Significance
Anemia is the predominant clinical manifestation of myelodysplastic syndromes (MDS). Genes that are aberrantly expressed and/or mutated that lead to the dysplastic erythroid morphology seen in −7/del(7q) MDS have not been identified. In this study, we show that reduced expression of dedicator of cytokinesis 4 (DOCK4) causes dysplasia by disrupting the actin cytoskeleton in developing red blood cells. In addition, our identification of the molecular pathway that leads to morphological defects in this type of MDS provides potential therapeutic targets downstream of DOCK4 that can be exploited to reverse the dysplasia in the erythroid lineage. Furthermore, we developed a novel single-cell multispectral flow cytometry assay for evaluation of disrupted F-actin filaments, which can be used for potential early detection of dysplastic cells in MDS.
Keywords: erythroid, MDS, DOCK4, signaling, actin
Abstract
Anemia is the predominant clinical manifestation of myelodysplastic syndromes (MDS). Loss or deletion of chromosome 7 is commonly seen in MDS and leads to a poor prognosis. However, the identity of functionally relevant, dysplasia-causing, genes on 7q remains unclear. Dedicator of cytokinesis 4 (DOCK4) is a GTPase exchange factor, and its gene maps to the commonly deleted 7q region. We demonstrate that DOCK4 is underexpressed in MDS bone marrow samples and that the reduced expression is associated with decreased overall survival in patients. We show that depletion of DOCK4 levels leads to erythroid cells with dysplastic morphology both in vivo and in vitro. We established a novel single-cell assay to quantify disrupted F-actin filament network in erythroblasts and demonstrate that reduced expression of DOCK4 leads to disruption of the actin filaments, resulting in erythroid dysplasia that phenocopies the red blood cell (RBC) defects seen in samples from MDS patients. Reexpression of DOCK4 in −7q MDS patient erythroblasts resulted in significant erythropoietic improvements. Mechanisms underlying F-actin disruption revealed that DOCK4 knockdown reduces ras-related C3 botulinum toxin substrate 1 (RAC1) GTPase activation, leading to increased phosphorylation of the actin-stabilizing protein ADDUCIN in MDS samples. These data identify DOCK4 as a putative 7q gene whose reduced expression can lead to erythroid dysplasia.
Myelodysplastic syndromes (MDS) are a group of clonal hematopoietic disorders that are characterized by cytopenias caused by ineffective hematopoiesis (1–3). Even though MDS may transform to acute leukemia in one-third of patients who have MDS, cytopenias drive morbidity for most patients (4). Most of the morbidity experienced by these patients is due to low red blood cell (RBC) counts; therefore, studies on the molecular pathogenesis of dysplastic erythropoiesis are critically needed. Cytogenetic studies have shown that stem and progenitor cells in MDS contain deletions in chromosomes 5, 7, 20, and others (5). Deletions of the chromosomal 7q region are seen in 10% of cases and are associated with significantly worse survival (6). These genomic deletions are usually large, and even though some candidate pathogenic genes have been postulated (7), it is not clear which of the deleted genes contribute to the pathogenesis of ineffective erythropoiesis and dysplasia in MDS.
In a previous study (8), we had observed that numerous 7q genes, including dedicator of cytokinesis 4 (DOCK4), were deleted or epigenetically silenced in MDS, thus prompting an examination of its role in erythropoiesis in the present study. DOCK4 belongs to a large family of proteins (CED5/DOCK180/MYOBLAST CITY class) that is well conserved in mammals (9–11). They are large proteins (>200 kb) that act as signaling intermediates and provide docking sites for many other signaling molecules. One of the well-described functions of DOCK4 is its ability to activate GTPases, such as ras-related C3 botulinum toxin substrate 1 (RAC1) and RAP1, in many cell types (12–14). Mutations in the DOCK4 gene have been identified in both prostate and ovarian cancers, and recent studies have demonstrated that DOCK4 can act as a tumor suppressor (12, 15). In the present study, we determined the functional role of DOCK4 depletion in RBC formation by using a zebrafish model (16) and an in vitro model of human erythropoiesis that recapitulates the erythroid differentiation program. The in vitro model we have developed uses human CD34+ stem/progenitor blood cells in which these cells are induced to commit to the erythroid lineage and then progressively differentiate into reticulocytes over a 2-wk period (17–19). The model takes advantage of erythropoietin (EPO) and stem cell factor (SCF), the two key cytokines responsible for driving erythropoiesis to sustain cell viability, proliferation, and terminal differentiation in an ordered fashion (20–22). Using this in vitro model and an established zebrafish model, we demonstrate a critical role of DOCK4 in maintaining the integrity of the erythrocyte cytoskeleton and implicate it as an important pathogenic gene in MDS. Furthermore, we established a novel single-cell–based assay to quantify the extent of actin filament disruptions in dysplastic erythroblasts from MDS patients.
Results
DOCK4 Expression Is Reduced in MDS Bone Marrow and Is Associated with Adverse Prognosis.
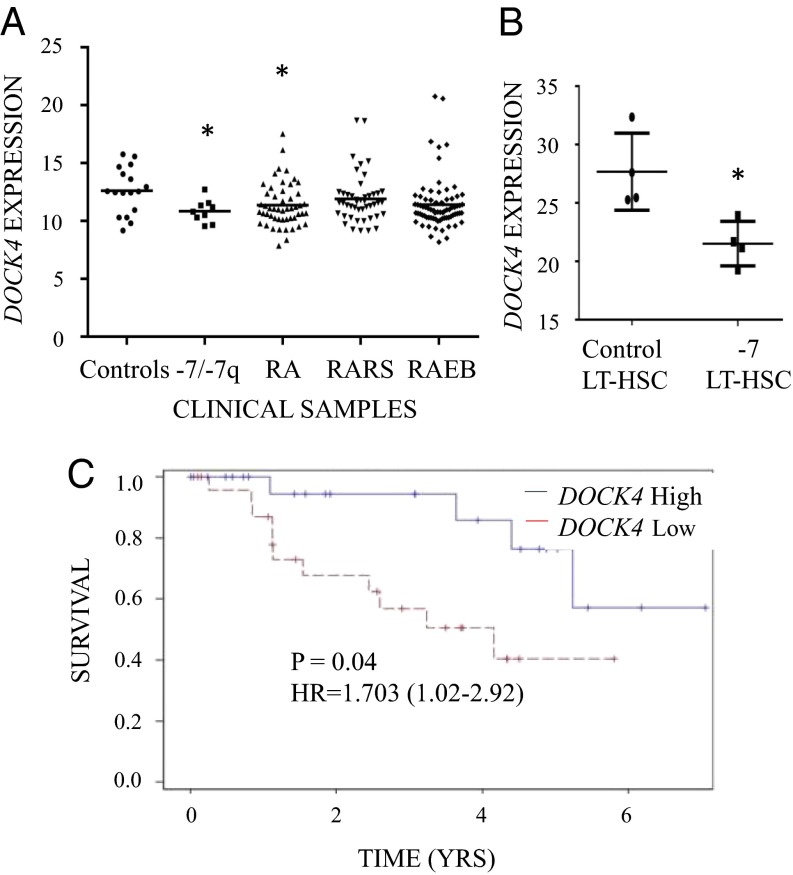
We examined expression levels for DOCK4 in a large gene expression dataset obtained from bone marrow CD34+ cells isolated from 183 MDS patients (23). Analysis of DOCK4 expression in the various subtypes of MDS found that DOCK4 expression is significantly reduced in MDS CD34+ samples that had deletion of chromosome 7/7q or belonged to the refractory anemia (RA) subtype compared with healthy controls (Fig. 1A). Determination of DOCK4 expression in an transcriptomic study (5) from an independent set of purified primitive hematopoietic stem cells (HSCs; CD34+, CD90+, Lin−, CD38−) also revealed significantly reduced levels in MDS/acute myeloid leukemia (AML) samples with deletion of chromosome 7 (Fig. 1B). Furthermore, the patients with low expression of DOCK4 within the RA subgroup of MDS had a significantly worse prognosis with a hazard ratio (HR) of 3.744 (range: 1.1–12.2) on univariate analysis (log rank P = 0.02). On multivariate analysis after adjusting for clinically relevant prognostic factors [International Prognostic Scoring System (IPSS)] (6), reduced expression of DOCK4 was also determined to be an independent adverse prognostic factor [HR = 1.703 (range: 1.02–2.91), P = 0.045] (Fig. 1C). Because RA patients present predominantly with anemia, this observation prompted us to examine the precise role played by DOCK4 in the pathogenesis of reduced erythropoiesis in MDS.
Fig. 1.
Reduced expression of DOCK4 in MDS correlates with anemia phenotype in patients. (A) Expression of DOCK4 is significantly reduced in −7/del(7q) MDS (n = 9) and in the RA (n = 55) subtype of MDS. Gene expression profiles were generated from CD34+ cells from MDS patients and controls (n = 17). RA with excess blasts (RAEB; n = 74) and RA with ringed sideroblasts (RARS; n = 48) are other subtypes of MDS (*P < 0.05, Student’s t test). (B) Comparison of gene expression data from sorted acute myeloid leukemia (AML)/MDS bone marrow samples (n = 4) with −7 with healthy controls (n = 4) revealed significantly decreased DOCK4 expression in long-term HSCs (LT-HSC) (*P = 0.01). (C) In the RA patients, lower expression of DOCK4 is significantly associated with worse overall survival [log rank P = 0.045, HR = 3.744 (1.1–12.2)]. Low expression < median expression, high expression > median expression.
In Vivo Suppression of DOCK4 in Zebrafish Embryos Generates Dysplastic Erythroid Cells.
To determine the impact of DOCK4 knockdown in vivo, we depleted Dock4 protein during zebrafish embryogenesis. Bioinformatic analysis indicated the presence of two duplicated orthologs of human DOCK4 in the zebrafish genome. We used in situ hybridization to compare the expression patterns of the two zebrafish dock4 genes. We found that dock4a maternal transcripts are present at the one-cell stage and that the gene is widely expressed during gastrulation, decreased in levels during early somatogenesis, but then again widely expressed from 24 to 48 h postfertilization (hpf) (SI Appendix, Fig. S1). Because dock4a appeared to encode the predominant embryonic isoform, we generated both translation-blocking (TB) and splice-blocking (SB) morpholino oligomers (MOs) to target the depletion of Dock4a expression. Titration of the SB MO showed that 5 ng was sufficient to deplete most of the normal message at 24 hpf (SI Appendix, Fig. S2). Injection of 5–10 ng of either the TB or SB MO generated embryos with similar morphant phenotypes at 48 hpf, with a predominant pericardial edema and a lack of blood circulation, with blood pooling on the yolk sac (SI Appendix, Fig. S3). Expression of markers of early erythroid transcriptional regulator gata1 appeared normal (SI Appendix, Fig. S4).
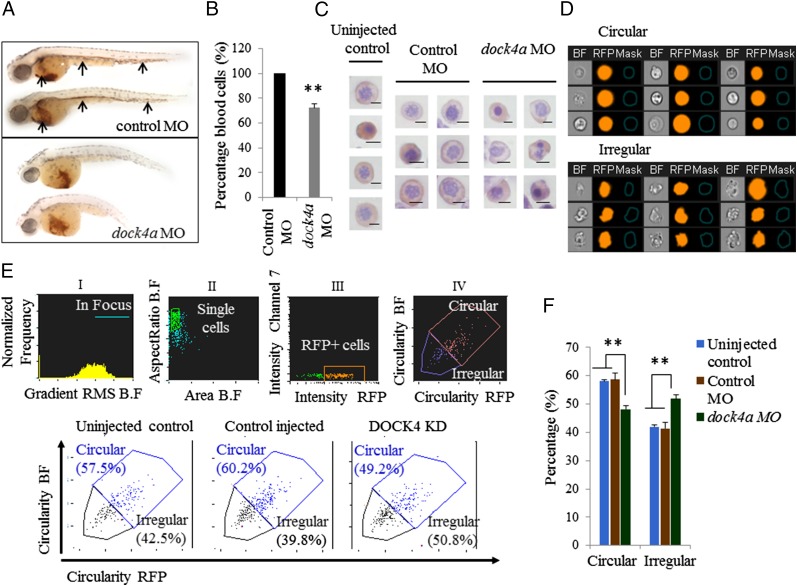
Although erythropoiesis appeared to initiate normally, when dock4a morphants were stained with o-dianisidine (Sigma) to visualize hemoglobin-staining cells, erythroid cells were missing from the circulation and the morphants were relatively depleted of RBCs in the yolk sac sinus compared with control injected embryos (Fig. 2A). To quantify erythropoiesis in the dock4a morphants better, we used a transgenic gata1:dsred reporter strain in which RBCs and erythroblasts express red fluorescent protein (RFP) and can be quantified by flow cytometry. When Dock4a was depleted in this model using either SB or TB MOs, the resulting embryos had ∼28% fewer erythroblasts/erythrocytes compared with the controls (Fig. 2B). We next focused on examining the morphology of erythroid cells in zebrafish embryos that were injected with either dock4a or control MOs by two approaches. First, we examined the overall morphology of benzidine- and hematoxylin-positive cells to identify erythroid cells on cytospun slides. We observed that erythroid cells from zebrafish embryos injected with dock4 MOs showed a large number of irregular shapes, whereas uninjected embryos or those embryos that received control MOs contained mostly cells that were circular in shape (Fig. 2C). We then developed a novel assay to distinguish cells with irregular shape from cells that were circular and to quantify the percentages of each population using multispectral flow cytometry (ImageStreamX; EMD-Millipore/Amnis). In developing this assay, we first examined Gata1+ erythroid cells (RFP+) from zebrafish embryos injected with dock4 MOs to capture cells with a circular shape and an irregular shape to identify the two different cell morphologies of interest (Fig. 2D). Applying these data to the ImageStreamX algorithm for circularity, we were able to differentiate between circular- and irregular-shaped cells (described in detail in Materials and Methods) to segregate the RFP+ erythroid cell compartment systematically based on the circularity feature (Fig. 2D). We used both bright-field (BF) and fluorescent channels, providing two independent measures of the circularity in our cell populations. The data from such an analysis showed increased numbers of irregular-shaped cells in dock4a morphant samples (Fig. 2E). We performed four independent experiments using zebrafish embryos depleted for Dock4a, along with three control replicates, and found that dock4a knockdown resulted in increased numbers of dysplastic erythroid cells in zebrafish embryos (Fig. 2F).
Fig. 2.
In vivo suppression of dock4a in a zebrafish model leads to dysplastic erythroid cells. (A) Shown are representative embryos following staining at 48 hpf with o-dianisidine to detect hemoglobin (red stain). (Top) Embryos were injected with 5 ng of standard control MO. Arrows indicate blood circulation through the caudal hematopoietic tissue, cardinal vein, and heart, with substantial staining in the sinus venosus on the yolk sac. (Lower) Embryos were injected with 5 ng of the dock4a SB MO, and at 48 hpf, they lack blood circulation and appear relatively pale with less blood in the yolk sac (n = 30 embryos for both panels). (B) Relative percentages of erythroid cells in zebrafish embryos were determined by flow cytometry comparing dock4a knockdown with control samples (with the later set to 100%). Use of gata1:dsred transgenic zebrafish embryos allowed direct assessment of erythroid cells in samples of dissociated whole embryos’ blood (n = 4 independent experiments, minimum of 200 embryos per experiment). (C) Photomicrographs of benzidine- and hematoxylin-positive erythroid cells from cytospins prepared using cells from dissociated zebrafish embryos. (Scale bars, 5 μm.) (D) BF and fluorescence images of RFP-expressing erythroid cells depicting circular- and irregular-shaped morphology harvested from embryos with reduced expression of Dock4a. (E) Quantitation of RFP-positive embryonic zebrafish blood cells that are circular or irregular in shape based on analysis of BF and RFP images using the ImageStreamX instrument. KD, knockdown. (F) Percentages of circular- and irregular-shaped erythroid cells harvested from dissociated zebrafish embryos 48 hpi of dock4a SB MO or control MO or from uninjected embryos (**P < 0.005, Student’s t test).
DOCK4 Knockdown in Primary Human Erythroblasts Results in Disruption of the F-Actin Network.
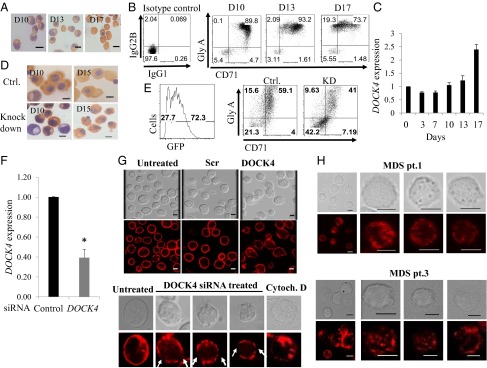
We next used an established in vitro human culture system to determine the mechanism by which DOCK4 depletion causes the generation of dysplastic erythrocytes that are observed in MDS patient samples and in our in vivo zebrafish knockdown studies. The in vitro human primary cell model uses cultured human CD34+ stem/early progenitors to promote lineage commitment and terminal differentiation into reticulocytes. The culture conditions are regulated by a strict cytokine feeding regimen at precise time intervals, allowing the cells to go through every major stage in the erythroid differentiation program and yielding large numbers of hemoglobinized reticulocytes by day 17 (Fig. 3A). These cells acquire Glycophorin A (eBioscience) and CD71 (BD Biosciences), two key surface markers that are highly up-regulated as the cells reach the reticulocyte stage (Fig. 3B). Using this dynamic model, we determined that DOCK4 is expressed throughout the differentiation program and gradual up-regulation occurs as the cells become terminally differentiated (Fig. 3C).
Fig. 3.
DOCK4 knockdown in primary human erythroblasts disrupts F-actin skeleton. (A) Photomicrographs depicting differentiating human primary erythroblasts on various days stained with benzidine and hematoxylin. D10, day 10 polychromatic erythroblasts; D13, day 13 orthochromatic erythroblasts; D17, day 17 reticulocytes. (B) Flow cytometry analysis of differentiating human primary erythroid progenitors for transferrin receptor (CD71) and Glycophorin A, showing gradual acquisition of surface markers that signify the erythroid program. (C) Quantitative RT-PCR (qRT-PCR) showing relative expression levels of DOCK4 transcripts during prelineage commitment (day 0) and postcommitted terminal differentiation into reticulocytes (P < 0.05, Student’s t test; n = 3). (D) Photomicrographs of differentiating human erythroblasts on various days stained with benzidine and hematoxylin following DOCK4 knockdown with shRNA against DOCK4 depicting multinucleated cells. Ctrl., control. (E) Flow cytometry analysis depicting the efficiency of DOCK4 knockdown (KD) (Left) and impeded differentiation as measured by the levels of glycophorin A (GlyA)/CD71 levels (Right). (F) qRT-PCR showing the extent of DOCK4 knockdown by siRNA on day 8 erythroblasts (*P < 0.05; Student’s t test). (G, Top) Immunofluorescence microscopy analysis showing disrupted actin filaments on day 8 erythroblasts after DOCK4 knockdown. Photomicrographs also depict cells that were treated with scrambled (Scr) or left untreated, with siRNAs along with corresponding differential interference contrast (DIC) images. (G, Bottom) Individual cells at high magnification showing gaps in the F-actin skeleton in a sample treated with DOCK4-specific siRNA and a sample treated with the actin-disrupting agent cytochalasin D (Cytoch. D) as a positive control. A cell from an untreated sample was used as the negative control. (Scale bars, 5 μm.) (H) DIC and immunofluorescence micrographs of day 10 (orthochromatic erythroblasts) derived from MDS patients showing disrupted F-actin skeleton. (Scale bars, 5 μm.)
To determine the functional significance of DOCK4 during erythroid differentiation, we knocked down its expression using high-titer lentiviral particles (with GFP reporter) carrying shRNA against DOCK4 and examined the cell morphology and levels of Glycophorin A and CD71. These experiments revealed that knockdown of DOCK4 resulted in a large number of late-stage erythroblasts (day 15) with multinucleate morphology compared with the control mock-transduced cells (Fig. 3D). Furthermore, we performed flow cytometry analysis on day 10 of culture, which showed reduced levels of Glycophorin A/CD71 expression in DOCK4-depleted cells compared with mock-transduced cells, suggesting that DOCK4 is important in promoting terminal differentiation of erythroblasts (Fig. 3E). Previous work had identified DOCK4 as a signaling intermediate capable of modulating RAC GTPase activity (12, 14). Because RAC-1 GTPases are known to regulate the actin cytoskeleton in erythroid cells (24), we reasoned that lack of DOCK4 expression might affect the F-actin skeleton in differentiating erythroblasts. Indeed, we found that suppression of DOCK4 resulted in disruption of F-actin skeleton as determined by immunofluorescence microscopy using Texas Red-conjugated phalloidin for detection (Fig. 3 F and G). DOCK4 siRNA-treated cells clearly exhibited gaps in the F-actin skeleton similar to when these cells were treated with cytochalasin D, an actin depolymerizing agent (Fig. 3G, Bottom). We examined erythroblasts derived from CD34+ cells from two MDS patients with −7/del(7q) lacking one allele of DOCK4, and found a phenotype similar to when we knocked down DOCK4 in control cells, where the F-actin skeleton was disrupted at numerous locations within the actin skeleton (Fig. 3H).
MDS Erythroblasts Exhibit Disrupted F-Actin Skeleton.
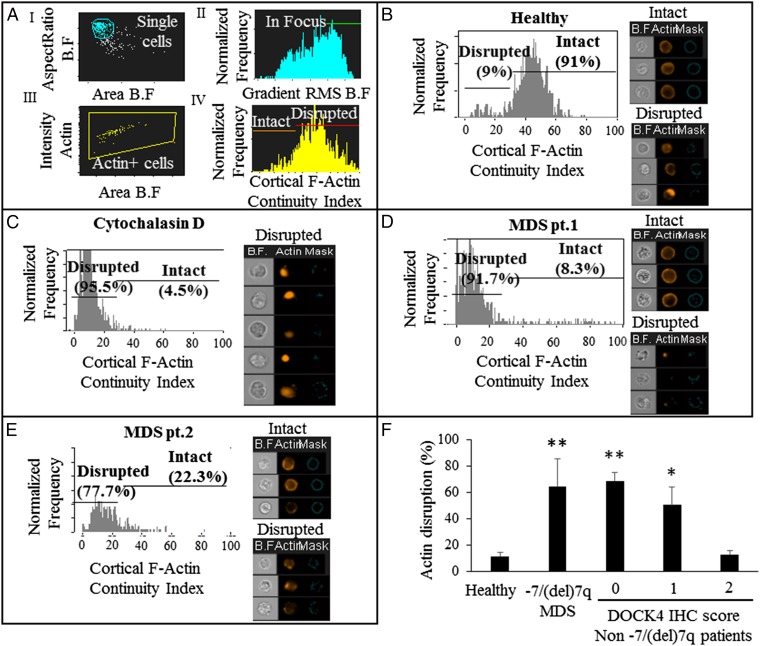
Although immunofluorescence microscopy enabled us to assess F-actin disruption, this approach did not provide us the means with which to assess the level of disruption objectively in a quantitative fashion, especially when using a large number of patient samples. To this end, we developed a novel assay capable of objectively quantifying the extent of F-actin disruption using multispectral flow cytometry (ImageStreamX). To quantify the extent of F-actin disruption in erythroblasts from −7/del(7q) MDS patients lacking DOCK4 expression, we purified CD34+ cells from bone marrow and/or blood samples obtained from 10 patients that included multiple subtypes of MDS (RA with excess blasts and refractory cytopenias with multilineage dysplasia). CD34+ cells were cultured for 10 d, and erythroblasts (polychromatic stage) were used to quantify the extent of F-actin disruption in these MDS samples. The multispectral flow cytometry assay we developed entailed first staining the cells for F-actin with fluorescently labeled phalloidin and using the ImageStreamX instrument to capture single cells in focus that were positive for actin (Fig. 4 A, i–iii). Using algorithms available on IDEAS 6.0 software (Amnis, Inc.), we defined the cortical F-actin continuity index, taking into account the fluorescence staining of actin filaments (Fig. 4 A, iv) (details are provided in Materials and Methods). Using this assay, we quantified the extent of intact and disrupted F-actin skeleton in erythroblasts from healthy individuals, as well as MDS patient samples (Fig. 4 B–E). As positive controls, we used samples from healthy donors treated with cytochalasin D or a RAC1 inhibitor (NSC23766) (Fig. 4C and SI Appendix, Fig. S5I). Both positive controls and −7/del(7q) MDS patient samples exhibited a clear pattern of major gaps in actin staining or complete absence of cortical actin staining, as seen in the image gallery for each sample, which was quantified by the histograms as depicted (Fig. 4 and SI Appendix, Fig. S5 A–H) On the other hand, erythroblasts from healthy individuals or a non-MDS anemia patient with chronic kidney disease (anemia) did not exhibit disrupted actin filaments (Fig. 4 and SI Appendix, Fig. S5J). Our earlier work had shown that DOCK4 can also be epigenetically silenced by aberrant DNA methylation in MDS (8). We examined MDS bone marrow samples without deletion of chromosome 7 and determined that reduced expression of DOCK4 can be seen in some samples and that this result correlates with actin filament disruption seen in erythroid cells generated from these samples (SI Appendix, Fig. S6). Taken together, these analyses reveal that MDS erythroblasts with reduced expression of DOCK4 had disrupted cortical actin filaments compared with those MDS erythroblasts from healthy individuals (Fig. 4F and SI Appendix, Fig. S6).
Fig. 4.
Quantitation of F-actin disruption in major MDS subtypes by multispectral flow cytometry (ImageStreamX). Day 10 erythroblasts (orthochromatic stage) from human primary cultured cells were labeled using fluorescently tagged phalloidin and used for the analysis. (A) Strategy used to develop the cortical F-actin continuity index for analysis and quantitation of actin filament disruption. (B) Histogram and image gallery of erythroblasts from a healthy individual showing percentages of intact and disrupted F-actin. (C) Histogram and image gallery of erythroblasts from a healthy individual after exposure to cytochalasin D used as a positive control. (D) Histogram and image gallery of erythroblasts from a patient with MDS [refractory cytopenias with multilineage dysplasia (RCMD)] showing percentages of intact and disrupted F-actin. (E) Histogram and image gallery of erythroblasts from a patient with MDS (RA) showing percentages of intact and disrupted F-actin. (F) Extent of actin disruption as measured by the cortical F-actin continuity index quantified from healthy controls (n = 5), patients with MDS (n = 10) with reduced or absence of DOCK4 expression, and patients without the −7/7q deletion with a DOCK4 immunohistochemistry (IHC) score of 0 (n = 4), 1 (n = 2), or 2 (n = 2) (**P < 0.00005, *P < 0.005; Student’s t test).
Reduced Expression of DOCK4 Leads to Reduction in RAC1 GTPase Activity.
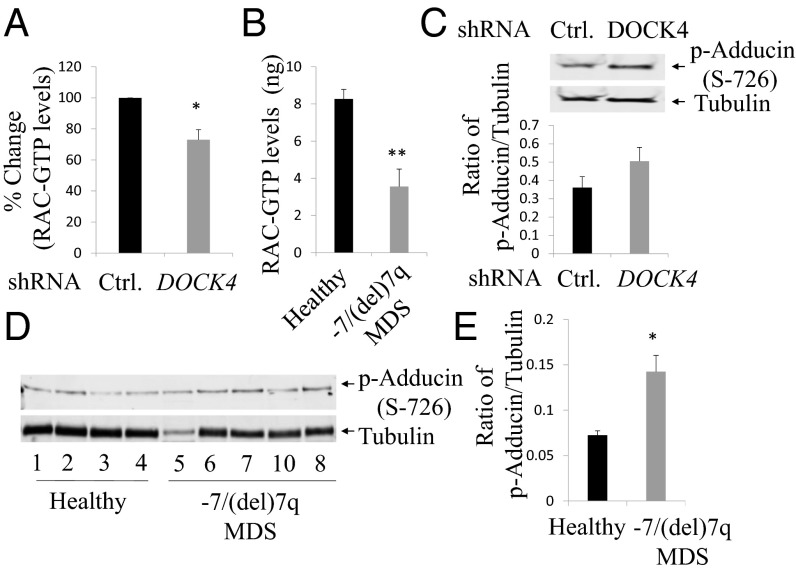
DOCK4 is a known guanine nucleotide exchange factor for the RAC GTPases (12). RAC1 and RAC2 are critical for maintaining proper actin filament length, which is pivotal for maintaining membrane stability, strength, and deformability of erythrocytes. Moreover, because actin is involved in the extrusion of nuclei during enucleation of late-stage erythroblasts, inhibition of RAC GTPase activity leads to reduction in reticulocyte formation (24, 25). Therefore, we investigated whether knockdown of DOCK4 in our ex vivo culture model reduced RAC GTPase activity. We infected human CD34+ early hematopoietic cells with high-titer lentiviral particles carrying shRNA against DOCK4 and selected with puromycin until day 6 of culture to select for cells (SI Appendix, Fig. S7A) that had suppressed DOCK4 expression (SI Appendix, Fig. S7B). We determined RAC GTPase activity (ELISA) in these cells and compared our findings against the activity in cells that were infected with scrambled control shRNAs. These experiments revealed that RAC GTPase activity was reduced when DOCK4 was suppressed, suggesting that actin disruption observed in DOCK4-deficient cells is mediated, at least in part, by RAC GTPases (Fig. 5A). We then examined levels of RAC GTPase in mature RBCs obtained from blood samples of −7/del(7q) MDS patients who were known to have deletion of one allele of DOCK4. We observed significantly lower RAC GTPase levels in MDS samples compared with RBCs from healthy volunteers (Fig. 5B).
Fig. 5.
DOCK4 suppression leads to reduction in RAC1 GTPase activity and increased ADDUCIN phosphorylation. (A) Levels of RAC1 GTPase activity were determined by ELISA in day 6 primary erythroblasts after suppression of DOCK4 expression. Percentage reduction of RAC1 GTPase activity is shown compared with samples infected with control shRNAs. The data are the mean of five biological replicates. (B) Levels of RAC1 GTPase activity in mature RBC ghosts prepared from blood samples of healthy donors and MDS patients. The data are the mean of seven biological replicates. (C, Top) Immunoblot analysis showing increased phosphorylation of ADDUCIN in day 6 erythroblasts after suppression of DOCK4 expression. Tubulin was used as a loading control. (C, Bottom) Quantitation of the levels of phospho-ADDUCIN after suppression of DOCK4 expression (n = 3). (D) Immunoblot (all lanes were run on the same gel but were noncontiguous) analysis showing phosphorylation of ADDUCIN in cultured human primary day 10 erythroblasts from healthy donors and MDS patients. Tubulin levels were used as a loading control. (E) Quantitation of the levels of phospho-ADDUCIN in day 10 erythroblasts from healthy donors and MDS patient samples (MDS patient 5 from D was excluded in this analysis). Data are mean values from four biological replicates (**P < 0.005, *P < 0.05; Student’s t test.)
Increased Phosphorylation of ADDUCIN in DOCK4-Deficient Erythroid Cells.
ADDUCIN is an actin-capping protein that is essential for maintaining the proper length of actin filaments (26, 27). Reduced RAC GTPase activity in erythrocytes leads to increased phosphorylation in the MARCKS domain of ADDUCIN (28, 29). In order for actin filaments to be capped at the fast-growing ends, enabling recruitment of SPECTRIN, ADDUCIN must be dephosphorylated at Ser726. These biochemical alterations occur in the RBC membranes in a dynamic fashion, which is critical for maintaining membrane stability and deformability. Therefore, we investigated whether DOCK4 suppression in primary human erythroblasts leads to increased phosphorylation of ADDUCIN. We suppressed DOCK4 expression by introducing DOCK4 shRNA into primary human erythroblasts and determined phospho-ADDUCIN levels, which showed that suppression of DOCK4 expression leads to increased phosphorylation of ADDUCIN (Fig. 5C). We next examined the status of ADDUCIN phosphorylation in erythroblasts from MDS patients and compared the levels with erythroblasts from healthy donors by immunoblot analysis (Fig. 5D). These studies revealed that −7/del(7q) MDS patient-derived erythroblasts had higher levels of ADDUCIN phosphorylation compared with healthy donor cells (Fig. 5E).
Reduced Terminal Differentiation and Abnormal Erythroid Morphology of DOCK4-Deficient Erythroid Cells.
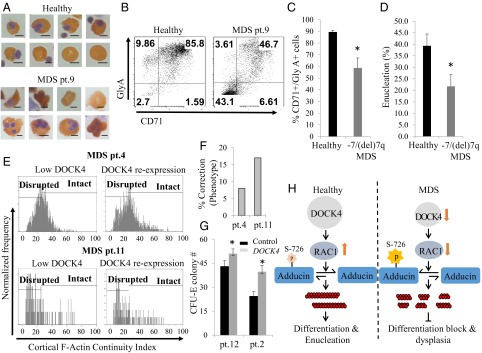
To determine the functional consequence of DOCK4 suppression in the erythroid lineage, we examined cells from healthy donors and MDS patients. Human CD34+ cells purified from blood or bone marrow samples from −7/del(7q) MDS patients were cultured to promote differentiation along the erythroid lineage. These cultures maintained growth of both healthy and −7/del(7q) cells, as confirmed by FISH analysis (SI Appendix, Fig. S8A). We then used cytospun slides of cultured cells from healthy donors and MDS patient samples to examine the morphology of orthochromatic cells (day 14), which showed large numbers of hyperlobulated and/or cells with a frayed cell membrane in −7/del(7q) MDS samples compared with the cells derived from healthy donors (Fig. 6A). Furthermore, analysis for expression of differentiation markers Glycophorin A/CD71 in our control and MDS cultures showed reduced levels of Glycophorin A/CD71 expression in MDS samples compared with cells from healthy donors (Fig. 6B and SI Appendix, Fig. S8 B and C). Examination of the extent of enucleation on each of the 10 MDS patient samples and comparing them with samples from five healthy donors also showed reduced levels of enucleation, further confirming an impact on terminal differentiation (Fig. 6D). We then enumerated the numbers of malignant cells (blast cell count) on FISH slides and correlated them with the size of the erythroid population that was devoid of the late-stage differentiation marker Glycophorin A, which showed that the malignant cells constituted the largest fraction of cells that did not gain positivity for Glycophorin A (SI Appendix, Fig. S9). In Table 1, we have tabulated the clinical features and patient information, along with the levels of F-actin disruption, that summarize our findings. Finally, to determine if reexpression of DOCK4 in −7/del(7q) MDS patient samples can reverse the extent of F-actin disruption, we subcloned the DOCK4 gene in-frame to the minicircle (MC) expression plasmid and transfected CD34+-derived erythroblasts from two −7/del(7q) MDS patients and examined the levels of actin disruption by multispectral flow cytometry. This approach not only allowed us to reexpress DOCK4 in patient samples (SI Appendix, Fig. S10) but also showed an improvement in the levels of intact actin filaments (Fig. 6 E and F). Furthermore, we found that reexpression of DOCK4 resulted in increased numbers of erythroid colonies in MDS patient samples (Fig. 6G). Altogether, our study demonstrated that reduced DOCK4 expression in −7/del(7q) MDS leads to RAC1 functional deficiency, resulting in aberrant ADDUCIN phosphorylation that disrupts proper formation of actin filaments (Fig. 6F).
Fig. 6.
Abnormal differentiation in DOCK4-deficient erythroid cells and reexpression of DOCK4 in DOCK4-deficient MDS patient erythroblasts. (A) Photomicrographs of benzidine- and hematoxylin-stained cells from a healthy donor and a patient (pt.) with MDS showing abnormal erythroid morphology present in erythroblasts derived from MDS CD34+ cells (Scale bars, 5 μm.) (B) Flow cytometry analysis of erythroid progenitors on day 14 of culture to enumerate the percentages of GlyA and CD71 expression as a measure of differentiation. (C) Quantitation of percentage of GlyA/CD71 double-positive cells on day 14 of culture from healthy donors (n = 6) and MDS patients (n = 10). (D) Quantitation of percentages of enucleated cells on day 14 of culture using cells from healthy donors (n = 5) and MDS patients (n = 10) as a measure of terminal differentiation. (E) Multispectral flow cytometry analysis for the levels of actin filament disruption in two MDS patient samples before and after reexpression of DOCK4 in DOCK4-deficient erythroblasts showing longer length actin filaments as reflected by the higher cortical actin staining index. (F) Percentage of correction of actin filament disruption in the two patients shown in E (patient 4, P = 0.04; patient 11, P = 0.007). (G) Increase in erythroid colony formation (CFU-E) after reexpression of DOCK4 in DOCK4-deficient MDS patients (*P < 0.05). (H) Schematic diagram of DOCK4 signaling in normal and MDS hematopoiesis.
Table 1.
MDS patient characteristics and summary of results
| Patient no. | Age, y | MDS subtype | Karyotype | Hemoglobin, g/dL | Clonal cells (FISH), % | Actin disruption, % | GlyA (day 14), % |
| MDS 1 | 60 | RCMD | −7 | 6 | 18.53 | 91.7 | 80.67 |
| MDS2 | 67 | RA | −7 | 8 | 2.96 | 77.7 | 98.7 |
| MDS 3 | 70 | RAEB | −7 | 6.6 | 66.67 | 93 | 2.7 |
| MDS 4 | 83 | RA | −7 | 7 | 12.26 | 50.2 | 90 |
| MDS 5 | 67 | RCMD | −7 | 6.7 | 10.15 | 24.9 | 97.9 |
| MDS 6 | 77 | RCMD | Complex −7 | 11.4 | 10 | 82.14 | 65.6 |
| MDS 7 | 63 | RCMD | Complex −7 | 12.7 | 35 | 52.8 | 55.5 |
| MDS 8 | 75 | RCMD | Complex −7 | 9.8 | 5.5 | 77.7 | 56.7 |
| MDS 9 | 84 | RAEB | Complex −7 | 7.6 | 6 | 50.37 | 59.9 |
| MDS 10 | 74 | RCMD | Complex −7 | 12 | 7.75 | 79.6 | 51 |
RAEB, RA with excess blasts; RCMD, refractory cytopenias with multilineage dysplasia.
Discussion
Anemia and dysplastic erythropoiesis are predominant clinical manifestations of myelodysplasia. Deletion of the chromosomal 7q segment is a common cytogenetic alteration in MDS, and it is associated with a poor prognosis. The commonly deleted segment contains a large number of genes, and studies have not identified the genes whose haploinsufficiency can lead to myelodysplastic phenotypes. Two recent studies identified CUX1 and MLL3 as 7q genes that can act as tumor suppressors in MDS, but these and other genes have not been implicated in erythropoietic defects that are a hallmark of MDS (30, 31). Another recent study using induced pluripotent stem (iPS) cells generated from −7 cases of MDS implicated reduced expression of HIPK2, ATP6V0E2, LUC7L2, and EZH2 in aberrant early hematopoiesis but did not specifically evaluate their role in erythropoiesis (7). We have identified DOCK4 as a candidate 7q pathogenic gene by demonstrating that it is reduced in MDS stem and progenitor cells and that its reduced expression leads to dysplastic erythropoiesis in vivo and in vitro.
Human RBCs are characterized by a biconcave shape that is essential for their functionality. These cells require the coordinated formation of actin cytoskeleton that relies on activation of RAC1 GTPases during erythropoiesis. Our work revealed that low DOCK4 expression in −7/del(7q) MDS RBCs, or in normal erythroblasts depleted of DOCK4, leads to diminished RAC1 GTPase activity, resulting in disruption of actin filaments and hyperphosphorylation of ADDUCIN, an actin protofilament-stabilizing protein. These changes in the erythroid membrane skeleton have a profound impact on the overall integrity of the cell, as seen by an inability to complete the terminal differentiation program and hemolysis of erythroid cells. Previous studies have shown that Rac1/Rac2−/− mice have impairment of skeletal stability and increased phosphorylation of ADDUCIN in the erythroid compartment (32). The significance of increased ADDUCIN phosphorylation at Ser-726 has far-reaching implications because this biochemical modification allows cleavage of ADDUCIN by caspase, yielding a 74-kDa fragment that does not dissociate from the cytoskeleton (33). Furthermore the Ser-726 is in the MARCKS domain, which acts as a substrate for PKA and PKC, therefore potentially providing us an opportunity to intervene and reverse hyperphosphorylation of ADDUCIN in −/del(7q) MDS (28, 29). Current findings provide mechanistic insight into how chromosomal deletion or epigenetic silencing of a signaling intermediate (DOCK4) can have far-reaching implications for cellular morphology and cell membrane stability, leading to dysplasia in the erythroid lineage, with possible therapeutic implications.
Our previous work has shown that DOCK4 can also be transcriptionally regulated by DNA methylation, and aberrant hypermethylation of the promoter can be seen in some cases of MDS (8). In fact, some of the samples examined by us without deletion of chromosome 7 did show reduced expression of DOCK4. The expression of DOCK4 increases during erythroid differentiation; in our studies, we observed a major role for DOCK4 in the postlineage commitment phase. Additionally, because DOCK4 can be epigenetically silenced (8) and numerous epigenetic marks are preserved between HSCs and differentiated cells (34, 35), it is possible that lower levels of DOCK4 in HSCs are a potential predictor of lower levels in differentiated cells in those samples, thus accounting for the higher RBC transfusion requirements in these cases. Future planned studies will evaluate the precise role of DOCK4 in stem cell dynamics. Erythroid cells generated from these samples also showed dysplastic actin filament disruption, further demonstrating the important role played by DOCK4 in myelodysplasia. Pathogenesis of MDS is multifactorial, and studies have shown that cooperative effects of different mutations and deletions can lead to the abnormal stem cell cycling, differentiation, and malignant clone expansions seen in this disease (4, 5). Because deleted segments of 7q contain large numbers of genes, with DOCK4 being one of them, our data suggest that reduction of this gene can contribute to erythroid dysplasia and contribute to the anemia phenotype seen in MDS.
MDS are one of the most common blood malignancies in the elderly and usually present as low blood counts in this population. Dysplasia is challenging to quantify and is often based on a subjective histological assessment. Our demonstration of F-actin network disruption in MDS erythroblasts, together with the development of a new approach to quantify the extent of disruption in the F-actin network (multispectral flow cytometry), raises the possibility that one could use this approach as a quantifiable measure of erythroid dysplasia. In summary, our study provides the first mechanistic basis, to our knowledge, of erythroid dysplasia due to reduced expression of a gene identified on chromosome 7q and demonstrates that disrupted F-actin filaments can be seen at the single-cell level in MDS.
Materials and Methods
Zebrafish dock4a Analysis.
Wild-type strain AB Tubingen (AB/TU hybrid) and transgenic zebrafish were maintained at 28.5 °C and staged as described (36). The Tg(gata1:dsRed) reporter line was used to examine RBC circulation (16). MOs were purchased from Gene Tools. Two distinct MOs were designed to target around the dock4a initiation ATG (5′-GTACCATCCTTCACATTTTT) or across the exon2/intron2 splice site (5′-AATAAAACAGCGATTACCTTCACAT). Blast analysis indicated the MOs are specific for dock4a. All morphants were compared with stage-matched embryos that were injected with a control MO (standard control MO; Gene Tools). Each MO was titrated by injection into one- to four-cell fertilized embryos to determine a minimal dose for the reproducible phenotype. Microinjection of MOs was performed using a PLI-100 Pico-Injector (Harvard Apparatus). Semiquantitative RT-PCR was used to measure the efficacy of the SB MO using the following primers: forward, 5′-TCGAGGAATGGTTCAGCATGGACT; reverse, 5′-TCTCTCCGTGGGACCAAATCCAAA. The same primers were used to generate a PCR fragment that was cloned into pCR-BluntII-TOPO plasmid (Life Technologies) as a clone used for generating the in situ hybridization probe, following digestion with Not1 and RNA synthesis using SP6 polymerase. Whole-mount in situ hybridization was performed as described (37). Embryos were treated with 0.003% phenylthiourea (PTU; Sigma) to block pigmentation. Following fixation in 4% (vol/vol) paraformaldehyde (Sigma), embryos were treated with 10 μg/mL proteinase K (Roche). Hybridization was performed at 68 °C in 57% (vol/vol) formamide buffer (Roche) with digoxigenin-labeled RNA probes (Roche).
Harvesting Cells from Zebrafish Embryos.
For each experiment, ∼200 dock4a morphant Tg(gata1:dsred) or control embryos were dechorionated at 48 hpf and placed into 1.5-mL tubes. Embryos were dissociated by manual agitation with a pellet pestle (Fisher) and trypsinized with prewarmed TrypLE (Life Technologies) at 32 °C for 20–30 min on a rotator. Trypsinized samples were pipetted through a 35-μm cell strainer into a 5-mL tube; trypsin was inhibited by addition of 4 mL of fluorescence-activated cell sorting (FACS) buffer (L-15 medium supplemented with 1% heat-inactivated FBS, 0.8 mM CaCl2, 50 U/mL penicillin, and 0.05 mg/mL streptomycin), followed by addition of FBS to a 7.5% (vol/vol) final concentration. Cells were pelleted at 300 RCF (relative centrifugal force) for 5 min and then washed with FACS buffer. Dissociated embryonic cells were resuspended in 1 mL of FACS buffer. FACS was performed on a Vantage cell sorter (Becton-Dickinson) into PBS. Alternatively, the cells were washed one more time and resuspended in PBS before analyzing with the ImageStreamX instrument.
HSC Culture.
CD34+ HSCs and progenitor cells were purified from G-mobilized peripheral blood of healthy donors purchased from AllCells, Inc. using a CliniMACS (Miltenyi Biotec, Inc.) instrument. Purified cells were cultured in Iscove’s modified Dulbecco’s medium (IMDM; Lonza) containing 15% (vol/vol) FBS (Life Technologies) and 15% (vol/vol) human serum, supplemented with 2 units/mL EPO, 50 ng/mL SCF, and 10 ng/mL IL-3 (R&D Systems). Throughout the entire 17-d culture duration, a strict cytokine feeding regimen was followed as described previously to promote lineage commitment and terminal differentiation (17, 19).
In experiments where CD34+ cells from peripheral blood or bone marrow samples from MDS patients were used, mononuclear cells (MNCs) were obtained by Ficoll-Hypaque separation and CD34+ cells were purified using an EasySep Human CD34 Positive Selection Kit (Stemcell Technologies, Inc.) according to the manufacturer’s protocol. In experiments where cryopreserved patient samples were used, initially thawed MNCs were cultured under short-term expansion conditions [IMDM with 20% (vol/vol) FBS, 20 ng/mL thrombopoietin (TPO), 20 ng/mL Fms-related tyrosine kinase-3 ligand (FLT3-L), 50 ng/mL SCF, and 50 ng/mL IL-6] for 2 d before CD34+ purification using the EasySep kit, and subsequently cultured under erythroid differentiation conditions. All of the MDS patient samples were obtained after obtaining informed consent and approval from the Institutional Review Boards (IRBs) of The University of Chicago and Albert Einstein College of Medicine.
Flow cytometry analysis and cytospun slides of cells were prepared on days 3, 7, 10, 13, and 17 for monitoring of terminal differentiation during the erythroid program. Benzidine and hematoxylin staining of cytospun slides was performed as described previously (38).
siRNA and Lentiviral Transductions.
In experiments where DOCK4 knockdown by siRNA was carried out for immunofluorescence microscopy studies, day 5 cells were plated at a density of 0.5 × 10^6 cells per milliliter of erythroid culture media in a 12-well plate and incubated at 37 °C for 30 min. The transfection mixture solution required per well was prepared in a 1.5-mL microcentrifuge tube by mixing 50 μL of X-vivo 15 (Lonza) with 3 μL of TransIT-siQUEST transfection reagent (Mirus) and 50 nM siRNA (scrambled control and DOCK4 siRNA; Thermo Scientific, Inc.). Each mixture was mixed well and incubated at room temperature for 15 min. The mixture was added to the 12-well plate and mixed gently. After incubating for 48 h, the cells were collected for immunofluorescence microscopy and RNA isolation.
Custom-made high-titer (1 × 109) lentiviral particles expressing shRNAs (control and DOCK4) were purchased commercially (Systems BioSciences, Inc.). The control and DOCK4 shRNA (5′ CTCAGTATTTGCAGATATA 3′) sequences were cloned into a pGreenPuro shRNA expression lentivector (Systems Biosciences, Inc.) with the H1 promoter driving shRNA and the EF1 promoter driving the GFP. Lentiviral transductions were carried out on 24-well plates coated with Retronectin (Takara Bio, Inc.). Day 0 CD34+ cells were thawed and prestimulated in X-vivo 10 base media (Lonza) supplemented with SCF (10 μg/mL), TPO (5 μg/mL), IL-3 (10 μg/mL), and FLT3-L (10 μg/mL) for 48 h (39). On the day of infection, an appropriate amount of viral soup (both control and DOCK4 shRNA) to infect 100,000 cells at a multiplicity of infection of 20 was added to 0.25 mL of complete X-vivo 10 media. This mixture was added to the Retronectin-coated well and incubated for 90 min. The plate with the virus was then centrifuged for 25 min at 4 °C at 800 × g. A total of 100,000 cells were then added to each well, along with polybrene at a concentration of 5 μg/mL, and the final volume was made up to 0.5 mL with X-vivo 10 complete media (Lonza). The virus/cells/polybrene mixture was incubated for 30 min at 37 °C, and the plate was centrifuged for 25 min at 27 °C at 400 × g. The plate was incubated for 20 h at 37 °C. On the following day (day 1), cultures were centrifuged and media containing polybrene and virus were removed before reculture in erythroid culture media. On the following day, puromycin was added at 2 μg/mL to select for transduced cells. On day 6, cells were collected for RAC activity assays, Western blotting, and RNA isolation. In addition on day 10, cytospins of cells followed by benzidine hematoxylin staining and flow cytometry for the detection of CD71 and Glycophorin A surface proteins were performed to monitor terminal differentiation of cells lacking DOCK4.
Immunofluorescence Microscopy.
CD34+ stem/progenitor cell-derived erythroblasts from healthy donors and MDS patients were immobilized on Alcian blue-coated coverslips and fixed with 2% (vol/vol) formaldehyde solution as previously described (19). Briefly, cells were permeabilized with PBS containing 0.5% Triton-X100 for 5 min, and the coverslips were washed once with PBS for 5 min and then blocked with 1% BSA for 20 min. Cells were subsequently stained for actin with phalloidin-Texas Red (1:160 dilution; Life Technologies) for 30 min in a humidifying chamber. Following multiple washes with PBS, the actin-stained cells on coverslips were mounted onto slides using Prolong gold antifade reagent (Life Technologies) and images were captured on a Leica STED-SP5 spectral confocal microscope using an oil immersion lens with a magnification of 63×.
Actin Disruption Quantitation by Multispectral Imaging Flow Cytometry.
An ImageStreamX (EMD-Millipore/Amnis) multispectral imaging flow cytometer was used to quantify the extent of actin filament disruption in erythroblasts derived from culturing CD34+ stem/progenitor cells (comparing healthy and MDS patient samples). Erythroblasts treated with Cytochalasin D (5 μg/mL for 5 h; Sigma) and Rac1 inhibitor (100 μM for 5 h; Calbiochem) were used as positive controls. Cells were fixed, permeabilized, and stained for F-actin using fluorescently labeled (Texas Red) phalloidin as described previously (40).
Acquisition of cellular images was performed on an ImageStreamX instrument using 50 mW of 561-nm laser light and an emission filter range of 594–660 nm. Both BF and fluorescence images were acquired at a magnification of 60×. A cell classifier was set on BF area to exclude small debris and system focus beads. Up to 10,000 events per sample were acquired.
Analysis to determine populations of cells with intact actin filaments and populations with disrupted actin was performed using the IDEAS 6.0 software package available through EMD Millipore. Intact single cells that were suitably in focus were gated using a hierarchical gating strategy starting with BF area and BF aspect ratio (single cells of appropriate size), and then by the BF gradient rms feature (cells in focus). Lastly, cells with no F-actin fluorescence were excluded from the downstream analysis (typically less than 10%). We used an algorithm available in the software package that enabled discrimination of defined fluorescence staining of the F-actin filaments, together with quantitation of the length (in microns) of the longest undisrupted actin filament staining seen by the custom-defined mask function of the software. The mask was defined using the built-in spot mask function. This option obtains bright regions from an image regardless of the intensity differences from one cell to another. The ability to extract bright objects is achieved using an image processing step that erodes the image and leaves only the bright areas. Once the mask properly captured the F-actin filament staining, the length feature was used to measure the longest part of the object defined by the mask, wherein cells with intact F-actin filaments would have a long length and cells with disrupted F-actin filaments would have a short length. A sample from a healthy donor for cells with intact F-actin and a Cytochalasin D-treated control with disrupted F-actin were used to set the cutoff level on the length parameter defining intact vs. disrupted F-actin based on fluorescent staining. The following sequential steps were taken in the analysis of populations from each of the samples that allowed us to develop the cortical F-actin staining index: (i) exclusion of aggregates and debris by gating cells with a moderate BF area and high BF aspect ratio, (ii) selection of a level of BF gradient rms to achieve well-focused cells (rms value >50), (iii) gating of cells with any level of F-actin staining, (iv) application of Spot Mask to the final subset, and (v) plotting of the length feature based on the F-actin Spot Mask (intact/disrupted cutoff value set at a unit of 25).
Quantifying Circular/Irregular-Shaped Erythroid Cells in Zebrafish Embryos by Multispectral Imaging Flow Cytometry (ImageStreamX Analysis).
The ImageStreamX instrument was also used for quantifying the ratio of circular/irregular cells in the erythroid (Gata1-expressing RFP cells) compartment of control and dock4a knockdown zebrafish embryos. A similar strategy as described above was used to eliminate the aggregates and debris. Well-focused RFP+ single cells were used in subsequent analysis. Using the built-in “Erode Mask,” a continuous mask was created on the periphery of the cell in both the BF channel and the RFP channel to define the shape of the cell. An IDEAS 6.0 built-in feature called circularity was then used to differentiate between the circular- and irregular-shaped cells. This feature measures the degree of the mask’s deviation from a circle. This measurement is based on the average distance of the object boundary from its center divided by the variation of this distance. Thus, the closer the object is to being a circle, the smaller is the variation, and the feature value will thus be high. On the other hand, the more the shape deviates from a circle, the higher is the variation, and the circularity value will thus be low. The following sequential steps were taken in the analysis of populations from each of the samples: (i) exclusion of aggregates and debris by gating cells with a moderate BF area and high BF aspect ratio, (ii) selection of a level of BF gradient rms to achieve well-focused cells (rms value >50), (iii) gating of cells with RFP signal, (iv) application of Erode Mask to the final subset, and (v) plotting a graph between the circularity features based on the BF Erode Mask and RFP Erode Mask and application of gates to separate out “circular” and “irregular” cells.
RAC1 G-LISA.
Levels of active GTP bound RAC1 were quantified using the G-LISA RAC1 Activation Assay Biochem Kit (Cytoskeleton, Inc.) according to the manufacturer’s protocol. In experiments where RBC ghosts or erythroblasts were used, frozen pellets from each preparation were lysed using reagents provided in the RAC1 activation kit.
RBC Ghost Preparation.
Fresh peripheral blood samples were collected from healthy volunteers and MDS patients with approval from the IRB. RBC ghosts were prepared according to published protocols (41) with minor modifications. Briefly, washed RBCs (devoid of the buffy coat) were suspended at 40% hematocrit containing hypotonic buffer [5 mM Tris⋅HCl, 5 mM KCl (pH 7.4)]. Cells were centrifuged at 20,000 × g for 5 min at 4 °C. The pellet obtained (white ghost) was washed two more times in hypotonic buffer. After washing, cells were transferred to microcentrifuge tubes and centrifuged at 14,000 × g for 10 min at 4 °C. The pellets consisting of erythroid ghosts were flash-frozen after discarding the supernatant and stored at −80 °C until subsequent use in the RAC1 activity assay.
FISH.
To quantify numbers of clonal malignant cells after in vitro culture, day 10 erythroblasts from MDS patients were cytospun (1,000–10,000 cells) for further analysis. Cells from the Mono7 cell line (42) were used as a positive control. FISH was performed according to the manufacturer’s instructions using the dual-color Vysis probe D7S486/CEP 7 (7q31 S.O./7p11.1-q11.1 Alpha Satellite DNA S. G.; Abbott Molecular, Inc.) enabling us to detect and quantify cells with loss of 7q. Analysis was performed using a Zeiss Axioplan Epifluorescence microscope.
Quantitative PCR.
To quantify DOCK4 expression levels, total RNA was isolated from erythroblasts at different stages of differentiation using an RNeasy Mini Kit (catalog no. 74104; Qiagen) following the manufacturer’s instructions. Isolated RNA was quantified, and cDNA was synthesized using a SuperScript VILO cDNA synthesis kit (catalog no. 11754050; Invitrogen). Control samples were prepared without an RT enzyme or RNA template. Specific DOCK4 primers were used (forward primer: 5′ GACCCACACACAGACTGCTTCA 3′ and reverse primer: 5′ GAGAGGGGGTGAAAGACTGC 3′), and the gene expression was quantified using the delta delta cycle threshold method utilizing Fast SYBR Green/Rox real-time PCR master mix (catalog no. 4385612; Invitrogen). 18S ribosomal RNA was used as the normalizing control. Primer specificity was verified using melting curve analysis.
Restoration of DOCK4 Expression in MDS Patient Samples.
To reexpress DOCK4 in MDS patient samples lacking DOCK4 expression, we subcloned the full-length DOCK4 cDNA (5,901 bp) into the MC (catalog no. MN530A1; System Biosciences) plasmid. The subcloned sequence was verified by DNA sequencing. The MC technology allows expression of large genes efficiently by only containing the circular DNA elements minimally required for sustained expression in cells once transfected into cells. The production of custom MC DNA was carried out by SBI, Inc., using an engineered Escherichia coli strain (ZYCY10P3S2T) that allows the production of the MCs. The MC DNA is conditionally produced by an expression of inducible integrase via intramolecular (cis)-recombination. Transfection of MCs into CD34+ patient-derived erythroid progenitors was performed on day 5 of culture, and reexpression of DOCK4 was verified by quantitative PCR on day 10 of culture from the same sample that was used in assaying for the restoration of F-actin filament length and cell shape. Erythroid colony-forming assays were done by plating 50,000 cells on day 7, following DOCK4 MC plasmid transfection on day 5. Enumeration of colonies was carried out on day 14.
Statistical Analyses.
The error bars are represented as mean ± SE. Paired Student’s t tests were performed to determine the statistical significance between the samples. Experiments were repeated at least three times, and values with P < 0.05 were considered statistically significant. Multivariate analysis for survival and DOCK4 expression was performed by adjusting for IPSS scores using SAS software.
Supplementary Material
Acknowledgments
We thank Dr. Velia M. Fowler at the Scripps Research Institute and Dr. Lucy A. Godley for their constructive comments; the personnel in the Flow Cytometry Facility for their expert advice; and Elizabeth M. Davis and Mathew Horch for technical assistance. This work was supported, in part, by NIH Grants HL16336 (to A.V. and A.W.), HL056183 (to T.E.), and CA40046 (to M.M.L.B.); by a University of Chicago Comprehensive Cancer Center Pilot Project Award, the Giving Tree Foundation, Harith Foundation, and Suhith Wickrema Memorial Cancer Fund (A.W.); and by Leukemia & Lymphoma Research of the United Kingdom (A.P. and J.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516394112/-/DCSupplemental.
References
- 1.Heaney ML, Golde DW. Myelodysplasia. N Engl J Med. 1999;340(21):1649–1660. doi: 10.1056/NEJM199905273402107. [DOI] [PubMed] [Google Scholar]
- 2.Bejar R, et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J Clin Oncol. 2012;30(27):3376–3382. doi: 10.1200/JCO.2011.40.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bejar R, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364(26):2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malcovati L, et al. Time-dependent prognostic scoring system for predicting survival and leukemic evolution in myelodysplastic syndromes. J Clin Oncol. 2007;25(23):3503–3510. doi: 10.1200/JCO.2006.08.5696. [DOI] [PubMed] [Google Scholar]
- 5.Will B, et al. Stem and progenitor cells in myelodysplastic syndromes show aberrant stage-specific expansion and harbor genetic and epigenetic alterations. Blood. 2012;120(10):2076–2086. doi: 10.1182/blood-2011-12-399683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenberg PL, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120(12):2454–2465. doi: 10.1182/blood-2012-03-420489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotini AG, et al. Functional analysis of a chromosomal deletion associated with myelodysplastic syndromes using isogenic human induced pluripotent stem cells. Nat Biotechnol. 2015;33(6):646–655. doi: 10.1038/nbt.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou L, et al. Aberrant epigenetic and genetic marks are seen in myelodysplastic leukocytes and reveal Dock4 as a candidate pathogenic gene on chromosome 7q. J Biol Chem. 2011;286(28):25211–25223. doi: 10.1074/jbc.M111.235028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasegawa H, et al. DOCK180, a major CRK-binding protein, alters cell morphology upon translocation to the cell membrane. Mol Cell Biol. 1996;16(4):1770–1776. doi: 10.1128/mcb.16.4.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nolan KM, et al. Myoblast city, the Drosophila homolog of DOCK180/CED-5, is required in a Rac signaling pathway utilized for multiple developmental processes. Genes Dev. 1998;12(21):3337–3342. doi: 10.1101/gad.12.21.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu YC, Horvitz HR. C. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature. 1998;392(6675):501–504. doi: 10.1038/33163. [DOI] [PubMed] [Google Scholar]
- 12.Yajnik V, et al. DOCK4, a GTPase activator, is disrupted during tumorigenesis. Cell. 2003;112(5):673–684. doi: 10.1016/s0092-8674(03)00155-7. [DOI] [PubMed] [Google Scholar]
- 13.Côté J-F, Vuori K. Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J Cell Sci. 2002;115(Pt 24):4901–4913. doi: 10.1242/jcs.00219. [DOI] [PubMed] [Google Scholar]
- 14.Upadhyay G, et al. Molecular association between beta-catenin degradation complex and Rac guanine exchange factor DOCK4 is essential for Wnt/beta-catenin signaling. Oncogene. 2008;27(44):5845–5855. doi: 10.1038/onc.2008.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kjeldsen E, Veigaard C. DOCK4 deletion at 7q31.1 in a de novo acute myeloid leukemia with a normal karyotype. Cell Oncol (Dordr) 2013;36(5):395–403. doi: 10.1007/s13402-013-0145-5. [DOI] [PubMed] [Google Scholar]
- 16.Traver D, et al. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol. 2003;4(12):1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- 17.Madzo J, et al. Hydroxymethylation at gene regulatory regions directs stem/early progenitor cell commitment during erythropoiesis. Cell Reports. 2014;6(1):231–244. doi: 10.1016/j.celrep.2013.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uddin S, Ah-Kang J, Ulaszek J, Mahmud D, Wickrema A. Differentiation stage-specific activation of p38 mitogen-activated protein kinase isoforms in primary human erythroid cells. Proc Natl Acad Sci USA. 2004;101(1):147–152. doi: 10.1073/pnas.0307075101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang J-A, et al. Osteopontin regulates actin cytoskeleton and contributes to cell proliferation in primary erythroblasts. J Biol Chem. 2008;283(11):6997–7006. doi: 10.1074/jbc.M706712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koury MJ, Bondurant MC. Erythropoietin retards DNA breakdown and prevents programmed death in erythroid progenitor cells. Science. 1990;248(4953):378–381. doi: 10.1126/science.2326648. [DOI] [PubMed] [Google Scholar]
- 21.Muta K, Krantz SB, Bondurant MC, Wickrema A. Distinct roles of erythropoietin, insulin-like growth factor I, and stem cell factor in the development of erythroid progenitor cells. J Clin Invest. 1994;94(1):34–43. doi: 10.1172/JCI117327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geslain R, et al. Distinct functions of erythropoietin and stem cell factor are linked to activation of mTOR kinase signaling pathway in human erythroid progenitors. Cytokine. 2013;61(1):329–335. doi: 10.1016/j.cyto.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 23.Pellagatti A, et al. Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia. 2010;24(4):756–764. doi: 10.1038/leu.2010.31. [DOI] [PubMed] [Google Scholar]
- 24.Konstantinidis DG, et al. Signaling and cytoskeletal requirements in erythroblast enucleation. Blood. 2012;119(25):6118–6127. doi: 10.1182/blood-2011-09-379263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji P, Jayapal SR, Lodish HF. Enucleation of cultured mouse fetal erythroblasts requires Rac GTPases and mDia2. Nat Cell Biol. 2008;10(3):314–321. doi: 10.1038/ncb1693. [DOI] [PubMed] [Google Scholar]
- 26.Franco T, Low PS. Erythrocyte adducin: A structural regulator of the red blood cell membrane. Transfus Clin Biol. 2010;17(3):87–94. doi: 10.1016/j.tracli.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhlman PA, Hughes CA, Bennett V, Fowler VM. A new function for adducin. Calcium/calmodulin-regulated capping of the barbed ends of actin filaments. J Biol Chem. 1996;271(14):7986–7991. doi: 10.1074/jbc.271.14.7986. [DOI] [PubMed] [Google Scholar]
- 28.Matsuoka Y, Hughes CA, Bennett V. Adducin regulation. Definition of the calmodulin-binding domain and sites of phosphorylation by protein kinases A and C. J Biol Chem. 1996;271(41):25157–25166. doi: 10.1074/jbc.271.41.25157. [DOI] [PubMed] [Google Scholar]
- 29.Matsuoka Y, Li X, Bennett V. Adducin is an in vivo substrate for protein kinase C: Phosphorylation in the MARCKS-related domain inhibits activity in promoting spectrin-actin complexes and occurs in many cells, including dendritic spines of neurons. J Cell Biol. 1998;142(2):485–497. doi: 10.1083/jcb.142.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNerney ME, et al. CUX1 is a haploinsufficient tumor suppressor gene on chromosome 7 frequently inactivated in acute myeloid leukemia. Blood. 2013;121(6):975–983. doi: 10.1182/blood-2012-04-426965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C, et al. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell. 2014;25(5):652–665. doi: 10.1016/j.ccr.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalfa TA, et al. Rac GTPases regulate the morphology and deformability of the erythrocyte cytoskeleton. Blood. 2006;108(12):3637–3645. doi: 10.1182/blood-2006-03-005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van de Water B, et al. Cleavage of the actin-capping protein alpha-adducin at Asp-Asp-Ser-Asp633-Ala by caspase-3 is preceded by its phosphorylation on serine 726 in cisplatin-induced apoptosis of renal epithelial cells. J Biol Chem. 2000;275(33):25805–25813. doi: 10.1074/jbc.M001680200. [DOI] [PubMed] [Google Scholar]
- 34.Polo JM, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28(8):848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Figueroa ME, et al. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood. 2009;114(16):3448–3458. doi: 10.1182/blood-2009-01-200519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 37.Alexander J, Stainier DY, Yelon D. Screening mosaic F1 females for mutations affecting zebrafish heart induction and patterning. Dev Genet. 1998;22(3):288–299. doi: 10.1002/(SICI)1520-6408(1998)22:3<288::AID-DVG10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 38.Wickrema A, Krantz SB, Winkelmann JC, Bondurant MC. Differentiation and erythropoietin receptor gene expression in human erythroid progenitor cells. Blood. 1992;80(8):1940–1949. [PubMed] [Google Scholar]
- 39.Millington M, Arndt A, Boyd M, Applegate T, Shen S. Towards a clinically relevant lentiviral transduction protocol for primary human CD34 hematopoietic stem/progenitor cells. PLoS One. 2009;4(7):e6461. doi: 10.1371/journal.pone.0006461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wickrema A, Koury ST, Dai CH, Krantz SB. Changes in cytoskeletal proteins and their mRNAs during maturation of human erythroid progenitor cells. J Cell Physiol. 1994;160(3):417–426. doi: 10.1002/jcp.1041600304. [DOI] [PubMed] [Google Scholar]
- 41.Andrews DA, Yang L, Low PS. Phorbol ester stimulates a protein kinase C-mediated agatoxin-TK-sensitive calcium permeability pathway in human red blood cells. Blood. 2002;100(9):3392–3399. doi: 10.1182/blood.V100.9.3392. [DOI] [PubMed] [Google Scholar]
- 42.Fujisaki H, et al. Establishment of a monosomy 7 leukemia cell line, MONO-7, with a ras gene mutation. Int J Hematol. 2002;75(1):72–77. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.