cells. Resulting mice were backcrossed onto a C57 Bl/6 background. Meox2-Cre and ERT-Cre mice were obtained from JAX mice (ref. #003755 and #004682, respectively) on C57BL/6 congenic background. The tamoxifen citrate diet was prepared by adding 5 g of tamoxifen citrate (TCI America) and 620 g of sucrose to 12 kg of standard diet base (Teklad Global 2016; Harlan Laboratories). Mice were housed and cared for in an Association for the Assessment and Accreditation of Laboratory Animal Care-accredited facility, and all animal experiments were conducted in accordance with Institutional Animal Care and Use Committee-approved protocols. Inquiries concerning Centrin1-GFP mice should be directed to S.M.E. (syevans@ucsd.edu).
DMBA-TPA Carcinogenic Treatment.
To generate animals overexpressing Plk4 in the epidermis, Plk4 OE animals were mated to K14-Cre (CD1) mice (33). DMBA/TPA experiments were performed as described previously (32).
Histology.
Animals were anesthetized with isoflurane and killed by cervical dislocation, after which necropsy was performed. Tissue and tumor specimens were fixed in 10% (vol/vol) formalin overnight and then incubated in 30% (wt/vol) sucrose at 4 °C before embedding and freezing. Tissues were embedded in TissueTek OCT (Sakura) and frozen in isopentane cooled to −40 °C with dry ice. After cryosectioning, 20-μm sections were directly mounted on Superfrost slides (Fisher Scientific), stained with DAPI, and mounted for imaging using ProLong Gold (Life Technologies). H&E-stained tissue sections were prepared at the University of California at San Diego’s Histology Core. All tumors were analyzed by a certified pathologist.
Quantitative Real-Time PCR.
Tissues were homogenized, and total RNA was isolated using TRIzol reagent (Invitrogen) and prepared for reverse transcription using the SuperScript First-Strand kit (Invitrogen). Quantitative real-time PCR for mouse Plk4 was performed using IQ SYBR Green Supermix (Bio-Rad) on IQ5 multicolor real-time PCR detection system (Bio-Rad). Analysis was performed using IQ5 optical system software (Bio-Rad). Reactions were carried out in duplicate using mouse Plk4 primers (forward, 5′-GGA GAG GAT CGA GGA CTT TAA GG-3′; reverse, 5′-CCA GTG TGT ATG GAC TCA GCT C-3′), as described previously (38). Expression values were normalized to two control genes, actin gamma 1 (ACTG1) and mouse ribosomal protein S9 (rps9). The fold changes in mRNA expression were calculated as described previously (48), and expression values were expressed as percentage of the average expression compared with nontransgenic tissues.
Cell Culture.
MEFs were derived from 14.5-d-old embryos using trypsin tissue dissociation as described previously with modifications (49). Skin fibroblasts were derived from ear punch tissue using the collagenase dispase dissociation method (50). Cells were maintained in a low-oxygen incubator with 3% (vol/vol) O2 and 7.5% CO2 at 37 °C. Cells were grown in DMEM containing 15% FBS, 0.1 mM nonessentials amino acids (Gibco), 1 mM sodium pyruvate (Gibco), 1 μM 2-mercaptoethanol (EMD-Millipore), 2 mM l-glutamine (Gibco), and 50 μg/mL penicillin-streptomycin (Gibco). To induce ERT-Cre excision at LoxP sites in culture, 600 nM of 4-hydroxytamoxifen (Sigma-Aldrich) was added to the culture medium for 2 d. For clonogenic assays, 400 cells were seeded into a 10-cm dish and cultured for 2 wk. Cells were fixed with methanol and stained with 0.1% (wt/vol) crystal violet solution.
Immunoblot and Immunofluorescence Analyses.
For immunoblot analysis, samples were separated by SDS/PAGE, transferred onto nitocellulose membranes using Mini Trans-Blot cells (Bio-Rad), and probed using the following antibodies: rabbit anti-RFP (a gift from Joo Seok Han, Ludwig Institute for Cancer Research), rabbit anti-GFP (Roche Life Science), and DM1A (mouse anti–α-tubulin; Sigma-Aldrich).
For immunofluorescence analysis, cells were grown on 18-mm glass coverslips coated with poly-l-lysine and fixed in ice-cold 100% methanol for 10 min at −20 °C. Cells were then blocked in 2.5% FBS, 0.1% (vol/vol) Triton X-100, and 200 mM glycine in PBS for 1 h at room temperature. Antibody incubation were carried out in blocking solution for 1 h at room temperature. DNA was labeled using DAPI, and coverslips were mounted in ProLong Gold (Life technologies). The following antibodies were used for immunofluorescence staining: DM1A (mouse anti–α-tubulin, Sigma-Aldrich), CEP192-Cy-5 (directly labeled rabbit antibody raised against CEP192 amino acids 1–211; a gift from K. Oegama, Ludwig Institute for Cancer Research), guinea pig anti-K5 (produced by the Fuchs group), and rabbit anti-Pericentrin (Covance).
Immunofluorescence images were collected at 0.2-μm Z-sections with an Olympus 100× 1.35 NA, 60× 1.42 NA or 100× 1.4 NA oil objective using a DeltaVision Core system (Applied Precision) with a CoolSnap camera (Roper) operated with SoftWoRx (Applied Precision). Images were deconvolved with SoftWoRx (Applied Precision), and maximum intensity 2D projections were assembled using Fiji (ImageJ, National Institutes of Health). Tissue immunofluorescence images were collected at 0.2-μm Z-sections with a Nikon 100× APO TIRF 1.49 NA objective using a Nikon A1 scanning confocal operated with NIS-Elements (Nikon). Maximum intensity 2D projections were assembled using Fiji.
Chromosome Spreads and FISH Analysis.
Control and Ad-Cre treated MEFs were treated with 10 μg/mL Karyomax Colcemid solution (1:100; Gibco) for 3 h. Cells were collected and treated with 0.075 mM KCl for 15 min at 37 °C, then fixed with Carnoy’s fixative (3:1 methanol:acetic acid). Cells were dropped onto slides and dried. Slides were dipped in 80% (vol/vol) ethanol and dried again. Probes (XMP1 green and XMP19 orange; Metasystems) were applied, and slides were sealed with Fixogum (Marabu). DNA and probes were denatured at 75 °C for 2 min, and hybridization was performed at 37 °C overnight.
Slides were washed with 0.4× SCC at 72 °C for 2 min and then with 2× SCC and 0.05% Tween-20 at room temperature for 30 s, stained with DAPI, and mounted using Prolong Gold antifade reagent (Molecular Probes). Slides were imaged using a Deltavision RT system (Applied Precision) with an Olympus 60× 1.42 NA oil lens using a DeltaVision Core system (Applied Precision) with a CoolSnap camera (Roper) operated with SoftWoRx (Applied Precision). Images were assembled and analyzed using Adobe Photoshop or Fiji software.
Live Cell Imaging.
Mouse ear fibroblasts expressing histone H2B-monomeric red fluorescent proteins were seeded 24 h before experiments on and eight-well chambered coverglass (ibiTreat uSlide; Ibidi). CO2-independent medium (Life Technologies) was added, and the cells were imaged at 37 °C for up to 12 h at 4- or 5-min intervals. Images were collected in 2-μm Z-sections, for seven sections, with an Olympus 60× 1.4 NA oil objective or 40× 1.35 NA oil objective using a DeltaVision Core system (Applied Precision) with a CoolSnap camera (Roper) operated with SoftWoRx (Applied Precision).
Statistical Analysis.
Statistical analyses were performed using GraphPad Prism software. The unpaired Student t test was performed where mentioned. Significance differences are denoted by asterisks: *P < 0.05; **P < 0.01; ***P < 0.005.
Acknowledgments
We thank Dr. Nissi Varki (University of California at San Diego Pathology Core) for histology sample preparation and analysis, and Melissa McAlonis-Downes (Ludwig Institute for Cancer Research) for valuable assistance in the management of the Cleveland laboratory mouse colony. We also thank N. Stokes, A. Aldeguer, and S. Hacker for valuable assistance with the DMBA/TPA treatments. B.V. was supported by a Human Frontier Science Program Long-Term Fellowship (LT000855/2010). A.J.H. was supported by a Leukemia Lymphoma Society Special Fellowship. A.K. was funded by a Jane Coffins Child postdoctoral fellowship. This work was supported by National Institutes of Health (NIH) Grants GM29513 (to D.W.C.) and GM114119 (to A.J.H), a Method to Extend Research in Time (MERIT) Award R37-AR27883 (to E.F.), and Grant DP1HL117649 from the NIH (to S.M.E.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1519388112/-/DCSupplemental.
References
- 1.Boveri T. Zellen-Studien: Über die Natur der Centrosomen. Gustav Fischer; Jena, Germany: 1900. [Google Scholar]
- 2.Strnad P, Gönczy P. Mechanisms of procentriole formation. Trends Cell Biol. 2008;18(8):389–396. doi: 10.1016/j.tcb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139(4):663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 4.Holland AJ, Cleveland DW. Boveri revisited: Chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10(7):478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vitre BD, Cleveland DW. Centrosomes, chromosome instability (CIN), and aneuploidy. Curr Opin Cell Biol. 2012;24(6):809–815. doi: 10.1016/j.ceb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460(7252):278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One. 2009;4(8):e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godinho SA, et al. Oncogene-like induction of cellular invasion from centrosome amplification. Nature. 2014;510(7503):167–171. doi: 10.1038/nature13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lingle WL, et al. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci USA. 2002;99(4):1978–1983. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giehl M, et al. Centrosome aberrations in chronic myeloid leukemia correlate with stage of disease and chromosomal instability. Leukemia. 2005;19(7):1192–1197. doi: 10.1038/sj.leu.2403779. [DOI] [PubMed] [Google Scholar]
- 11.Chan JY. A clinical overview of centrosome amplification in human cancers. Int J Biol Sci. 2011;7(8):1122–1144. doi: 10.7150/ijbs.7.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabino D, et al. Moesin is a major regulator of centrosome behavior in epithelial cells with extra centrosomes. Curr Biol. 2015;25(7):879–889. doi: 10.1016/j.cub.2015.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castellanos E, Dominguez P, Gonzalez C. Centrosome dysfunction in Drosophila neural stem cells causes tumors that are not due to genome instability. Curr Biol. 2008;18(16):1209–1214. doi: 10.1016/j.cub.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 14.Basto R, et al. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133(6):1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holland AJ, et al. The autoregulated instability of Polo-like kinase 4 limits centrosome duplication to once per cell cycle. Genes Dev. 2012;26(24):2684–2689. doi: 10.1101/gad.207027.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marthiens V, et al. Centrosome amplification causes microcephaly. Nat Cell Biol. 2013;15(7):731–740. doi: 10.1038/ncb2746. [DOI] [PubMed] [Google Scholar]
- 17.Boveri T. 1914. Zur Frage der Entstehung Maligner Tumoren (Gustav Fischer, Jena, Germany)
- 18.O’Connell KF, et al. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 2001;105(4):547–558. doi: 10.1016/s0092-8674(01)00338-5. [DOI] [PubMed] [Google Scholar]
- 19.Habedanck R, Stierhof Y-D, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7(11):1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 20.Bettencourt-Dias M, et al. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005;15(24):2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 21.Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol. 2007;17(10):834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. “Green mice” as a source of ubiquitous green cells. FEBS Lett. 1997;407(3):313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 23.Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: A tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26(2):113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: A tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244(2):305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 25.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 26.Kulukian A, et al. Epidermal development, growth control, and homeostasis in the face of centrosome amplification. Proc Nat Acad Sci USA. 2015;112:E6311–E6320. doi: 10.1073/pnas.1518376112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin SC, et al. Somatic mutation of p53 leads to estrogen receptor alpha-positive and -negative mouse mammary tumors with high frequency of metastasis. Cancer Res. 2004;64(10):3525–3532. doi: 10.1158/0008-5472.CAN-03-3524. [DOI] [PubMed] [Google Scholar]
- 28.Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude GF. Abnormal centrosome amplification in the absence of p53. Science. 1996;271(5256):1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- 29.Donehower LA, et al. Deficiency of p53 accelerates mammary tumorigenesis in Wnt-1 transgenic mice and promotes chromosomal instability. Genes Dev. 1995;9(7):882–895. doi: 10.1101/gad.9.7.882. [DOI] [PubMed] [Google Scholar]
- 30.French JE, et al. Loss of heterozygosity frequency at the Trp53 locus in p53-deficient (+/−) mouse tumors is carcinogen-and tissue-dependent. Carcinogenesis. 2001;22(1):99–106. doi: 10.1093/carcin/22.1.99. [DOI] [PubMed] [Google Scholar]
- 31.Jonkers J, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29(4):418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 32.Abel EL, Angel JM, Kiguchi K, DiGiovanni J. Multi-stage chemical carcinogenesis in mouse skin: Fundamentals and applications. Nat Protoc. 2009;4(9):1350–1362. doi: 10.1038/nprot.2009.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: Genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci USA. 1999;96(15):8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boveri T. Concerning the Origin of Malignant Tumours. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 2008. [Google Scholar]
- 35.Sato N, et al. Correlation between centrosome abnormalities and chromosomal instability in human pancreatic cancer cells. Cancer Genet Cytogenet. 2001;126(1):13–19. doi: 10.1016/s0165-4608(00)00384-8. [DOI] [PubMed] [Google Scholar]
- 36.Pihan GA, Wallace J, Zhou Y, Doxsey SJ. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003;63(6):1398–1404. [PubMed] [Google Scholar]
- 37.Cuomo ME, et al. p53-driven apoptosis limits centrosome amplification and genomic instability downstream of NPM1 phosphorylation. Nat Cell Biol. 2008;10(6):723–730. doi: 10.1038/ncb1735. [DOI] [PubMed] [Google Scholar]
- 38.Holland AJ, et al. Polo-like kinase 4 controls centriole duplication but does not directly regulate cytokinesis. Mol Biol Cell. 2012;23(10):1838–1845. doi: 10.1091/mbc.E11-12-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong YL, et al. Cell biology: Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 2015;348(6239):1155–1160. doi: 10.1126/science.aaa5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lambrus BG, et al. p53 protects against genome instability following centriole duplication failure. J Cell Biol. 2015;210(1):63–77. doi: 10.1083/jcb.201502089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duncan AW, et al. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467(7316):707–710. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitelman F, Johansson B, Mertens F. 2012 Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. Available at cgap.nci.nih.gov/Chromosomes/Mitelman. Accessed May 1, 2012.
- 43.Davoli T, de Lange T. The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol. 2011;27(1):585–610. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]
- 44.Davoli T, Denchi EL, de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141(1):81–93. doi: 10.1016/j.cell.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujiwara T, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437(7061):1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka H, et al. Cytokinetic failure-induced tetraploidy develops into aneuploidy, triggering skin aging in phosphovimentin-deficient mice. J Biol Chem. 2015;290(21):12984–12998. doi: 10.1074/jbc.M114.633891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelly-Spratt KS, Gurley KE, Yasui Y, Kemp CJ. p19Arf suppresses growth, progression, and metastasis of Hras-driven carcinomas through p53-dependent and -independent pathways. PLoS Biol. 2004;2(8):E242. doi: 10.1371/journal.pbio.0020242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conner DA. 2001. Mouse embryo fibroblast (MEF) feeder cell preparation. Curr Protoc Mol Biol Chap. 23, Unit 23.2.
- 50.Shao C, et al. Mitotic recombination produces the majority of recessive fibroblast variants in heterozygous mice. Proc Natl Acad Sci USA. 1999;96(16):9230–9235. doi: 10.1073/pnas.96.16.9230. [DOI] [PMC free article] [PubMed] [Google Scholar]