Abstract
Bacterial isolates from 30 farmed bullfrogs (Lithobates catesbeianus) weighing 500-600 g at Johore, Malaysia with external clinical signs of ulcer, red leg and torticollis were tested for their antibiograms and heavy metal tolerance patterns. A total of 17 bacterial species with 77 strains were successfully isolated and assigned to 21 antibiotics and 4 types of heavy metal (Hg2+, Cr6+, Cd2+, Cu2+). Results revealed that bacteria were resistant against lincomycin (92%), oleandomycin (72.7%) and furazolidone (71.4%) while being susceptible to chloramphenicol and florfenicol at 97.4%. The multiple antibiotic resistance (MAR) index for C. freundii, E. coli and M. morganii was high with the value up to 0.71. Bacterial strains were found to exhibit 100 % resistance to chromium and mercury. High correlation of resistance against both antibiotics and heavy metals was found (71.4 to 100%) between bullfrog bacteria isolates, except bacteria that were resistant to kanamycin showed only 25% resistance against Cu2+. Based on the results in this study, bacterial pathogens of bullfrog culture in Johore, Malaysia, were highly resistant to both antibiotics and heavy metals.
Keywords: Antibiotics, Heavy metal, Bullfrog, Bacteria
Introduction
Overdosing of antibiotics in feed and excessive use of chemicals in prophylaxis has caused bacteria to become antibiotic-heavy metal resistant. Their residue may stay in the environment and could transfer to other bacteria via antibiotic resistance genes, which are often located in plasmids and transposons (Gillings et al., 2008). Horizontal gene transfer among microorganisms is an important pathway for acquisition of antibiotic and heavy metal resistance in bacterial pathogens. Interaction by the co-resistance of the specific genes can confer resistance to both antibiotics and heavy metals (Baker-Austin et al., 2006; Stepanauskas et al., 2006). Resistance to antibiotics and heavy metals in frogs (Rana ridibunda) and rice frogs (Fejervarya limnocharis) have been reported (Vogiatzis and Loumbourdis, 1998; Othman et al., 2009).
The hazards present in frog farm, mainly due to use of chemicals for treatment of diseases, stay inherently in the farmed products, and remain a health risk in public concerns (Boyd and Massaut, 1999).
According to the statistical data by FAO (2010), the global production of bullfrogs in 2009 was 1439 tons with the estimated value of 6,007,000 USD, with a 15.4% increase compared to the previous year. Besides, The United Nations’ Commodity Trade Statistic Database (United Nations Statistics Division, 2008) reported that major exporting frog legs countries of bullfrog were Indonesia, China, Belgium and Luxembourg. The present study investigated the antibiogram and heavy metal tolerances of bullfrog bacteria in Malaysia.
Materials and Methods
Samples
Thirty bullfrogs (Lithobates catesbeianus) weighing 500-600 g with external clinical signs such as ulcer, red leg and torticollis were brought from a bullfrog farm located at Johore (02o15.549’ N, 102o39.261’ E). Bullfrogs were euthanized by transdermal exposure to 1.0% solution of buffered MS-222 (McDaniel et al., 2008). Internal organs (liver, kidney, spleen, heart, intestine, lung, ovary and gall bladder) were aseptically – excised and homogenized for 15 min in distilled physiological saline. Two-fold serial dilutions were plated in triplicates on Glutamate Starch Phenol Red Agar (GSP agar), MacConkey agar, Xylose Lysine Deoxycholate Agar (XLD agar), Thiosulfate-Citrate-Bile Salts-Sucrose Agar (TCBS agar), Baird-parker agar and Trypticase Soy Agar (TSA) (Oxoid, England). Plates were incubated at 28oC for 24 to 48 h and counted for colony forming units (cfu) per gram. A total of 17 bacterial species with 77 strains were identified by indole, oxidase, hemolysis tests on horse blood agar and commercial biochemical test, BBL Crystal TM Enteric/Nonfermenter Identification System (Becton Dickinson, USA).
They were Acinetobacter lwoffii, Aeromonas hydrophila, Aeromonas caviae, Chryseobacterium indologenes, Citrobacter freundii, Citrobacter amalonaticus, Edwardsiella tarda, Elizabethkingia meningoseptica, Escherichia coli, Escherichia hermannii, Morganella morganii, Pantoea agglomerans, Plesiomonas shigelloides, Pseudomonas aeruginosa, Serratia liquefaciens, Shewanella putrefaciens and Stenotrophomonas maltophilia, as described by Cappuccino and Sherman (2002).
Antibiogram
A total of 21 commercial antibiotic discs were used as follows: ampicillin (AMP 25 µg), amoxicillin (AML 10 µg), chloramphenicol (C 30 µg), colistin sulphate (CT 25 µg), doxycycline (DO 30 µg), erythromycin (E 15 µg), florfenicol (FFC 30 µg), flumequine (UB 30 µg), fosfomycin (FOS 50 µg), furazolidone (FR 15 µg), kanamycin (K 30 µg), lincomycin (MY 15 µg), nalidixic acid (NA 30 µg), nitrofurantoin (F 50 µg), novobiocin (NV 30 µg), oleandomycin (OL 15 µg), oxolinic acid (OA 2 µg), oxytetracycline (OT 30 µg), spiramycin (SP 100 µg), tetracycline (TE 30 µg), and sulphamethoxazole (RL 25 µg) (Oxoid, England). Bacterial suspensions were adjusted to 0.5 McFarland. The antibiotic discs were placed on the surface of the medium by using sterile forceps and incubated at 28°C for 24 h. Diameter of inhibition zones around the discs were measured in millimeter (mm) and characterized as Sensitive (S), Intermediate (I) and Resistance (R) according to Clinical and Laboratory Standard Institute (CLSI, 2006).
Multiple Antibiotic Resistance (MAR) Test
The multiple antibiotic resistance (MAR) index of bacterial strains against antibiotics was calculated based on method used by Krumperman (1983) as follow: MAR index= X/(Y x Z). Where, X: Total bacteria resistant to antibiotics; Y: Total antibiotic used and Z: Total isolates. MAR index value less than 0.20 indicated that the antibiotics are seldom and never used, whereas a value greater than 0.20 suggests that the antibiotics are exposed to the bacteria.
Heavy Metal Tolerance Test
In heavy metal studies, bacterial cultures were grown for 24 h at 37°C on plates containing Trypticase Soy Agar (Oxoid, England) supplemented with Mercuric Chloride (HgCl2) (Amresco, Ohio) at 2.5 µg/ml, 5.0 µg/ml, 10.0 µg/ml and 20.0 µg/ml; Potassium Dichromate (K2Cr2O7) (Hamburg, Germany) and Cadmium Chloride Anhydrous (CdCl) (Fluka, USA) at 25 µg/ml, 50 µg/ml, 100 µg/ml, 200 µg/ml and 400 µg/ml; Cooper II Sulphate (CuSO4) (Nacalai Tesque, Japan) at 150 µg/ml, 300 µg/ml, 600 µg/ml, 1200 µg/ml and 2400 µg/ml.
Heavy metal resistant indicative values were: 10 µg/ml for mercury; 100 µg/ml for cadmium and chromium; and 600 µg/ml for copper, respectively (Miranda and Castillo, 1998).
Results
Antimicrobial resistance patterns
Out of the 77 bacterial isolates tested, 71 isolates were resistant to lincomycin (92%), 56 isolates and 55 isolates were resistant to oleandomycin (72.7%) and furazolidone (71.4%), respectively. On the other hand, there were only two isolates (2.6%) resistant to chloramphenicol and florfenicol (Figure 1).
Fig. 1.
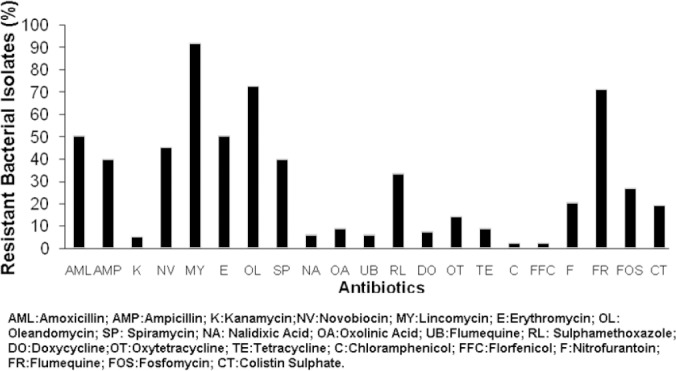
Percentage of bacterial strains resistance to antibiotics.
The majority of the bacteria such as E. meningoseptica, C. freundii, E. coli, E. hermanii, E. tarda, M. morganii, P. agglomerans, P. shigelloides, P. aeruginosa and S. maltophilia were found to be resistant to lincomycin, followed by oleandomycin (Table 1).
Table 1.
Percentage of bacterial resistance to various antibiotics.
| Antibiotic Resistance profile (%) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Organism | NA | CT | AMP | DO | OL | SP | K | OA | NV | FFC | TE | FR | FOS | OT | AML | F | UB | MY | C | E | RL |
| A. Freundii (n=1) | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 100 |
| A.hydrophila(n=9) | 0 | 0 | 67 | 11 | 89 | 22 | 11 | 11 | 67 | 0 | 0 | 78 | 0 | 11 | 67 | 11 | 11 | 89 | 0 | 30 | 11 |
| A.caviae (n=1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 100 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 |
| C. indologenes(n=2) | 0 | 0 | 50 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 50 | 0 | 0 | 0 | 0 | 0 | 100 |
| E.meningoseptica(n=4) | 0 | 50 | 75 | 0 | 50 | 0 | 50 | 50 | 25 | 0 | 50 | 100 | 50 | 50 | 100 | 50 | 0 | 100 | 0 | 0 | 25 |
| C. amalonaticus(n=3) | 0 | 33 | 0 | 0 | 33 | 33 | 0 | 0 | 0 | 0 | 0 | 67 | 33 | 0 | 0 | 0 | 0 | 67 | 0 | 67 | 0 |
| C. freundii(n=17) | 12 | 6 | 18 | 6 | 82 | 41 | 0 | 6 | 71 | 0 | 6 | 82 | 12 | 12 | 35 | 12 | 6 | 100 | 6 | 41 | 29 |
| E. coli(n=4) | 50 | 50 | 75 | 50 | 100 | 75 | 0 | 50 | 50 | 0 | 25 | 75 | 25 | 50 | 75 | 25 | 50 | 100 | 0 | 100 | 75 |
| E. hermanii(n=1) | 0 | 0 | 100 | 0 | 100 | 100 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 100 | 0 |
| E. tarda(n=8) | 13 | 25 | 13 | 13 | 100 | 75 | 0 | 0 | 25 | 0 | 13 | 88 | 0 | 13 | 25 | 0 | 13 | 100 | 0 | 88 | 0 |
| M. morganii(n=10) | 0 | 70 | 90 | 10 | 90 | 90 | 0 | 0 | 40 | 0 | 10 | 80 | 90 | 10 | 90 | 70 | 0 | 100 | 0 | 90 | 90 |
| P. agglomerans(n=4) | 0 | 0 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | 50 | 0 | 25 | 0 | 0 | 100 | 0 | 0 | 0 |
| P. shigeloides(n=3) | 0 | 0 | 33 | 0 | 100 | 0 | 0 | 0 | 33 | 0 | 0 | 33 | 0 | 33 | 67 | 0 | 0 | 100 | 0 | 0 | 33 |
| P. aeruginosa(n=3) | 0 | 0 | 33 | 0 | 100 | 33 | 0 | 33 | 100 | 67 | 0 | 100 | 67 | 0 | 100 | 67 | 0 | 100 | 33 | 67 | 33 |
| S. liquefaciens(n=2) | 0 | 0 | 0 | 0 | 50 | 0 | 0 | 0 | 50 | 0 | 0 | 50 | 50 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 50 |
| S. putrefaciens(n=1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| S. maltophilia (n=4) | 0 | 0 | 25 | 0 | 25 | 25 | 25 | 0 | 25 | 0 | 0 | 50 | 25 | 0 | 25 | 25 | 0 | 100 | 0 | 25 | 25 |
AMP: ampicillin (25 µg); AML: amoxicillin (10 µg), C: chloramphenicol (30 µg), CT: colistin sulphate (25 µg), DO: doxycycline (30 µg), E: erythromycin (15 µg), FFC: florfenicol (30 µg), UB: flumequine (30 µg), FOS: fosfomycin (50 µg), FR: furazolidone (15 µg), K: kanamycin (30 µg), MY: lincomycin (15 µg), NA: nalidixic acid (30 µg), F: nitrofurantoin (50 µg), NV: novobiocin (30 µg), OL: oleandomycin (15 µg), OA: oxolinic acid (2 µg), OT: oxytetracycline (30 µg), SP: spiramycin (100 µg), TE: tetracycline (30 µg), RL: sulphamethoxazole (25 µg) (Oxoid, England).
Among the effective antibiotics against bacteria tested were chloramphenicol, florfenicol, kanamycin, doxycycline, nalidixic acid, colistin sulphate and oxolinic acid.
Antibiogram showed that P. agglomerans and S. liquefaciens were susceptible to up to 16 and 15 antibiotics, respectively. While P. aeruginosa was 100% resistant against 5 antibiotics namely oleandomycin, novobiocin, furazolidone, amoxicillin and lincomycin. Besides, it was susceptible to nalidixic acid, colistin sulphate, doxycycline, kanamycin, tetracycline, oxytetracycline and flumequine. S. putrefaciens was susceptible to all antibiotics except fosfomycin (Table 2).
Table 2.
Antibiogram patterns of bullfrog bacteria.
| Bacteria Species | Antibiotic Resistance Profiles |
|---|---|
| Acinetobacter lwoffiii | OL, E,RL |
| Aeromonas hydrophila 1 | OL, NV, MY, E |
| Aeromonas hydrophila 2 | AMP, OL, SP, NV, FR, AML, MY |
| Aeromonas hydrophila 3 | AMP, DO, OL, SP, NV, FR, OT, AML, MY |
| Aeromonas hydrophila 4 | AMP, OL, FR, AML, MY |
| Aeromonas hydrophila 5 | NV, FR, MY, E, RL |
| Aeromonas hydrophila 6 | AMP, OL, NV, FR, AML, MY |
| Aeromonas hydrophila 7 | AMP, OL, NV, FR, AML, MY |
| Aeromonas hydrophila 8 | AMP, OL, K, AML, MY |
| Aeromonas hydrophila 9 | OL, OA, FR, F, UB,E |
| Aeromonas caviae | TE, FR, MY |
| Chryseobacterium indologenes 1 | RL |
| Chryseobacterium indologenes 2 | AMP, AML, RL |
| Elizabethkingia meningoseptica 1 | OL, NV, FR, AML, MY,E |
| Elizabethkingia meningoseptica 2 | CT, AMP, K, OA, TE, FOS, OT, AML, F, MY |
| Elizabethkingia meningoseptica 3 | CT, AMP, OL, K, OA, TE, FR, FOS, OT, AML, F, MY |
| Elizabethkingia meningoseptica 4 | AMP, FR, AML, MY, RL |
| Citrobacter amalonaticus 1 | CT, OL, SP, FR, MY, E |
| Citrobacter amalonaticus 2 | FOS, MY |
| Citrobacter amalonaticus 3 | FR,E |
| Citrobacter freundii 1 | OL, SP, MY |
| Citrobacter freundii 2 | OL, SP, NV, FR, AML, MY, E |
| Citrobacter freundii 3 | AMP, OL, SP, NV, FR, AML, MY,E |
| Citrobacter freundii 4 | NA, AMP, DO, OL, SP, OA, NV, TE, FR, OT, AML, UB, MY, E, RL |
| Citrobacter freundii 5 | CT, AMP, OL, SP, FR, FOS, AML, F, MY, E, RL |
| Citrobacter freundii 6 | OL, SP, NV, FR, OT, MY, E, RL |
| Citrobacter freundii 7 | FR, F, MY |
| Citrobacter freundii 8 | MY |
| Citrobacter freundii 9 | OL, SP, NV, FR, AML, MY |
| Citrobacter freundii 10 | NA, OL, NV, FR, MY |
| Citrobacter freundii 11 | NV, FR, MY |
| Citrobacter freundii 12 | OL, NV, FR, MY |
| Citrobacter freundii 13 | OL, FR, AML, MY, E |
| Citrobacter freundii 14 | OL, NV, FR, FOS, MY, C |
| Citrobacter freundii 15 | OL, NV, FR, MY, RL |
| Citrobacter freundii 16 | OL, NV, FR, MY |
| Citrobacter freundii 17 | OL, NV, MY, E, RL |
| Escherichia coli 1 | NA, AMP, DO, OL, SP, OA, NV, TE, FR, OT, AML, UB, MY, E, RL |
| Escherichia coli 2 | NA, AMP, DO, OL, SP, OA, NV, FR, OT, AML, F, UB, MY, E, RL |
| Escherichia coli 3 | CT, OL, FR, MY, E |
| Escherichia coli 4 | CT, AMP, OL, SP, FR, FOS, AML, MY, E, RL |
| Escherichia hermanii | AMP, OL, SP, NV, AML, MY, E |
| Edwardsiella tarda 1 | OL, SP, NV, FR, AML, MY |
| Edwardsiella tarda 2 | NA, AMP, DO, OL, SP, TE, FR, OT, AML, UB, MY, E |
| Edwardsiella tarda 3 | OL, SP, FR, MY, E |
| Edwardsiella tarda 4 | CT, OL, SP, FR, MY, E |
| Edwardsiella tarda 5 | CT, OL, MY, E |
| Edwardsiella tarda 6 | OL, SP, FR, MY, E |
| Edwardsiella tarda 7 | OL, SP, FR, MY, E |
| Edwardsiella tarda 8 | OL, NV, FR, MY, E |
| Morganella morganii 1 | CT, AMP, OL, SP, NV, FR, FOS, AML, MY, E, RL |
| Morganella morganii 2 | CT, AMP, OL, SP, FR, FOS, AML, F, MY, E, RL |
| Morganella morganii 3 | CT, OL, SP, MY,E |
| Morganella morganii 4 | CT, AMP, DO, OL, SP, NV, TE, FR, FOS, OT, AML, F, MY, E, RL |
| Morganella morganii 5 | CT, AMP, OL, SP, FR, FOS, AML, F, MY, E, RL |
| Morganella morganii 6 | CT, AMP, OL, SP, FR, FOS, AML, F, MY, E, RL |
| Morganella morganii 7 | AMP, OL, SP, NV, FR, FOS, AML, F, MY, E, RL |
| Morganella morganii 8 | AMP, OL, SP, FR, FOS, AML, F, MY, E, RL |
| Morganella morganii 9 | CT, AMP, OL, SP, FR, FOS, AML, F, MY, E, RL |
| Morganella morganii 10 | AMP, NV, FOS, AML, MY, RL |
| Pantoea agglomerans 1 | FOS, MY |
| Pantoea agglomerans 1 | MY |
| Pantoea agglomerans 1 | FOS, MY |
| Pantoea agglomerans 1 | AMP, FR, AML, MY |
| Plesiomonas shigeloides 1 | OL, OT, MY, RL |
| Plesiomonas shigeloides 2 | AMP, OL, NV, FR, AML, MY |
| Plesiomonas shigeloides 3 | OL, AML, MY |
| Pseudomonas aeruginosa 1 | AMP, OL, SP, OA, NV, FFC, FR, FOS, AML, F, MY, C, E |
| Pseudomonas aeruginosa 2 | OL, NV, FR, AML, MY, RL |
| Pseudomonas aeruginosa 3 | OL, NV, FFC, FR, FOS, AML, F, MY, E |
| Serratia liquefaciens 1 | FOS, MY |
| Serratia liquefaciens 2 | OL, NV, FR, MY, RL |
| Shewanella putrefaciens | OT |
| Stenotrphomonas maltophilia 1 | FOS, MY |
| Stenotrphomonas maltophilia 2 | FR, MY |
| Stenotrphomonas maltophilia 3 | MY |
| Stenotrphomonas maltophilia 4 | AMP, OL, SP, K, NV, FR, AML, F, MY, E, RL |
AMP: ampicillin (25 µg); AML: amoxicillin (10 µg), C: chloramphenicol (30 µg), CT: colistin sulphat (25 µg), DO: doxycycline (30 µg), E: erythromycin (15 µg), FFC: florfenicol (30 µg), UB: flumequine (30 µg), FOS: fosfomycin (50 µg), FR: furazolidone (15 µg), K: kanamycin (30 µg), MY: lincomycin (15 µg), NA: nalidixic acid (30 µg), F: nitrofurantoin (50 µg), NV: novobiocin (30 µg), OL: oleandomycin (15 µg), OA: oxolinic acid (2 µg), OT: oxytetracycline (30 µg), SP: spiramycin (100 µg), TE: tetracycline (30 µg), RL: sulphamethoxazole (25 µg) (Oxoid, England).
Multiple Antibiotic Resistant (MAR) Index
The lowest MAR index value was seen with S. putrefaciens, and the highest was found in C. freundii, E. coli and M. morganii as high as 0.71 (Table 3).
Table 3.
MAR Index for bullfrog bacteria.
| Bacteria Species | Multiple Antibiotic Resistance Index |
|---|---|
| Acinetobacter lwoffii (n=1) | 0.14 |
| Aeromonas hydrophila (n=9) | 0.19-0.43 |
| Aeromonas caviae (n=1) | 0.14 |
| Chryseobacterium indologenes (n=2) | 0.05-0.14 |
| Elizabethkingia meningoseptica (n=4) | 0.24-0.57 |
| Citrobacter amalonaticus (n=3) | 0.10-0.29 |
| Citrobacter freundii (n=17) | 0.05-0.71 |
| Escherichia coli (n=4) | 0.24-0.71 |
| Escherichia hermanii (n=1) | 0.33 |
| Edwardsiella tarda (n=8) | 0.19-0.57 |
| Morganella morganii (n=10) | 0.24-0.71 |
| Pantoea agglomerans (n=4) | 0.10-0.19 |
| Plesiomonas shigeloides (n=3) | 0.14-0.29 |
| Pseudomonas aeruginosa (n=3) | 0.29-0.62 |
| Serratia liquefaciens (n=2) | 0.10-0.24 |
| Shewanella putrefaciens (n=1) | 0.05 |
| Stenotrphomonas maltophilia (n=4) | 0.05-0.52 |
The MAR value for E. meningoseptica, E. coli, E. hermanii, M. morganii and P. aeruginosa isolates was higher than 0.20.
Heavy Metal Tolerance of Bacteria
In bullfrog farm, antibiotic multiple-resistance in isolates was distinctly associated with tolerance among heavy metals (Hg2+, Cr6+, Cd2+, Cu2+).
Isolates were found to be tolerant to different concentrations of heavy metals, ranging from 2.5 to 2400 µg/ml. In our study, heavy metal resistance varies as in the pattern of Hg-Cr>Cd>Cu (Table 4). All the isolates showed 100% resistant to mercury and chromium. There were 89.6% and 76.6% isolates resistance to cadmium and copper, respectively. The maximum heavy metal tolerance of bacteria was found at > 400 μg/ml for copper, and minimum for mercury (20 µg/ml).
Table 4.
Incidence of heavy metal tolerance in bacteria from bullfrog farm.
| Heavy metal | n | Number of isolates with heavy metal tolerance (µg/ml) | Resistancea | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2.5 | 5 | 10 | 20 | 40 | 25 | 50 | 100 | 200 | 400 | 150 | 300 | 600 | 1200 | 2400 | n | % | ||
| Cadmium | 77 | - | - | - | - | - | 0 | 5 | 8 | 9 | 30 | - | - | - | - | - | 69 | 89.6 |
| Cromium | 77 | - | - | - | - | - | 0 | 0 | 0 | 0 | 0 | - | - | - | - | - | 77 | 100 |
| Copper | 77 | - | - | - | - | - | - | - | - | - | - | 3 | 7 | 18 | 42 | 77 | 59 | 76.6 |
| Mercury | 77 | 0 | 0 | 0 | 0 | 12 | - | - | - | - | - | - | - | - | - | - | 77 | 100 |
Resistance Concentration:: Hg2+ (10µg/ml); Cd2+ and Cr6+ (100µg/ml); Cu2+ (600µg/ml). n : Number of total isolates; - : Not Tested
Mercury was found to be the most toxic heavy metal with the inhibition concentration of 40 μg/ml for 12 bacterial isolates.
High percentages of resistant patterns among heavy metals and antibiotics were observed. Isolates resistant to heavy metals were also resistant to nalidixic acid, flumequine, doxycycline, chloramphenicol and florfenicol.
The 100% of double-resistant strains were: mercury and chromium to all antibiotics; cadmium to NV, SP, NA, OA, UB, DO, OT, C, FFC, F and CT; and lastly copper to NA, UB, DO, C and FFC (Table 5).
Table 5.
Expression of antimicrobial activity between antibiotics and heavy metal resistance of bacterial isolates from bullfrogs.
| Antibiotic | TNo. | Heavy metal | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Cd | Cr | Cu | Hg | ||||||
| No. | % | No. | % | No. | % | No. | % | ||
| AML | 39 | 38 | 97.4 | 39 | 100 | 33 | 84.6 | 39 | 100 |
| AMP | 31 | 30 | 96.8 | 31 | 100 | 25 | 80.6 | 31 | 100 |
| K | 4 | 3 | 75 | 4 | 100 | 1 | 25 | 4 | 100 |
| NV | 35 | 35 | 100 | 35 | 100 | 32 | 91.4 | 35 | 100 |
| MY | 71 | 65 | 91.5 | 71 | 100 | 56 | 78.9 | 71 | 100 |
| E | 39 | 37 | 94.9 | 39 | 100 | 35 | 89.7 | 39 | 100 |
| OL | 56 | 54 | 96.4 | 56 | 100 | 49 | 87.5 | 56 | 100 |
| SP | 31 | 31 | 100 | 31 | 100 | 29 | 93.5 | 31 | 100 |
| NA | 5 | 5 | 100 | 5 | 100 | 5 | 100 | 5 | 100 |
| OA | 7 | 7 | 100 | 7 | 100 | 6 | 85.7 | 7 | 100 |
| UB | 5 | 5 | 100 | 5 | 100 | 5 | 100 | 5 | 100 |
| RL | 26 | 24 | 92.3 | 26 | 100 | 21 | 80.8 | 26 | 100 |
| DO | 6 | 6 | 100 | 6 | 100 | 6 | 100 | 6 | 100 |
| OT | 11 | 11 | 100 | 11 | 100 | 8 | 72.7 | 11 | 100 |
| TE | 7 | 6 | 85.7 | 7 | 100 | 5 | 71.4 | 7 | 100 |
| C | 2 | 2 | 100 | 2 | 100 | 2 | 100 | 2 | 100 |
| FFC | 2 | 2 | 100 | 2 | 100 | 2 | 100 | 2 | 100 |
| F | 16 | 16 | 100 | 16 | 100 | 13 | 81.3 | 16 | 100 |
| FR | 55 | 53 | 96.4 | 55 | 100 | 46 | 83.6 | 55 | 100 |
| FOS | 21 | 20 | 95.2 | 21 | 100 | 18 | 85.7 | 21 | 100 |
| CT | 15 | 15 | 100 | 15 | 100 | 13 | 86.7 | 15 | 100 |
AML: Amoxicillin; AMP: Ampicillin; K: Kanamycin; NV: Novobiocin; My: Lincomycin; E: Erythromycin; OL: Oleandomycin; SP:Spiramycin: NA: Nalidixic acid; OA: Oxolinic Acid; UB: Flumequine; RL: Sulphamethoxazole; DO: Doxycycline; OT: Oxytetracycline; TE: Tetracycline; C: Chloramphenicol; FFC: Florfenicol; F:Nitrofurantoin; FR: Furazolidone; FOS: Fosfomycin; CT: Colistin Sulphate; TNo: Number of isolates resistant to particular antibiotic; No: Number of isolates resist to heavy metal and antibiotic; %: Percentage of isolate resistance to antibiotic and heavy metal.
Discussion
Intensive farming of Lithobates catesbeianus are always risked with bacterial infection, which is mostly due to the environmental factors (Mauel et al., 2002). In order to reduce bacterial infection, the farmers utilize antibiotics to control and prevent diseases. This has been reported to result in bacterial resistance to various antibiotics and heavy metals (Miranda and Castillo, 1998; Mauel et al., 2002; Akinbowale et al., 2007). In this study, bullfrog bacteria isolated from a local farm in Johore were tested for their antibiotics susceptibility and heavy metal tolerance patterns. Antibiogram results in this study showed that an impressive abundance of bacteria isolated from the diseased bullfrogs were resistant to antibiotic and heavy metals.
In this study, resistance to lincomycin was found to be in around 92% of total bacteria tested, while a total of 72 isolated bacterial strains were all susceptible to chloramphenicol, florfenicol and flumequine. These findings were in contrast with Akinbowale et al. (2007), reporting chloramphenicol and florfenicol resistance in Pseudomonas spp. isolates. Studies from Mauel et al. (2002) were similar to the present results that A. hydrophila isolated from bullfrog were resistant to ampicillin, erythromycin and oxytetracycline. However E. meningoseptica and C. indologenes from the previous study were resistant to erythromycin, in contrast with our results. The difference could be due to types of antibiotics applied in different farms. The continuous use of antibiotics with a high dosage in the farming areas is highly associated with the occurrence of resistant microorganism, probably by the transferring resistant plasmids or intergons (Kümmerer, 2004).
MAR index value was high (>0.2) for many bacterial strains such as E. meningoseptica, E. coli, E. hermanii, M. morganii and P. aeruginosa. This indicates that antibiotics were commonly used by bullfrog farm at Johore. Furthermore, MAR index value for E. coli in the present study was 0.24 to 0.71. Similarly, 0.25 to 0.69 were achieved for E. coli isolated from seawater, sediment and shrimp from the south coast of Turkey where the contamination level of domestic waste was high (Fatih et al., 2008). Nevertheless, multiple antibiotic resistance up to 15 types of antibiotics were of special concern. Many of the antibiotics present in the aquaculture area are extrinsic. It is likely driven to the contamination either by run-off or the off-label used (Akinbowale et al., 2007). This may explain the antibiotic resistance problems in the present study.
A heavy metal resistance patterns of Hg-Cr>Cd>Cu in bacterial isolates was observed in the present study, which is different from the heavy metal resistance pattern as Cd>Cu>Hg>Cr for the isolates in a different pollution level in various freshwater sources reported by Miranda and Castillo (1998). Resistance pattern of Pseudomonas spp. and Aeromonas spp. isolated from rainbow trout farms in Australia was Cu>Cr>Cd (Akinbowale et al., 2007). The differences could be due to standard stain E. coli K12 used. The MIC for copper and chromium were 200 and 800 µg/ml, respectively, in the study by Akinbowale et al. (2007), while it was 600 and 200 µg/ml in the present study. All the bacteria were resistant to copper at the concentration of 200 µg/ml in the study by Akinbowale et al. (2007), but all bullfrog bacteria were only resistant at the concentration of 2400 µg/ml. Large amount of copper used in bullfrog farm for the treatment of red leg diseases, of this study, could be a reason leading to the high resistance patterns of the bacterial strains in our study. Furthermore, bacterial isolates which are resistant to heavy metals tend to be also resistant to antibiotics. This may be due to the co-location of resistance determinants where specific plasmids carried the resistance genes as defense mechanisms (Stepanauskas et al., 2006).
Usage of antibiotics and chemicals in prophylaxis and treatment of bullfrog culture is becoming problematic. Multiple antibiotic resistances in microorganism arise mainly due to injudicious use of antibiotics in disease treatments. Besides, high antibiotic resistance in bacteria isolated from aquaculture organisms could pose a risk to human health. Therefore, the antibiograms are important to review and revise the empirical disease management used in the aquaculture farm or as indicator of the dissemination of antibiotic elements.
It is well-known that the use of chemotherapeutics in the treatment of bacterial diseases represents a public health hazard. In particular, heavy metals are easily accumulated in the food chain and remain in the muscle tissue. The judicious use of antibiotics and heavy metals by the adoption of best management practices (BMPs) by aquaculturists is essential to reduce the risk of bacterial resistance (Boyd and Massaut, 1999). Dosage, withdrawal period, proper use, storage, disposal, and other constraints on the chemicals including environmental, human and food safety precautions should be followed stringently in reducing those problems.
Acknowledgement
We would like to thank members from Fish Disease Laboratory, Faculty of Agrotechnology and Food Science for their supports in this project.
References
- Akinbowale O.L, Peng H.H, Grant P, Barton M.D. Antibiotic and heavy metal resistance in motile aeromonads and pseudomonads from rainbow trout (Oncorhynchus mykiss) farms in Australia. Int. J. Antimicrob. Agents. 2007;30:177–182. doi: 10.1016/j.ijantimicag.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Baker-Austin C, Wright M.S, Stepanauskas R, McArthur J.V. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006;14(4):176–182. doi: 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Boyd C.E, Massaut L. Risks associated with the use of chemicals in pond aquaculture. Aquacult. Eng. 1999;20:113–132. [Google Scholar]
- Cappuccino J.G, Sherman N. 6th Edition. San Francisco, CA: Benjamin Cummings; 2002. Microbiology: a laboratory manual; pp. 1–195. [Google Scholar]
- Clinical and Laboratory Standard Institute (CLSI) Wayne (PA): CLSI; 2006. Performance standard for antimicrobial susceptibility testing;sixteenth information supplement, M-100-S 16. [Google Scholar]
- Fatih M, Aysenur K, Sadik D. Antibacterial agents and heavy metal resistance in Gram-negative bacteria isolated from seawater, shrimp and sediment in Iskenderun Bay, Turkey. Sci. Total Environ. 2008;407:279–285. doi: 10.1016/j.scitotenv.2008.08.014. [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organization of United Nation. Fisheries and aquaculture information and statistics service. 2010. [Accessed 16 January 2010]. http://www.fao.org/figis/servlet/SQServlet?ds=Aquaculture&k1=SPECIES&k1v=1&k1s=10048&outtype=html .
- Gillings M, Boucher Y, Labbate M, Holmes A, Krishnan S, Holley M, Stokes H.W. The Evolution of Class 1 Integrons and the Rise of Antibiotic Resistance. J. Bacteriol. 2008;190(14):5095–5100. doi: 10.1128/JB.00152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumperman P.H. Multiple antibiotic resistance indexing of E. coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983;46(1):165–170. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kümmerer K. Resistance in the environment. J. Antimicrob. Chemother. 2004;54:311–320. doi: 10.1093/jac/dkh325. [DOI] [PubMed] [Google Scholar]
- Mauel M.J, Miller D.L, Frazier K.S, Hines II M.E. Bacterial pathogens isolated from cultured bullfrogs (Rana castesbeiana) J. Vet. Diagn. Invest. 2002;14:431–433. doi: 10.1177/104063870201400515. [DOI] [PubMed] [Google Scholar]
- McDaniel T.V, Martin P.A, Struger J, Sherry J, Marvin C.H, McMaster M.E, Clarence S, Tetreault G. Potential endocrine disruption of sexual development in free ranging male northern leopard frogs (Rana pipiens) and green frogs (Rana clamitans) from areas of intensive row crop agriculture. Aquat. Toxicol. 2008;88:230–242. doi: 10.1016/j.aquatox.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Miranda C.D, Castillo G. Resistance to antibiotic and heavy metals of motile aeromonads from Chilean freshwater. Sci. Total Environ. 1998;224:167–176. doi: 10.1016/s0048-9697(98)00354-4. [DOI] [PubMed] [Google Scholar]
- Othman M.S, Khonsue W, Kitana J, Thirakhupt K, Robson M.G, Kitana N. Cadmium Accumulation in Two Populations of Rice Frogs (Fejervarya limnocharis) Naturally Exposed to Different Environmental Cadmium Levels. Bull. Environ. Contam. Toxicol. 2009;83:703–707. doi: 10.1007/s00128-009-9845-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanauskas R, Glenn T.C, Jagoe C.H, Tuckfield R.C, Lindell A.H, King C. J, McArthur J.V. Coselection for microbial resistance to metals and antibiotics in freshwater microcosms. Environ. Microbiol. 2006;8(9):1510–1514. doi: 10.1111/j.1462-2920.2006.01091.x. [DOI] [PubMed] [Google Scholar]
- United Nations Statistics Division. Trade of Goods, US$, HS. 1992, 02 meat and edible meat offal. 2008. [Accessed on 21 July 2011]. http://data.un.org/Data.aspx?q=frog+leg&d=ComTrade&f=_l1Code%3a3%3bcmdCode%3a020820 .
- Vogiatzis A.K, Loumbourdis N.S. Cadmium accumulation in liver and kidneys and hepatic metallothionein and glutathione levels in Rana ridibunda after exposure to CdCl2. Arch. Environ. Contam. Toxicol. 1998;34:64–68. doi: 10.1007/s002449900286. [DOI] [PubMed] [Google Scholar]


