Abstract
Introduction
Macromolecular X-ray crystallography has been the primary methodology for determining the three-dimensional structures of proteins, nucleic acids and viruses. Structural information has paved the way for structure-guided drug discovery and laid the foundations for structural bioinformatics. However, X-ray crystallography still has a few fundamental limitations, some of which may be overcome and complemented using emerging methods and technologies in other areas of structural biology.
Areas covered
This review describes how structural knowledge gained from X-ray crystallography has been used to advance other biophysical methods for structure determination (and vice versa). This article also covers current practices for integrating data generated by other biochemical and biophysical methods with those obtained from X-ray crystallography. Finally, the authors articulate their vision about how a combination of structural and biochemical/biophysical methods may improve our understanding of biological processes and interactions.
Expert opinion
X-ray crystallography has been, and will continue to serve as, the central source of experimental structural biology data used in the discovery of new drugs. However, other structural biology techniques are useful not only to overcome the major limitation of X-ray crystallography, but also to provide complementary structural data that is useful in drug discovery. The use of recent advancements in biochemical, spectroscopy and bioinformatics methods may revolutionize drug discovery, albeit only when these data are combined and analyzed with effective data management systems. Accurate and complete data management is crucial for developing experimental procedures that are robust and reproducible.
Keywords: data management, hybrid methods, protein crystallography, reproducibility, structural data interpretation, target-based drug discovery
1. Macromolecular X-ray crystallography: the past, current and the near future
The past decade has witnessed enormous methodological developments in X-ray crystallography, with a special emphasis on advancements in protocols and software, accompanied by rapid increases in computational power and storage capacity. The contributions of X-ray crystallography to modern science cannot be overstated. Nine of the top 100 most cited scientific publications of all time [1] are structural biology papers. Many of these papers describe fundamental theories and techniques in the field that will (or already have) considerably advance structure-based rational drug discovery.
1.1 Recent progress in pipelines for structure determination – the high-throughput (high-output) era
The remarkable progress in methods for recombinant protein production [2,3] and crystallization in recent years is in part due to the success of various protein structure and function initiatives [4]. Methodologies and strategies that make protein production more cost-effective and robust include ligation-independent cloning, recombinant protein expression and purification on metal affinity resins [4–9]. Automated approaches to protein crystallization, both for initial crystallization screening and hits optimization, are now routinely used by structural biology laboratories. The use of 96-well screening plates containing 0.2 μL of protein solution (or less) in each drop is now commonplace, further minimizing the amount of sample and consumables used and time per trial. The mining of crystallization data has resulted in the creation of more efficient crystallization cocktail screens [10] and novel approaches for sample preparation have been developed. These include in situ proteolysis [11] and reductive methylation [12] for improving the probability of crystallization of macromolecules, as well as fluorescence-based thermal shift assays (TSAs) [13,14], and dynamic light scattering [15] for choosing the best protein constructs, buffers and ligands for co-crystallization and/or soaking.
Recent developments in X-ray data collection protocols, low noise detectors, sample handling robotics, synchrotron beam-lines and associated software also significantly contributed to progress in X-ray crystallography. However, perhaps, the most dramatic changes that have led to faster structure determination are in structure solution, refinement and validation software. SHELX was the first software package to revolutionize efficient and robust experimental phasing for macromolecules [16]. HKL-3000 has implemented a complete structure determination pipeline that can generate a complete atomic model from raw diffraction images in a matter of minutes, both by molecular replacement and anomalous diffraction [17]. PHENIX has also provided a framework for automated protocols in structure determination and refinement processes [18]. Refmac has significantly improved structure refinement by highly optimizing global refinement algorithms, including jelly-body refinement, non-crystallographic symmetry restraints and translation/libration/screw analysis [19]. COOT provides state-of-the-art visualization of structures and integrates numerous tools that facilitate interactive manual refinement and validation [20]. Auto-Rickshaw incorporates decision making to fully automate the selection of crystal structure determination protocols [21]. The availability of these modern tools has made the determination of many macromolecular structures ‘low-hanging fruit’ even for novice structural biologists. However, even in the most straightforward cases, fundamental errors may occur and expert oversight of the structure solution process should not be replaced by blind use of computational tools. Moreover, there are still many important structures that require both high-level expertise and sometimes many years of effort.
1.2 The growth of the Protein Data Bank and issues of reproducibility
Technology development and large ‘high-throughput’ structural biology initiatives have led to the availability of a large collection of high- and medium-resolution structures. The number of macromolecular structures deposited in the Protein Data Bank (PDB) has recently exceeded 100,000 [22], with 80% of them determined during the last decade. This avalanche of structural data has extended our understanding of macromolecular structures in general and increased the overall structural coverage of proteins up to 40% [23], which forms a solid foundation for homology modeling and threading approaches to generate structural models for proteins recalcitrant to crystallization [24].
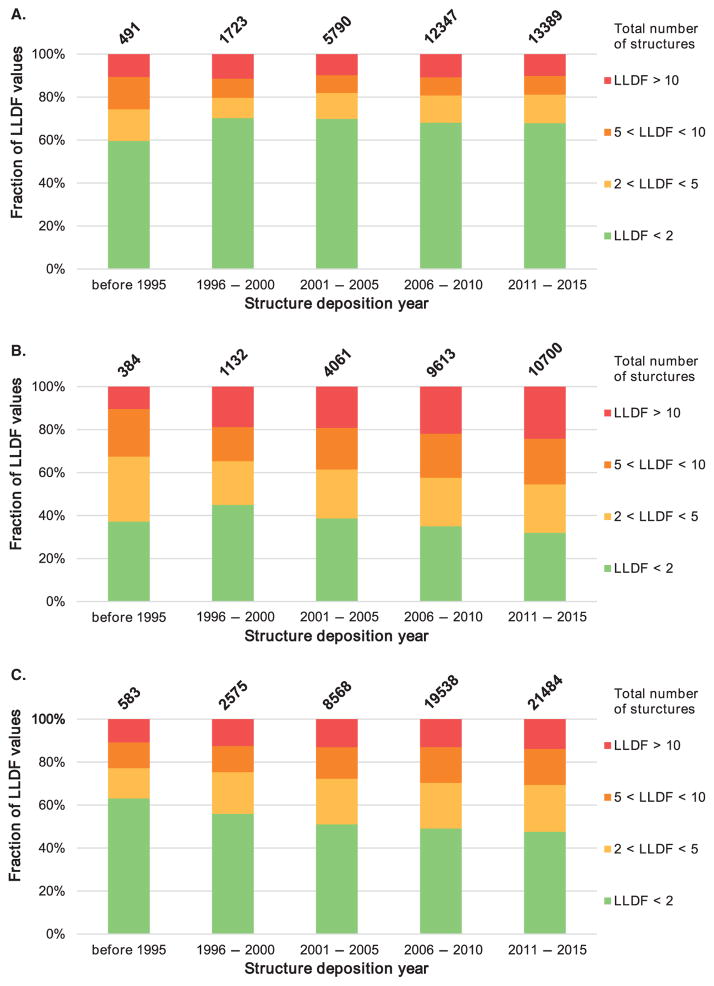
Unfortunately, a small number of crystal structures deposited in the PDB are of suboptimal quality [25–30]. Automatic re-refinement has been demonstrated to significantly improve overall structure quality [31]; however, these advances do not necessarily improve the biological interpretation of structures, since the actual atomic model is not changed and questionable regions are not re-modeled. The errors most relevant to drug discovery are the incorrect identification and modeling of ligands in protein–ligand complexes [32]. In recent years, increased awareness of potential problems in structures deposited in the PDB has led to the development of sophisticated general validation tools [30,33], and tools for evaluating the quality of ligands in protein–ligand complexes [34–37]. However, despite progress in methods and software, the number of macromolecular structures with suboptimal ligand quality has not decreased with time, as measured by LLDF (which represents electron density fit quality for ligands; Figure 1). If anything, the fit of complex ligands to electron density is getting worse over time, which may be partly due to overreliance on automation in structure determination and limited availability of ligand validation tools. Furthermore, old structures are reprocessed with the newest technology only in rare cases [38,39]. The consequences for drug discovery are that the substantial number of structures with incorrectly modeled or misidentified ligands may not only result in erroneous conclusions and non-reproducible results published in literature, but also pollute the databases that are used for computational biology and chemistry. An additional complication arises from inconsistencies between metadata and reduced data. For example, out of approximately 14,000 structures in the PDB reported to use anomalous signal, only 2359 structures include the associated anomalous data in their deposited structure factors. To summarize, despite overall progress towards higher quality (and presumably, greater reliability) of structures and their interpretations, the issue of data reproducibility and consistency is still a major concern [40]. Recent initiatives from the PDB, International Union of Crystallography and individual scientists [26,27,39] have led to proposals for the mandatory deposition of unmerged intensity data with the intent of providing a means for future reanalysis of the data. There are also pilot programs that plan to accept deposition of raw diffraction data and develop tools that will enhance and sustain macromolecular diffraction data and provide metadata for raw X-ray diffraction images. The public availability of raw diffraction images for all PDB deposits is critical for making the results of X-ray crystallography wholly reproducible [39].
Figure 1. The quality of small molecule ligands in crystal structures of macromolecule-ligand complexes during the past two decades, evaluated by the agreement of modeled ligand with local electron densities using LLDF.
Each chart shows the distribution of structures containing (A) ligand(s) with a single non-hydrogen atom (excluding water molecules), (B) ligand(s) with 2 – 5 non-hydrogen atoms, and (C) ligand(s) with 6 or more non-hydrogen atoms. The structures are then classified by the highest LLDF score (i.e., poorest fit) among the ligands in the respective ligand-size category in each structure. The distributions are also subdivided by year of deposition; the numbers atop the bars count all structures in the time range with at least one ligand in the respective size category.
LLDF: Local Ligand Density Fit.
1.3 Unmet limitations of macromolecular crystallography
While techniques for macromolecular X-ray crystallography have led to advancements in the field, problems that may not be so easily resolved in X-ray crystallography still persist and stand out more prominently. As large-scale structure determination efforts have moved the boundaries closer to the limitations of X-ray crystallography [41], the application of complementary alternative biophysical and/or biochemical methods became very important. These limitations include; i) difficulties in protein crystallization due to limited solubility; ii) unresolved protein dynamics (conformational diversity); and iii) limited detection of chemical heterogeneity (such as post-translational modifications [PTM]).
First, solubility is a key bottleneck in macromolecular X-ray crystallography for drug discovery because many drug targets are anchored on the cell membrane and/or are insoluble. For example, there are ~ 20,000 proteins in the human genome that may be expressed in different tissues [42], with ~ 13,000 soluble proteins that may be modeled based on a known fold. The remaining ~ 7000 insoluble proteins are primarily associated with the membrane, including more than 800 GPCR proteins that may be modeled with comparative confidence. This leaves ~ 6000 insoluble proteins with structures yet to be explored. While it is potentially possible to crystallize membrane proteins via the use of either detergents or lipid cubic phases, at the moment high-output platforms for determination of integral membrane protein structures by X-ray crystallography are still far from optimal [43,44], and the selection of detergents for protein production and successful crystallization of membrane proteins remains highly challenging.
Recent advancements in cryo-electron microscopy (cryo-EM) that allow structure determination with near-atomic resolution (higher than 3.5 Å) [45,46] provide an alternative approach to determine macromolecular structure without crystallization and is less dependent on issues of solubility [47,48] but still require sample homogeneity. The progress in solid-state nuclear magnetic resonance (NMR) methods for biological macromolecules provides another approach for the determination of insoluble protein structures [49].
X-ray crystallography, like many other biophysical methods, averages out many comparatively static molecules which is biased towards the most populous conformation and leaves most protein dynamics unresolved. Solution NMR, however, provide information about the dynamics of the molecules studied, represented as an ensemble of structures with varying conformations. The incorporation of NMR data to evaluate the flexibility of various regions of a macromolecule, especially at substrate binding sites, is of particular interest for examining drug binding and release mechanisms [50]. EPR also provides additional information about different conformation states of the protein [51,52].
Detection and characterization of structural variations in proteins not encoded in their DNA, including post-translation modifications such as phosphorylation, ubiquiti-nation, methylation, acetylation and glycosylation, presents the third limitation for X-ray crystallography. Although most epigenetic modifications are thought to be correlated with the ‘histone code’, glycosylation usually takes place within the ER and Golgi complex. X-ray crystallography typically requires a homogeneous fraction of macromolecules to obtain diffraction quality crystals (and/or the process of crystallization naturally selects for a homogenous fraction of the molecule). Therefore, crystallography is not a suitable biophysical method for the investigation of multiple species of epigenetic modifications in the same sample unless the fractions are separated. As this comprises a great deal of information relevant to cell regulation that is not encoded in the protein sequence, structural information of such epigenetic modifications are invaluable, especially for the understanding of the functions of eukaryotic macromolecules and their drug targetability [53,54]. Emerging techniques for the analysis of PTM in proteomics rely predominantly upon mass spectrometry (MS) [55]. Further advancements in single-particle cryo-EM also may enable one to observe heterogeneous structural details of large covalent modifications on the same macromolecule [56].
Target-based drug discovery has to date relied heavily on high-resolution structures determined by X-ray crystallography, such as in the cases of the design of HIV protease inhibitors [57,58], influenza virus neuraminidase inhibitors [59] and L. mexicana glyceraldehyde-3-phosphate dehydrogenase inhibitors [60]. However, the structural knowledge of protein and nucleic acids we have gained from X-ray crystallography has also been used to develop methodologies for other biophysical techniques. We expect that in the near future, the integration of X-ray crystallography with other biophysical methods will become more prevalent. As a result, the use of multiple biophysical techniques will become increasingly important to tackle different aspects of macromolecular structures for the purpose of drug discovery.
2. Integration of X-ray crystallography with other biophysical structure determination methods – applications in the investigation of structural aspects of drug targets
Besides traditional X-ray crystallography, other forms of X-ray experiments can also provide essential structural information at different stages of structure-guided drug discovery. Serial femtosecond crystallography uses highly focused X-ray free electron lasers to collect single-shot diffraction data on streams of nanocrystals, which has been shown to be useful for the investigation of structures where well-diffracting mesoscopic crystals cannot be grown [61]. Small angle X-ray scattering provides data about average particle size, shape and surface-to-volume ratio. Small angle X-ray scattering may be used to highlight large-scale structural changes that may accompany drug binding [62].
This section will highlight the complementary roles played by non-X-ray-based biophysical structure determination methods. Cryo-EM and NMR spectroscopy, like X-ray crystallography, provide high-resolution structural information. Mass spectrometry (MS) and electron paramagnetic resonance (EPR) experiments also provide complementary information aiding in the interpretation of crystallographic data. The complementary use of X-ray crystallography with these other methods are further illustrated by references to developed protocols that involve the use of a combination of > 1 biophysical structure determination method. Various techniques used to perform ligand binding assays in a structure-guided context will also be discussed. Recent advancements in these techniques call for successful integration of structural data that present new opportunities in rational drug design.
2.1 The complementarity between cryo-EM and X-ray crystallography
Recent advancement in single-particle cryo-electron microscopy (cryo-EM) sample preparation techniques, detectors and reconstruction algorithms have produced > 30 near-atomic resolution structures (3.5 Å or higher), and the 3.0 Å barrier was broken even for particles with no or low symmetry [45–48,63–67]. However, despite these great advancements, the typical resolution of cryo-EM structures is much lower as compared to X-ray structures. Nevertheless, cryo-EM has several advantages when compared with X-ray crystallography, making these two techniques highly complementary to each other [63,67,68]. First, cryo-EM allows the visualization of very large macromolecules and their complexes, such as the ribosome or the nuclear pore complex. These giant structures are usually hard to crystallize [69], and even if crystallized, the resolutions of structures solved by X-ray diffraction are comparable to that of cryo-EM [63–66,70]. Second, in cryo-EM experiments data are collected from single molecules or complexes with molecular conformations unaffected by potentially ‘unnatural’ interactions due to the formation of crystal contacts. In addition, X-ray crystallography can struggle with obtaining phases for structure solution even when high-resolution data are available [68], and cryo-EM can be of help. By using phases derived from cryo-EM density maps, after applying various density modification techniques, X-ray structures may be determined by molecular replacement [71,72]. For example, the X-ray structure of the reovirus core at 3.6 Å resolution was determined using a 27 Å cryo-EM envelope of the core as a starting point for solving the phase problem [73]. Cryo-EM may also be used for cross verification of results to ensure data consistency, such as the study of mechanism of the adhesive function of cadherins, where the high-resolution structure was solved by X-ray crystallography and the probable biological assembly in vivo was confirmed by cryo-EM [74].
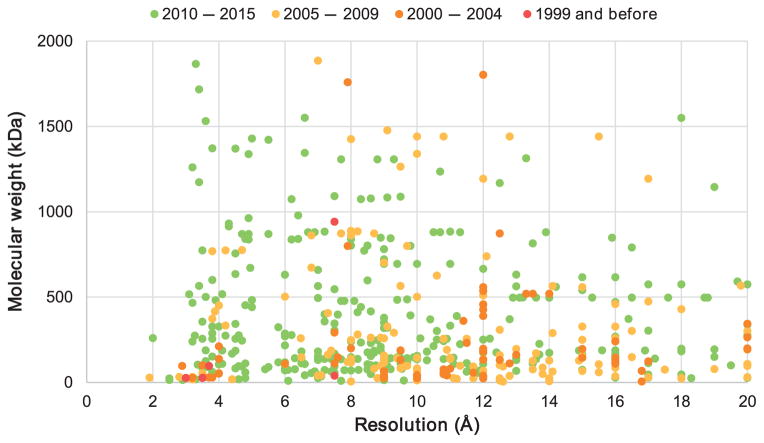
The resolution of cryo-EM has been historically insufficient to confidently place amino acids/nucleotides in the cryo-EM map (Figure 2), even though there has been substantial recent progress in de novo model-building approaches [46]. Fortunately, this problem may be solved if structures of smaller components of a macromolecular assembly (single proteins, nucleic acids or their domains) in higher resolution are available [67]. Structural coordinates of complex components deposited in the PDB can be fitted directly into three-dimensional (3D) maps obtained by cryo-EM, or used as a template for homology modeling if the structure of a particular component is not available. For example, the 3D structure of poliovirus receptor bound to poliovirus (PDBID: 1DGI) was solved by cryo-EM to 22 Å resolution. Obviously, this resolution precluded the identification of specific amino acid chains in the density, but fitting a combination of X-ray crystallographic models and custom-built homology models led to a plausible atomic model of the overall structure [75,76]. A similar strategy has been used to fit structures determined by X-ray crystallography (1 – 3 Å) into lower resolution EM maps (7 – 10 Å) in order to investigate the overall arrangement of subunits in filaments [77]. The vast library of X-ray crystal structures accumulated in the PDB may be useful for solution of novel structures by cryo-EM even where the positions and identities of individual components are unknown, as was the case with the yeast mitochondrial large ribosomal subunit [70,78]. In addition, the use of various software developed for X-ray crystallography (MOLREP, BALBES [79], COOT [20] and REFMAC [19]) are being adopted for cryo-EM structure determination with significant modifications [78,80]. However, single-particle cryo-EM is still a rapidly evolving field that requires significant improvement in both experimental and computational protocols to achieve the higher resolutions needed to characterize of the binding mode of small ligand molecules in structure-guided drug discovery.
Figure 2.
Molecular weight versus resolution for structures determined using cryo-electron microscopy.
The employment of novel classification algorithms together with recent advancements in cryo-EM equipment also allows the separation of different populations of single particles for the determination of multiple distinct molecules, or similar molecules in different conformational states – although currently only for very large complexes such as ribosomes [81]. For samples with low homogeneity which are thus difficult to organize into a crystal lattice for diffraction, cryo-EM single-particle analysis, in conjunction with advanced classification algorithms, may in the future permit investigation of their structural arrangement. Even if samples with heterogeneous molecules manage to arrange into a productive crystal lattice, the diffraction pattern produced by X-ray crystallography would only be able to differentiate heterogeneity in terms of occupancy, while cryo-EM has the potential in the future to selectively average the different species of the target molecule separately, especially if major conformational differences are present or if the modifications are very large.
2.2 Solution NMR, Solid-state NMR and X-ray crystallography
NMR provides spin relaxations, J-couplings and chemical shifts, which can measure the dynamics of macromolecular structure in solution. However, the ambiguity of NMR restraints still requires the incorporation of additional geometrical restraints from fundamental knowledge about stereo-chemical structure at atomic resolution generated by X-ray crystallography [50]. Therefore, the determination of NMR structures has been highly dependent on the integration of prior structural knowledge by computational methods. A key question about the application of NMR structure in drug discovery is that whether or not the timescale for the measured ensemble is long enough to cover all conformations relevant to the process of binding of a substrate. The past decade has seen significant improvements in the computational algorithms used for the integration of prior structural knowledge with experimental restraints, such as the CNS, PHENIX, and Rosetta structural biology software systems [82]. Despite size limits and high concentration requirement, NMR is particularly strong for determining structures of very flexible proteins which are resistant to forming diffraction quality crystals.
Solid-state NMR, on the other hand, offers many desirable features including the ability to overcome the molecular tumbling size limit in solution NMR [83]. For large membrane proteins or biomolecular assemblies that only diffract weakly due to internal dynamics, solid-state NMR is an attractive method to provide vital structural data to improve the quality of structural models. Hybrid methods have been developed to combine data from both solid-state NMR data and X-ray crystallography. For example, using a 3.7 Å crystal structure of the disulfide bond-forming enzyme complex as a starting point, the incorporation of solid-state NMR data was demonstrated to enhance the transmembrane backbone precision to 0.92 Å and to improve structural quality overall [84]. However, solid-state NMR alone still suffers from low-spectral resolution due to a low signal-to-noise ratio.
NMR is capable of determining important functional data about flexible elements of macromolecules not revealed in X-ray studies. The combination of NMR with X-ray crystallography provides more complete structural information for the investigation of important drug targets. In the case of the voltage-dependent anion channel, the most abundant mitochondrial outer membrane protein, the crystal [85] and solution NMR structures [86] were determined independently, then conjointly by combined NMR/X-ray refinement to reveal different aspects of the structure [87]. The use of NMR has also been reported to monitor changes in protein conformation due to changes in pH [88]. In addition to its use in structure determination, NMR spectroscopy has also been used in ligand-binding assays. For example, Abbott used fragment-based NMR screening in part to discover high-affinity ligands for Bcl-xL [89].
2.3 EPR supplements X-ray crystallography with mobility information
Pulsed EPR (electron paramagnetic resonance, or ESR, electron spin resonance) has been used to detect anisotropic motion using proteins with spin labels [90]. When used in conjunction with X-ray crystallography, EPR is particularly useful to determine the conformational state of structures [52]. For example, the EPR spectrum has been used to determine nitroxide motion using the T4 lysozyme crystal structure as a framework [91,92]. Knowledge of conformational information in ligand binding is useful for the elucidation of the mechanism of action of proteins, essential for rational design of drug-like entities.
EPR is especially suitable for the evaluation of metal binding sites in metalloproteins, especially transition metals. For example, EPR was used as the key methodology for the characterization of Fe-S cluster reaction mechanism during diphthamide biosynthesis [93,94]. When combined with X-ray crystallography, EPR has also been used to study conformational states of ion channels [95] and metal incorporation in proteins [96].
Besides the investigation of the motion of small molecules, other applications of EPR would include the discovery of multiple conformational states for multi-domain proteins in the solution. For example, the co-chaperone DnaJ crystal structure shows multiple domains; and EPR was used to detect different domain arrangements exhibited by DnaJ [51]. In combination with X-ray crystallography, EPR may provide insightful information on the mobility of structural features of membrane proteins and large proteins with multiple domains.
2.4 Mass spectrometry supports testing of hypotheses imposed by crystal structures
Although mass spectrometry lacks the capability for de novo tertiary structure determination, it is, however, an excellent tool to verify or falsify testable hypotheses imposed by structures determined by X-ray crystallography. Experimental data used to determine the crystal structures can be subjectively overinterpreted, and this can bias hypotheses and follow-up experiments [41]. In some situations where the presence of a covalently-linked feature is uncertain or disordered in an electron density map produced by X-ray crystallography, mass spectrometry provides higher precision data in terms of chemical identity. A recent example of such hypothesis testing is the application of mass spectrometry in PTM analysis for shotgun proteomics [55].
Chemical cross-linking with subsequent mass spectrometry (CXMS) is another emerging analytical method that can provide experimental restraints for other techniques [97,98]. CXMS introduces covalent links between two spatially close amino acids as a chemical constraint that will remain intact through the digestion prior to mass spectrometry analysis. This provides experimental evidence about relative arrangements between two domains or two subunits that would otherwise be difficult to obtain by X-ray crystallography alone, especially when obtaining homogeneous, high-concentration samples for crystallization is infeasible. CXMS particularly complements X-ray crystallography by providing data on interactions within protein complexes or between multiple proteins.
2.5 Biochemical and spectroscopic experiments used to extend the functional implications of X-ray crystal structures
The identification of metabolic ligands of proteins with unknown (or partially unknown) functions by crystallographic screening would not be possible without recent advances in macromolecular crystallography techniques. For example, previously unknown functions of two protein families (PF01256 and YjeF_N) were determined by crystallographic screening of a metabolite library [99]. The project required soaking of crystals of three proteins with 11 cocktails comprising 87 commercially available natural metabolites, resulting in over 500 crystal structures which were determined and the bound ligand identified using the observed electron density. This number of crystals and structures could be handled only once a very efficient structure determination pipeline [17], coupled with comprehensive database for experiment management, was available [100]. The same approach can be used to screen for lead compound identification in drug discovery [101,102]. Further advancements in high-throughput structure determination coupled with efficient data handling could revolutionize drug discovery.
Another method for ligand identification using a compound library that has come to active use in the last decade is fluorescence-based TSA. In this method, the thermal stability of protein (unfolding as measured by critical melting temperature – Tm) is tested, and the best ligand chosen assuming that the protein stability will increase upon ligand binding [103]. There is a broad spectrum of environmentally sensitive dyes available which monitor protein unfolding by binding to exposed hydrophobic parts of protein and emitting fluorescence [104]. This technique is useful not only for physiological ligand [105] and drug hit compound identification [106], but also for selection of stable protein constructs and buffer conditions for biomedical experiments including crystallization. Multiple studies report increased chances of getting crystals with enhanced thermal stability [13]. Taking into account the broad range of applications, low cost and high-throughput setup, TSA is used both in academia and industry as a routine practice [107].
Even if TSA does provide some information about ligand binding, data crucial for drug characterization, the accuracy of the method may not be sufficient for reliably ranking the potency of identified compounds [106]. For many years, isothermal titration calorimetry (ITC) has been used to establish reaction stoichiometry and ligand affinity. ITC measures the direct heat exchanged between interacting molecules [108]. In its standard use, this technique requires a significant amount of protein and long-duration titrations. However, high-throughput developments in ITC have been aimed at reducing the amount of sample used as well as improving the measurement speed [109]. ITC is still, however, used as a secondary screening technique in high-throughput setups.
Another approach suitable for efficient estimation of ligand binding parameters is fluorescence-based methods like tryptophan fluorescence quenching (TQ) or fluorescence polarization/anisotropy. In the former, binding affinity of ligand is expressed as a function of decreasing tryptophan fluorescence with respect to increasing ligand concentration. The applicability and success of a TQ experiment is dependent on several parameters of each particular system: the presence of tryptophan, the inner-filter effect, collision quenching, and the distance of the ligand binding site from tryptophan [110]. Fluorescence polarization (FP) derives from the fact that FP has an inverse dependence with molecular mobility [111]. FP experiments require large changes of molecular volume during formation of the protein-ligand complex and the lifetime of the fluorophore has to be longer than the time needed for the molecule to tumble in solution [112].
Optical biosensor technology, such as surface plasmon resonance (SPR), has been recently exploited in the field of drug discovery, both in pharmaceutical companies and in basic research. SPR measures changes in the optical reflectivity of thin metal (e.g., gold) films, which occur when molecules bind the film or targets that have been precoated on its surface. Measurements are carried out in real time and do not require additional labeling steps. SPR can be used to effectively monitor interactions among a variety of molecules, including protein, DNA, RNA and small molecules [113]. In particular, SPR is being adapted for the study of membrane proteins that remain principal targets for drug discovery [114]. SPR may be applied to drug discovery in a variety of applications: to screen for potential binding to target proteins, to monitor reaction rates of different substrates, and to establish cofactors required for protein activity [115]. SPR has long been used as a secondary screening technique in drug discovery; however, recent advances in methodology and instruments (such as the possibility of using 384-well plates [116]) in combination with the wealth of data output by the method have elevated its importance.
2.6 From bench to bedside
Within the past few decades, the time and cost of drug development have significantly increased. Today it takes around 10 – 15 years to get new drugs on the market, and the process can cost up to $1 billion [117]. These costs reflect the complex and highly regulated process of drug development. In an effort to decrease the prolonged development time and streamline the process, high throughput screening systems have been developed for X-ray crystallography and NMR that range from semi-automated to fully automated robots, which have been used successfully by industry for lead identification and optimization.
Among FDA-approved new molecular entities [118], there are several examples of successful crystallography-based drug discovery stories that have led to clinical trials [119]. Astex Technology, for example, has demonstrated the use of crystallographic fragment-based lead discovery methods to identify low-affinity ligand hits (>100 μM). In this approach, single crystals of the target protein are soaked with cocktails of 2 – 8 molecules from a ligand fragment library composed of small compounds (100 – 250 Da) and the bound entities are identified by structure determination [120]. Crystallographic fragment-based screening has been applied for the development of a cyclin-dependent kinase 2 (cdk2) inhibitor, AT7519 [121]. CDKs are a family of serine-threonine protein kinases having key roles in cell cycle regulation and have been linked to many types of cancer [122]. The screening set of about 500 compounds that was tested consisted of kinase-oriented drugs, known drug fragments and molecules selected as potential cdk2 binders based on virtual screening. After initial crystallographic screening, four low-molecular-weight hits were selected and one-indazole was further optimized, leading to the development of AT7519. Currently, AT7519 has passed the second phase of clinical trials and is being tested for the treatment of patients with chronic lymphocytic leukemia and mantle cell lymphoma [123].
Abbott used fragment-based NMR screening in combination with parallel synthesis to discover high-affinity ligands for Bcl-xL. Bcl-xL is an anti-apoptotic protein that plays a key role in maintaining cellular homeostasis. However, its overexpression can lead to oncogenic transformation and is responsible for drug resistance in certain types of cancer making it an attractive candidate as a drug target [89]. Abbott found an initial biaryl ligand for Bcl-xL of which the affinity was increased by four orders of magnitude after subsequent optimization. The solubility issues and inadvertent binding to serum albumin were also overcome using structure-guided synthesis.
3. Expert opinion
3.1 Further automation and collaborative data management systems
Solving macromolecule structure with X-ray crystallography is a multistep and labor intensive process. Recently, many automated tools have been developed to accelerate many steps of crystal preparation. Construct preparation and cloning have been accelerated as a result of development of ligation independent cloning. Multi-channel liquid chromatography systems such as AKTA Express (GE Healthcare) have been introduced for automated protein purification. Preparation of crystallization plates has become much easier as a result of extensive development of liquid handlers, screen design tools and crystallization plate setup robots (Mosquito from TTP Labtech, NT8 from Formulatrix, Gryphon from ARI, etc.). Automated plate observation systems such as the Minstrel (Rigaku) speed up the process of reviewing crystallization results.
We expect further advancement of ligand screening methods in the near future. There is already quite extensive use of absorption and fluorescence-based screening plate readers, but methods like ITC or dynamic light scattering are also moving in the direction of multi-well plates, small experimental volumes, and the ability to test many different protocols with minimal human intervention (MicroCal™ Auto-iTC200, DynaPro™ Plate Reader II, etc.). Crystallographic fragment-based screening will become more widely used as the compound libraries become better optimized, semi-automated structure solutions will be more widely utilized, and the laboratory management systems and databases will become of even greater help in managing experiments and data.
The coming years will bring further automation in harvesting raw data, curation of data already deposited in databases, and automated integration of results originating from different sources. Algorithms will integrate data from many biochemical tests and will be able to return ligands with significant affinity to a target protein as reported by multiple different techniques. Information obtained in this way will be used to design subsequent crystallization experiments or select a group of promising compounds for additional rational drug development.
Recent concerns about reproducibility of biomedical experiments trend to force the storage and public availability of raw data after its publication. This will require development of databases and laboratory-management systems that can handle large amounts of data and automatically update databases upon completion of experiments. These systems will allow researchers to share information between all labs involved in the project and will send notifications about all recent changes in the project to all members of a team. In addition, there will be significant development of methods and algorithms for automated integration of results from different experiments within one database. The seamless integration of raw data from different structural biology technologies will require further advances in structure analysis and interpretation tools in addition to the availability of sophisticated data management systems.
The direction of most modern techniques used to extend functional knowledge about proteins of interest should not only be towards increasing throughput or even efficiency, but also to increase reliability and reproducibility of functional results. The replacement of 24-well plates by 96- and 384-well ones would increase the speed of data harvesting and decrease the costs of a single experiment, but does not necessarily decrease the cost of reliable functional assignment. This brute force approach has its limitations, especially when applied to science. For that reason, the ‘high throughput’ paradigm should be replaced by a ‘high output’ concept. However, this approach requires more thinking than just automation and advanced data management techniques.
3.2 Increased use of hybrid biophysical methods to identify and characterize potent drug target
For years, we have been able to visualize elements of the cell, but even the most powerful microscopes cannot show the 3D structure of proteins and other macromolecules directly in the living organism. Structural biology has overcome these limitations by determining protein structures in vitro and opening the path to rational structure-based drug design. A thorough understanding of the structural features of a drug target is multi-faceted, including its 3D structure, its dynamic conformations, and chemical modification patterns. The main technique used to determine 3D structures of target protein is X-ray crystallography, which provided the basis for development of techniques and software used for structure solution. However, many drug targets (especially in their actionable conformation) are located in or on the membrane, and thus these targets may not be easily crystallizable. Therefore, other structural biology techniques, such as cryo-EM, NMR and solid-state NMR, have become indispensable alternative approaches for structure determination.
In the future, X-ray crystallography will continue to serve as the central source of experimental structural biology data used in novel drug discovery. However, other structural biology techniques are going to be used more extensively not only to overcome the major limitation of X-ray crystallography, that is the availability of diffraction quality crystals, but also to provide complementary structural data that is useful in drug discovery. Cryo-EM is expected to bring major breakthroughs in structural biology as further technological and software developments increase the achievable resolution, as well as the accessibility of cryo-EM experiments for more general users [78,80].
It is often insufficient to determine 3D structures of protein and characterize its biochemical properties in vitro. A continuous challenge lies in the interplay between proteins in macromolecular assemblies or even various metabolic pathways in the whole cell. As more challenging targets, assemblies, and pathways await our elucidation, data derived from just one protein using a single technique are often insufficient. In the midst of the increasing demand of using multiple structure determination techniques, we expect to see more and more labs specializing in >1 structural biology technique (e.g., both cryo-EM and X-ray crystallography) to tackle difficult structural biology questions.
Recent years have shown the maturation of experimental structural biology techniques, led by X-ray crystallography. With the maturity and increased automation of structural biology techniques lowering the effort and cost of determining macromolecular structures, researchers can focus more on the drug targets of their interest than on the structural biology techniques used to determine them. We expect to see the trend that as experimental structural biology techniques mature, they are/will gradually become routine procedures and/or commercialized.
3.3 Enhancement in reproducibility
In the near future, we should see an increase in the reliability of fundamental research due to development of better databases with automatic data curation and improvement of policies in academia and industry. Various factors which positively affect reproducibility include improved validation tools, stricter procedures for deposition of data to databanks such as PDB and EMDB [30], creating and developing tools for raw data deposition which will enable validation and/or re-interpretation of published hypotheses, novel open publishing models [124] and last but not least, stricter NIH regulations [40]. The establishment of consortia to repeat the most important experiments will further assess the reproducibility of results obtained from various techniques. Further development of automated data harvesting will eliminate censoring of negative results and reduce cases of inaccurate descriptions of experimental results. Elimination of the human factor from the data harvesting process will eliminate the effect of ‘hidden variables’, variables which are not recorded during the experiment, such as the batches of chemicals used, uncontrolled laboratory conditions, undocumented details of the protocol, and so on, but might be important for data interpretation. An illustration of the importance of hidden parameters is this: one can describe the procedure for preparing a cup of sweet tea in extraordinary detail, such as the shape and size of the teapot, the preparation of the tea leaves, the volume and purity of the water, the size and type of cup, how the beverage should be stirred, and so on. Yet regardless how comprehensive the instructions may be, if they fail to note one detail–that sugar should also be added–anyone who tries to follow them will be disappointed with the results. This problem is magnified as people have a tendency to focus on the final steps, as one might try to stir the tea in different ways in order to make it sweet.
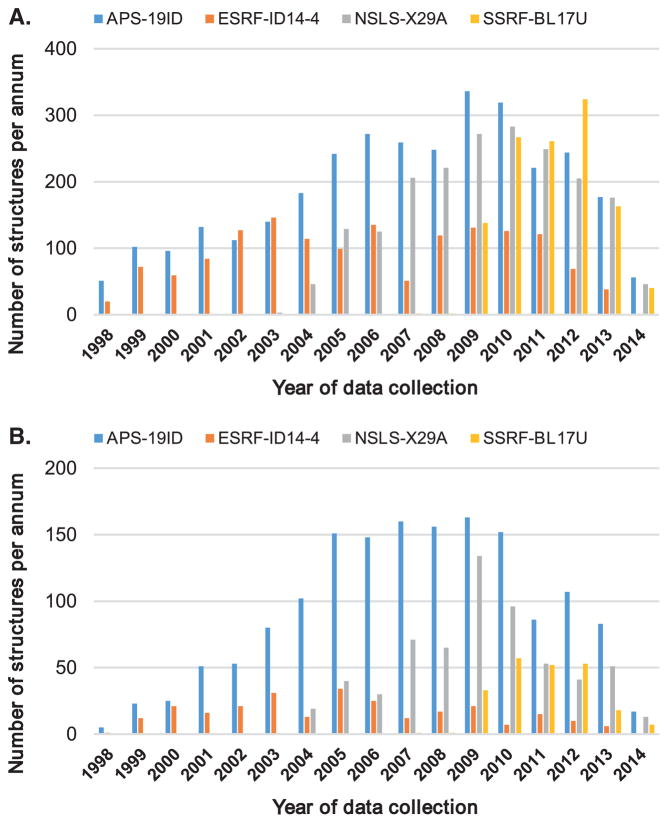
A similar story may be told of microfocus synchrotron beamlines. It was necessary for these beamlines to develop or acquire high quality sample alignment systems that work for both very small and large samples. Many synchrotron datasets collected with use of obsolete sample alignment systems (where the camera angle is not coincident with the collimator) are affected by poor sample alignment. The strong variability in the productivity of beamlines [41] depends strongly on the ability of beamline personnel to provide protocols that would allow researchers to use complicated beamline hardware and software systems optimally. It is somewhat surprising that one of the most productive beamlines in 2012 (NSLS-X19A) was a station of an old, ‘last generation’ synchrotron that recently closed, and used an old detector of which production was terminated in order to make a supposedly better, faster and much more expensive state-of-the-art detector (Figure 3). The two examples show that sometimes one can significantly improve a drug discovery pipeline with better protocols rather than multiple millions of dollars on new equipment. Despite advances in automation, the expert judgement of experienced scientists is the most valuable part of the drug discovery process.
Figure 3.
Productivity of a selected set of the most productive synchrotron beamlines around the world for all structures (A); and for structures phased using Single-wavelength anomalous dispersion (SAD)/Multiple-wavelength anomalous dispersion (MAD) (B).
Article highlights.
X-ray crystallography will continue to serve as the central source of experimental structural data used in novel drug discovery.
The fundamental limitations of X-ray crystallography may be complemented using methods and technologies in other areas of structural biology.
Integration of data from different biophysical methods for structure determination provides a panoply of information invaluable for target-based drug discovery.
Single-particle cryo-EM is expected to bring major breakthroughs in structural biology by achieving near crystallographic resolution.
Accurate and integrated data management solutions are crucial for developing experimental procedures that are robust and reproducible.
This box summarizes key points contained in the article.
Footnotes
Declaration of interests
The authors’ research was supported with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272201200026C, by NIH grants GM093342, GM094585, GM094662, and HG008424. The authors note that they have also been involved in the development of state-of-the-art software, data management and mining tools; some of them were commercialized by HKL Research and are mentioned in the paper. W Minor is co-founder of HKL Research and member of the board. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Van Noorden R, Maher B, Nuzzo R. The top 100 papers. Nature. 2014;514(7524):550–3. doi: 10.1038/514550a. [DOI] [PubMed] [Google Scholar]
- 2.Almo SC, Garforth SJ, Hillerich BS, et al. Protein production from the structural genomics perspective: Achievements and future needs. Curr Opin Struct Biol. 2013;23(3):335–44. doi: 10.1016/j.sbi.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almo SC, Love JD. Better and faster: Improvements and optimization for mammalian recombinant protein production. Curr Opin Struct Biol. 2014;26:39–43. doi: 10.1016/j.sbi.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson WF. Structural genomics and drug discovery for infectious diseases. Infect Disord Drug Targets. 2009;9(5):507–17. doi: 10.2174/187152609789105713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson WF, editor. Structural genomics and drug discovery. 1. Humana Press; New York City: 2014. [Google Scholar]
- 6.Chen YW, editor. Structural genomics. Humana Press; New York City: 2014. [Google Scholar]
- 7.Grabowski M, Chruszcz M, Zimmerman MD, et al. Benefits of structural genomics for drug discovery research. Infect Disord Drug Targets. 2009;9(5):459–74. doi: 10.2174/187152609789105704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joachimiak A. High-throughput crystallography for structural genomics. Curr Opin Struct Biol. 2009;19(5):573–84. doi: 10.1016/j.sbi.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grabowski M, Joachimiak A, Otwinowski Z, et al. Structural genomics: Keeping up with expanding knowledge of the protein universe. Curr Opin Struct Biol. 2007;17(3):347–53. doi: 10.1016/j.sbi.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newman J, Egan D, Walter TS, et al. Towards rationalization of crystallization screening for small- to medium-sized academic laboratories: The PACT/JCSG+ strategy. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 10):1426–31. doi: 10.1107/S0907444905024984. [DOI] [PubMed] [Google Scholar]
- 11.Dong A, Xu X, Edwards AM, et al. In situ proteolysis for protein crystallization and structure determination. Nat Methods. 2007;4(12):1019–21. doi: 10.1038/nmeth1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y, Quartey P, Li H, et al. Large-scale evaluation of protein reductive methylation for improving protein crystallization. Nat Methods. 2008;5(10):853–4. doi: 10.1038/nmeth1008-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupeux F, Rower M, Seroul G, et al. A thermal stability assay can help to estimate the crystallization likelihood of biological samples. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 11):915–19. doi: 10.1107/S0907444911036225. [DOI] [PubMed] [Google Scholar]
- 14•.Seabrook SA, Newman J. High-throughput thermal scanning for protein stability: Making a good technique more robust. ACS Comb Sci. 2013;15(8):387–92. doi: 10.1021/co400013v. Fluorescence-based thermal shift assays are a cost-effective high-throughput method routinely employed in drug discovery. [DOI] [PubMed] [Google Scholar]
- 15.Wilson WW. Light scattering as a diagnostic for protein crystal growth–a practical approach. J Struct Biol. 2003;142(1):56–65. doi: 10.1016/s1047-8477(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 16.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–22. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 17.Minor W, Cymborowski M, Otwinowski Z, et al. HKL-3000: The integration of data reduction and structure solution - from diffraction images to an initial model in minutes. Acta Crystallogr D. 2006;62:859–66. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 18.Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Cryst D. 2010;66(Pt 2):213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murshudov GN, Skubak P, Lebedev AA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D. 2011;67(Pt 4):355–67. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emsley P, Lohkamp B, Scott WG, et al. Features and development of coot. Acta Crystallogr D. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panjikar S, Parthasarathy V, Lamzin VS, et al. Auto-rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 4):449–57. doi: 10.1107/S0907444905001307. [DOI] [PubMed] [Google Scholar]
- 22.Berman HM, Coimbatore Narayanan B, Di Costanzo L, et al. Trendspotting in the protein data bank. FEBS Lett. 2013;587(8):1036–45. doi: 10.1016/j.febslet.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khafizov K, Madrid-Aliste C, Almo SC, et al. Trends in structural coverage of the protein universe and the impact of the protein structure initiative. Proc Natl Acad Sci USA. 2014;111(10):3733–8. doi: 10.1073/pnas.1321614111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webb B, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2014;1137:1–15. doi: 10.1007/978-1-4939-0366-5_1. [DOI] [PubMed] [Google Scholar]
- 25•.Chruszcz M, Wlodawer A, Minor W. Determination of protein structures–a series of fortunate events. Biophys J. 2008;95(1):1–9. doi: 10.1529/biophysj.108.131789. A review discussing the difficulties presented by each step of X-ray crystallography that may affect the overall experimental productivity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Wlodawer A, Minor W, Dauter Z, et al. Protein crystallography for aspiring crystallographers or how to avoid pitfalls and traps in macromolecular structure determination. FEBS J. 2013;280(22):5705–36. doi: 10.1111/febs.12495. A paper discussing all aspects of protein crystallography, including structure quality and limitations of the technique. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wlodawer A, Minor W, Dauter Z, et al. Protein crystallography for non-crystallographers, or how to get the best (but not more) from published macromolecular structures. FEBS J. 2008;275(1):1–21. doi: 10.1111/j.1742-4658.2007.06178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng H, Chruszcz M, Lasota P, et al. Data mining of metal ion environments present in protein structures. J Inorg Biochem. 2008;102(9):1765–76. doi: 10.1016/j.jinorgbio.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chruszcz M, Domagalski M, Osinski T, et al. Unmet challenges of structural genomics. Curr Opin Struct Biol. 2010;20(5):587–97. doi: 10.1016/j.sbi.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Read RJ, Adams PD, Arendall WB, III, et al. A new generation of crystallographic validation tools for the protein data bank. Structure. 2011;19(10):1395–412. doi: 10.1016/j.str.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Majorek KA, Kuhn ML, Chruszcz M, et al. Double trouble-buffer selection and his-tag presence may be responsible for nonreproducibility of biomedical experiments. Protein Sci. 2014;23(10):1359–68. doi: 10.1002/pro.2520. A discussion of reproducibility issues and validity of results obtained in functional studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Cooper DR, Porebski PJ, Chruszcz M, et al. X-ray crystallography: Assessment and validation of protein-small molecule complexes for drug discovery. Expert Opin Drug Discov. 2011;6(8):771–82. doi: 10.1517/17460441.2011.585154. An overview of recent approaches for validation of results obtained by X-ray crystallography as used in drug discovery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen VB, Arendall WB, III, Headd JJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weichenberger CX, Pozharski E, Rupp B. Visualizing ligand molecules in twilight electron density. Acta Crystallogr F. 2013;69(Pt 2):195–200. doi: 10.1107/S1744309112044387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pozharski E, Weichenberger CX, Rupp B. Techniques, tools and best practices for ligand electron-density analysis and results from their application to deposited crystal structures. Acta Crystallogr D. 2013;69:150–67. doi: 10.1107/S0907444912044423. [DOI] [PubMed] [Google Scholar]
- 36.Debreczeni JE, Emsley P. Handling ligands with coot. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):425–30. doi: 10.1107/S0907444912000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng H, Chordia MD, Cooper DR, et al. Validation of metal-binding sites in macromolecular structures with the CheckMyMetal web server. Nat Protoc. 2014;9(1):156–70. doi: 10.1038/nprot.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chruszcz M, Chapman MD, Vailes LD, et al. Crystal structures of mite allergens der f 1 and der p 1 reveal differences in surface-exposed residues that may influence antibody binding. J Mol Biol. 2009;386(2):520–30. doi: 10.1016/j.jmb.2008.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Shabalin IG, Dauter Z, Jaskolski M, et al. Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin. Acta Crystallogr D. 2015:D71. doi: 10.1107/S139900471500629X. A paper illustrating the importance of metal-binding chemistry in X-ray crystallography, using platinum-based drugs as examples. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collins FS, Tabak LA. Policy: NIH plans to enhance reproducibility. Nature. 2014;505(7485):612–13. doi: 10.1038/505612a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41••.Zheng H, Hou J, Zimmerman MD, et al. The future of crystallography in drug discovery. Expert Opin Drug Discov. 2014;9(2):125–37. doi: 10.1517/17460441.2014.872623. A review discussing how the limitations of crystal structures should be addressed in the context of drug discovery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ezkurdia I, Juan D, Rodriguez JM, et al. The shrinking human protein coding complement: Are there now fewer than 20,000 genes? ArXiv e-Prints. 2013.arXiv:1312.7111. Available from: http://adsabs.harvard.edu/abs/2013arXiv1312.7111E.
- 43.Mancia F, Love J. High throughput platforms for structural genomics of integral membrane proteins. Curr Opin Struct Biol. 2011;21(4):517–22. doi: 10.1016/j.sbi.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kloppmann E, Punta M, Rost B. Structural genomics plucks high-hanging membrane proteins. Curr Opin Struct Biol. 2012;22(3):326–32. doi: 10.1016/j.sbi.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DiMaio F, Song Y, Li X, et al. Atomic-accuracy models from 4.5-A cryo-electron microscopy data with density-guided iterative local refinement. Nat Methods. 2015;12(4):361–5. doi: 10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46••.Wang RY, Kudryashev M, Li X, et al. De novo protein structure determination from near-atomic-resolution cryo-EM maps. Nat Methods. 2015;12(4):335–8. doi: 10.1038/nmeth.3287. Describes a method to determine atomic protein structure from near-crystallographic resolution electron microscopy maps. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fischer N, Neumann P, Konevega AL, et al. Structure of the E. coli ribosome-EF-tu complex at <3 A resolution by C-corrected cryo-EM. Nature. 2015;520(7548):567–70. doi: 10.1038/nature14275. [DOI] [PubMed] [Google Scholar]
- 48.Kudryashev M, Wang RY, Brackmann M, et al. Structure of the type VI secretion system contractile sheath. Cell. 2015;160(5):952–62. doi: 10.1016/j.cell.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Linser R, Bardiaux B, Higman V, et al. Structure calculation from unambiguous long-range amide and methyl 1H-1H distance restraints for a microcrystalline protein with MAS solid-state NMR spectroscopy. J Am Chem Soc. 2011;133(15):5905–12. doi: 10.1021/ja110222h. [DOI] [PubMed] [Google Scholar]
- 50.Torchia DA. NMR studies of dynamic biomolecular conformational ensembles. Prog Nucl Magn Reson Spectrosc. 2015;84–85C:14–32. doi: 10.1016/j.pnmrs.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barends TR, Brosi RW, Steinmetz A, et al. Combining crystallography and EPR: Crystal and solution structures of the multidomain cochaperone DnaJ. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 8):1540–52. doi: 10.1107/S0907444913010640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fleissner MR, Cascio D, Hubbell WL. Structural origin of weakly ordered nitroxide motion in spin-labeled proteins. Protein Sci. 2009;18(5):893–908. doi: 10.1002/pro.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y, Liu K, Qin S, et al. Epigenetic targets and drug discovery: Part 1: Histone methylation. Pharmacol Ther. 2014;143(3):275–94. doi: 10.1016/j.pharmthera.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 54.Liu K, Liu Y, Lau JL, et al. Epigenetic targets and drug discovery part 2: Histone demethylation and DNA methylation. Pharmacol Ther. 2015;151:121–40. doi: 10.1016/j.pharmthera.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 55.Ren RJ, Dammer EB, Wang G, et al. Proteomics of protein post-translational modifications implicated in neurodegeneration. Transl Neurodegener. 2014;3(1):23. doi: 10.1186/2047-9158-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song F, Chen P, Sun D, et al. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344(6182):376–80. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- 57.Srivastava HK, Bohari MH, Sastry GN. Modeling anti-HIV compounds: The role of analogue-based approaches. Curr Comput Aided Drug Des. 2012;8(3):224–48. doi: 10.2174/157340912801619085. [DOI] [PubMed] [Google Scholar]
- 58.Wlodawer A. Rational approach to AIDS drug design through structural biology. Annu Rev Med. 2002;53:595–614. doi: 10.1146/annurev.med.53.052901.131947. [DOI] [PubMed] [Google Scholar]
- 59.Feng E, Ye D, Li J, et al. Recent advances in neuraminidase inhibitor development as anti-influenza drugs. ChemMedChem. 2012;7(9):1527–36. doi: 10.1002/cmdc.201200155. [DOI] [PubMed] [Google Scholar]
- 60.Callens M, Hannaert V. The rational design of trypanocidal drugs: Selective inhibition of the glyceraldehyde-3-phosphate dehydrogenase in trypanosomatidae. Ann Trop Med Parasitol. 1995;89(Suppl 1):23–30. doi: 10.1080/00034983.1995.11813011. [DOI] [PubMed] [Google Scholar]
- 61.Kern J, Hattne J, Tran R, et al. Methods development for diffraction and spectroscopy studies of metalloenzymes at X-ray free-electron lasers. Philos Trans R Soc Lond B Biol Sci. 2014;369(1647):20130590. doi: 10.1098/rstb.2013.0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Faruqi AR, Cross RA, Kendrick-Jones J. Small angle X-ray scattering studies on myosin. J Cell Sci Suppl. 1991;14:23–6. doi: 10.1242/jcs.1991.supplement_14.5. [DOI] [PubMed] [Google Scholar]
- 63.Zhou ZH. Atomic resolution cryo electron microscopy of macromolecular complexes. Adv Protein Chem Struct Biol. 2011;82:1–35. doi: 10.1016/B978-0-12-386507-6.00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoenger A. High-resolution cryo-electron microscopy on macromolecular complexes and cell organelles. Protoplasma. 2014;251(2):417–27. doi: 10.1007/s00709-013-0600-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiang J, Pentelute BL, Collier RJ, et al. Atomic structure of anthrax protective antigen pore elucidates toxin translocation. Nature. 2015;521(7553):545–9. doi: 10.1038/nature14247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nannenga BL, Shi D, Leslie AG, et al. High-resolution structure determination by continuous-rotation data collection in MicroED. Nat Methods. 2014;11(9):927–30. doi: 10.1038/nmeth.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schroder GF. Hybrid methods for macromolecular structure determination: Experiment with expectations. Curr Opin Struct Biol. 2015;31:20–7. doi: 10.1016/j.sbi.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 68.Grimes JM, Fuller SD, Stuart DI. Complementing crystallography: The role of cryo-electron microscopy in structural biology. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 10):1742–9. doi: 10.1107/s0907444999009956. [DOI] [PubMed] [Google Scholar]
- 69.Tao Y, Zhang W. Recent developments in cryo-electron microscopy reconstruction of single particles. Curr Opin Struct Biol. 2000;10(5):616–22. doi: 10.1016/s0959-440x(00)00139-1. [DOI] [PubMed] [Google Scholar]
- 70.Amunts A, Brown A, Bai XC, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343(6178):1485–9. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiong Y. From electron microscopy to X-ray crystallography: Molecular-replacement case studies. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 1):76–82. doi: 10.1107/S090744490705398X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dodson EJ. Using electron-microscopy images as a model for molecular replacement. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 10):1405–9. doi: 10.1107/s0907444901013415. [DOI] [PubMed] [Google Scholar]
- 73.Reinisch KM, Nibert ML, Harrison SC. Structure of the reovirus core at 3.6 A resolution. Nature. 2000;404(6781):960–7. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- 74.Miyaguchi K. Direct imaging electron microscopy (EM) methods in modern structural biology: Overview and comparison with X-ray crystallography and single-particle cryo-EM reconstruction in the studies of large macromolecules. Biol Cell. 2014;106(10):323–45. doi: 10.1111/boc.201300081. [DOI] [PubMed] [Google Scholar]
- 75.Belnap DM, McDermott BM, Jr, Filman DJ, et al. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc Natl Acad Sci USA. 2000;97(1):73–8. doi: 10.1073/pnas.97.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.He Y, Bowman VD, Mueller S, et al. Interaction of the poliovirus receptor with poliovirus. Proc Natl Acad Sci USA. 2000;97(1):79–84. doi: 10.1073/pnas.97.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Montabana EA, Agard DA. Bacterial tubulin TubZ-bt transitions between a two-stranded intermediate and a four-stranded filament upon GTP hydrolysis. Proc Natl Acad Sci U S A. 2014;111(9):3407–12. doi: 10.1073/pnas.1318339111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brown A, Long F, Nicholls RA, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71(Pt 1):136–53. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Long F, Vagin AA, Young P, et al. BALBES: A molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 1):125–32. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80••.Cheng Y. Single-particle cryo-EM at crystallographic resolution. Cell. 2015;161(3):450–7. doi: 10.1016/j.cell.2015.03.049. Excellent review introducing recent breakthroughs in single-particle cryo-EM techniques that make the determination of atomic structures from near-crystallographic resolution EM maps possible. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bai XC, McMullan G, Scheres SH. How cryo-EM is revolutionizing structural biology. Trends Biochem Sci. 2015;40(1):49–57. doi: 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 82.Adams PD, Baker D, Brunger AT, et al. Advances, interactions, and future developments in the CNS, phenix, and rosetta structural biology software systems. Annu Rev Biophys. 2013;42:265–87. doi: 10.1146/annurev-biophys-083012-130253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weingarth M, Baldus M. Solid-state NMR-based approaches for supramolecular structure elucidation. Acc Chem Res. 2013;46(9):2037–46. doi: 10.1021/ar300316e. [DOI] [PubMed] [Google Scholar]
- 84.Tang M, Sperling LJ, Berthold DA, et al. High-resolution membrane protein structure by joint calculations with solid-state NMR and X-ray experimental data. J Biomol NMR. 2011;51(3):227–33. doi: 10.1007/s10858-011-9565-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ujwal R, Cascio D, Colletier JP, et al. The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating. Proc Natl Acad Sci USA. 2008;105(46):17742–7. doi: 10.1073/pnas.0809634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hiller S, Garces RG, Malia TJ, et al. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321(5893):1206–10. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bayrhuber M, Meins T, Habeck M, et al. Structure of the human voltage-dependent anion channel. Proc Natl Acad Sci USA. 2008;105(40):15370–5. doi: 10.1073/pnas.0808115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gregory SM, Harada E, Liang B, et al. Structure and function of the complete internal fusion loop from ebolavirus glycoprotein 2. Proc Natl Acad Sci USA. 2011;108(27):11211–16. doi: 10.1073/pnas.1104760108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Petros AM, Dinges J, Augeri DJ, et al. Discovery of a potent inhibitor of the antiapoptotic protein bcl-xL from NMR and parallel synthesis. J Med Chem. 2006;49(2):656–63. doi: 10.1021/jm0507532. [DOI] [PubMed] [Google Scholar]
- 90.Prisner T, Rohrer M, MacMillan F. Pulsed EPR spectroscopy: Biological applications. Annu Rev Phys Chem. 2001;52:279–313. doi: 10.1146/annurev.physchem.52.1.279. [DOI] [PubMed] [Google Scholar]
- 91.Guo Z, Cascio D, Hideg K, et al. Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme. Protein Sci. 2007;16(6):1069–86. doi: 10.1110/ps.062739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guo Z, Cascio D, Hideg K, et al. Structural determinants of nitroxide motion in spin-labeled proteins: Solvent-exposed sites in helix B of T4 lysozyme. Protein Sci. 2008;17(2):228–39. doi: 10.1110/ps.073174008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dong M, Su X, Dzikovski B, et al. Dph3 is an electron donor for Dph1-Dph2 in the first step of eukaryotic diphthamide biosynthesis. J Am Chem Soc. 2014;136(5):1754–7. doi: 10.1021/ja4118957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang Y, Zhu X, Torelli AT, et al. Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme. Nature. 2010;465(7300):891–6. doi: 10.1038/nature09138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cieslak JA, Focia PJ, Gross A. Electron spin-echo envelope modulation (ESEEM) reveals water and phosphate interactions with the KcsA potassium channel. Biochemistry. 2010;49(7):1486–94. doi: 10.1021/bi9016523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lagerstedt JO, Petrlova J, Hilt S, et al. EPR assessment of protein sites for incorporation of gd(III) MRI contrast labels. Contrast Media Mol Imaging. 2013;8(3):252–64. doi: 10.1002/cmmi.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Singh P, Panchaud A, Goodlett DR. Chemical cross-linking and mass spectrometry as a low-resolution protein structure determination technique. Anal Chem. 2010;82(7):2636–42. doi: 10.1021/ac1000724. [DOI] [PubMed] [Google Scholar]
- 98•.Merkley ED, Cort JR, Adkins JN. Cross-linking and mass spectrometry methodologies to facilitate structural biology: Finding a path through the maze. J Struct Funct Genomics. 2013;14(3):77–90. doi: 10.1007/s10969-013-9160-z. A review that shows the experimental considerations for successful application of chemical cross-linking coupled with mass spectrometry, illustrated by examples. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99•.Shumilin IA, Cymborowski M, Chertihin O, et al. Identification of unknown protein function using metabolite cocktail screening. Structure. 2012;20(10):1715–25. doi: 10.1016/j.str.2012.07.016. Describes the use of metabolite cocktail soaking methods to characterize binding compounds. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100••.Zimmerman MD, Grabowski M, Domagalski MJ, et al. Data management in the modern structural biology and biomedical research environment. Methods Mol Biol. 2014;1140:1–25. doi: 10.1007/978-1-4939-0354-2_1. Highlights the importance of data management in a new era of productive and reproducible structural biology research and structure-guided drug discovery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nienaber VL, Richardson PL, Klighofer V, et al. Discovering novel ligands for macromolecules using X-ray crystallographic screening. Nat Biotechnol. 2000;18(10):1105–8. doi: 10.1038/80319. [DOI] [PubMed] [Google Scholar]
- 102.Badger J. Crystallographic fragment screening. Methods Mol Biol. 2012;841:161–77. doi: 10.1007/978-1-61779-520-6_7. [DOI] [PubMed] [Google Scholar]
- 103.Zhang R, Monsma F. Fluorescence-based thermal shift assays. Curr Opin Drug Discov Devel. 2010;13(4):389–402. [PubMed] [Google Scholar]
- 104.Sasse J, Gallagher SR. Staining proteins in gels. Curr Protoc Immunol. 2004;Chapter 8(Unit 8):9. doi: 10.1002/0471142735.im0809s58. [DOI] [PubMed] [Google Scholar]
- 105.Giuliani SE, Frank AM, Collart FR. Functional assignment of solute-binding proteins of ABC transporters using a fluorescence-based thermal shift assay. Biochemistry. 2008;47(52):13974–84. doi: 10.1021/bi801648r. [DOI] [PubMed] [Google Scholar]
- 106.Lo MC, Aulabaugh A, Jin G, et al. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal Biochem. 2004;332(1):153–9. doi: 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 107.Ciulli A, Abell C. Fragment-based approaches to enzyme inhibition. Curr Opin Biotechnol. 2007;18(6):489–96. doi: 10.1016/j.copbio.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Freyer MW, Lewis EA. Isothermal titration calorimetry: Experimental design, data analysis, and probing macromolecule/ligand binding and kinetic interactions. Methods Cell Biol. 2008;84:79–113. doi: 10.1016/S0091-679X(07)84004-0. [DOI] [PubMed] [Google Scholar]
- 109.Torres FE, Recht MI, Coyle JE, et al. Higher throughput calorimetry: Opportunities, approaches and challenges. Curr Opin Struct Biol. 2010;20(5):598–605. doi: 10.1016/j.sbi.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.van de Weert M, Stella L. Fluorescence quenching and ligand binding: A critical discussion of a popular methodology. J Mol Struct. 2011;998(1–3):144–50. [Google Scholar]
- 111••.Lea WA, Simeonov A. Fluorescence polarization assays in small molecule screening. Expert Opin Drug Discov. 2011;6(1):17–32. doi: 10.1517/17460441.2011.537322. A broad overview of fluorescence polarization assays and their novel application to various drug targets, as well as recent advances in the field. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lakowicz JR, editor. Principles of fluorescence spectroscopy. 3. Springer; New York City: 2006. [Google Scholar]
- 113.Campbell CT, Kim G. SPR microscopy and its applications to high-throughput analyses of biomolecular binding events and their kinetics. Biomaterials. 2007;28(15):2380–92. doi: 10.1016/j.biomaterials.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 114.Patching SG. Surface plasmon resonance spectroscopy for characterisation of membrane protein-ligand interactions and its potential for drug discovery. Biochim Biophys Acta. 2014;1838(1 Pt A):43–55. doi: 10.1016/j.bbamem.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 115.Myszka DG, Rich RL. Drug Discovery World. 2003. Spring. SPR’s high impact on drug discovery: Resolution, throughput, and versatility; pp. 1–5. [Google Scholar]
- 116.SPR instruments [Internet] 2015 Available from: http://www.sprpages.nl/instruments [Cited 28 May 2015]
- 117.Khan MTH, editor. Recent trends on QSAR in the pharmaceutical perceptions. Bentham Science Publishers; Sharjah, United Arab Emirates: 2012. [Google Scholar]
- 118.Kinch MS, Haynesworth A, Kinch SL, et al. An overview of FDA-approved new molecular entities: 1827–2013. Drug Discov Today. 2014;19(8):1033–9. doi: 10.1016/j.drudis.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 119•.Alex AA, Millan DS. Contribution of structure-based drug design to the discovery of marketed drugs. In: Livingstone DJ, Davis AM, editors. Drug Design Strategies: Quantitative Approaches. Royal Society of Chemistry Publishing; Cambridge, UK: 2011. pp. 108–63. A review that highlights the impact and limitations of various structural biology methods, particularly when assessing the factors to consider in structure-based drug design during lead generation and subsequent optimization. [Google Scholar]
- 120.Hartshorn MJ, Murray CW, Cleasby A, et al. Fragment-based lead discovery using X-ray crystallography. J Med Chem. 2005;48(2):403–13. doi: 10.1021/jm0495778. [DOI] [PubMed] [Google Scholar]
- 121.Wyatt PG, Woodhead AJ, Berdini V, et al. Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H-pyrazole-3-carboxamide (AT7519), a novel cyclin dependent kinase inhibitor using fragment-based X-ray crystallography and structure based drug design. J Med Chem. 2008;51(16):4986–99. doi: 10.1021/jm800382h. [DOI] [PubMed] [Google Scholar]
- 122.Sharma PS, Sharma R, Tyagi R. Inhibitors of cyclin dependent kinases: Useful targets for cancer treatment. Curr Cancer Drug Targets. 2008;8(1):53–75. doi: 10.2174/156800908783497131. [DOI] [PubMed] [Google Scholar]
- 123.Astex pharmaceuticals. clinical pipeline products [Internet] 2015 Available from: http://astx.com/pipeline/products/clinical [Cited 28 April 2015]
- 124.The center for open science [Internet] 2015 Available from: http://centerforopenscience.org [Cited 28 April 2015]