Abstract
Recent advances in genomic sequencing technologies now allow results from deep next-generation sequencing to be obtained within clinically meaningful timeframes, making this an attractive approach to better guide personalized treatment strategies. No multiple myeloma-specific gene panel has been established so far; we therefore designed a 47-gene-targeting gene panel, containing 39 genes known to be mutated in ≥3%of multiple myeloma cases and eight genes in pathways therapeutically targeted in multiple myeloma (MM). We performed targeted sequencing on tumor/germline DNA of 25 MM patients in which we also had a sequential sample post treatment. Mutation analysis revealed KRAS as the most commonly mutated gene (36 % in each time point), followed by NRAS (20 and 16 %), TP53 (16 and 16 %), DIS3 (16 and 16 %), FAM46C (12 and 16 %), and SP140 (12 and 12 %). We successfully tracked clonal evolution and identified mutation acquisition and/or loss in FAM46C, FAT1, KRAS, NRAS, SPEN, PRDM1, NEB, and TP53 as well as two mutations in XBP1, a gene associated with bortezomib resistance. Thus, we present the first longitudinal analysis of a MM-specific targeted sequencing gene panel that can be used for individual tumor characterization and for tracking clonal evolution over time.
Keywords: Multiple myeloma, Targeted sequencing
Introduction
Initial therapy in multiple myeloma (MM) consistently induces high-quality remission, including CR, in the majority of patients. However, relapses occur in almost all patients over time, best explained by the existence of tumor clone heterogeneity already existent at initial diagnosis with different drug susceptibilities, leading to clonal selection and evolution over time [1–3]. Successful treatment of the disease therefore needs a broad target that includes minor subclones, from which a final, ultimately resistant clone could arise [4]. Consequently, awareness of the individual genetic profile of the tumor cell population and the surveillance of changes over time under therapeutic selective pressure is needed to assess treatment efficacy. Today, whole genome/exome sequencing data of more than 300 MM patients are publically available from recent large sequencing studies [5–7, 3] describing genomic complexity of the disease including baseline clonal heterogeneity [4], linear and branching evolution [3], and therapeutic selection of clones and subclones resulting in clonal tides [2]. However, whereas genetic diagnostics in MM as cytogenetics, fluorescence-in-situ-hybridization and gene expression profiling are well established [8], individual mutation profiling has not yet been adopted to the routine risk assessment. In this work, we report on an innovative gene panel investigating 25 MM patients at sequential time points before and after therapy. We employed semiconductor sequencing technology that provides rapid mutation analysis at reasonable costs, needing low sample input (10 ng) and a sample turnaround time in clinically meaningful timeframes (hours).
Material and methods
We obtained DNA of 22 newly diagnosed and 3 pretreated MM patients, including a later time point sample and corresponding germline from peripheral blood mononuclear cells from the German MM trial group (DSMM). All samples were collected with informed consent according to the Declaration of Helsinki. Plasma cells were enriched using CD138+ beads (median purity 95 %). The purity of the samples was assessed cytologically. DNA was subsequently extracted using the Qiagen AllPrep DNA/RNA Mini Kit according to the manufacturer’s recommendations. The time between the sample biopsies ranged from 63 to 2054 days, with an average of 374 days. Treatment information and response data were not available. Baseline FISH was available in 23 of 25 pairs and revealed a increased risk cohort defined by 70 % gain of 1q21 (16/23), 48 % of del13q (11/23), 35 % of t(4;14) (8/23), 9 % of t(11;14) (2/23) and t(14;16) (2/23), and 26 % (6/23) of del17p.
The 47 gene multiple myeloma mutation panel (M3P)
We established a MM-specific 47 gene mutation panel [9] including a selection of 39 genes expressed in MM (by analyzing gene expression profiling public datasets) with nonsynonymous mutations found in ≥3 % of published MM genomes [6, 10]. To this, we added eight genes targeted by the most commonly used MM therapies, associated with resistance to IMiDs (CRBN, CUL4A, CUL4B, DDB1, and IRF4), proteasome inhibitors (PSMG2, PSMB5) and glucocorticoid therapies (NR3C1) (Table 1). We employed the Ion Torrent semiconductor sequencing platform (PGM, Life Technologies, Carlsbad, CA, USA), using 20 ng of starting DNA for each sample (10 ng per primer pool). The coding regions of the 47 genes were amplified in 200-bp libraries using customized oligos (Ion AmpliSeq Designer, Life Technologies). Overall, 2875 amplicons, covering 96 % of the M3P exons, were analyzed per sample, multiplexed in two library preparations (Ion AmpliSeq Library Kit 2.0, Life Technologies). Template preparation and enrichment of DNA libraries was done on the Ion OneTouch2 and Ion OneTouch ES (Life Technologies) automated system, respectively. Batches of four samples were barcoded (Ion Xpress Barcode Adapters, Life Technologies), pooled, and sequenced using Ion 318 and 318v2 chips and the Ion Sequencing 200 Kit v2 (Life Technologies). Sequencing data were analyzed using the Ion Reporter Software v1.6 (protocols applied: “TumorNormalTemplate 1.6.2” and “Ion QC protocol”, Life Technologies, Carlsbad, CA, USA), visualized, and manually reviewed using the Integrative Genomics Viewer (IGV, Broad Institute, Cambridge, MA, USA). Variants were analyzed using SIFT, Provean (J. Craig Venter Institute) [11, 12], PolyPhen-2 (Harvard University) [13], and the Catalogue of Somatic Mutations in Cancer (COSMIC, Welcome Trust Sanger Institute, UK) [14]. Mutation calls were considered positive when called by ≥10 % variant reads and >20 times sequencing coverage depth in the tumor sample. In already characterized cancer-related mutations (COSMIC database), the threshold was reduced to 3 %. We additionally considered mutations called below threshold if a matching variant above the threshold was found in the corresponding tumor sample.
Table 1.
The multiple myeloma mutation panel (M3P) v1.0
| ADAMTS9 | DIS3 | IRF4 | NRAS | TP53 |
| ANK2 | DNAH5 | KRAS | PRDM1 | TRAF3 |
| ATM | EGFR | LRP1B | PSMB5 | TRIP12 |
| BRAF | EGR1 | LTN1 | PSMG2 | VCAN |
| CCND1 | FAM46C | LYST | PTPRD | XBP1 |
| CRBN | FAT1 | MECOM | RASA2 | ZFHX3 |
| CUL4A | FAT3 | MLL3 | RB1 | ZFHX4 |
| CUL4B | FAT4 | NBPF1 | SP140 | |
| CYLD | FRYL | NEB | SPEN | |
| DDB1 | HECW1 | NR3C1 | TIAM1 |
Results
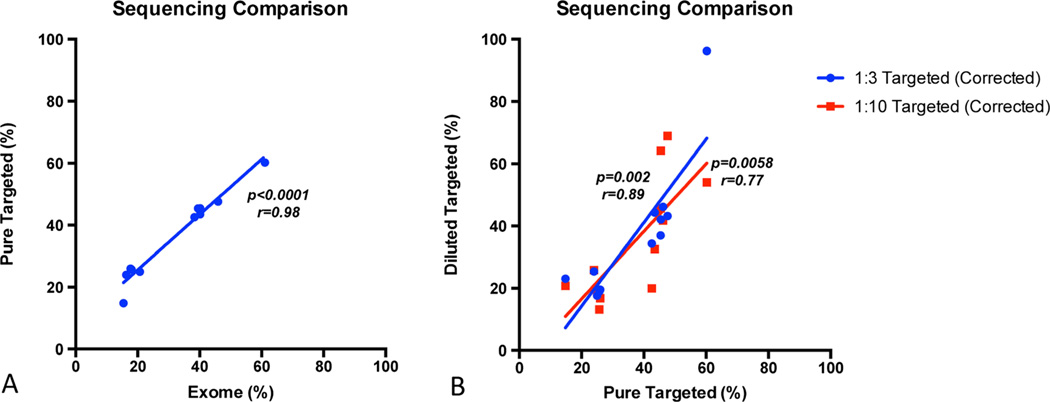
First we validated the targeted sequencing technology by investigating a MM tumor sample previously analyzed by whole exome sequencing (WES) and thus known to be harboring 12 mutations included in the M3P panel [15]. To determine the sequencing accuracy, especially in low allele frequency mutations, additional targeted sequencing was performed at 1:3 and 1:10 dilutions of the tumor sample. The sample was diluted using a commercially available lymphoproliferative cell line (GM19240, Coriell Cell Repositories, Camden, NJ, USA) for which sequencing results were publicly available from the HapMap project.
We obtained a mean coverage of × 278 read depth for the M3P panel. All 12 mutations initially found by WES were also reported using targeted sequencing. The mutation allelic frequencies, ranging between 15 and 60 % by WES, correlated very well with the results obtained by targeted sequencing (R=0.98, p<0.0001). A good correlation was also found in the 1:3 (p=0.0002, R=0.84) and 1:10 (p=0.006, R=0.75) dilution (Fig. 1), and all mutations were found in both dilutions, including a mutation with a lowest non reference allele read frequency of 1.32 % in the 1:10 dilution.
Fig. 1.
Correlation between Illumina exome and Torrent targeted semiconductor sequencing technology. a Correlation between techniques on the frequency of non reference allele reads of the undiluted tumor sample. b Correlation between WGS and the 1:3 and the 1:10 dilutions (dilution corrected)
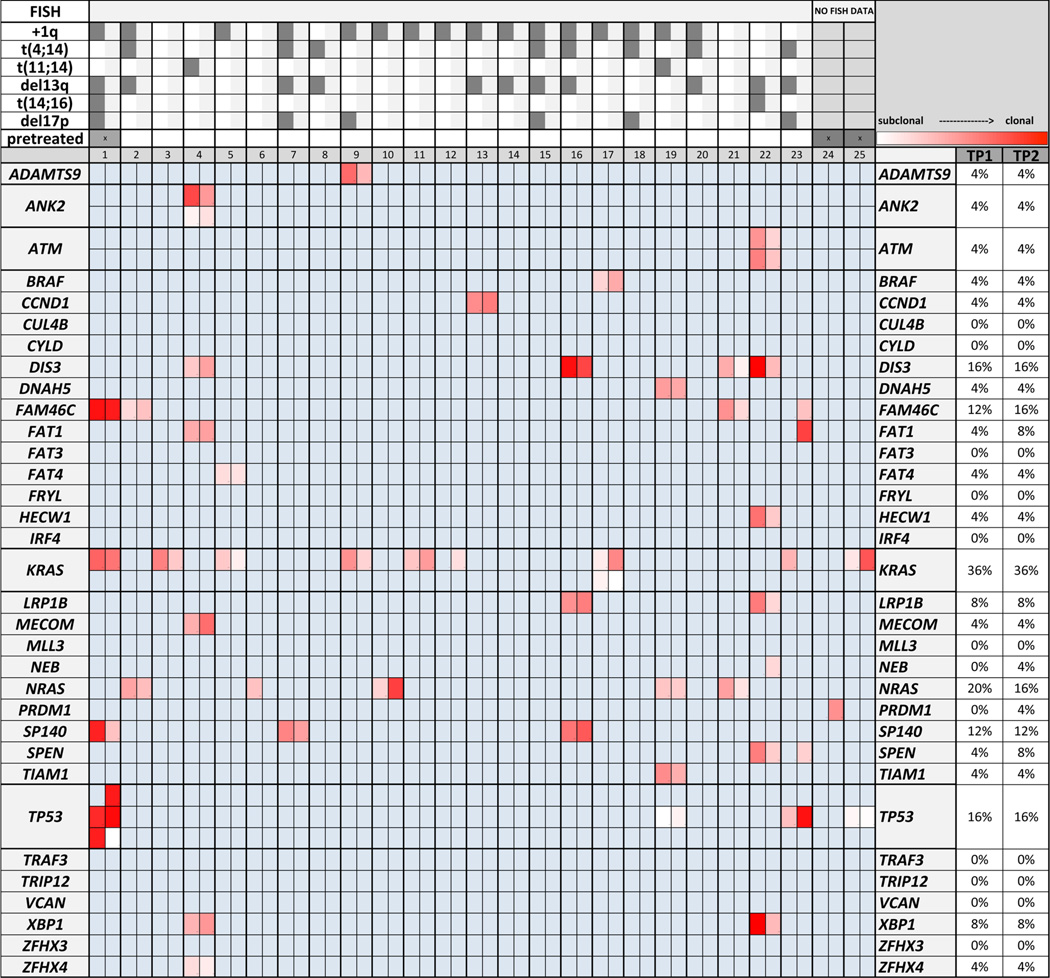
An average read sequencing depth of × 280 (median × 255, amplicons with at least 20 reads in >90 %) was achieved across the 25 paired samples. In 20 patients (80 %) and for 23 of the 47 M3P genes (49 %), mutations were identified at the cutoffs employed. The number of mutations per patient varied from 0 to 8, with a total of 101 mutations identified of which 82 (81.2 %) were predicted damaging/deleterious by Polyphene-2, Provean, or SIFT. In the earlier time point, an average of 1.92 variants was found per patient. In time point 2, the mean mutation prevalence increased slightly to 2.12 mutations per patient. Changes in mutation abundance exceeding 20 % were seen in 52 % of the patients (measured by purity corrected variant reads). Six genes were mutated in more than 10 % of the patients at both time points, with KRAS as the most prevalent (36 % in each time point), followed by NRAS (20 %, 16 %), TP53 (20 %, 24 %), DIS3 (16 %, 16 %), FAM46C (12 %, 16 %), and SP140 (12 %, 12 %) (Fig. 2). Acquisition of mutations over time was seen in several genes, including FAM46C (p.Ile276Thr), FAT1 (p.His3512Asn), KRAS (p.Gln61His, known activating), SPEN (p.Thr2747Ala), PRDM1 (p.Lys620Arg), NEB (p.Thr3677Ser), and TP53 (p.Tyr163Cys). Disappearance of mutations was found in KRAS (p.Gly12Val, known activating) and NRAS (p.Pro54Leu). Probable parallel evolution [3] in the RAS/MAPK pathway was observed in two patients with one patient presenting two mutations in one gene (KRAS, p.Tyr64Asn, p.Ala146Thr) and one patient harboring mutations in different genes of the pathway (KRAS and BRAF). Furthermore, multiple mutations on one gene were found in ANK2 (p.Glu2857Lys, p.His923Tyr), ATM (truncating p.Glu431*, p.Leu2945Met), and TP53 (p.Tyr163Cys, p.Arg181His, p.Met246Ile). Of interest, only the mutations in ATM showed concordant changes of abundance over time, whereas the mutations in ANK2, KRAS, and TP53 had opposing changes, indicating that these mutations are present in different subclones of the tumor cell population. Accordingly, in 8 of 12 patients with more than one mutation, the existence of coexisting tumor (sub)clones could be determined by opposing changes in variant read abundance over time. Most strikingly, in one relapsed del(17p) patient, we detected a TP53, KRAS, and FAM46C mutations in a pleural effusion of a pretreated patient at time point A while 5 months later, this patient developed a plasma cell leukemia in which loss of this TP53-mutated subclone and emergence of a second subclone with a different TP53 mutation was seen. Notably, a third clonal TP53 mutation was found to be shared by both subclones (Table 2).
Fig. 2.
Track of clonal evolution in the longitudinal analysis and mutation prevalence in 22 untreated and 3 pretreated MM patients. Clonal evolution is evident in the majority of patients, with changes in clonal size (indicated by the heatmap), acquisition of a TP53, FAM46C, FAT1, SPEN, PRDM1, and NEB and a gain and loss of a NRAS and KRAS mutation over time, representing individual change of mutational profile under therapeutic selection pressure. Multiple mutations in one gene at the same time point were identified in ANK2, ATM, KRAS, and TP53; each mutation is represented by an individual line. FISH: Analysis was performed on time point 1 in patients 1–23
Table 2.
This table illustrates the individual change of the mutational profile over time tracked by M3P
| TP 1 | TP 2 | |
|---|---|---|
| FAM46C | 87 % | 83 % |
| KRAS | 57 % | 51 % |
| SP140 | 81 % | 22 % |
| TP53 | 78 % | 90 % |
| 82 % | 0 % | |
| 0 % | 84 % |
Percentage in box represents purity corrected variant reads. In this patient one TP53 mutation gets eradicated over time, whereas a second TP53 mutation expands between the time points, suggesting the presence of at least two different clones. Additionally, a SP140 mutation is decreased but not eradicated, suggesting the existence of a third clone. Furthermore, the mutation frequency of a FAM46C, a KRAS and a third TP53 mutation stay unchanged over time and are not affected by the clonal evolution tracked by M3 P, providing evidence that these mutations are clonal
Damaging mutations in drug resistance related genes were rare, even when we checked below the chosen threshold of significance. However, variants in XBP1, a gene related to proteasome inhibitor resistance [16], were found in two patients. While one of the mutations did not change significantly between the time points (p.Glu99Lys), one was reduced in abundance by therapy by 74% of the reads (p.Arg94Gln) over time.
Discussion
The investigation of tumor samples on multiple time points allows insights in the pathomechanisms that lead to tumor progression and the development of drug resistance in MM patients. Whole genome/whole exome sequencing data as well as single cell analyses on sequential MM samples have been performed, confirming baseline clonal heterogeneity, linear and branching evolution, and the selection of clones and subclones under selective pressure of drug therapy resulting in clonal tides [3, 7, 17, 2, 4]. We established a MM-specific next-generation targeted sequencing panel (M3P) to investigate the most commonly mutated genes in MM as well as genes for which a targetable drug is available or that are related to drug resistance [9]. This approach allows to obtain results faster (within clinically meaningful timeframes), cheaper, and with far less sample demand than established WES/WGS sequencing technologies. We investigated 22 untreated and 3 pretreated tumor normal pairs and a subsequent time point sample by M3P. We observed clonal evolution in the majority of patients including clonal expansion or contraction, as well as complete extinction of subclones (KRAS, TP53) and the emergence of new subclones (FAM46C, FAT1, SPEN, TP53). Of interest, we found the majority of mutations in our cohort to be present in both patient time points and true extinction of clones or subclones by therapy was uncommon. Baseline FISH data indicate increased incidence of high-risk markers in our cohort, including del17p or t(4;14), explaining the increased incidence of TP53 mutations in our cohort(16 %). DIS3 incidences matched t(4;14)/t(11;14) restricted cohorts [18] with four patients harboring a mutations in this gene. DIS3, located on chromosome 13, is one of the most commonly mutated genes in MM, but its role in the pathophysiology of MM remains undiscovered. DIS3 is component of the RNA exosome complex and may be involved in Ig class switch recombination and Ig variable region somatic hypermutation in human B lymphocytes [19]. Mutations of DIS3 have been described in other malignant diseases such as medulloblastoma [20], acute myeloid leukemia [21], and nodular melanoma [22]. In MM, DIS3 mutations have been associated to t(4;14), t(11;14) and a dependency on del(13q) was reported [18]. In our cohort, four DIS3 mutations were identified prior to treatment of which two were clonal (p.Pro412Leu and p.Asp784His) and two subclonal (p.Arg780Lys and p.Arg780Thr). Of interest, mutations in DIS3 in MM at amino acid position 780 have been described to alter gene function by causing significant aberrations of hDIS3 exoribonucleolytic activity [23]. We could see clonal evolution caused by therapy-induced selective pressure in all DIS3 mutations in our cohort: In three of them, we observed a significant decrease of variant read (VR) abundance over time (p.Asp784His −73 %, p.Pro412Leu −34 %, and p.Arg780Lys −29 %), whereas in one patient an increase was seen (p.Arg780Thr +17 %).
Another gene, frequently mutated in our cohort, was SP140. This gene is expressed in mature B cells and on plasma cell lines. It is the lymphoid-restricted homolog of SP100 which contributes to EBV-mediated B cell immortalization [24]. It is involved in the pathogenesis of chronic lymphocytic leukemia [25] and squamous cell carcinoma [26]. However, the clinical impact of mutations in SP140 in MM is not yet determined. Truncating mutations of SP140 have been recently described in MM [7], and indeed, we also saw two truncating SP140 mutations (p.Arg576* and p.Glu75*) and one missense mutation (p.Glu856Lys) in our cohort. We observed a 72 % decrease in abundance of the missense mutation after treatment; however, the truncating mutations remained stable over time.
Of interest, mutations in genes related to drug resistance were rare. We identified mutations in XBP1 in two patients, a gene reported to be associated with resistance against proteasome inhibition. Mutation taster predicts splice site changes by both mutations and it has been shown that only spliced XBP1s is transcriptionally active and other mutations affecting the XBP1 splicesite have been demonstrated to cause bortezomib resistance [16]. Furthermore, both mutations occurred in highly conserved regions, affecting the bzip domain and the Leucine zipper region of the gene and both were predicted as damaging or possibly damaging by Polyphene-2 or SIFT. Thus, both mutations might alter sensitivity to proteasome inhibition; however, no data on treatment are available; therefore, the effect of these mutations remains speculative.
We believe that targeted mutation profiling will likely become part of the clinical workup in MM in the near future. M3P is the first MM-specific targeted sequencing panel so far and using this targeted sequencing panel, we were not only able to characterize the individual mutational tumor profiles of the 25 investigated patients, but also identified clonal heterogeneity and tracked successfully clonal evolution over time. Investigation in larger cohorts of clinically well annotated patients are needed; however, our study could already illustrate how M3P may serve as a practical tool to provide information needed to more precisely and more efficiently conduct individualized therapy concepts.
Supplementary Material
Acknowledgments
Funding source supporting this work This work is supported by grants R01 CA83724, CA167511, and CA183968, ECOG CA 21115 T, Predolin Foundation, Mayo Clinic Cancer Center, the Mayo Foundation, and the DFG (Ko 4604/1-1 to KMK, BU 1339/7-2 and BU 1339/3-1 to LBu, and LA 2414/2-1 to CL); EB has support by the Henry Predolin Foundation, the Marriott Specialized Workforce Development Awards in Individualized Medicine and the Fraternal Order of Eagles.
RF is a Clinical Investigator of the Damon Runyon Cancer Research Fund and received a patent for the prognostication of MM based on genetic categorization of the disease. He has received consulting fees from Medtronic, Otsuka, Celgene, Genzyme, BMS, Lilly, Onyx, Binding Site, Millennium, and AMGEN. He also has sponsored research from Cylene and Onyx.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00277-015-2344-9) contains supplementary material, which is available to authorized users.
Conflict of interests KMK, CL, JM, LBr, YXZ, CXS, PJ, JBE, JO, LBu, MK, GA, LR, SK, HE, AKS, and EB declare that they have no conflict of interest.
Contributor Information
KM Kortüm, Division of Hematology - Oncology, Mayo Clinic, Scottsdale, AZ, USA.
C Langer, Department of Internal Medicine III, University Hospital of Ulm, Ulm, Germany.
J Monge, Division of Hematology - Oncology, Mayo Clinic, Scottsdale, AZ, USA.
L Bruins, Division of Hematology - Oncology, Mayo Clinic, Scottsdale, AZ, USA.
YX Zhu, Division of Hematology - Oncology, Mayo Clinic, Scottsdale, AZ, USA.
CX Shi, Division of Hematology - Oncology, Mayo Clinic, Scottsdale, AZ, USA.
P Jedlowski, Division of Hematology - Oncology, Mayo Clinic, Scottsdale, AZ, USA.
JB Egan, Comprehensive Cancer Center, Mayo Clinic, Scottsdale, AZ, USA.
J Ojha, Division of Hematology - Oncology, Mayo Clinic, Scottsdale, AZ, USA.
L Bullinger, Department of Internal Medicine III, University Hospital of Ulm, Ulm, Germany.
M Kull, Department of Internal Medicine III, University Hospital of Ulm, Ulm, Germany.
G Ahmann, Division of Hematology - Oncology, Mayo Clinic, Scottsdale, AZ, USA.
L Rasche, Department of Internal Medicine II, University Hospital of Würzburg, Würzburg, Germany.
S Knop, Department of Internal Medicine II, University Hospital of Würzburg, Würzburg, Germany.
R Fonseca, Division of Hematology - Oncology, Mayo Clinic, Scottsdale, AZ, USA.
H Einsele, Department of Internal Medicine II, University Hospital of Würzburg, Würzburg, Germany.
AK Stewart, Division of Hematology - Oncology, Mayo Clinic, Scottsdale, AZ, USA.
Esteban Braggio, Email: Braggio.Esteban@mayo.edu, Division of Hematology - Oncology, Mayo Clinic, Scottsdale, AZ, USA.
References
- 1.Brioli A, Melchor L, Cavo M, Morgan GJ. The impact of intraclonal heterogeneity on the treatment of multiple myeloma. Br J Haematol. 2014;165(4):441–454. doi: 10.1111/bjh.12805. doi: 10.1111/bjh.12805. [DOI] [PubMed] [Google Scholar]
- 2.Egan JB, Shi CX, Tembe W, Christoforides A, Kurdoglu A, Sinari S, Middha S, Asmann Y, Schmidt J, Braggio E, Keats JJ, Fonseca R, Bergsagel PL, Craig DW, Carpten JD, Stewart AK. Whole-genome sequencing of multiple myeloma from diagnosis to plasma cell leukemia reveals genomic initiating events, evolution, and clonal tides. Blood. 2012;120(5):1060–1066. doi: 10.1182/blood-2012-01-405977. doi: 10.1182/blood-2012-01-405977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melchor L, Brioli A, Wardell CP, Murison A, Potter NE, Kaiser MF, Fryer RA, Johnson DC, Begum DB, Hulkki Wilson S, Vijayaraghavan G, Titley I, Cavo M, Davies FE, Walker BA, Morgan GJ. Single-cell genetic analysis reveals the composition of initiating clones and phylogenetic patterns of branching and parallel evolution in myeloma. Leukemia. 2014;28(8):1705–1715. doi: 10.1038/leu.2014.13. doi: 10.1038/leu.2014.13. [DOI] [PubMed] [Google Scholar]
- 4.Keats JJ, Chesi M, Egan JB, Garbitt VM, Palmer SE, Braggio E, Van Wier S, Blackburn PR, Baker AS, Dispenzieri A, Kumar S, Rajkumar SV, Carpten JD, Barrett M, Fonseca R, Stewart AK, Bergsagel PL. Clonal competition with alternating dominance in multiple myeloma. Blood. 2012;120(5):1067–1076. doi: 10.1182/blood-2012-01-405985. doi: 10.1182/blood-2012-01-405985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lonial S, Yellapantula VD, Liang W, Kurdoglu A, Aldrich J, Legendre CM, Stephenson K, Adkins J, McDonald J, Helland A, Russell M, Christofferson A, Cuyugan L, Rohrer D, Blanski A, Hodges M, CoMMpass Network M, Derome M, Auclair D, Kidd PG, Jewell S, Craig D, Carpten J, Keats JJ. Interim analysis of the Mmrf Commpass Trial: identification of novel rearrangements potentially associated with disease initiation and progression. Blood. 2014;124(21):722–722. [Google Scholar]
- 6.Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, Sougnez C, Knoechel B, Gould J, Saksena G, Cibulskis K, McKenna A, Chapman MA, Straussman R, Levy J, Perkins LM, Keats JJ, Schumacher SE, Rosenberg M, Multiple Myeloma Research C. Getz G, Golub TR. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25(1):91–101. doi: 10.1016/j.ccr.2013.12.015. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, Dawson KJ, Iorio F, Nik-Zainal S, Bignell GR, Hinton JW, Li Y, Tubio JM, McLaren S, O’ Meara S, Butler AP, Teague JW, Mudie L, Anderson E, Rashid N, Tai YT, Shammas MA, Sperling AS, Fulciniti M, Richardson PG, Parmigiani G, Magrangeas F, Minvielle S, Moreau P, Attal M, Facon T, Futreal PA, Anderson KC, Campbell PJ, Munshi NC. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997. doi: 10.1038/ncomms3997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faiman B. Myeloma Genetics and Genomics: Practice Implications and Future Directions. Clin Lymphoma Myeloma Leuk. 2014 doi: 10.1016/j.clml.2014.07.008. doi: 10.1016/j.clml.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Kortum KM, Langer C, Monge J, Bruins L, Egan JB, Zhu YX, Shi CX, Jedlowski P, Schmidt J, Ojha J, Bullinger L, Liebisch P, Kull M, Champion MD, Van Wier S, Ahmann G, Rasche L, Knop S, Fonseca R, Einsele H, Stewart AK, Braggio E. Targeted sequencing using a 47 gene multiple myeloma mutation panel (MP) in-17p high risk disease. Br J Haematol. 2014 doi: 10.1111/bjh.13171. doi: 10.1111/bjh.13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, Anderson KC, Ardlie KG, Auclair D, Baker A, Bergsagel PL, Bernstein BE, Drier Y, Fonseca R, Gabriel SB, Hofmeister CC, Jagannath S, Jakubowiak AJ, Krishnan A, Levy J, Liefeld T, Lonial S, Mahan S, Mfuko B, Monti S, Perkins LM, Onofrio R, Pugh TJ, Rajkumar SV, Ramos AH, Siegel DS, Sivachenko A, Stewart AK, Trudel S, Vij R, Voet D, Winckler W, Zimmerman T, Carpten J, Trent J, Hahn WC, Garraway LA, Meyerson M, Lander ES, Getz G, Golub TR. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. doi: 10.1038/nature09837. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 12.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7(10):e46688. doi: 10.1371/journal.pone.0046688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, Kok CY, Jia M, Ewing R, Menzies A, Teague JW, Stratton MR, Futreal PA. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38(Database issue):D652–D657. doi: 10.1093/nar/gkp995. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egan JB, Kortuem KM, Kurdoglu A, Izatt T, Aldrich J, Reiman R, Phillips L, Baker A, Shi CX, Schmidt J, Liang WS, Craig DW, Carpten JD, Stewart AK. Extramedullary myeloma whole genome sequencing reveals novel mutations in Cereblon, proteasome subunit G2 and the glucocorticoid receptor in multi drug resistant disease. Br J Haematol. 2013;161(5):748–751. doi: 10.1111/bjh.12291. doi: 10.1111/bjh.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung-Hagesteijn C, Erdmann N, Cheung G, Keats JJ, Stewart AK, Reece DE, Chung KC, Tiedemann RE. Xbp1s-negative tumor B cells and pre-plasmablasts mediate therapeutic proteasome inhibitor resistance in multiple myeloma. Cancer Cell. 2013;24(3):289–304. doi: 10.1016/j.ccr.2013.08.009. doi: 10.1016/j.ccr.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walker BA, Wardell CP, Melchor L, Brioli A, Johnson DC, Kaiser MF, Mirabella F, Lopez-Corral L, Humphray S, Murray L, Ross M, Bentley D, Gutierrez NC, Garcia-Sanz R, San Miguel J, Davies FE, Gonzalez D, Morgan GJ. Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia. 2014;28(2):384–390. doi: 10.1038/leu.2013.199. doi: 10.1038/leu.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker BA, Wardell CP, Melchor L, Hulkki S, Potter NE, Johnson DC, Fenwick K, Kozarewa I, Gonzalez D, Lord CJ, Ashworth A, Davies FE, Morgan GJ. Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4;14) and t(11;14) myeloma. Blood. 2012;120(5):1077–1086. doi: 10.1182/blood-2012-03-412981. doi: 10.1182/blood-2012-03-412981. [DOI] [PubMed] [Google Scholar]
- 19.Basu U, Meng FL, Keim C, Grinstein V, Pefanis E, Eccleston J, Zhang T, Myers D, Wasserman CR, Wesemann DR, Januszyk K, Gregory RI, Deng H, Lima CD, Alt FW. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011;144(3):353–363. doi: 10.1016/j.cell.2011.01.001. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, Gallia GL, Jallo GI, Binder ZA, Nikolsky Y, Hartigan J, Smith DR, Gerhard DS, Fults DW, VandenBerg S, Berger MS, Marie SK, Shinjo SM, Clara C, Phillips PC, Minturn JE, Biegel JA, Judkins AR, Resnick AC, Storm PB, Curran T, He Y, Rasheed BA, Friedman HS, Keir ST, McLendon R, Northcott PA, Taylor MD, Burger PC, Riggins GJ, Karchin R, Parmigiani G, Bigner DD, Yan H, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331(6016):435–439. doi: 10.1126/science.1198056. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, McMichael JF, Wallis JW, Lu C, Shen D, Harris CC, Dooling DJ, Fulton RS, Fulton LL, Chen K, Schmidt H, Kalicki-Veizer J, Magrini VJ, Cook L, McGrath SD, Vickery TL, Wendl MC, Heath S, Watson MA, Link DC, Tomasson MH, Shannon WD, Payton JE, Kulkarni S, Westervelt P, Walter MJ, Graubert TA, Mardis ER, Wilson RK, DiPersio JF. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506–510. doi: 10.1038/nature10738. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rose AE, Poliseno L, Wang J, Clark M, Pearlman A, Wang G, Vega Y, Saenz deMiera EC, Medicherla R, Christos PJ, Shapiro R, Pavlick A, Darvishian F, Zavadil J, Polsky D, Hernando E, Ostrer H, Osman I. Integrative genomics identifies molecular alterations that challenge the linear model of melanoma progression. Cancer Res. 2011;71(7):2561–2571. doi: 10.1158/0008-5472.CAN-10-2958. doi: 10.1158/0008-5472.CAN-10-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomecki R, Drazkowska K, Kucinski I, Stodus K, Szczesny RJ, Gruchota J, Owczarek EP, Kalisiak K, Dziembowski A. Multiple myeloma-associated hDIS3 mutations cause perturbations in cellular RNA metabolism and suggest hDIS3 PIN domain as a potential drug target. Nucleic Acids Res. 2014;42(2):1270–1290. doi: 10.1093/nar/gkt930. doi: 10.1093/nar/gkt930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ling PD, Peng RS, Nakajima A, Yu JH, Tan J, Moses SM, Yang WH, Zhao B, Kieff E, Bloch KD, Bloch DB. Mediation of Epstein-Barr virus EBNA-LP transcriptional coactivation by Sp100. EMBO J. 2005;24(20):3565–3575. doi: 10.1038/sj.emboj.7600820. doi: 10.1038/sj.emboj.7600820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Bernardo MC, Crowther-Swanepoel D, Broderick P, Webb E, Sellick G, Wild R, Sullivan K, Vijayakrishnan J, Wang Y, Pittman AM, Sunter NJ, Hall AG, Dyer MJ, Matutes E, Dearden C, Mainou-Fowler T, Jackson GH, Summerfield G, Harris RJ, Pettitt AR, Hillmen P, Allsup DJ, Bailey JR, Pratt G, Pepper C, Fegan C, Allan JM, Catovsky D, Houlston RS. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2008;40(10):1204–1210. doi: 10.1038/ng.219. doi: 10.1038/ng.219. [DOI] [PubMed] [Google Scholar]
- 26.Zhou JR, Fu ZX, Wei LZ, Li YP, Li JC. Identification of tumor-associated proteins in laryngeal squamous cell carcinoma by proteomics. Chin J Otorhinolaryngol Head Neck Surg. 2007;42(12):934–938. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.