Significance
Activation of G protein-coupled receptors (GPCRs) by agonists is the first step of eukaryotic cellular signal transduction. Because GPCRs are expressed in almost all human tissues and play a key role in human physiology, they are the targets for more than 30% of pharmaceutical drugs. Binding of ligands on the extracellular surface of a GPCR induces a conformational change on the cytoplasmic surface, which is recognized by G proteins or other cellular effectors. Here we show that the β2-adrenergic receptor, a prototypical GPCR, naturally fluctuates between inactive and active conformations, and that agonist or inverse agonist ligands modulate the conformational exchange kinetics in distinct ways, explaining their different pharmacological efficacies. These insights should assist in the design of improved GPCR-targeting drugs.
Keywords: signal transduction mechanisms, agonists and inverse agonists, conformational polymorphism, single-molecule fluorescence spectroscopy, phospholipid nanodiscs
Abstract
Binding of extracellular ligands to G protein-coupled receptors (GPCRs) initiates transmembrane signaling by inducing conformational changes on the cytoplasmic receptor surface. Knowledge of this process provides a platform for the development of GPCR-targeting drugs. Here, using a site-specific Cy3 fluorescence probe in the human β2-adrenergic receptor (β2AR), we observed that individual receptor molecules in the native-like environment of phospholipid nanodiscs undergo spontaneous transitions between two distinct conformational states. These states are assigned to inactive and active-like receptor conformations. Individual receptor molecules in the apo form repeatedly sample both conformations, with a bias toward the inactive conformation. Experiments in the presence of drug ligands show that binding of the full agonist formoterol shifts the conformational distribution in favor of the active-like conformation, whereas binding of the inverse agonist ICI-118,551 favors the inactive conformation. Analysis of single-molecule dwell-time distributions for each state reveals that formoterol increases the frequency of activation transitions, while also reducing the frequency of deactivation events. In contrast, the inverse agonist increases the frequency of deactivation transitions. Our observations account for the high level of basal activity of this receptor and provide insights that help to rationalize, on the molecular level, the widely documented variability of the pharmacological efficacies among GPCR-targeting drugs.
G protein-coupled receptors (GPCRs) mediate a multitude of physiological functions and are the targets for a myriad of drugs (1), many of which elicit different functional outcomes through the same receptor (2). It remains to be rationalized at the molecular level why some drugs stimulate the signaling activity of a GPCR (full or partial agonists), whereas others either repress the receptor (inverse agonists) or have no effect on the intrinsic signaling activity (neutral antagonists). Moreover, the existence of a high basal activity of some GPCRs (3) suggests that the conformational transitions leading to activation may occur spontaneously, even in the absence of ligands, which in turn raises questions about the mechanistic roles of GPCR ligands. Understanding the mechanisms and pathways of receptor activation or deactivation, and how these are linked to the binding of ligands with different chemical structures and pharmacological efficacies, will aid in design of new GPCR-targeted drugs with tailored pharmacological responses and fewer side effects. To attain these goals, new methods are required to visualize the conformational dynamics of GPCRs in the presence and absence of drugs.
The β2-adrenergic receptor (β2AR) has been extensively investigated in crystals (4, 5), by NMR in solution (6–11), by bulk fluorescence spectroscopy in solution (12–14) and in cells (2), by single-molecule fluorescence spectroscopy (15–17), and by molecular dynamics simulations (18). Despite the availability of high-resolution crystal structures of β2AR in inactive (4) and active (5) conformations, it remains unknown how ligands regulate transitions between the two states and why β2AR exhibits a significant level of ligand-independent, basal signaling activity. To address these questions, we use single-molecule fluorescence spectroscopy to monitor activation-linked conformational transitions of individual receptor molecules in real time over extended time periods. Our results highlight the intrinsically dynamic character of β2AR and provide insights into the mechanism of receptor activation and the roles of β2AR ligands.
Results
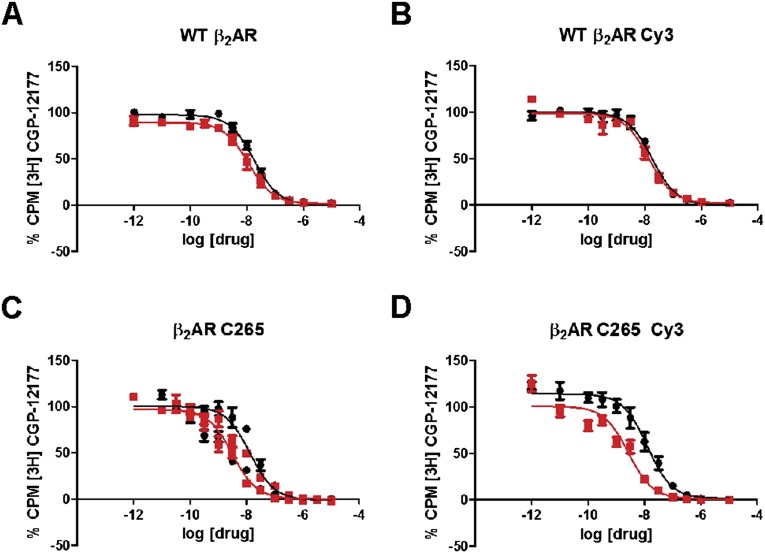
To visualize individual human β2AR molecules, we labeled the receptor with a bright fluorescent dye (Cy3) and reconstituted the receptor in phospholipid nanodiscs, under conditions that favor incorporation of just a single receptor per nanodisc (19). The receptor–nanodisc complexes were tethered to a quartz surface and monitored over time for an average period of 70 s by total internal reflection fluorescence (TIRF) microscopy (Fig. 1A). The Cy3 label was attached to Cys265 near the cytoplasmic end of helix VI (Fig. 1B). Comparison of crystal structures of β2AR in inactive (4) and active (5) states reveals that the cytoplasmic end of helix VI moves outward by 14 Å and rotates during receptor activation, a conformational change that is expected to alter the fluorophore environment (Fig. 1B). Using a radioligand binding assay, we found that the Cy3-labeled receptor reconstituted in nanodiscs is fully functional in ligand binding (Fig. S1). The Kd value for the radioligand [3H]CGP-12177 (Table S1) is similar to that reported for native β2AR in membranes (20), confirming that the nanodisc environment preserves the functionality of the receptor and that the receptor mutations required for site-specific labeling and the attached Cy3 label do not disrupt ligand binding. Control experiments confirmed that the receptor–nanodisc complexes were specifically bound to the surface and that individual complexes contained just one labeled receptor molecule, as intended (Fig. S2 A–C). A small fraction of receptor–nanodisc aggregates were also present on the surface, but these were readily identified by their high fluorescence intensity (Fig. S2A) and were excluded from further analysis.
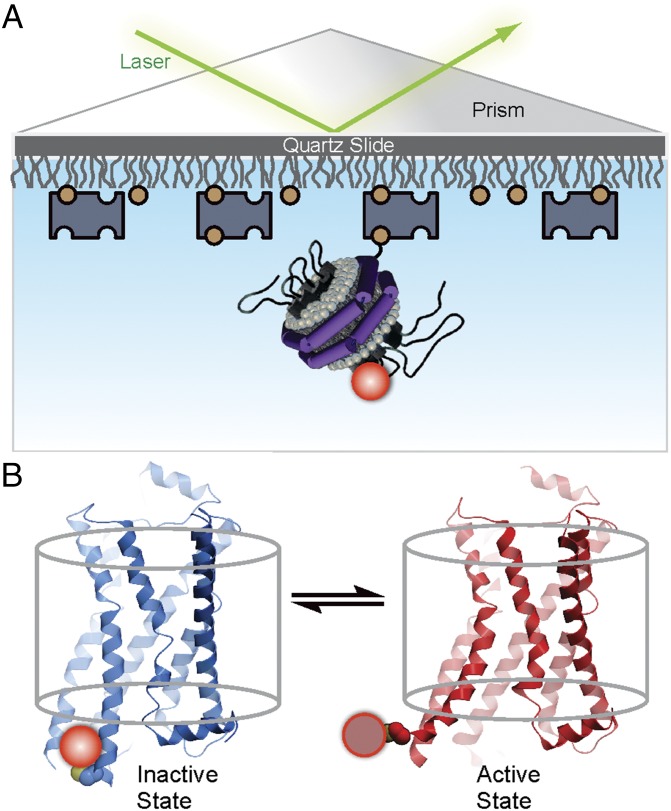
Fig. 1.
Experimental system to monitor conformational transitions of β2AR at the single-molecule level. (A) An individual receptor molecule (black) labeled with Cy3 (red sphere) incorporated in a phospholipid nanodisc (with the belt protein MSP1 shown as purple cylinder and phospholipids shown with gray head groups) is tethered to a quartz surface coated with polyethylene glycol (wavy lines) via biotin (orange circles) and streptavidin (dark blue rectangles). The labeled receptor is illuminated in the evanescent field of a totally internally reflected 532-nm laser beam (green). The cartoon of the receptor–nanodisc complex is adapted from ref. 21. (B) Expanded view of a single receptor–nanodisc complex, showing the receptor exchanging between inactive (blue) and active (red) conformations, with corresponding changes in the local environment of the Cy3 probe attached to Cys265 (light or dark red spheres, respectively). The two structures correspond to crystal structures of β2AR in active (red, PDB ID code 3SN6; ref. 5) and inactive (blue, PDB ID code 2RH1; ref. 4) states, with residues Lys263 to Glu268 in the latter structure modeled in α-helical conformation, as observed in the closely related β1AR receptor structure (PDB ID code 4AMJ). The transparent cylinder represents an abstraction of the lipid bilayer.
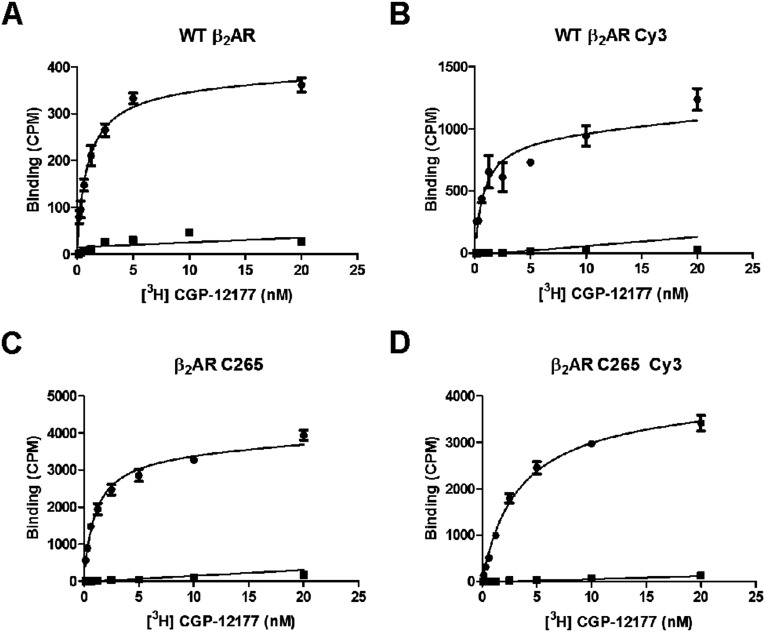
Fig. S1.
Saturation binding of radioligand [3H]CGP-12177 to various β2AR constructs reconstituted in nanodiscs. (A) WT β2AR. (B) WT β2AR labeled with Cy3. (C) β2AR C265 construct. (D) β2AR C265 construct labeled with Cy3. In all cases, the square symbols represent nonspecific binding of [3H]CGP-12177 to nanodisc samples in the presence of a competitve reference ligand (10 µM alprenolol), and the circular symbols represent total binding of [3H]CGP-12177 to nanodisc samples in the absence of reference ligand. The generated curves were used to determine the affinity of [3H]CGP-12177 for the above receptor constructs (corresponding Kd values are shown in Table S1).
Table S1.
Ligand binding to β2AR constructs reconstituted in nanodiscs
| Sample | Saturation binding with [3H]CGP-12177 (Kd,* nM) | Competition binding with Form (Ki, † nM) | Competition binding with ICI (Ki, † nM ) |
| WT | 1.0 ± 0.1 | 5.6 ± 0.8 | 8.9 ± 1.0 |
| Unlabeled | |||
| WT | 0.8 ± 0.2 | 6.1 ± 0.9 | 8.1 ± 1.0 |
| Cy3-labeled | |||
| C265 construct | 1.0 ± 0.1 | 1.6 ± 0.2 | 6.6 ± 1.1 |
| Unlabeled | |||
| C265 construct | 3.2 ± 0.3 | 2.0 ± 0.5 | 9.6 ± 1.8 |
| Cy3-labeled |
Dissociation constants (Kd) were determined using a saturation binding assay. The data are shown as the mean ± SEs, which were determined through three independent experiments.
Inhibition constants (Ki) were determined from radioligand competition binding assays in the presence of [3H]CGP-12177 (1 nM concentration). IC50 was determined from the competition binding and converted to Ki by the following equation: Ki = IC50/(1 + [CGP-12177]/Kd), where Kd is the dissociation constant for CGP-12177. The data are shown as the mean ± SEs, which were determined through three or more independent experiments.
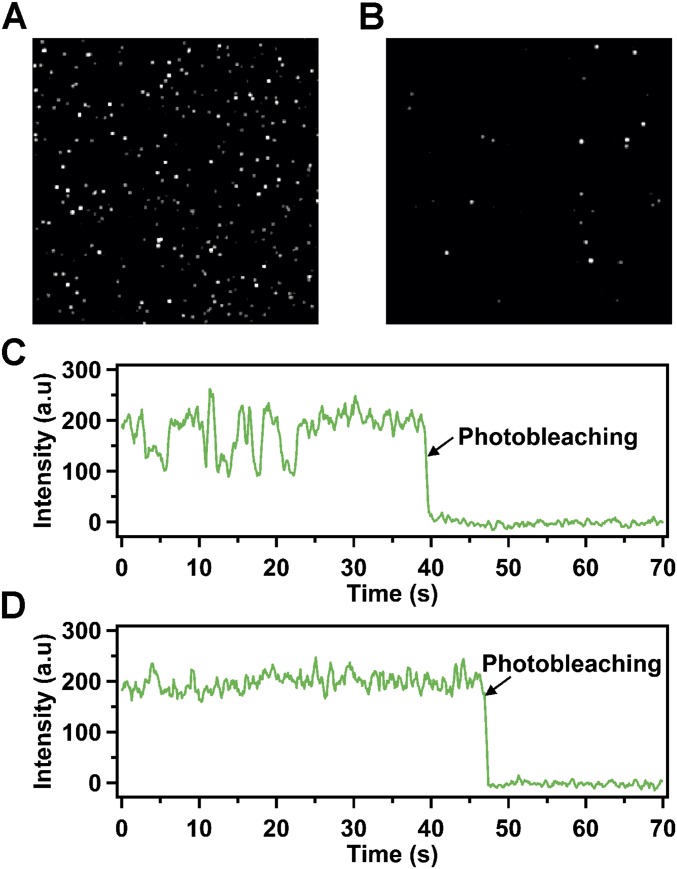
Fig. S2.
TIRF images of immobilized receptor–nanodisc complexes and photobleaching of single receptor–nanodisc complexes. (A) Typical TIRF image showing a 5- × 5-μm region (approximate dimensions) of a streptavidin-coated coverslip decorated with biotin-labeled nanodiscs containing β2AR labeled with Cy3 at Cys265 in helix VI. Each spot is due to the fluorescence emitted from an individual receptor–nanodisc complex. The very bright spots are due to a small fraction of receptor–nanodisc aggregates, which were excluded from the data analysis. (B) Corresponding TIRF image for a control surface lacking streptavidin. The number of fluorescent spots is greatly reduced relative to A, indicating a negligible level of nonspecific adsorption of receptor–nanodisc complexes. (C) Representative fluorescence intensity trajectory from a single nanodisc-bound receptor, showing repeated intensity jumps before an irreversible photobleaching event. Raw intensity values are shown (arbitrary units, not normalized). The single-step photobleaching transition indicates that a single receptor molecule is contained within the nanodisc. (D) Representative fluorescence intensity trajectory for a static receptor molecule. Other details as in C.
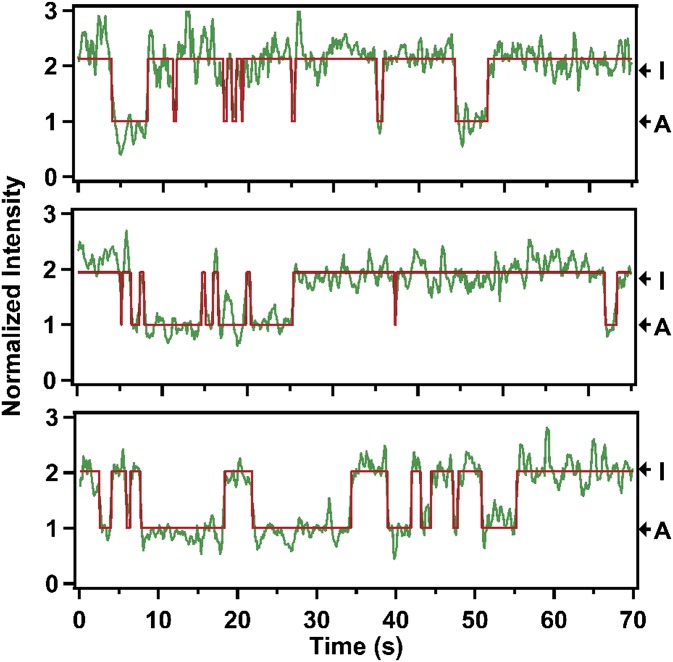
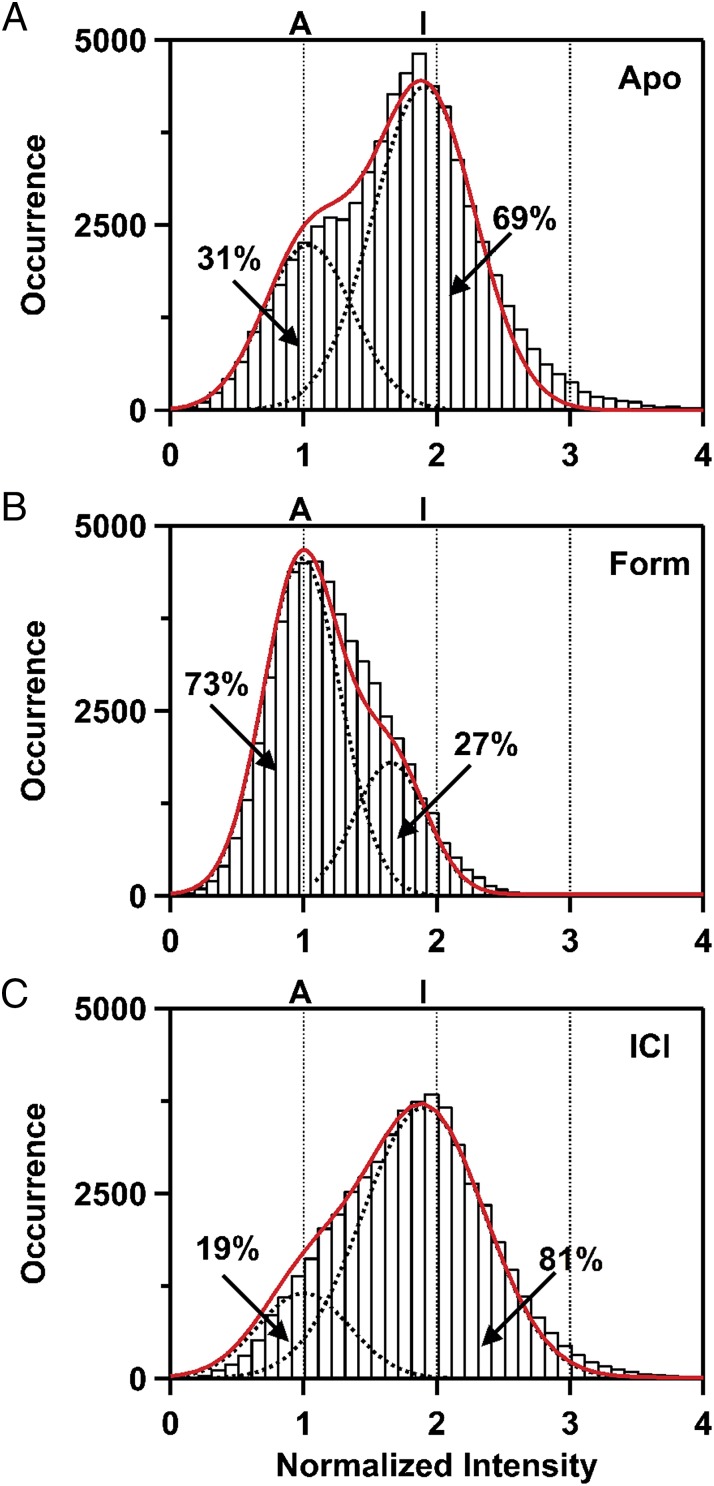
Fluorescence time trajectories from a collection of individual apo β2AR molecules were recorded simultaneously. Most trajectories reveal abrupt transitions between two distinct intensity states (three examples are shown in Fig. 2), whereas ∼30% of the trajectories exhibit relatively constant fluorescence intensity (Fig. S2D and Table S2). These static trajectories may represent a population of inactive receptors, consistent with a previous report that β2AR reconstituted in nanodiscs retained ∼60% of the starting activity (21). The static receptor population was excluded from further analysis. The state of higher fluorescence intensity in the fluctuating trajectories likely arises from protein-induced fluorescence enhancement (PIFE) of Cy3 (22, 23). Accordingly, for each single molecule, we normalized the fluorescence intensities at each time point to the mean intensity of the lower intensity state (Fig. 2). A histogram compiled from 94 individual apo receptors reveals two distinct peaks, with relative areas of 31 and 69% (Fig. 3A). The occurrence of two distinct peaks is analogous to fluorine NMR spectra recorded with a 19F label attached to Cys265 of β2AR, and the relative peak areas are also similar (8). As we explain below, the high-intensity state can be assigned to an inactive conformation of helix VI (state I), whereas the low-intensity state likely represents an active-like conformation (state A).
Fig. 2.
Representative fluorescence intensity trajectories of Cy3 in three individual β2AR molecules in the apo form recorded under identical conditions. The trajectories are shown in green and the red lines are best fits obtained from hidden Markov modeling. The intensity values are normalized to the mean intensity of the lower-intensity state (defined as 1). The intensity levels corresponding to states I and A are indicated (see text for details).
Table S2.
Percentages of fluctuating and static intensity trajectories
Fluctuating trajectories require two fluorescence intensity states for best fit during hidden Markov analysis.
Static trajectories are adequately fitted with a single fluorescence intensity state.
Fig. 3.
Fluorescence intensity histograms compiled from a collection of individual β2AR molecules. Fluorescence intensity is normalized to the mean intensity of the low-intensity state. (A) Histogram compiled from 94 receptor molecules in the apo form. The dotted black lines show fits to two Gaussian functions, corresponding to states I and A, and the composite fit is shown by the solid red line. The percentage populations of each state, calculated from the areas enclosed by the respective peaks, are indicated. (B) Histogram compiled from 89 receptor molecules in the presence of the full agonist Form. Other details as in A. (C) Histogram compiled from 101 receptor molecules in the presence of the inverse agonist ICI. Other details as in A.
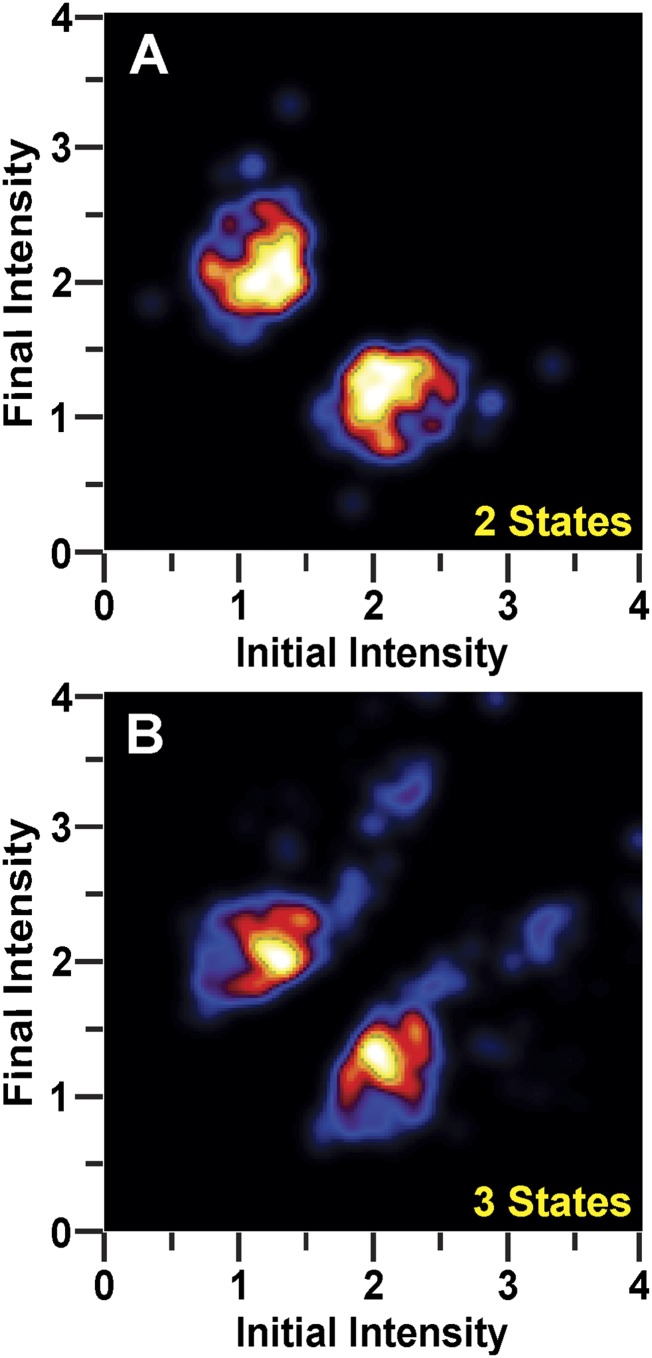
We next fitted the set of trajectories using hidden Markov modeling, allowing for two intensity states, and constructed a 2D plot of transition probability density (24). The prominent cross-peaks reflect frequent transitions between states I and A (Fig. S3A). To determine whether additional states were present, the trajectories were refitted, allowing for three intensity states, and the transition probability density plot was recalculated. The resulting plot revealed the same two major cross-peaks and no additional prominent features (Fig. S3B), indicating that the majority of intensity jumps observed across all datasets are adequately represented by transitions between two states with distinct fluorescence intensities (states I and A).
Fig. S3.
Transition probability density plots presented as heat maps, with regions of high probability shown in lighter colors and regions of low probability shown in darker colors. Emission intensity values are normalized to the mean intensity of the low-intensity state. (A) Constructed from results of hidden Markov analysis allowing for two fluorescence intensity states. (B) Constructed from results of hidden Markov analysis allowing for three fluorescence intensity states.
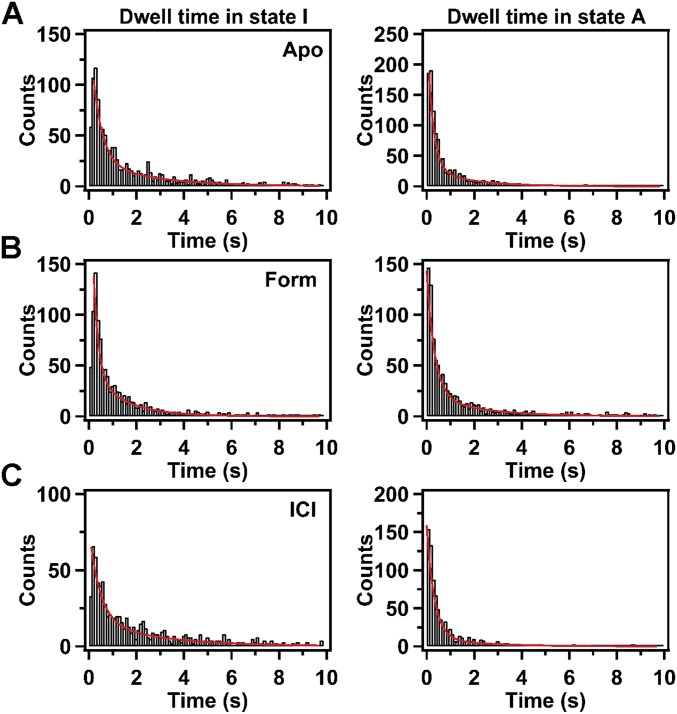
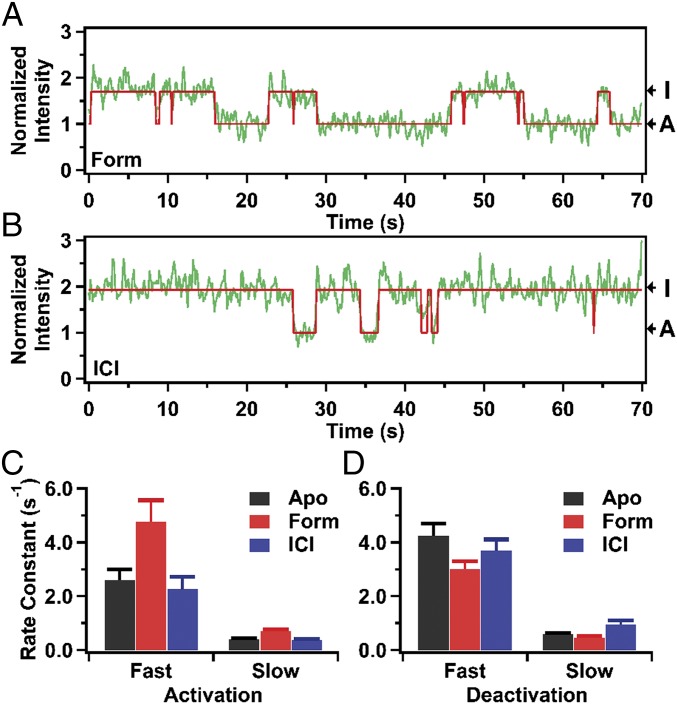
Individual receptor molecules exhibited a range of dwell times in one state before transition to the other state (three examples are shown in Fig. 2). We compiled histograms of the dwell times in each state from the set of 94 receptor molecules (Fig. S4A). For a stochastic system composed of two exchanging states, the dwell-time distributions of either state should be described by a single exponential function. However, the dwell-time histograms of state I or state A do not fit well to a single exponential function and instead require a biexponential function for best fit (reduced χ2 values for monoexponential and biexponential fits are given in Table S3). The majority of transitions from state I to state A (84 ± 23%) occur with high frequency, described by a rate constant of 2.6 ± 0.4 s−1, whereas the remaining transitions are less frequent, with a rate constant of 0.40 ± 0.04 s−1. Similarly, the majority of transitions from state A to state I (89 ± 18%) are described by a rate constant of 4.2 ± 0.4 s−1, whereas the remaining transitions are described by a rate constant of 0.58 ± 0.05 s−1. It is unlikely that the second kinetic phase is due to a population of misfolded or otherwise defective receptors, because short dwell times (corresponding to the rapid phase) and very long dwell times (corresponding to the slow phase) are observed within the same molecule (Fig. 2). Rather, it seems that state I consists of two kinetically distinct substates that share the same fluorescence intensity. The same argument applies to state A.
Fig. S4.
Dwell-time histograms. (A) Dwell-time histograms compiled from 94 individual receptor molecules in the apo form. Solid red lines are the best fits to a double exponential function. The corresponding rate constants, amplitudes, and reduced χ2 values are presented in Table S3. (B) Dwell-time histograms compiled from 89 individual receptor molecules in the presence of the full agonist Form. Other details as in A. (C) Dwell-time histograms compiled from 101 individual receptor molecules in the presence of the inverse agonist ICI. Other details as in A.
Table S3.
Kinetic parameters
| Ligand | Transition | Fitting function* | k1, s−1 | k2, s−1 | A1, † % | A2, † % | Reduced χ2 |
| None | I to A | Monoexponential | 0.70 ± 0.03 | 100 | 1.98 | ||
| I to A | Biexponential | 2.6 ± 0.4 | 0.40 ± 0.04 | 84 ± 23 | 16 ± 5 | 1.07 | |
| A to I | Monoexponential | 1.47 ± 0.06 | 100 | 2.27 | |||
| A to I | Biexponential | 4.2 ± 0.5 | 0.58 ± 0.05 | 89 ± 18 | 11 ± 3 | 0.84 | |
| Form | I to A | Monoexponential | 1.09 ± 0.05 | 100 | 1.71 | ||
| I to A | Biexponential | 4.8 ± 0.8 | 0.71 ± 0.05 | 72 ± 16 | 28 ± 7 | 1.01 | |
| A to I | Monoexponential | 1.22 ± 0.05 | 100 | 1.98 | |||
| A to I | Biexponential | 3.0 ± 0.3 | 0.47 ± 0.05 | 85 ± 14 | 15 ± 4 | 0.88 | |
| ICI | I to A | Monoexponential | 0.60 ± 0.03 | 100 | 1.60 | ||
| I to A | Biexponential | 2.3 ± 0.5 | 0.37 ± 0.04 | 74 ± 19 | 26 ± 8 | 1.06 | |
| A to I | Monoexponential | 2.0 ± 0.1 | 100 | 1.50 | |||
| A to I | Biexponential | 3.7 ± 0.4 | 0.94 ± 0.15 | 84 ± 28 | 16 ± 14 | 0.93 |
Dwell-time histograms were fitted with a mono- or biexponential function, , where N is the number of counts at dwell time t, fi is the amplitude of component i, ki is the rate constant for component i, and i = 1 or 2.
Percentage amplitudes Ai were calculated from the fitted amplitudes fi, according to the formula .
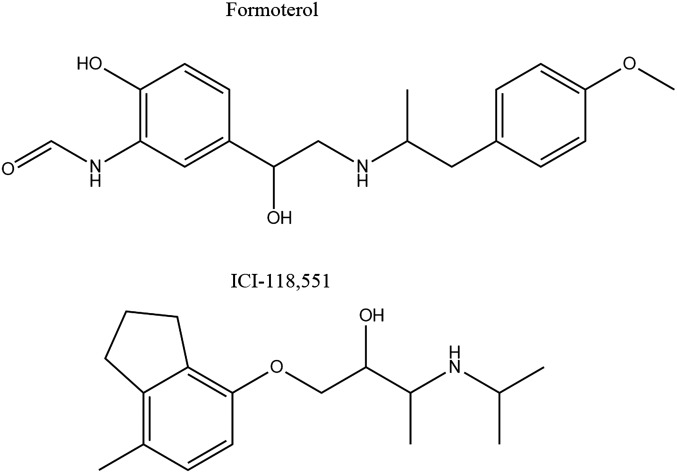
We then examined the effects of two prototypical β2AR ligands on the receptor’s behavior, the full agonist formoterol (Form) and the inverse agonist ICI-118,551 (ICI), whose chemical structures are shown in Fig. S5. Radioligand competition binding measurements showed that both ligands bind tightly to the receptor in nanodiscs (Fig. S6), with Ki values that are similar to the affinities reported for these ligands binding to β2AR on membranes (20, 25), and that the Cy3 label does not perturb binding of either ligand (Table S1). Single-molecule fluorescence measurements performed in the presence of saturating concentrations (1 mM) of either ligand show that Cy3 fluctuates between the same two major intensity states that were observed in the apo form (Fig. 4 A and B), indicating that the ligands do not induce new conformations at the probe location on helix VI. However, relative to the apo receptor, binding of Form shifted the equilibrium distribution toward state A (Fig. 3B), whereas binding of ICI shifted the distribution toward state I (Fig. 3C).
Fig. S5.
Chemical structures of Formoterol and ICI-118,551.
Fig. S6.
Competition binding results. Competition binding curves were generated as described in the main text to determine Ki values of ICI (black symbols) and Form (red symbols) in various receptor constructs reconstituted in nanodiscs: (A) WT β2AR, (B) WT β2AR labeled with Cy3, (C) β2AR C265 construct, and (D) β2AR C265 construct labeled with Cy3. The corresponding Ki values for each ligand and receptor construct are listed in Table S1. The ligand affinity is similar for all conditions.
Fig. 4.
Single-molecule data for receptor–ligand complexes. (A) Representative fluorescence trajectory for an individual β2AR molecule in the presence of the full agonist Form. The trajectory is in green and the red line is a best fit from hidden Markov modeling. The intensity levels corresponding to states I and A are indicated. Intensity values are normalized to the mean intensity of the lower intensity state, as in Fig. 2. (B) Representative fluorescence trajectory for an individual β2AR molecule in the presence of the inverse agonist ICI. Other details as in A. (C) Rate constants describing the activation transition (state I to state A). The two rate constants obtained from biexponential fits to dwell-time histograms are shown (see Table S3 for full details). The error bars represent the uncertainties in the fitted parameters. (D) Rate constants describing the deactivation transition (state A to state I). Other details as in C.
To gain further insights into the population shifts induced by these ligands, we analyzed the kinetics of conformational exchange between states I and A, in the presence of either Form or ICI. Dwell-time histograms of states I and A display biexponential kinetics in the presence of these ligands (Fig. S4 B and C), as before, and the amplitudes of each phase are also similar to those observed in the apo form (Table S3). However, relative to the apo receptor, transitions from state I to state A occur more frequently when Form is bound (Fig. 4C) and deactivation transitions occur less frequently (Fig. 4D). The major effect of ICI binding is to increase the frequency of deactivation transitions, evident in the slow kinetic phase (Fig. 4D).
Discussion
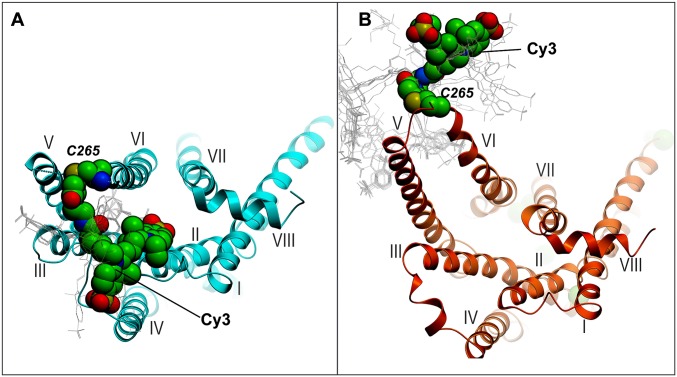
Cy3 exhibits an enhanced fluorescence quantum yield when it is located in a protein environment, a phenomenon referred to as PIFE (22, 23). Recently, it was shown that PIFE is specifically due to a reduction in excited-state cis–trans isomerization of Cy3, which normally competes with fluorescence emission (26). Hence, the emission intensity of Cy3 is determined by the degree of steric restriction imposed on the fluorophore by the local environment: Highly restricted environments that inhibit cis–trans isomerization will produce relatively bright emission, whereas unrestricted environments that permit isomerization will lead to relatively weak emission. We constructed a model of the Cy3-β2AR conjugate based on the crystal structure of β2AR in an inactive conformation (bound to inverse agonist; ref. 4) and determined the most likely location of the Cy3 moiety after 106 rounds of Monte Carlo energy minimization. The resulting model shows that Cy3 lies in a channel surrounded by helices III, IV, and V (Fig. 5A), an environment that is expected to restrict cis–trans isomerization. Accordingly, the state of higher fluorescence intensity (state I) likely corresponds to an inactive conformation of helix VI. In contrast, a model of the Cy3-β2AR conjugate based on the crystal structure of β2AR in an active conformation (bound to agonist and G protein; ref. 5) shows that the Cy3 moiety is fully solvent-exposed and free to undergo facile cis–trans isomerization (Fig. 5B). Hence, the low-intensity state (state A) likely corresponds to an active or active-like conformation of helix VI. The fluorescence intensity of Cy3 in state I is approximately twofold higher than in state A, reflecting a significant PIFE effect (22). It should be noted that the assignments of the states I and A are fully consistent with the observed effects of the β2AR ligands. The full agonist Form shifts the conformational distribution in favor of state A, whereas the inverse agonist ICI shifts the balance toward state I (Fig. 3). Indeed, Form and ICI are known to favor active and inactive functional states of β2AR, respectively (20, 27).
Fig. 5.
Models of Cy3-β2AR conjugates, looking into the transmembrane helical bundle from the cytoplasmic side. (A) β2AR-Cy3 conjugate modeled in the inactive-state conformation based on PDB ID code 2RH1 (with the α-helical part of helix VI extended by one turn). Cy3 is surrounded by helices III, IV, and V, which are expected to inhibit cis–trans isomerization of the fluorophore and thereby increase the fluorescence quantum yield. (B) β2AR-Cy3 conjugate modeled in the active-state receptor conformation as in PDB ID code 4SN6. Cy3 is located in an unrestricted solvent-exposed environment and is expected to exhibit relatively low fluorescence intensity due to facile cis–trans isomerization. In both cases, receptor models are shown as a cartoon, the predicted lowest-energy position of the Cy3 label is shown in CPK presentation, and the alternative conformations within 4 kJ/mol from the lowest energy are shown as thin gray lines. The conformational sampling of the conjugated Cy3 label and the receptor residues of the intracellular region were performed in internal coordinates by a Monte Carlo minimization procedure implemented in ICM-Pro (Molsoft LLC).
The active-like receptor conformation was the major species (73%) populated in the presence of the full agonist Form, although a significant population of the inactive conformation remained (Fig. 3B). These observations are consistent with previous studies showing that binding of a full agonist to β2AR does not produce a homogeneous population of active-like receptor conformations (7, 8, 11). Notably, our results are in close agreement with a previous fluorine NMR study using a 19F label attached to Cys265 of β2AR, which showed that ∼60% of receptors occupied an active-like conformation in the presence of Form (8).
The kinetic information obtained from dwell-time analysis of the single-molecule trajectories is consistent with previous estimates of the conformational exchange rates or activation times of β2AR and also reveals previously unidentified kinetic properties of β2AR. Fluorine NMR saturation transfer experiments have established an upper limit of 10 s−1 for the overall rate of exchange between inactive and active-like conformations of β2AR (10), which is consistent with our estimates of the rate constants for transitions between states I and A in the apo receptor (based on the data in Table S3, the total exchange rates are 6.8 ± 0.9 s−1 and 0.98 ± 0.09 s−1 for the fast and slow kinetic phases, respectively). Another study based on a 19F-NMR probe attached to Cys265 reported a lifetime of 660 ms for an agonist-bound state of β2AR (11), which is consistent with our estimate of the rate constant (rapid component) for the deactivation transition of helix VI in the presence of Form (Fig. 4D). A fluorescence-based (FRET) study using a β2AR construct fused to CFP and YFP reporter proteins revealed activation times of 48 ms or 128 ms in intact cells perfused with epinephrine or norepinephrine agonists, respectively (2). For comparison, we used the rate constants of the fast kinetic phase (Table S3) to calculate the time required to establish an equilibrium distribution of states I and A after addition of the full agonist Form to the apo receptor. The resulting value of 128 ± 18 ms is similar to the activation times measured in intact cells, confirming that the nanodiscs used in our experiments provide a native-like environment for the receptor. In addition to the rapid conformational transitions, which are in accord with previous studies, our single-molecule observations also reveal that helix VI spends prolonged periods in either the inactive or active-like conformation. These long dwell times are readily evident in the fluorescence trajectories of individual receptors (Figs. 2 and 4) and give rise to a long tail in the dwell-time histograms compiled from a population of receptors (Fig. S4), necessitating the use of a biexponential function for best fit (Table S3). The long periods in either conformation have not been observed before. We speculate that as helix VI initially adopts an active-like conformation, additional interactions are formed within the receptor core, without any change in fluorophore emission intensity, further stabilizing this conformation and giving rise to the long dwell times in the low-intensity state (state A). Similarly, we speculate that the long dwell times in the high-intensity state (state I) are due to the formation of additional stabilizing interactions while helix VI is in the inactive conformation.
Our results clearly show that β2AR in the apo form (i.e., in the absence of either agonist ligands or G proteins) can spontaneously transition between inactive and active-like conformations. In fact, the active-like conformation of helix VI is significantly populated under these conditions (Fig. 3A). In the cellular context, where G proteins are present, our results imply that the receptor can access the active conformation and initiate a signaling response in the absence of extracellular ligand binding. In fact, β2AR exhibits a significant level of ligand-independent (basal) signaling activity (27). In addition, the results of our study also provide insights into the mechanisms by which ligands regulate the signaling activity of β2AR. The full agonist Form increases the frequency of activation transitions and also reduces the frequency of deactivation transitions (Fig. 4 C and D), both of which are expected to enhance signaling activity. In contrast, the inverse agonist ICI increases the frequency of deactivation transitions (Fig. 4D), explaining why this ligand reduces signaling activity. Overall, our results show that agonist and inverse agonist ligands regulate the signaling activity of β2AR by fine-tuning the kinetics of transitions between two intrinsic receptor conformations.
In a related study, individual detergent-solubilized β2AR molecules labeled with tetramethylrhodamine at Cy265 were observed in solution while confined in an electrokinetic trap (17). A broad distribution of fluorescence intensity states with dwell times in the millisecond time range were observed. In contrast, we have observed slower transitions between two discrete intensity states and presented strong evidence that these correspond to inactive and active-like conformations of β2AR. Our ability to detect spontaneous transitions between two intrinsic receptor conformations is likely due to the unique photophysical properties of Cy3, the native-like environment of the phospholipid nanodiscs, and the relatively long time scale of our observations. Recently, single-molecule fluorescence spectroscopy was used to visualize conformational transitions in the extracellular ligand-binding domain of the metabotropic glutamate receptor, either as an isolated protein dimer (28) or as part of the full-length receptor (29). In contrast, our observations of β2AR in nanodiscs reveal the dynamics of the membrane-embedded region of a GPCR.
Single-molecule fluorescence (SMF) spectroscopy in nanodiscs complements and significantly extends the insights available from other biophysical approaches. Similar to 19F NMR spectroscopy (8), SMF analysis is able to resolve inactive and active-like receptor conformations and to quantify their relative populations in the presence or absence of drug ligands. However, SMF is the only technique capable of directly visualizing spontaneous transitions between these receptor conformations in real time. Moreover, whereas 19F NMR saturation transfer experiments have established an upper limit for the overall rate of exchange between inactive and active-like conformations of β2AR (10), dwell-time analysis of the two fluorescence intensity states observed by SMF can determine rate constants for the individual conformational transitions. The resulting kinetic information provides previously unidentified insights, showing that the full agonist Form affects both the activation and deactivations steps, whereas the inverse agonist ICI primarily affects deactivation (Fig. 4 C and D). Understanding the specific effects of drug ligands on the activation/deactivation processes in the molecular detail described here can be expected to provide new opportunities for improved design of GPCR-targeting drugs. Moreover, we can now use these advances in SMF spectroscopy of GPCRs in nanodiscs to explore how interactions with G proteins and/or addition of specific membrane components influence receptor activation.
Materials and Methods
Expression and Purification of Cy3-Labeled β2AR.
The human β2AR construct used in this study has been described previously (8). The construct contains the thermostabilizing E122W mutation (25), is truncated at residue 348, and a portion of intracellular loop 3 (ICL3) not required for G protein binding (residues 245–249) is removed. For simplicity, this construct is referred to here as WT. It has been shown that only three cysteine residues within this construct (Cys265, Cys327, and Cys341) are available for labeling with sulfhydryl-reactive reagents (8). The construct used for site-specific fluorophore labeling, referred to as the C265 construct, also contains C327S and C341A mutations. All receptor constructs were expressed in Sf9 cells and extracted as described (8), before incubating with cobalt-based immobilized metal affinity chromatography beads (Talon) overnight. The receptor-loaded Talon beads were washed extensively with wash buffer 1 [50 mM Hepes (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 1 mM n-dodecyl-β-d-maltoside (DDM), 0.2 mM cholesteryl hemisuccinate (CHEMS), 20 mM imidazole, and 8 mM ATP] and wash buffer 2 [50 mM Hepes (pH 7.5), 150 mM NaCl, 1 mM DDM, 0.2 mM CHS, and 20 mM imidazole) and then exchanged into 10 mL labeling buffer [50 mM Hepes (pH 7.5), 150 mM NaCl, 1 mM DDM, and 0.2 mM CHS]. A 10-fold molar excess of Cy3 maleimide (GE Healthcare) (20 μL of a 5 mg⋅mL−1 solution in DMSO) was then added and the reaction mixture incubated in the dark for 1 h. After Cy3 labeling, β2AR was further purified using the standard protocol (8).
Nanodisc Preparation.
Biobeads were added to a mixture of labeled β2AR, membrane scaffold protein 1 (MSP1), and phospholipids (1:10:700) in cholate buffer, following the reconstitution procedure described previously (30). The phospholipid mixture contained POPC, POPS, and biotinyl CAP PE (67.5:27.5:5%) (Avanti Polar Lipids). The receptor, MSP1, and lipid mixture was incubated overnight at 4 °C, after which the biobeads were removed and the nanodisc–receptor complex was purified by size-exclusion chromatography. The reconstituted receptor–nanodisc complexes were further separated from empty nanodiscs by purifying His-tag-containing complexes using Talon columns.
Radioligand Binding Assays.
Radioligand binding assays were carried out as described in the Assay Protocol Book (version II) of the National Institute of Mental Health Psychoactive Drug Screening Program (available at https://pdspdb.unc.edu/pdspWeb/content/PDSP%20Protocols%20II%202013-03-28.pdf). Briefly, the Kd value of radioligand [3H]CGP-12177 was determined immediately after nanodisc reconstitution for each sample by a saturation binding assay. This was carried out in a 96-well plate at a final volume of 125 μL per well. Twenty-five microliters of radioligand was added to each well (ranging from 0.16–20 nM), followed by the addition of either 25 μL binding buffer (total binding) or 25 μL of 10 μM alprenolol (to assess nonspecific binding). Receptor–nanodisc complexes were added and incubated for 1 h before vacuum filtration onto cold 0.3% polyethyleneimine-soaked glass fiber filter mats. Wax scintillation mixture was melted on the filter mat and radioactivity was counted in a Microbeta2 counter (Perkin-Elmer). Competition binding measurements were performed to determine the Ki values of the full agonist Form and the inverse agonist ICI 118,551. Nanodisc samples were incubated with 25 μL of 1 nM of [3H]CGP-12177 and 25 μL of 0–10 μM test ligand. All conditions were done in triplicate at least three times. Kd and Ki values were calculated using Prism GraphPad software according to established data analysis protocols (https://pdspdb.unc.edu/pdspWeb/content/PDSP%20Protocols%20II%202013-03-28.pdf).
Single-Molecule Fluorescence Measurements.
Single-molecule data collection was performed using an inverted Axiovert 200 microscope (Zeiss) modified for prism-based TIRF imaging (TIRF Labs Inc.) under oxygen-scavenging conditions, as described (31, 32). Quartz slides were cleaned, passivated with polyethylene glycol, and coated with streptavidin, as described (33). Biotin conjugated receptor–nanodisc complexes in imaging buffer [50 mM Hepes (pH 7.5), 150 mM NaCl, and 2 mM trolox] were introduced into the sample chamber and allowed to bind to the streptavidin-coated surface, after which unbound nanodiscs were washed away and the imaging buffer was enriched with glucose oxidase and catalase as an oxygen scavenging system was introduced. Immobilized nanodisc–receptor complexes were excited using a green laser (532 nm) and the Cy3 emission intensity was recorded over time on an intensified CCD camera (Andor Technology) with a 100-ms frame time. Binary complexes were formed by incubating a saturating concentration (1 mM) of a desired ligand with the receptor on the slide surface before excitation and data recording. All measurements were performed at 298 K. A custom-written single-molecule data acquisition package was used in combination with IDL software (ITT VIS, version 8.1) to record CCD camera data and generate fluorescence intensity time traces.
Single-Molecule Data Analysis.
Trajectories displaying reversible intensity fluctuations before a single-step photobleaching event were selected for analysis. Each selected emission intensity trajectory was corrected for background, smoothed by three-point sliding averaging, and truncated before photobleaching. Binned fluorescence intensity histograms were compiled from multiple trajectories and fitted with two Gaussian functions using Igor Pro software (version 6; Wavemetrics). Individual trajectories were fitted with a hidden Markov model using the program HaMMy (24). All trajectories were fitted adequately with two distinct intensity states. The intensity levels observed before and after each transition were assigned to state I or state A, based on a chosen threshold value. The dwell times spent in each state before transition to the other state were compiled in the form of histograms, using data from multiple individual receptor molecules. The resulting histograms were fitted with both single and double exponential functions to determine the rate constant(s) for the corresponding conformational transition and the associated uncertainty in the rate constant(s) using Igor Pro software. The quality of the resulting fits was judged from the reduced χ2 values. The data points were weighted according to their variance (square root of the observed value).
Modeling of the Fluorescent Dye-Receptor Conjugates.
Energy-based conformational modeling of Cy3-β2AR conjugates was performed using the ICM molecular modeling suite (Molsoft, LLC). Modeling of the inactive state was based on the crystal structure of the β2AR complex with inverse agonist carazolol (PDB ID code 2RH1). After removal of the T4 lysozyme fusion in intracellular loop 3, the conformation of Cys265 was modeled by extension of the α-helical conformation in helix VI for residues Lys263–Lys267. The α-helical conformation at these residue positions is supported by previous observations of this conformation in other β2AR structures in both active and inactive states (PDB ID codes 3SN6 and 4GBR), as well as in several structures of the closely related β1AR receptor (e.g., PDB ID code 4AMJ). The active-state model was based on the crystal structure of β2AR complex with agonist BI-167107 and G protein heterotrimer (PDB ID code 3SN6). The Cy3 dye molecule was built and conjugated to the corresponding cysteine thiol moiety in both models, with its covalent geometry optimized using the MMFF force field. The conformation of the conjugated dye was thoroughly sampled using more than 106 steps of a Monte Carlo minimization procedure in internal coordinates, and assuming flexibility of all side chains at the intracellular interface. For both inactive and active states, the lowest energy conformation of the Cy3 dye as well as all nonredundant conformations within 4 kJ/mol from the lowest energy conformation were collected (Fig. 5).
Acknowledgments
We thank Katya Kadyshevskaya for assistance in preparing the figures and Angela Walker for her comments on the manuscript. We also thank the Sligar laboratory at the University of Illinois at Urbana–Champaign for generously sharing their protocols for nanodisc reconstitution of membrane proteins. This research was supported by NIH Road Map Initiative Grant P50 GM073197.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1519626112/-/DCSupplemental.
References
- 1.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1(9):727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 2.Reiner S, Ambrosio M, Hoffmann C, Lohse MJ. Differential signaling of the endogenous agonists at the β2-adrenergic receptor. J Biol Chem. 2010;285(46):36188–36198. doi: 10.1074/jbc.M110.175604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milligan G. Constitutive activity and inverse agonists of G protein-coupled receptors: A current perspective. Mol Pharmacol. 2003;64(6):1271–1276. doi: 10.1124/mol.64.6.1271. [DOI] [PubMed] [Google Scholar]
- 4.Cherezov V, et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318(5854):1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasmussen SG, et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477(7366):549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kofuku Y, et al. Efficacy of the β₂-adrenergic receptor is determined by conformational equilibrium in the transmembrane region. Nat Commun. 2012;3(1045):1045. doi: 10.1038/ncomms2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nygaard R, et al. The dynamic process of β(2)-adrenergic receptor activation. Cell. 2013;152(3):532–542. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu JJ, Horst R, Katritch V, Stevens RC, Wüthrich K. Biased signaling pathways in β2-adrenergic receptor characterized by 19F-NMR. Science. 2012;335(6072):1106–1110. doi: 10.1126/science.1215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim TH, et al. The role of ligands on the equilibria between functional states of a G protein-coupled receptor. J Am Chem Soc. 2013;135(25):9465–9474. doi: 10.1021/ja404305k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horst R, Liu JJ, Stevens RC, Wüthrich K. β2-adrenergic receptor activation by agonists studied with 19F NMR spectroscopy. Angew Chem Int Ed Engl. 2013;52(41):10762–10765. doi: 10.1002/anie.201305286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manglik A, et al. Structural insights into the dynamic process of β2-adrenergic receptor signaling. Cell. 2015;161(5):1101–1111. doi: 10.1016/j.cell.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghanouni P, Steenhuis JJ, Farrens DL, Kobilka BK. Agonist-induced conformational changes in the G-protein-coupling domain of the β 2 adrenergic receptor. Proc Natl Acad Sci USA. 2001;98(11):5997–6002. doi: 10.1073/pnas.101126198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghanouni P, et al. Functionally different agonists induce distinct conformations in the G protein coupling domain of the β 2 adrenergic receptor. J Biol Chem. 2001;276(27):24433–24436. doi: 10.1074/jbc.C100162200. [DOI] [PubMed] [Google Scholar]
- 14.Yao XJ, et al. The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc Natl Acad Sci USA. 2009;106(23):9501–9506. doi: 10.1073/pnas.0811437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peleg G, Ghanouni P, Kobilka BK, Zare RN. Single-molecule spectroscopy of the β2 adrenergic receptor: Observation of conformational substates in a membrane protein. Proc Natl Acad Sci USA. 2001;98(15):8469–8474. doi: 10.1073/pnas.151239698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whorton MR, et al. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc Natl Acad Sci USA. 2007;104(18):7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bockenhauer S, Fürstenberg A, Yao XJ, Kobilka BK, Moerner WE. Conformational dynamics of single G protein-coupled receptors in solution. J Phys Chem B. 2011;115(45):13328–13338. doi: 10.1021/jp204843r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dror RO, et al. Activation mechanism of the β2-adrenergic receptor. Proc Natl Acad Sci USA. 2011;108(46):18684–18689. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bayburt TH, Sligar SG. Membrane protein assembly into nanodiscs. FEBS Lett. 2010;584(9):1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker JG. The selectivity of β-adrenoceptor agonists at human β1-, β2- and β3-adrenoceptors. Br J Pharmacol. 2010;160(5):1048–1061. doi: 10.1111/j.1476-5381.2010.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leitz AJ, Bayburt TH, Barnakov AN, Springer BA, Sligar SG. Functional reconstitution of β2-adrenergic receptors utilizing self-assembling nanodisc technology. Biotechniques. 2006;40(5):601–602, 604, 606 passim. doi: 10.2144/000112169. [DOI] [PubMed] [Google Scholar]
- 22.Hwang H, Kim H, Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc Natl Acad Sci USA. 2011;108(18):7414–7418. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang H, Myong S. Protein induced fluorescence enhancement (PIFE) for probing protein-nucleic acid interactions. Chem Soc Rev. 2014;43(4):1221–1229. doi: 10.1039/c3cs60201j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91(5):1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roth CB, Hanson MA, Stevens RC. Stabilization of the human β2-adrenergic receptor TM4-TM3-TM5 helix interface by mutagenesis of Glu122(3.41), a critical residue in GPCR structure. J Mol Biol. 2008;376(5):1305–1319. doi: 10.1016/j.jmb.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stennett EM, Ciuba MA, Lin S, Levitus M. Demistifying PIFE: The photophysics behind the protein-induced fluorescence enhancement of Cy3. J Phys Chem Lett. 2015;6(10):1819–1823. doi: 10.1021/acs.jpclett.5b00613. [DOI] [PubMed] [Google Scholar]
- 27.Bond RA, et al. Physiological effects of inverse agonists in transgenic mice with myocardial overexpression of the β 2-adrenoceptor. Nature. 1995;374(6519):272–276. doi: 10.1038/374272a0. [DOI] [PubMed] [Google Scholar]
- 28.Olofsson L, et al. Fine tuning of sub-millisecond conformational dynamics controls metabotropic glutamate receptors agonist efficacy. Nat Commun. 2014;5:5206. doi: 10.1038/ncomms6206. [DOI] [PubMed] [Google Scholar]
- 29.Vafabakhsh R, Levitz J, Isacoff EY. Conformational dynamics of a class C G-protein-coupled receptor. Nature. 2015;524(7566):497–501. doi: 10.1038/nature14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ritchie TK, et al. Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berezhna SY, Gill JP, Lamichhane R, Millar DP. Single-molecule Förster resonance energy transfer reveals an innate fidelity checkpoint in DNA polymerase I. J Am Chem Soc. 2012;134(27):11261–11268. doi: 10.1021/ja3038273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamichhane R, Berezhna SY, Gill JP, Van der Schans E, Millar DP. Dynamics of site switching in DNA polymerase. J Am Chem Soc. 2013;135(12):4735–4742. doi: 10.1021/ja311641b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamichhane R, Solem A, Black W, Rueda D. Single-molecule FRET of protein-nucleic acid and protein-protein complexes: Surface passivation and immobilization. Methods. 2010;52(2):192–200. doi: 10.1016/j.ymeth.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]