Significance
Myosins use ATP hydrolysis to power movement, but how they do this remains enigmatic. If we understood exactly how ATP’s chemical energy is coupled to force generation, we would gain insight into the fundamental biophysics of these fascinating and medically important proteins. We combined subnanosecond time-resolved FRET with millisecond-resolved transient kinetics to monitor subnanometer structural transitions in myosin during the actin-activated myosin power stroke. Our results show that the power stroke precedes phosphate release, thus contradicting the dominant model of myosin function, which argues that phosphate release should precede the power stroke. Our work prompts future studies investigating how this coordination is altered by disease-causing mutations and therapies designed to treat these diseases.
Keywords: FRET, myosin, power stroke, phosphate release, structural kinetics
Abstract
A principal goal of molecular biophysics is to show how protein structural transitions explain physiology. We have developed a strategic tool, transient time-resolved FRET [(TR)2FRET], for this purpose and use it here to measure directly, with millisecond resolution, the structural and biochemical kinetics of muscle myosin and to determine directly how myosin’s power stroke is coupled to the thermodynamic drive for force generation, actin-activated phosphate release, and the weak-to-strong actin-binding transition. We find that actin initiates the power stroke before phosphate dissociation and not after, as many models propose. This result supports a model for muscle contraction in which power output and efficiency are tuned by the distribution of myosin structural states. This technology should have wide application to other systems in which questions about the temporal coupling of allosteric structural and biochemical transitions remain unanswered.
Myosin family proteins use ATP hydrolysis to generate force and movement required for normal physiology. They drive muscle contraction, help control cell division and cellular motility, move organelles through the cytoplasm, and are important elements of the cellular mechanical-sensing machinery (1, 2). The key to understanding how myosin and related enzymes function in cells, and how to modulate their activity to treat disease, is to determine how the protein’s structural dynamics and biochemical kinetics are coupled. Although high-resolution crystal structures provide best-guess snapshots of protein structure over a range of biochemical states, determining the physiological relevance of these snapshots remains one of the central challenges of structural biophysics.
How myosin generates force remains debated despite more than 50 y of intense research (1–3). The most popular current model (2, 4) proposes that after ATP hydrolysis, myosin interacts weakly with actin and this interaction initiates an ordered series of structural and biochemical transitions that culminate in the dissociation of hydrolyzed phosphate, followed by the isomerization of the actin-binding interface to a state that binds actin with nanomolar affinity and then the rotation of the myosin light-chain domain (LCD) toward the plus end of the actin filament. This rotation converts the thermodynamic energy of phosphate release and actin binding into mechanical energy that performs work. A number of results question this model, however, including spectroscopic data showing that a structural transition in the myosin relay helix, hypothesized to be coupled to LCD rotation, precedes Pi release (5) and force development precedes Pi release in muscle fibers (6).
Determining how these events take place in solution and in cells is an important question, because (i) differences in the mechanics of different myosins likely reflect differences in how the biochemical and structural transitions described above are coordinated (4); (ii) disease-causing mutations in the myosin heavy chain should alter this coordination (7, 8); (iii) recent studies show that modulating mechanochemical coupling in myosin is a viable therapeutic approach to treat myosin-associated diseases (9, 10); and (iv) myosin remains a model system for mechanochemical coupling in general, so what we learn about the myosin power stroke will inform work on related enzymes (3). Understanding the coordination of actin binding, phosphate release, and LCD rotation will address each of these goals. However, direct detection of the kinetics of both LCD rotation and phosphate release, the key to understanding mechanochemical coupling in myosin, has not been achieved (4).
We have developed a spectroscopic approach that allows us to detect angstrom-scale structural changes in protein samples using time-resolved FRET (TR-FRET) measured on the submillisecond time scale. We call this approach transient time-resolved FRET [(TR)2FRET] (5, 11) and use it to determine the structural kinetic mechanism of the myosin power stroke. The results show that actin induces LCD rotation before phosphate dissociation, suggesting a more nuanced interpretation of recent high-resolution crystal structures of myosins complexed with ADP and phosphate (4). They also prompt a general model for tuning myosin force generation where the nonequilibrium distribution of pre- and postpower stroke LCD orientations controls power output and muscle efficiency.
Results
TR-FRET Detects ADP- and ATP-Sensitive Structural States of Myosin.
Numerous experimental approaches have been used to investigate how myosin biochemistry is coupled to myosin structure and force generation, including high-resolution X-ray crystallography, EM, cellular mechanics, EPR, fluorescence, and single-molecule studies (reviewed in 2, 3, 12). However, none of these approaches has directly resolved how LCD rotation is coupled to actin-induced phosphate release during the weak-to-strong actin-binding transition.
We previously examined the structural dynamics of the myosin relay helix (5, 11, 13), a conserved structural element downstream from switch-2 in myosins. The structure of this helix is allosterically coupled to interactions between switch-2 and bound ATP, ADP, and hydrolyzed phosphate (14, 15). Movement of the relay helix is hypothesized to be tightly coupled to LCD rotation (14–16). We showed that in Dictyostelium myosin II, actin binding in the presence of ATP straightens the relay helix before dissociation of hydrolyzed phosphate (5). If the straight structural state of the relay helix is tightly coupled to the postpower stroke orientation of the LCD as proposed (14, 15), then actin should induce LCD rotation before phosphate release. This hypothesis disagrees with several existing proposals (2, 4) but agrees with recent studies performed in isolated muscle fibers (6).
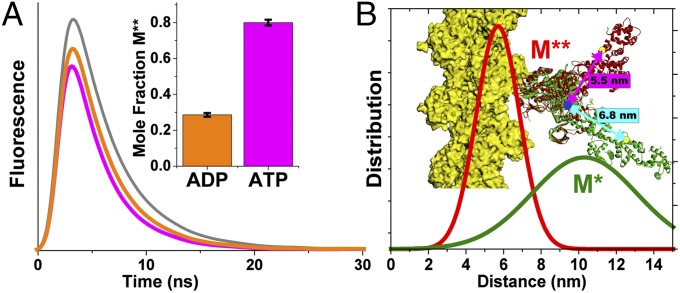
To test this hypothesis directly, we designed a TR-FRET biosensor of two-headed skeletal muscle heavy meromyosin (HMM; Fig. 1 and preparation described in SI Appendix). The biosensor (Fig. 1) contains a fluorescently labeled (Alexa Fluor 488, the FRET donor; Invitrogen) chicken-gizzard smooth muscle regulatory light chain (cgRLC) exchanged onto skeletal muscle HMM (SI Appendix) and a fluorescent nucleotide (Cy3-ATP, the FRET acceptor) bound to the myosin nucleotide binding pocket (17–19). We used the cgRLC because it contains a single reactive Cys and can be exchanged efficiently with the native nonlabeled RLC under conditions that do not affect ATPase activity (SI Appendix, Fig. S1). The characterization of this construct and the validation of FRET between the probes are described in SI Appendix. Spectral overlap between the donor and acceptor gives a Förster distance (R0) of 6.7 nm (SI Appendix). Based on high-resolution crystal structures [summarized in SI Appendix, Table S1; Protein Data Bank (PDB) ID codes 2MYS (20), shown in Fig. 1 as a representative postpower stroke structure, and 1Br1 (16) with added RLC based on homology to scallop myosin RLC structures, shown as a representative prepower stroke state], we predicted that the distance between the C-lobe of the RLC (residue C108 of the exchanged light chain) and the Cy3-labeled ribose will be 5.5 nm when myosin is in the prepower stroke state and between 6.8 and 9.0, indicated by the range of predicted distances in rigor-like X-ray structures (SI Appendix, Table S1), when myosin is in the postpower stroke state.
Fig. 1.
TR-FRET detects nucleotide-dependent LCD orientation. (A) Representative fluorescence waveforms (gray, donor only; orange, donor plus acceptor, saturating Cy3-ADP; magenta, donor plus acceptor, saturating Cy3-ATP). (Inset) Mole fraction of the M** structural state obtained from a two-component fit to the waveforms (n = 9; SEM). (B) Distance probability distributions from the above-mentioned fit, corresponding to the two resolved structural states (M*, green line; M**, red line). The cartoon depicts predicted distances from 2MYS (green ribbon; distance, cyan arrow) and 1Br1 (green ribbon; distance, magenta arrow) with modeled RLC. Actin is shown in yellow.
We measured TR-FRET of the labeled HMM in the presence of saturating bound Cy3-ADP or Cy3-ATP without actin (Fig. 1). Data from these experiments were analyzed using programs written in MATLAB (MathWorks) and with software used in our previous publications (5, 11), and they are described in SI Appendix. The observed time-resolved fluorescence (TRF) waveform of the Alexa Fluor donor probe decays faster in the presence of Cy3-ADP than when the acceptor is not present or when the samples are incubated with actin (Fig. 1A). Cy3-ATP binding decreases the decay time further (Fig. 1A). Thus, the donor’s excited state is quenched by the presence of the fluorescent acceptors, and the degree of quenching increases when the γ-phosphate is present. We fit a structure-based TR-FRET model to the observed data (SI Appendix and SI Appendix, Fig. S2) and evaluated the validity of the fit based on χ2 minimization (SI Appendix, Fig. S2), the geometry of the χ2 error surface for each parameter in the fit (SI Appendix, Fig. S4), and the upper and lower confidence intervals for each parameter obtained from the χ2 error surface (SI Appendix, Table S3).
In the best-fit model, the mole fraction of two TR-FRET interprobe distance distributions depended on the nucleotide-binding conditions (Fig. 1A, Inset). This dependence was consistent with the structural models (Fig. 1 and SI Appendix, Table S3). ATP increased FRET compared with ADP by increasing in the mole fraction of a 5.7-nm distance distribution (denoted M**) and decreasing a 10.3-nm distance (denoted M*; Fig. 1B, Inset). These distances are consistent with the expected values from the range of LCD orientations seen in myosin crystal structures (SI Appendix, Table S3). The short 5.7-nm distance state is also detected in the presence of ADP, although at a lower relative mole fraction (Fig. 1A, Inset). These results indicate that the LCD isomerizes between pre- and postpower stroke orientations in the presence of saturating ATP and ADP, and that the γ-phosphate shifts the [M**]/[M*] equilibrium constant for this isomerization by a factor of 10 (0.4 with ADP and 4.0 with ATP). This equilibrium constant is remarkably consistent with the measured equilibrium constant for ATP hydrolysis at 20 °C and 30 °C in rabbit skeletal myosin S1 (21).
We measured the spectroscopic determinants of FRET, including the absorbance, excitation, and emission spectra; fluorescence time-resolved anisotropy of the donor; and steady-state anisotropy of the acceptor. The spectra were measured to ensure that the overlap integral [J(λ), SI Appendix, Eq. 11] does not change during the ATPase cycle, and the anisotropy controls were performed to estimate the orientation-sensitive term, κ2 (SI Appendix, Eq. 11), and the resulting uncertainty in FRET R0. These controls showed that the donor and acceptor absorption and excitation spectra do not change with ATP binding or with ATP hydrolysis (SI Appendix, Fig. S3). The acceptor (Cy3) fluorescence emission is enhanced upon binding to myosin, indicating a change in quantum yield, but this quantum-yield change does not affect FRET (SI Appendix, Eq. 11), and enhanced Cy3 emission does not contaminate the measured fluorescence of the donor emission detected at 520 nm (SI Appendix, Fig. S3). The anisotropies of the probes do not change during the experiment (SI Appendix, Table S4), and the upper and lower limits for R0 determined from κ2 are 6.5 nm and 8.0 nm, respectively (SI Appendix, Table S4). These limits are smaller than the magnitude of change in distance detected by structure-based TR-FRET modeling (5.7–10.2 nm). Thus, changes in TR-FRET report changes in interprobe distance and LCD rotation.
Structural Kinetics of the Power Stroke Detected by (TR)2FRET.
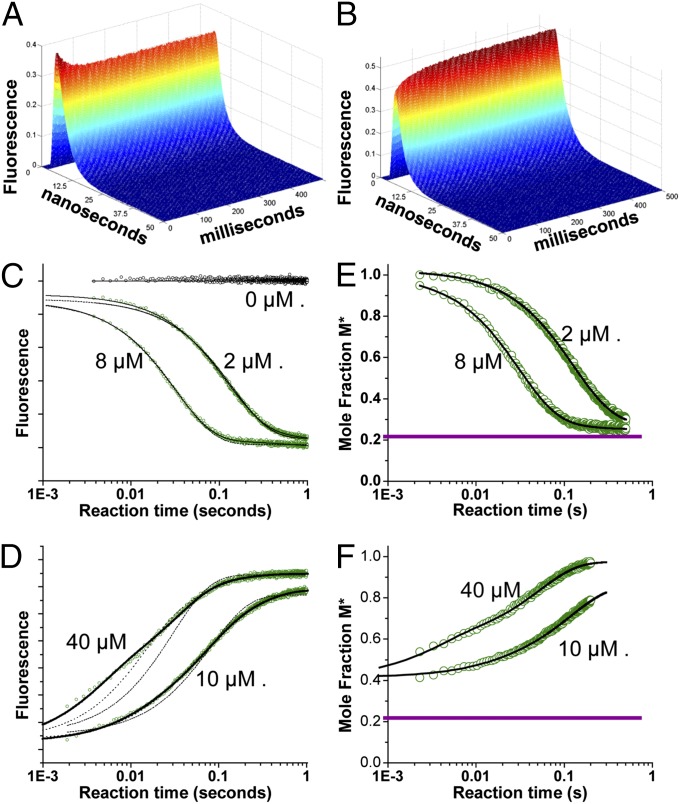
Armed with the biosensor described above, we performed (TR)2FRET experiments during ATP binding and actin-activated single ATP turnover to determine how the molar fractions of the resolved structural states change during the force generating the weak-to-strong actin-binding transition. We acquired TR-FRET waveforms every 0.2 ms after mixing in a stopped-flow device (SI Appendix). Representative waveforms during Cy3-ATP binding, and the subsequent myosin recovery stroke and hydrolysis steps (Fig. 2A), and during actin-activated single ATP turnover (Fig. 2B) show robust changes in TR-FRET.
Fig. 2.
(TR)2FRET was used to detect actomyosin structural kinetics. (A) Recovery stroke following Cy3-ATP binding: TR-FRET waveforms were acquired every 0.2 ms (1-ms average shown) upon mixing 0.2 μM donor-labeled myosin HMM with 2 μM Cy3-ATP (0.1 μM and 1.0 μM final concentrations, respectively). (B) Power stroke: TR-FRET waveforms starting from the steady state of A, after mixing with actin (40 μM after mixing). (C) Total fluorescence of Alexa Fluor 488-labeled HMM mixed with [Cy3-ATP]. (D) Total fluorescence of an HMM Cy3-ATP steady state as in C, mixed with [actin] containing 1 mM ATP. Best fit (two-exponential fit for C and E and three-exponential fit for D and F, solid black line), nonfitting (C and D only, dotted lines), one-exponential fit (C), and one- and two-exponential fits (D) are shown. (E) Mole fractions of M* structural state during Cy3-ATP binding determined by fitting (TR)2FRET data obtained as in A to a structural state model. (F) Mole fractions of M* structural state during the power stroke determined by fitting (TR)2FRET data obtained as in B. The dotted purple line indicates M* mole fraction during steady-state ATPase cycling in the absence of actin (n = 9). Best-fit functions: and (C) and and (E) for 2 μM and 8 μM Cy3ATP, respectively. Best fit functions: and (D) and and (F) for 10 μM and 40 μM actin, respectively.
We analyzed the data by first determining each sample’s total fluorescence (Fig. 2 C and D), calculated by integrating the TRF waveforms over the nanosecond decay time (SI Appendix). The donor total fluorescence in the presence of acceptor provides an indication of the average energy transfer and distance between probes (22), reflecting changes in the average structural state of the protein. By fitting the TR-FRET waveforms to a structure-based model (described below), we resolved the structural states of myosin and measured the changes in their mole fractions with millisecond kinetic resolution.
Cy3-ATP binding to labeled HMM induces a single-exponential time-dependent change in the total fluorescence (Fig. 2B) of Alexa Fluor donor, consistent with Cy3-ATP binding and formation of a high-FRET state. The binding is specific and is blocked by 1 mM MgATP in the presence of excess actin (SI Appendix, Fig. S1). The rate constant for this exponential transient increased linearly with [Cy3-ATP] (SI Appendix, Fig. S5) and exhibited an apparent second-order rate constant of 4.0 μM−1⋅s−1 and a dissociation rate constant <0.1 s−1 (SI Appendix, Table S2), consistent with the kinetics of ATP binding and ATPase cycling (17). The preexponential amplitude did not change with increasing [Cy3-ATP] (SI Appendix, Fig. S5); thus, the binding saturated at concentrations the 1 μM concentration used.
We measured actin-induced structural changes by preparing a steady-state of 0.2 μM Alexa Fluor-labeled HMM with 2 μM excess Cy3-ATP in syringe A of the stopped flow, and then mixed this steady-state complex, which is stable for 5 min (SI Appendix, Fig. S1), with varied concentrations of actin (10–40 μM final concentrations after mixing) in syringe B containing 1 mM MgATP after mixing. We added the 1 mM MgATP to prevent multiple actin-activated Cy3-ATP turnover reactions to ensure that changes in total fluorescence (Fig. 2D) reflect actin-induced changes in FRET during a single weak-to-strong transition (23) and not multiple turnovers.
Actin induced a three-exponential increase in total fluorescence (Fig. 2D), consistent with a multistep process for actin-activated turnover of the Cy3-ATP and a transition from a high-FRET state, before actin binding, to a no-FRET state with dissociation of the Cy3-labeled acceptor at the end of the reaction transient. The slowest phase exhibited a rate constant of 3.4 s−1 (Fig. 3D) and was less than 10% of the total amplitude (SI Appendix, Fig. S5), consistent with slow flux through actin-bound ATP hydrolysis (21).
Fig. 3.
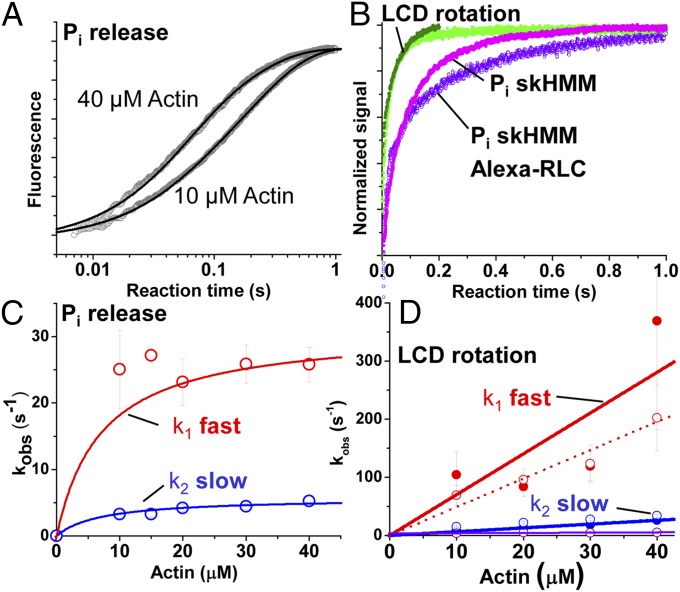
Comparison of phosphate release and LCD structural kinetics. (A) Representative fluorescence of phosphate-binding protein after mixing 1 μM HMM containing 2 μM ATP with [actin]. Best fit functions: and at 10 μM and 40 μM actin, respectively. (B) Direct comparison between phosphate release (magenta, skHMM with native RLC; purple, skHMM with exchanged Alexa fluor-RLC), total myosin fluorescence (light green), and M* mole fraction (dark green). (C) Observed rate constants (kobs) for phosphate release (red, fast phase; blue, slow phase). (D) Observed rate constants for actin-induced LCD rotation (total fluorescence, open circles; M* mole fractions, solid circles) fit as in Fig. 2 D and F. Triple-exponential fit (red, phase 1; blue, phase 2; purple, phase 3).
The observed rate constants for the dominant phases [phase 1 (red symbols) and phase 2 (blue symbols) in Fig. 3D] of the actin-activated fluorescence transients increased linearly with increasing [actin] with a rate constant at 40 μM actin of 200 s−1 and 34 s−1, respectively (SI Appendix, Table S3). The amplitudes of the pre–dead-time increase in M* increased linearly with increasing actin (SI Appendix, Fig. S5). This increase is consistent with the expected Kd of >100 μM for weak actin-binding states of myosin that contain either ATP or posthydrolysis ADP and phosphate in the nucleotide-binding pocket. The amplitudes of the actin-induced exponential transitions that occur after the rapid pre–dead-time phase did not change with increasing actin (SI Appendix, Fig. S5), indicating that the partitioning between each exponential phase after mixing is determined by the equilibrium constants for the recovery stroke and ATP hydrolysis in the absence of actin, and that the multiexponential character of the transient represents flux through a linear or branched kinetic mechanism after the formation of the actomyosin collision complex.
The kinetics of phase 1 during the actin-induced FRET transients were distinct from and faster than actin-induced fluorescent ATP (Cy3-ATP and mant-ATP) turnover (SI Appendix, Fig. S1), whereas phase 2 (35 s−1 at 40 μM actin) is identical to the maximum turnover rate constants in SI Appendix, Fig. S1, indicating that phase 1 of the actin-induced change in FRET reflects structural transitions that precede ADP release after the myosin binds actin.
Structure-Based TR-FRET Modeling Reveals the Structural Kinetics and Thermodynamics of the Power Stroke.
We determined the structural underpinnings for the changes in FRET described above by fitting a structure-based FRET model (described in SI Appendix) to the (TR)2FRET waveforms. The best-fit model was selected by χ2 minimization (SI Appendix, Fig. S2), evaluated by the geometry of χ2 error surface (SI Appendix, Fig. S4) and the error surface confidence intervals (SI Appendix, Fig. S4).
The best-fit model reveals two distinct populations of interprobe distance distributions, indistinguishable from the populations in Fig. 1. These distributions (M* and M**; SI Appendix, Table S3) are in apparent equilibrium under steady-state ATPase cycling conditions, with a [M**]/[M*] equilibrium constant (Keq) of 4 consistent with measurements of the ATP hydrolysis reaction equilibrium constant (21). The mole fraction of the M* and M** states exchanged exponentially during Cy3-ATP binding (Fig. 2E) and actin-activated turnover (Fig. 2F), with the M* fraction decreasing during ATP binding from 1.0 to 0.2 (Fig. 2E) and increasing during actin-activated turnover from 0.2 to 1.0 (Fig. 2F). During actin-activated turnover, the M* fraction increased from 0.2 to ∼0.4 within the dead time of the stopped-flow mix, indicating that the initial stages of LCD rotation occur on the microsecond time scale. This pre–dead-time change is uncertain in the total fluorescence signal depicted in Fig. 2D after normalization, due to uncertainty in the total fluorescence at the start of the transient, but is well resolved when the TRF of the waveforms is analyzed directly, because the time-resolved lifetime is independent of the total concentration of the sample, and thus independent of the ∼10% variation in total fluorescence between successive stopped flow mixes. We fit the observed time dependence after mixing (2 ms onward) of the M* mole fraction with one, two, and three exponential functions, as described for the total fluorescence transients in Fig. 2D. The observed rate constants from these fits (Fig. 3D) were indistinguishable from those rate constants obtained by fitting the total fluorescence transients. Thus, the changes in total fluorescence reflect exponential time-dependent changes in the mole fractions of M* and M** structural states identified by the structure-based TR-FRET model. Because the structure-based model identifies the mole fractions of the M* and M** structural states, it reveals the apparent equilibrium constants separating the states during ATP binding and actin-activated single ATP turnover; these equilibrium constants are not resolved in the analysis of total fluorescence. These equilibrium constants directly reveal the structural kinetic mechanism of the myosin power stroke during the weak-to-strong actin-binding transition. This analysis showed that actin induces the increase in the M* mole fraction, at 40 μM [actin] from 0.2 before formation of the weak-binding actomyosin interface; to 0.38 in the pre–dead-time phase; and then to 0.53 at the completion of phase 1, 0.95 at the completion of phase 2, and 1.0 at the completion of phase 3, with each corresponding to an increase in the apparent M*/M** equilibrium constant from 0.25 to 0.63, 1.13, 19, and >100, respectively.
Actin-Initiated Phosphate Release Is Slower than the Power Stroke.
We compared the structural kinetics of the myosin power stroke, described above, with the kinetics of actin-activated phosphate release using fluorescent-labeled phosphate-binding protein (SI Appendix) (21). The observed rate constant for actin-activated phosphate release (Fig. 3 and SI Appendix, Table S2) increased hyperbolically with increasing [actin], with a K0.5 of 8 μM and a maximum rate constant of 31 s−1 (Fig. 3C) for skeletal HMM (skHMM) with the native RLC and 38 s−1 for skHMM with the exchanged Alexa Fluor-labeled cgRLC. These results, at 6 mM ionic strength, are consistent with previous reports at a lower ionic strength of 1.2 mM (21), which predict 120 s−1 at 25 °C, and 12 mM, which showed 10 s−1 (24) at 10 °C. Direct comparison of FRET and Pi release transients at 40 μM actin (Fig. 3B) and the apparent rate constants for the fast phase of each process, 30–40 s−1 for Pi release and >350 s−1 for the M* mole fraction (Fig. 3D and SI Appendix, Table S2), show that LCD rotation precedes biochemical dissociation of inorganic phosphate in skeletal muscle myosin.
Blebbistatin and Vanadate Block Actin-Induced LCD Rotation.
To test the structural and biochemical requirements for actin-induced changes in LCD orientation, we performed experiments similar to those experiments shown in Fig. 2, in the presence of saturating blebbistatin or saturating vanadate. Both reagents are potent inhibitors of the weak-to-strong actin-binding transition and actin-activated phosphate release. We incubated the labeled HMM with either blebbistatin (10–100 μM) and excess Cy3-ATP or vanadate (10 μM to 1 mM) and excess Cy3-ADP, and then mixed these complexes with varied [actin]. We analyzed the results from experiments performed in the presence of blebbistatin by evaluating the total fluorescence: Blebbistatin itself is fluorescent, having a longer lifetime than the Alexa Fluor donor, and interferes with the TR-FRET data fitting. Blebbistatin did not prevent Cy3-ATP binding (SI Appendix, Fig. S6A, magenta) but did prevent actin-induced increases in fluorescence (SI Appendix, Fig. S6A, purple) and slowed Cy3-ATP turnover in the absence of actin as well (SI Appendix, Fig. S6B), consistent with blebbistatin binding and inhibition of ATPase cycling. We analyzed experiments performed with vanadate by fitting the TR-FRET waveforms to a structure-based model. Mixing 100 μM vanadate with labeled HMM in the presence of excess Cy3-ADP slowly stabilizes the M** state (SI Appendix, Fig. S6D) and prevents the actin-induced M**-to-M* transition on the 0- to 1-s time scale (SI Appendix, Fig. S6C). At longer times, actin weakens vanadate binding and drives nucleotide exchange with nonlabeled ATP included in the actin syringe (SI Appendix, Fig. S6D), evident by the [vanadate] dependence of the observed transients. These results indicate that actin does not induce LCD rotation when these inhibitors are bound.
Structural Kinetics Modeling of FRET Confirms That Actin Initiates LCD Rotation Before Phosphate Dissociation.
We modeled the LCD structural kinetics by fitting three different structural kinetic mechanisms to the Pi release and the M* and M** transients acquired during ATP binding and actin-induced ATP turnover (SI Appendix, Fig. S7). In mechanism 1 (SI Appendix, Eq. 24), actin binding is followed by phosphate release and then LCD rotation; in mechanism 2 (SI Appendix, Eq. 25), actin binding is followed by reversible rotation of the LCD and then Pi release; and in mechanism 3 (SI Appendix, Eq. 26), mechanisms 1 and 2 are combined into a branched pathway. The χ2 value from these fits showed that mechanisms 2 and 3 fit the data (SI Appendix, Fig. S7), whereas mechanism 1 exhibits a χ2 value nearly 10-fold greater than either mechanism 2 or 3 and does not fit the data.
Discussion
Our results show that actin can induce rotation of the myosin LCD before dissociation of hydrolyzed Pi. This finding contradicts the popular model in which LCD rotation follows Pi release (2, 4). The best-fit structural modeling of the TR-FRET (Fig. 1) and (TR)2FRET (Fig. 2) waveforms reveals the structural, thermodynamic, and kinetic underpinnings of the actin-induced LCD rotation, showing that the LCD partitions between two orientations under steady-state ATPase cycling conditions. The first structural state (M*; LCD oriented down in Fig. 1) is indicated by an interprobe distance distribution centered at 10.3 nm (SI Appendix, Table S1), consistent with rigor-like crystal structures predicting distances between 6.8 and 9.0 nm (distance of PDB ID code 2MYS is 6.8 nm). The second structural state, (M**; LCD oriented up in Fig. 1) is indicated by an interprobe distance distribution centered at 5.7 nm, consistent with the predicted prepower stroke orientation of the LCD based on the 1Br1 crystal structure homology model (5.5 nm). Actin binding shifts the [M*]/[M**] equilibrium constant distribution toward the M* state before Pi release, 0.25–6.3 as determined by kinetic modeling (SI Appendix, Table S2). Thus, the difference in the apparent Gibbs free energy (ΔΔG) at 25 °C associated with this distribution change is 8.0 kJ/mol, which is greater than the thermal background (2.4 kJ/mol) but much lower than the energy available from ATP hydrolysis (50 kJ/mol). Once the LCD has rotated, phosphate release is accelerated to 30–40 s−1 under our experimental conditions (6 mM ionic strength). Pi release is predicted to be faster at 1.2 mM ionic strength (21) and slower at 12 mM (24), presumably reflecting the ionic strength dependence of actomyosin binding. After Pi release, the M*/M** ratio is >100 and myosin is trapped in the postpower stroke structural state until ATP rebinds.
Relation to High-Resolution Structural Studies.
Recent crystallographic studies (4) propose a detailed structural mechanism for actin-induced phosphate dissociation from myosin. In this work, Llinas et al. (4) concluded that (i) actin induces the transit of phosphate from the active site down a phosphate release pathway and toward the inner actin-binding cleft before phosphate dissociation and (ii) this transit follows switch-2 opening but precedes LCD rotation and the weak-to-strong actin-binding transition. Our results are consistent with the first conclusion but do not support the second, because we clearly resolve LCD rotation before Pi dissociation and formation of the strong binding interface, which forms more slowly than Pi release (4).
Llinas et al. (4) reach these conclusions by showing that phosphate can bind to the phosphate-release tube when the LCD is in the prepower stroke orientation but not the postpower stroke orientation. An alternate interpretation, supported by the data presented in that study, is that phosphate does not bind to the release tube when myosin is in the postpower stroke crystal state because the binding is too weak. This interpretation is consistent with our data, with our previous FRET studies performed on the myosin relay helix (5), with tryptophan fluorescence studies performed in the absence of actin (14), with studies performed on skinned muscle fibers (6), and with optical trapping studies showing the single-molecule force develops faster than Pi release (25).
Coordination of LCD Rotation with Switch-2 Opening.
Although the LCD rotates before Pi dissociation from myosin, blebbistatin and vanadate inhibit the actin-activated M**-to-M* transition in the LCD (SI Appendix, Fig. S6). Crystal structures of myosin bound by blebbistatin with ADP and vanadate (26) or ADP and vanadate without blebbistatin (27) show switch-2 in a closed conformation. Furthermore, spectroscopic studies suggest that blebbistatin (28) and vanadate (13) stabilize the closed state of switch-2 in the presence of ADP without Pi or Pi analogs. Thus, switch-2 is likely predominantly closed in the presence of blebbistatin and vanadate, and because actin-induced LCD rotation is inhibited by both, we infer that the power stroke requires acceleration of switch-2 opening just as predicted by previous studies (5, 14) and by crystallographic models (4, 16).
LCD Rotation and the Power Stroke.
ATP-driven molecular motors, myosin included, couple the chemical energy of ATP hydrolysis with structural transitions that sustain piconewton-scale forces, and thus perform mechanical work. The force/velocity dependence and subsequent power output of these proteins are determined by their structural kinetic mechanism operating over a range of nonequilibrium and steady-state biochemical conditions. This mechanism defines the distribution of structural states during ATPase cycling, how these states evolve with time, and how they respond to strain.
LCD rotation is one of the primary determinants of force, velocity, and power output in myosins because it acts as an energy-transducing lever arm that converts changes in structure into work. Based on kinetic modeling (SI Appendix, Fig. S7 and Table S2), we find that the apparent Gibbs free energy change for LCD rotation before Pi release is low, −4.5 kJ/mol, suggesting that nonequilibrium diffusion through the transition state is driven by the downstream kinetics of Pi release rather than the free energy change of the transition itself. Thus, this result predicts that power is generated by LCD rotation, not as the result of the free energy change of the rotation but because after the LCD diffuses from the M** state to the M* state, phosphate release inhibits the reversal of the transition while simultaneously facilitating the formation of the strong actin-binding interface. This mechanism is analogous to a thermally activated “ratchet” that is driven by the energetics and kinetics of actin-activated phosphate dissociation. It will be interesting to test this hypothesis in conjunction with high-speed single-molecule laser trap experiments (25) and in high-resolution muscle fiber mechanics studies (6) to determine how the apparent equilibrium constant for LCD rotation is coupled to load and the development of muscle force.
Lastly, our results are nicely consistent with the long-held proposal, first made by Huxley and Simmons in 1971 (29), that the myosin power stroke occurs in at least two thermodynamically distinct steps: a fast elastic transition followed by a slower viscoelastic transition (29). As discussed above, the >500-s−1 actin-induced LCD rotation that we observe is highly reversible, with a predicted change in free energy of −4.5 kJ⋅mol−1, which is only twofold greater than thermal background at the temperature of our experiments (25 °C). The later stages in the FRET-detected LCD rotation are consistent with the kinetics and energetics of phosphate release, which exhibits a much larger change in free energy and is less readily reversed. Reversible LCD rotation is consistent with the elastic process observed by Huxley and Simmons (29), whereas the slower, less reversible phase in our data is consistent with the viscoelastic processes.
Conclusions
We have used (TR)2FRET and transient phosphate release assays to determine the coordination of LCD rotation and actin-induced dissociation of hydrolyzed Pi in fast skeletal muscle myosin. The detection of nanosecond-resolved TR-FRET in real time during the millisecond-resolved biochemical transient allowed direct and quantitative measurement of the mole fraction of pre- and postpower stroke LCD structural states, and thus the transient state distribution of these states during the critical actin-activated power stroke. The major change in Gibbs free energy is associated with phosphate release and the weak-to-strong actin binding transition, and not with the large structural reorientation of the LCD. These results provide direct and vivid insight into one of the most enigmatic aspects of myosin enzymology. Our work also serves as an important primer for future studies of other enzymes, such as kinesins, dyneins, small GTPases, nucleotide polymerases, and kinases, where questions regarding the coupling of allosteric structural and biochemical transitions remain unanswered.
Methods
Protein Purification and Labeling.
Skeletal myosin (rabbit) was purified as described by Margossian et al. (30) and is described in detail in SI Appendix. Purified HMM was generated by α-chymotrypsin digestion stopped by addition of pefabloc (Roche). The cgRLC with a single reactive Cys at position 108 was expressed in Escherichia coli, as described in detail in SI Appendix, and labeled with Alexa Fluor 488 (Invitrogen). Actin was purified from rabbit skeletal muscle by acetone dehydration followed by extraction into ice cold water, as described in detail in SI Appendix.
Spectroscopy.
TR-FRET experiments were carried out as described in our previous studies (5), and are explained in detail in SI Appendix.
Transient Kinetics.
Stopped-flow experiments were performed on a Biologic SFM-20 instrument equipped with a transient TRF spectrophotometer, as described in our previous work (5).
Supplementary Material
Acknowledgments
This study was supported by NIH Grant AR32961 (to D.D.T.), American Heart Association Grant 14SDG20480032 (to J.M.M.), and NIH Grant 5 P30 AR05722), and by the Paul and Sheila Wellstone Foundation. J.A.R. was supported by the Graduate Excellence Fellowship from the University of Minnesota.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1514859112/-/DCSupplemental.
References
- 1.Houdusse A, Sweeney HL. Myosin motors: Missing structures and hidden springs. Curr Opin Struct Biol. 2001;11(2):182–194. doi: 10.1016/s0959-440x(00)00188-3. [DOI] [PubMed] [Google Scholar]
- 2.Sweeney HL, Houdusse A. Structural and functional insights into the Myosin motor mechanism. Annu Rev Biophys. 2010;39:539–557. doi: 10.1146/annurev.biophys.050708.133751. [DOI] [PubMed] [Google Scholar]
- 3.Málnási-Csizmadia A, Kovács M. Emerging complex pathways of the actomyosin powerstroke. Trends Biochem Sci. 2010;35(12):684–690. doi: 10.1016/j.tibs.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llinas P, et al. How actin initiates the motor activity of Myosin. Dev Cell. 2015;33(4):401–412. doi: 10.1016/j.devcel.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muretta JM, Petersen KJ, Thomas DD. Direct real-time detection of the actin-activated power stroke within the myosin catalytic domain. Proc Natl Acad Sci USA. 2013;110(18):7211–7216. doi: 10.1073/pnas.1222257110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caremani M, Melli L, Dolfi M, Lombardi V, Linari M. The working stroke of the myosin II motor in muscle is not tightly coupled to release of orthophosphate from its active site. J Physiol. 2013;591(Pt 20):5187–5205. doi: 10.1113/jphysiol.2013.257410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bloemink M, et al. The hypertrophic cardiomyopathy myosin mutation R453C alters ATP binding and hydrolysis of human cardiac β-myosin. J Biol Chem. 2014;289(8):5158–5167. doi: 10.1074/jbc.M113.511204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sommese RF, et al. Molecular consequences of the R453C hypertrophic cardiomyopathy mutation on human β-cardiac myosin motor function. Proc Natl Acad Sci USA. 2013;110(31):12607–12612. doi: 10.1073/pnas.1309493110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malik FI, et al. Cardiac myosin activation: A potential therapeutic approach for systolic heart failure. Science. 2011;331(6023):1439–1443. doi: 10.1126/science.1200113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, White HD, Belknap B, Winkelmann DA, Forgacs E. Omecamtiv Mecarbil modulates the kinetic and motile properties of porcine β-cardiac myosin. Biochemistry. 2015;54(10):1963–1975. doi: 10.1021/bi5015166. [DOI] [PubMed] [Google Scholar]
- 11.Nesmelov YE, et al. Structural kinetics of myosin by transient time-resolved FRET. Proc Natl Acad Sci USA. 2011;108(5):1891–1896. doi: 10.1073/pnas.1012320108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas DD, Kast D, Korman VL. Site-directed spectroscopic probes of actomyosin structural dynamics. Annu Rev Biophys. 2009;38:347–369. doi: 10.1146/annurev.biophys.35.040405.102118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agafonov RV, et al. Structural dynamics of the myosin relay helix by time-resolved EPR and FRET. Proc Natl Acad Sci USA. 2009;106(51):21625–21630. doi: 10.1073/pnas.0909757106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gyimesi M, et al. The mechanism of the reverse recovery step, phosphate release, and actin activation of Dictyostelium myosin II. J Biol Chem. 2008;283(13):8153–8163. doi: 10.1074/jbc.M708863200. [DOI] [PubMed] [Google Scholar]
- 15.Kintses B, Yang Z, Málnási-Csizmadia A. Experimental investigation of the seesaw mechanism of the relay region that moves the myosin lever arm. J Biol Chem. 2008;283(49):34121–34128. doi: 10.1074/jbc.M805848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominguez R, Freyzon Y, Trybus KM, Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: Visualization of the pre-power stroke state. Cell. 1998;94(5):559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 17.Kuhlman PA, Bagshaw CR. ATPase kinetics of the Dictyostelium discoideum myosin II motor domain. J Muscle Res Cell Motil. 1998;19(5):491–504. doi: 10.1023/a:1005304408812. [DOI] [PubMed] [Google Scholar]
- 18.Conibear PB, Kuhlman PA, Bagshaw CR. Measurement of ATPase activities of myosin at the level of tracks and single molecules. Adv Exp Med Biol. 1998;453:15–26; discussion 26–27. doi: 10.1007/978-1-4684-6039-1_3. [DOI] [PubMed] [Google Scholar]
- 19.Chaen S, Shirakawa I, Bagshaw CR, Sugi H. Measurement of ATP turnover during shortening and lengthening of rabbit psoas myofibrils using a fluorescent ATP analog. Adv Exp Med Biol. 1998;453:569–576. doi: 10.1007/978-1-4684-6039-1_62. [DOI] [PubMed] [Google Scholar]
- 20.Rayment I, et al. Three-dimensional structure of myosin subfragment-1: A molecular motor. Science. 1993;261(5117):50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 21.White HD, Belknap B, Webb MR. Kinetics of nucleoside triphosphate cleavage and phosphate release steps by associated rabbit skeletal actomyosin, measured using a novel fluorescent probe for phosphate. Biochemistry. 1997;36(39):11828–11836. doi: 10.1021/bi970540h. [DOI] [PubMed] [Google Scholar]
- 22.Lakowicz JR. Principles of Fluorescent Spectroscopy. 3rd Ed Springer; New York: 2006. [Google Scholar]
- 23.De La Cruz EM, Ostap EM. Kinetic and equilibrium analysis of the myosin ATPase. Methods Enzymol. 2009;455:157–192. doi: 10.1016/S0076-6879(08)04206-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brune M, Hunter JL, Corrie JE, Webb MR. Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry. 1994;33(27):8262–8271. doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- 25.Capitanio M, et al. Ultrafast force-clamp spectroscopy of single molecules reveals load dependence of myosin working stroke. Nat Methods. 2012;9(10):1013–1019. doi: 10.1038/nmeth.2152. [DOI] [PubMed] [Google Scholar]
- 26.Allingham JS, Smith R, Rayment I. The structural basis of blebbistatin inhibition and specificity for myosin II. Nat Struct Mol Biol. 2005;12(4):378–379. doi: 10.1038/nsmb908. [DOI] [PubMed] [Google Scholar]
- 27.Smith CA, Rayment I. X-ray structure of the magnesium(II).ADP.vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 A resolution. Biochemistry. 1996;35(17):5404–5417. doi: 10.1021/bi952633+. [DOI] [PubMed] [Google Scholar]
- 28.Takács B, et al. Myosin complexed with ADP and blebbistatin reversibly adopts a conformation resembling the start point of the working stroke. Proc Natl Acad Sci USA. 2010;107(15):6799–6804. doi: 10.1073/pnas.0907585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huxley AF, Simmons RM. Proposed mechanism of force generation in striated muscle. Nature. 1971;233(5321):533–538. doi: 10.1038/233533a0. [DOI] [PubMed] [Google Scholar]
- 30.Margossian SS, Lowey S. Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzymol. 1982;85(Pt B):55–71. doi: 10.1016/0076-6879(82)85009-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.