Abstract
Reversibly-stabilized DNA nanoparticles (rSDN) were prepared by coating reducible polycation/DNA complexes with multivalent N-(2-hydroxypropyl)methacrylamide (HPMA) copolymers. RGD targeted rSDN were formulated by linking cyclic c(RGDyK) to the surface layer of rSDN. Cellular uptake in B16F10 mouse melanoma cells, human endothelial (HUVEC) cells, and THLE immortalized hepatic cells was quantified by real-time PCR. RGD-targeted rSDN exhibited ~2 fold higher cell uptake in integrin-positive cells: B16F10 and HUVEC compared to THLE cells with low integrin content. RGD targeting mediated increased transfection activity in B16F10 cells but not in THLE cells. Overall, the studies show that rSDN can be effectively targeted with RGD while exhibiting reduced non-specific cell interactions and favorable stability. As such, these gene delivery vectors have the potential to permit targeting therapeutic genes to tumors by systemic delivery. In addition, the study shows that real-time PCR could be used effectively for quantification of cellular uptake of gene delivery vectors.
Keywords: RGD, HPMA, Gene delivery, Gene therapy, Tumor targeting, Real-Time PCR
1. Introduction
Safe and efficient systemic delivery of therapeutic nucleic acids remains a major challenge compromising successful application of gene therapy. The first major obstacle is the fast plasma clearance of almost all currently available delivery vectors. Naked plasmid DNA degrades in rat plasma with a half-life of 1–3 minutes (Houk et al., 1999, Houk et al., 2001). DNA polyplexes typically are prone to aggregation and a wide range of non-specific interactions (Lai and van Zanten, 2001, Burak et al., 2003) that contribute to their fast elimination from blood circulation after intravenous injection (Wiethoff et al., 2001, Ruponen et al., 1999). DNA lipoplexes show similarly unfavorable pharmacokinetics with less than 3% of DNA remaining in the plasma after 2–5 minutes after tail vein injection in mice (Niven et al., 1998, Osaka et al., 1996). In addition to non-specific interactions, susceptibility to disassembly was identified as a major contributing factor to the fast plasma clearance of polyplexes. Multivalent HPMA copolymers were successfully used in improving the stability against disassembly (Oupicky et al., 2002a, Dash et al., 2000). Using the optimized formulation of HPMA-coated stabilized DNA polyplexes (SDN) led to circulation half-lives of almost 90 minutes in mice, a significant improvement over the typical 3–5 min observed for control non-stabilized polyplexes (Oupicky et al., 2002b). The irreversible nature of the HPMA stabilization in SDN, which compromised transfection activity, was overcome by using reducible polycations that allowed easy intracellular reversal of the stabilization. These reversibly stabilized DNA polyplexes (rSDN) also were shown to exhibit improved plasma circulation, similar to SDN (Zhou et al., 2005) (Oupicky et al., 2002b).
The second challenge for gene delivery is the targeted delivery to the specific disease tissues or cells. Numerous reports describe the use of specific ligands ranging from peptides to monoclonal antibodies to target polyplexes to tumors. Despite substantial efforts, effective formulations of tumor-targeted polyplexes remain elusive. Peptide-based targeting ligands appear advantageous over protein-based ligands due to lower possibility of immune reactions and easier conjugation to the delivery vectors. Peptides containing the RGD motif have become among the most investigated targeting ligands due to their specific binding to integrins in tumor neovasculature (Baillie et al., 1995, Ruoslahti, 2002, Arap et al., 1998). The RGD motif binds many members of the integrin family in the cell membrane, including αvβ3, αvβ5, αvβ6, αvβ8, α5β1, α8β1 and αIIbβ3 (Hynes, 2002). An additional desirable feature of the RGD ligands is their ability to bind to both the tumor vasculature and the neoplastic cells (Rokhlin and Cohen, 1995, Kim et al., 2000) (Rokhlin and Cohen, 1995, Kim et al., 2000, Lang et al., 1997, Romanov and Goligorsky, 1999, Enns et al., 2005, Brenner et al., 2000). RGD peptides have been previously used to target DNA polyplexes (Kim et al., 2005, Kunath et al., 2003, Aris and Villaverde, 2000, Schiffelers et al., 2004) and lipoplexes (Harvie et al., 2003, Colin et al., 1998). While not always successful, enhanced gene expression of about one or two orders of magnitude was reported (Kunath et al., 2003, Kim et al., 2005).
The objective of this study was to demonstrate the utility of RGD targeting in combination with rSDN exhibiting low levels of non-specific cellular interactions (Fig. 1). An additional goal of this study was to evaluate the suitability of real-time PCR methodology for quantitative determination of the polyplexes that would be applicable for analyzing in vivo disposition of non-degraded DNA. The cyclic version of the RGD peptide used here provides the benefit of a better control of the spatial arrangement of the peptide compared with linear analogues and has been well established in animal and human imaging and targeting studies (Haubner et al., 2001, Mitra et al., 2005).
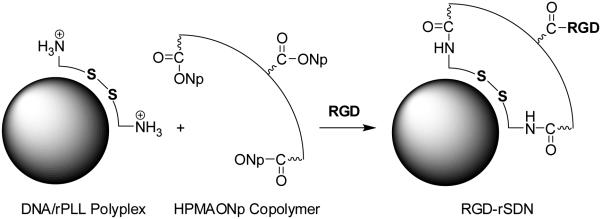
Figure 1.
Preparation of RGD-targeted rSDN. rPLL/DNA polyplexes are prepared by mixing rPLL with plasmid DNA and their surface is coated with multivalent HPMA copolymer. Targeting ligand c(RGDyK) is then covalently attached to the surface layer of HPMA-modified polyplex.
2. Materials and methods
2.1 Materials
Copolymer of HPMA with 9.3 mol% of methacryloylglycylglycine 4-nitrophenyl ester (Mw 4.1×104) was synthesized as described previously (Oupicky et al., 2000). Reducible PLL (rPLL, Mw 3.47×104) was synthesized by polymerization of CK10C peptide in 50% DMSO in Hepes buffer (pH 7.4) using previously described general protocol (Oupicky et al., 2002b). Average molecular weights of the synthesized polymers were determined by size exclusion chromatography equipped with a refractive index detector and multi-angle laser light scattering detector using either CATSEC-300 (rPLL) or Polymer Labs PL gel 5 µm mixed C column (HPMA copolymer). Targeting peptides, c(RGDyK) and control c(RADyK), were purchased from Peptides International, Inc. (Louisville, KY). gWiz™ High-Expression Luciferase (gWIZLuc) plasmid was purchased from Aldevron. Immortalized hepatic cell line THLE-3 (CRL-11233) was from ATCC (Rockville, MD) and maintained in BEGM® Bullet Kit (Lonza Group Ltd, catalog CC-3170), which includes 500 mL basal medium supplemented with 10% fetal bovine serum (FBS), 5 ng/mL EGF and separate frozen additives from which gentamycin/amphotericin and epinephrine were discarded. Human umbilical vein endothelial cells (HUVEC) were purchased from Cambrex Biosciences and cultured in EGM-2 medium with 5% FBS. The B16F10 mouse melanoma cells were cultured in DMEM supplemented with 10% FBS. Real-time PCR (RT-PCR) lysis buffer was made of 0.5 mg/mL Proteinase K, 2 mg/mL poly-L-aspartic acid (PAA; molecular weight 35,000) and 20 mM dithiothreitol (DTT). Anti mouse/human β-actin antibody (cat. no A2228) was from Sigma and anti mouse/human αv integrin antibody (cat. no 611012) was from BD-Pharmagen. Secondary antibody goat anti-mouse HRP-Ig conjugate (cat # 170-6516) was from Bio-Rad Corp.
2.2 Formulation of polyplexes
rPLL/DNA polyplexes were prepared by fast addition of rPLL solution to DNA solution (32 µg/mL) to achieve N:P ratio 2. The polyplexes were immediately vortexed for 30 s and incubated for 30 min before use. To coat the rPLL/DNA polyplexes, HPMA copolymer solution was added to the polyplex solution to achieve final copolymer concentration 2 mg/mL. The pH of the reaction mixture was adjusted to 7.8 with 500 mM Hepes buffer as previously described (Zhou et al., 2005). RGD-targeted and control RAD-containing rSDN were prepared by adding 200 µg/mL of the respective peptide (c(RGDyK) or c(RADyK)) to the coating reaction, 5 min after addition of HPMA copolymer. Unreacted polymer and peptides were removed after 24 h reaction at room temperature by VivaSpin20 centrifugal concentrator (molecular weight cut-off 1×105). The DNA recovery after purification was measured by gel electrophoresis after incubating the polyplexes with 20 mM DTT and 2 mg/mL PAA to ensure complete release of DNA. DNA recovery after concentration was typically >90% with only minimal changes in particle size (Oupicky et al., 2002a). The content of the RGD and RAD peptides in the polyplexes was determined by amino acid analysis at Texas A&M University Protein Chemistry Lab.
2.3 Determination of size and zeta potential of the polyplexes
The determination of hydrodynamic diameters and zeta potentials of polyplexes was performed using ZetaPlus Particle Size and Zeta Potential analyzer equipped with 35 mW solid state laser (658 nm) (Brookhaven Instruments Inc.) as described previously(Zhou et al., 2005). Three independent determinations were performed for each experiment.
2.4 Polyplex stability against polyelectrolyte exchange reactions
The stability of polyplexes against polyelectrolyte exchange reactions with PAA (MW 35,000) was evaluated by monitoring the release of free DNA using agarose gel electrophoresis. The polyplexes were incubated with 2 mg/mL PAA at 37 °C for 8 h in the presence or absence of 10 mM DTT and analyzed on 0.8% agarose gel with 50 μL of Ethidium Bromide (1 mg/mL) added to 100 mL gel. The amount of free DNA was determined using Kodak 1D Image Analysis Software. Three independent determinations were performed for each experiment.
2.5 Western blot
B16F10, HUVECs and THLE-3 were rinsed three times with PBS and lysed in the RIPA cell lysis buffer including 0.5% sodium deoxycholate, 1% NP40, 50 mM Tris (pH 7.6), 2 mM EGTA, 2 mM EDTA, 150 mM NaCl, 2 mM sodium vanadate, 1 mM phenylmethylsulfonyl fluoride, 10 µg/mL leupeptin, and 10 µg/mL aprotinin (Ustach et al., 2004). The cell lysates were centrifuged for 15 min and the protein content in supernatant was quantified using BCA™ Protein Assay Reagent Kit (PIERCE, Rockford, IL). Equal amounts of total protein from the cell lysates were run on Bio-Rad Criterion™ XT 10% Bis-Tris gel PAGE and electro-transferred onto nitrocellulose membrane (1.5 h, 4 °C, 100 V constant voltage) and blotted with αv integrin and β-actin antibodies to determine levels of αv integrin and β-actin. Secondary antibody used was goat anti-mouse HRP-Ig conjugate with dilution 1:10000. The immunoreactive bands were developed with Millipore Immobilon Western Chemiluminescent HRP Substrate.
2.6 Cytotoxicity evaluation
The cytotoxicity of control PLL (Mw =3.20×104 Da, Sigma) and rPLL (Mw =3.47×104 Da) were measured by MTS assay in the following cell lines: Eahy, Hela, MCF7, B16F10, HUVEC and THLE. Briefly, the cells were incubated with 0, 5, 10, 12.5, 15, 20, 40, 60 µg/mL of PLL and 0, 10, 20, 50, 100, 150, 200, 400 µg/mL of rPLL for 16 h. Then, the cells were washed with PBS and mixed with CellTiter96® AQueous Assay reagent (Promega Inc.) for 1 h and absorbance was read at the λ=505nm. The cytotoxicity of hydrolyzed HPMA copolymer was tested in the same cell lines. The cytotoxicity of the RGD and RAD peptides was tested in B16F10, HUVEC and THLE cells. Half lethal concentration (LC50) was calculated for each sample by survival analysis using StatPlus® software. Each experiment was performed four times.
2.7 Transfection efficiency in vitro
All transfection studies were performed in 96-well plates with cells plated 24 h before transfection at the seeding density of 60,000 cells/ml, 200 µL per well. (Soundara Manickam et al., 2005). The cells were incubated for 3 h with the different polyplexes (non-targeted polyplex, rSDN, RAD-rSDN, RGD-rSDN) in 150 µL of medium at 2 µg DNA/mL in the absence or presence of 100 μM chloroquine. All transfections were conducted in the presence of 10% FBS. For RGD competition experiments, 50 µg/mL of free c(RGDyK) was added to the wells 30 min before the incubation with polyplexes. The transfection results are expressed as Relative Light Units (RLU) per milligram of cellular protein ± SD, four independent determinations were performed for each transfection experiment (n=4).
2.8 Cellular uptake
The cell uptake of plasmid DNA (gWIZLuc) in the polyplexes was measured with RT-PCR (ABI Prism® 7300). The following probe and primers were designed in BioSearch online RealTimeDesign™ software: FAM-BHQ probe TCAGGATTACAAGATTCAAAGTGCGCT, forward primer GAAGAGCTGTTTCTGAGG, reverse primer CGAAGAAGGAGAATAGGGT. The 18S rRNA probe and the master mix were purchased from Eurogentec (cat. # RT-CKFT-18S, RT-QP2X-03). B16F10, THLE-3 and HUVEC cells were incubated with the polyplexes under conditions used in the transfection studies. After incubation, the cells were washed four times with medium using MultiWasher III (TriContinent Scientific, Inc.) and lysed with 25 μL of RT-PCR lysis buffer overnight. The lysate was then incubated for 10 min at 95 °C to deactivate the Proteinase K in the lysis buffer and diluted with 200 μL RNase-free water. During the RT-PCR cycle, 5 μL of DNA template, 500 nM probe and 100 nM primer pairs were added. To determine 18S rDNA, 600 nM probe and 125 nM primer pairs were added. The PCR cycle was run for 2 min at 50 °C, 10 min at 95 °C, 40 cycles for 15 s at 95 °C, and 1 min at 60 °C. Under these conditions, the PCR efficiency of the luciferase and 18SDNA is close to 1 (data not shown). The normalized content of the luciferase gene per 18S rRNA was calculated by the comparative CT method for relative quantification. The formula of normalized content was: 2^(CT(18sr) − CT(luc)), where CT(18sr) is the threshold cycle for 18SDNA and CT (luc) is threshold cycle for luciferase plasmid DNA. The results are presented as a mean ± S.D. obtained from four samples (n=4).
2.9 Statistical analysis
Statistical significance of the results was evaluated using two-tailed heteroscedastic Student’s t-test in Microsoft Excel. A p-value of < 0.05 was considered to be significant.
3. Results and Discussion
3.1. Biophysical properties of RGD-targeted rSDN
Biophysical properties such as size and surface charge are well known determinants of pharmacokinetic properties and transfection efficiency of DNA polyplexes. Typical DNA polyplexes exhibit highly positive surface charge and thus bind indiscriminately to all negatively charged cell membranes. The nonspecific binding, while acceptable for in vitro transfections, makes such polyplexes unsuitable for selective targeting in vivo. Direct conjugation of specific targeting ligands (especially small molecule ligands) to positively charged polyplexes often fails to improve their cellular uptake and transfection activity because of the overwhelming effect of the nonspecific, charge-mediated cell binding. While RGD-targeting was successfully achieved in some previous reports when the ligand was conjugated directly to the polycation, other reports found that RGD could not increase the cell uptake of PEI/DNA polyplexes because of the already high cell binding of the parent PEI/DNA which masked any potential effects of the RGD (Clements et al., 2006). For successful targeting of polyplexes, nonspecific interactions with cells must be minimized by using polyplexes with neutral or slightly negative surface charge.
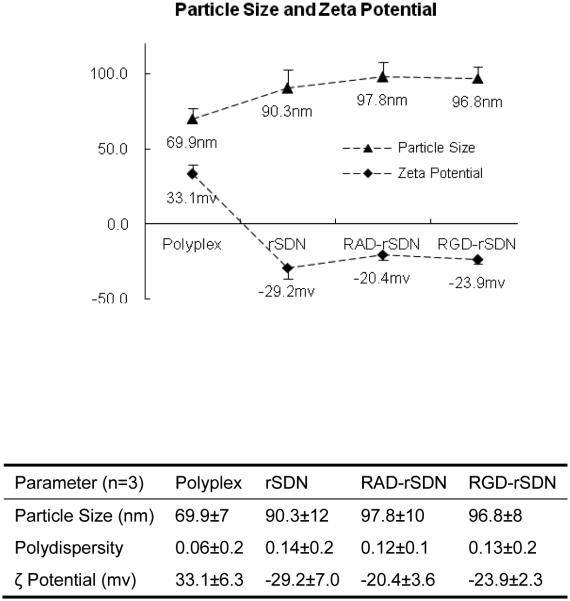
Fig. 2 shows the effect of coating the surface of rPLL/DNA polyplexes with the HPMA copolymers and RGD or RAD peptides on the size and ζ potential of the resulting rSDN. As expected, coating with the HPMA copolymers (Fig. 1) increased the size from ~70 nm to 90–97 nm. The increase in the size of the polyplexes is due to the surface layer of the HPMA copolymer. Importantly, addition of c(RGDyK) and c(RADyK) had no adverse effect on the size of rSDN. rPLL/DNA polyplexes exhibited the expected positive charge because of the excess of polycation. After coating with the HPMA copolymer, the positive charge was reversed to a negative charge ranging from −20 mV to −30 mV. The negative charge is a result of partial hydrolysis of the p-nitrophenyl ester groups of the HPMA copolymers attached to the surface of the polyplexes. The rSDN prepared here show size and surface charge suitable for minimized nonspecific cellular binding and effective targeting.
Figure 2.
Hydrodynamic diameters (triangles) and ζ potentials (diamonds) of the polyplex formulations used in the study. (n=3) (Polyplex: rPLL/DNA (N:P = 2); rSDN: reversibly-stabilized DNA nanoparticles; RAD-rSDN: control RAD-targeted rSDN; RGD-rSDN: RGD-targeted rSDN)
3.2. Stability of RGD-rSDN against polyelectrolyte exchange reactions
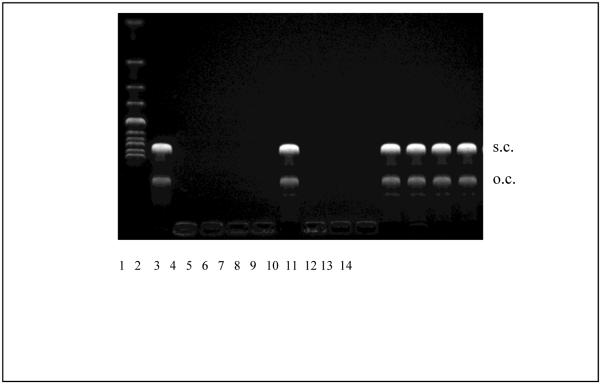
Polyelectrolyte exchange reactions are believed to play an essential role in gene delivery (Varga et al., 2001, Bronich et al., 2000). Although both monovalent and multivalent types of surface modification can endow DNA polyplexes with steric stability, using multivalent HPMA copolymers leads to an additional lateral stabilization, which is linked to improved pharmacokinetics of polyplexes (Oupicky et al., 2002b). The lateral stabilization can be demonstrated for example by the resistance of polyplexes to polyelectrolyte exchange reactions. As shown in Fig. 3, rSDN exhibit significantly improved stability against exchange reactions with competing polyanions. While rPLL/DNA polyplexes are completely destabilized and release free DNA in the presence of 2 mg/mL poly-L-aspartic acid (PAA), rSDN exhibit full stabilization under the experimental conditions indicated by undetectable free DNA. Incubation of rSDN with 10 mM DTT leads to a complete reversal of the lateral stabilization as indicated by an almost complete release of DNA from the polyplexes. Attachment of c(RGDyK) or c(RADyK) has no observable effect on the lateral stability of the polyplexes as both RGD-rSDN and RAD-rSDN resist the exchange reaction with PAA. Similar to the non-targeted rSDN, the lateral stability of both RGD-rSDN and RAD-rSDN is reversed by the addition of a reducing agent after which the treatment with PAA leads to a complete release of the DNA. These results confirm easy redox reversibility of the lateral stabilization of polyplexes based on reducible polycations (Oupicky et al., 2002b), and suggest that RGD-rSDN should exhibit a good stability in vivo.
Figure 3.
Resistance of rSDN against polyelectrolyte exchange reactions. Lane (1) DNA ladder; (2) luciferase plasmid DNA; (3) rPLL/DNA polyplex; (4) rSDN; (5) RAD-rSDN; (6) RGD-rSDN; (7) rPLL/DNA polyplex + 2 mg/mL PAA; (8) rSDN + 2 mg/mL PAA; (9) RAD-rSDN + 2 mg/mL PAA; (10) RGD-rSDN + 2 mg/mL PAA; (11) rPLL/DNA polyplex + 2 mg/mL PAA + 10 mM DTT; (12) rSDN + 2 mg/mL PAA + 10 mM DTT; (13) RAD-rSDN + 2 mg/mL PAA + 10 mM DTT; (14) RGD-rSDN + 2 mg/mL PAA + 10 mM DTT.
3.3. Cellular levels of αv integrins (move this section before the toxicity)
Levels of integrin expression were determined by Western blot in the three cell lines selected to represent endothelial cells, tumor cells, and liver cells (HUVEC, B16F10, THLE). HUVEC cells are known to express high levels of αvβ3, αVβ5 (Patrycja et al., 2005, Suh et al., 2002), while B16F10 melanoma cells show high levels of αvβ3 (Shiras et al., 2002, Ray et al., 1999, Smolarczyk et al., 2006). The cells were chosen to represent the neovasculature cells and tumor cells respectively. The THLE human hepatic cell line, which was reported to show only low levels of αv compared with hepatocellular carcinoma cells, was chosen as a control to represent cells with low levels of integrin receptors (Mayoral et al., 2005). The rationale for selecting the above cell lines was to verify enhanced cell uptake in the endothelial and tumor cells while minimizing uptake and transfection in the low-integrin hepatic cells.
The αvβ3 and αvβ5 are the two most important integrins in the RGD-mediated targeting and tumor angiogenesis. HUVEC and THLE are of human origin while B16F10 is of mouse origin. Unfortunately, there are no commercially available antibodies that react specifically with both human and mouse αvβ3 and αvβ5 integrins. Therefore, we used commercially available αv antibody which has cross reactivity with the human and mouse cells to perform the Western blot. Since both αvβ3 and αvβ5 share the αv part of all known αv dimers (αvβ1, αvβ3, αvβ5, αvβ6, αvβ8) bind RGD motif (Hynes, 2002), the expression of αv should provide a sufficient measure of the RGD binding capacity in each corresponding cell line. Fig. 4 demonstrates that HUVEC and B16F10 cells express substantially higher levels of αv integrin than THLE cells, confirming the suitability of the selected panel of cells for the subsequent studies.
Figure 4.
Analysis of αv integrin in B16F10, THLE-3 and HUVEC cells by Western blot. (molecular weight of αv integrin is 125kDa and β-actin 42kDa)
3.4 Cytoxicity
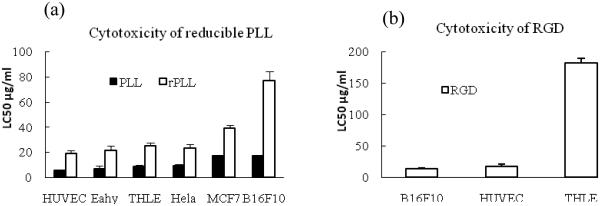
Measuring cytotoxicity of the individual components that constitute the particles provides a more comprehensive understanding of the toxic effects due to the possibility to measure toxicity over a wide concentration range, which is experimentally difficult to achieve with the nanoparticles. It should be pointed out, however, that none of the nanoparticle formulations show any significant signs of cytotoxicity in the concentration range used in the in vitro transfection experiments described below. The rPLL used these studies display lower cytotoxicity than control PLL in all the cell lines tested. The LC50 of rPLL is about 3 times higher than the control PLL (Fig 5a). The LC50 of PLL ranges from ~5 µg/mL to 20 µg/mL, while the LC50 of rPLL ranges from ~20 µg/mL to 70 µg/mL. As expected, the HPMA copolymer shows no signs of cytotoxicity up to at least 1 mg/mL in all the tested cell lines (not shown). Similarly, the control RAD peptide doesn’t display any obvious signs of cytotoxicity up to 1 mg/mL (not shown). On the other hand, it was previously reported that unlikely RAD, RGD peptide and RGD antagonist can induce an apoptosis in HUVEC {Aguzzi, 2004 #100} and human erythroleukemia cell line K562 stably transfected with αvβ3 (Meerovitch et al., 2003). The previous findings are confirmed in the cells used in this study (Fig. 5b). The RGD peptide displays higher cytotoxicity in cells with high levels of integrin receptors (B16F10 and HUVEC) than in THLE cells with low levels of integrin expression. The toxicity of RGD should not adversely affect studies of the RGD-targeted polyplexes since the RGD concentration in the cell uptake and transfection experiments described here is less than 0.2 µg/ml (i.e. far less than LC50 of RGD).
Figure 5.
Cytotoxicity (a) of rPLL and PLL in different cell lines and (b) of RGD in THLE, HUVEC, B16F10 cells. (n=4)
3.5. Increased cell uptake of RGD-targeted rSDN
The two main features that distinguish rSDN from typical PEGylated polyplexes are the negative surface charge and resistance against disassembly. The negative surface charge is expected to reduce non-specific cellular uptake and thus increase the selectivity of RGD-mediated delivery. Cellular uptake of DNA polyplexes can be measured by a variety of analytical methods, including the use of fluorescently or radioactively labeled DNA. Neither of the two methods, however, can easily distinguish inactive (i.e. degraded) from intact DNA. Furthermore, methods relying on fluorescently labeled DNA usually do not lend themselves readily for quantitative analysis of in vivo distribution of polyplexes. Real-time PCR, on the other hand, is not only highly sensitive and requires no chemical modification of DNA but also is suitable for in vivo analysis and allows quantification of only the active, non-degraded form of DNA.
Several commercial qPCR lysis buffers were tested that allow determination of the target DNA from crude cell lysates without the need for purification. None of the evaluated lysis buffers we tested such as one step lysis buffer by Strategene Co. Ltd. could quantitatively release plasmid DNA from the rSDN. A modified lysis buffer was therefore formulated by supplementing a standard buffer with DTT to reduce disulfide bonds in the rSDN and with PAA to dissociate the polyplexes.
The content of the RGD and RAD peptides in the polyplex formulations was determined by amino acid analysis. Polyplexes were prepared at 32 μg DNA/mL and RGD and RAD contents were determined from the content of tyrosine residues. It was found that the average RGD content was 5.0 ± 0.2 nmol/mL(n=3) and average RAD content was 4.8 ± 0.1 nmol/mL) (n=3). Assuming that molecular weight of DNA (6732 bp) is 4.38×106 Da, this equals to an average 680 ± 30 RAD and 660 ± 20 RGD molecules for every DNA molecule.
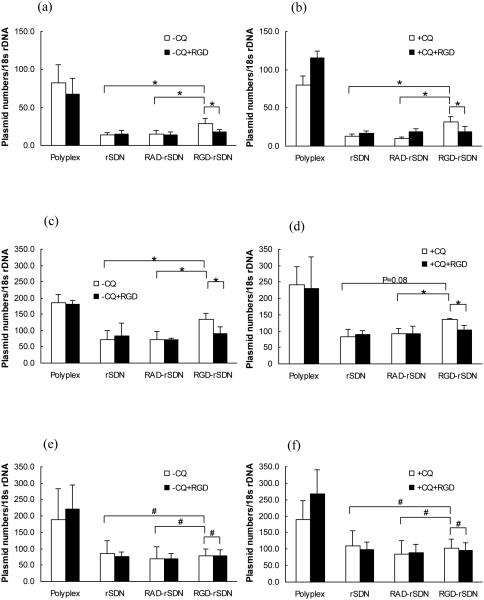
Cellular uptake was determined in the panel of the three cell lines using RT-PCR (Fig. 6). The uptake is expressed as number of plasmid DNA copies normalized to the content of 18srDNA. The average uptake of rPLL/DNA polyplexes in the integrin-negative THLE cells was about 200 plasmid copies per every 18SrDNA (Fig. 6e, f). Coating with the HPMA reduced the cell uptake almost 4-fold, due to minimization of non-specific uptake of the negatively charged rSDN. No increase in the cellular uptake was observed for RGD-rSDN in the THLE cells compared to the non-targeted or control RAD-targeted rSDN. Addition of the lysosomotropic agent chloroquine did not affect the cellular uptake. Cellular uptake of the parent rPLL/DNA polyplexes in the integrin-positive B16F10 cells was lower than that observed in THLE cells. About 80 copies of plasmid DNA per 18SrDNA were taken up by B16F10 and the uptake was further decreased after coating with HPMA to 15 copies plasmid per 18SrDNA. RGD-rSDN exhibited almost 2-fold increase in the cell uptake to 30 copies of plasmid DNA per 18SrDNA, while the control RAD-rSDN had no effect on the cell uptake (Fig. 6a, b). The increased cellular uptake of RGD-rSDN was reduced to the original levels observed for rSDN in competitive experiments with excess of free c(RGDyK) (50 μg/mL). The specific concentration of free c(RGDyK) was selected because it was the maximum concentration at which no significant signs of cytotoxicity were observed. In HUVEC cells, the rPLL/DNA polyplexes showed maximum cell uptake of 200 copies of plasmid DNA per 18SrDNA. Coating with HPMA decreased the uptake to ~60 copies of plasmid per 18SrDNA, while the cell uptake of the RGD-targeted rSDN increased 2-fold (130 plasmid copies per 18SrDNA) (Fig. 6c, d). Control RAD-rSDN was taken up by the HUVEC cells to the same extent as the parent rSDN polyplexes. Similar to the B16F10, the increased cell uptake of RGD-rSDN in HUVEC can be inhibited by the addition of free RGD. In conclusion, cyclic RGD ligand successfully enhanced cell uptake of rSDN in integrin expressing cells, HUVEC and B16F10, which points to the suitability of RGD-rSDN for tumor targeted delivery in vivo. On the other hand, it is obvious that the non-specific charge-mediated cellular uptake is more efficient than RGD-mediated uptake as evidenced by the higher levels of cellular uptake of the control rPLL/DNA polyplexes compared to the RGD-targeted ones. As shown in our experiments, the rSDN shows 2-fold increase in cell uptake in the cells over-expressing integrins. Nonetheless, the rSDN nanoparticles have a highly negative surface charge (zeta potential < −20 mV) as shown in Figure 2. We predict that reducing the negative surface charge to a less negative value will have a positive effect on RGD-mediated cell uptake due to decreasing effect of charge repulsion between the particles and cell surface.
Figure 6.
Normalized cell uptake of rSDN. (a) uptake in B16F10 cells in the absence of chloroquine (−CQ); (b) uptake in B16F10 cells in the presence of 100 µM chloroquine (+CQ); (c) uptake in HUVEC cells in the absence of chloroquine (−CQ); (d) uptake in HUVEC cells in the presence of 100 µM chloroquine (+CQ); (e) uptake in THLE-3 cells in the absence of chloroquine (−CQ); (f) uptake in THLE-3 cells in the presence of 100 µM chloroquine (+CQ). Cell uptake was measured in the presence (black bars) or absence (white bars) of competing free c(RGDyK) by real time PCR. The uptake is expressed as the number of plasmid DNA copies per 18s rDNA. (# p-value > 0.05; * p-value < 0.05, n=4)
3.6. Enhanced transfection activity of the RGD-targeted rSDN in B16F10 tumor cells
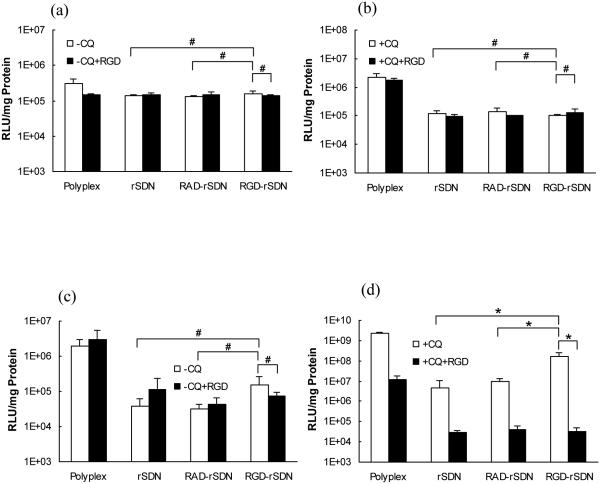
Transfection activity of RGD-rSDN was measured to determine whether the increased cell uptake translates into enhanced transgene expression in vitro (Fig. 7). The transfection activity of RGD targeted rSDN increased almost 10-fold in our study compared with non-targeted rSDN in the B16F10 tumor cells in the presence of chloroquine. This modest level of transfection increase is fully comparable to previously published results. For example, it was reported that PEI-g-PEG-RGD increased transfection about 2-fold compared with PEI in angiogenic human dermal microvascular endothelial cells (Kim et al., 2005). When c(RGDfK)-PEG-PLLL was used as the delivery vector, a three-fold increase in luciferase transfection was observed compared with control PEG-PLL polyplexes in HeLa cells (Oba et al., 2007). Only low levels of transfection were observed in THLE cells (Fig. 7a, b) even in the presence of chloroquine. In contrast, transfection activity was significantly higher and strongly dependent on the presence of chloroquine in B16F10 cells (Fig. 7c, d). Coating with HPMA decreased the transfection in B16F10 almost 100-fold, which correlated well with the reduced cellular uptake (Fig. 6).
Figure 7.
Transfection activity of rSDN in B16F10 and THLE-3 cells. (a) THLE-3 cells without chloroquine; (b) THLE-3 cells with 100 µM chloroquine; (c) B16F10 without chloroquine; (d) B16F10 cells with 100 µM chloroquine. Transfection was measured in the presence (black bar) or absence of (white bar) of competing free RGD. (# p-value > 0.05; * p-value < 0.05, n=4)
Transfection activity of the RGD-targeted rSDN increased more than 10-fold compared to non-targeted rSDN and control RAD-rSDN. Although the transfection activity of RGD-rSDN in B16F10 did not reach levels observed for rPLL/DNA polyplexes, it was significantly higher than transfection of the parent rPLL/DNA in THLE cells. These results also confirm previous observation by Pan et al. (Pan et al., 2006) which showed that only in the presence of chloroquine can RGD increase transfection. Despite comparable levels of cellular uptake, no transfection was observed in the HUVEC cells, confirming the importance of subcellular trafficking in addition to absolute levels of cellular uptake for effective transfection. Inefficient endosomal escape and nuclear entry are the most likely factors that contributed to the poor transfection observed in the slow growing HUVEC cells. Addition of free excess RGD ligand during the incubation of B16F10 cells with the polyplexes counteracts the enhanced transfection activity of RGD-rSDN in the presence of chloroquine (Fig. 7d). Overall, targeting rSDN with RGD allows increasing cellular uptake and transfection activity in integrin-expressing cancer cells. However, overcoming the reliance of the rSDN transfection on external lysosomotropic agents is required to increase the potential of rSDN for in vivo tumor-targeted gene delivery (Zhou et al., 2005). The next step of this study is to verify the findings in vivo. Because B16F10 tumors retain the high levels of integrin expression in vivo in C57B6 mice, it should be possible to achieve increased accumulation and transfection in these tumors in vivo.
4. Conclusion
RGD peptides have been widely used for polyplex targeting, however, most of the published literature deals with vectors stabilized by monovalent attachment of PEG (Schiffelers et al., 2004, Oba et al., 2007, Kunath et al., 2003, Kim et al., 2005, Harvie et al., 2003). The gene delivery vectors reported here are stabilized by crossing linking of the surface of the polyplex with multivalent hydrophilic polymer. We successfully used the cyclic RGD ligand in this stabilized system to increase cell uptake and transfection. In addition, we report the development of a convenient real-time PCR method for measuring DNA accumulation in the cells. This is a simple method using a one-step lysis with several advantages over the conventional fluorescent or radioactive labeling methods to quantify cell uptake: (i) there is no need for labeling of DNA, which may change the properties of the nucleic acids, (ii) it allows to easily distinguish between intact active form of DNA and inactive degraded DNA, which is difficult to achieve with the labeling methods. The results presented in this study therefore demonstrate the suitability of RT-PCR for analyzing cellular uptake of reversibly-stabilized polyplexes based on reducible polycations. The results further show that rSDN can be effectively targeted with RGD while exhibiting reduced non-specific cell interactions and favorable stability. As such, these gene delivery vectors have the potential to permit targeting therapeutic genes to tumors by systemic delivery. In vivo disposition studies of the RGD-rSDN vectors are underway.
Acknowledgement
This research was financially supported by the National Institutes of Health (CA 109711). We thank Dr. Amjad and Dr. Cha of the Anti-Infective Research Laboratory of Wayne State University for help with the real-time PCR.
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper
References
- ARAP W, PASQUALINI R, RUOSLAHTI E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science. 1998;279:377–80. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]
- ARIS A, VILLAVERDE A. Molecular organization of protein-DNA complexes for cell-targeted DNA delivery. Biochem Biophys Res Commun. 2000;278:455–61. doi: 10.1006/bbrc.2000.3824. [DOI] [PubMed] [Google Scholar]
- BAILLIE CT, WINSLET MC, BRADLEY NJ. Tumour vasculature--a potential therapeutic target. Br J Cancer. 1995;72:257–67. doi: 10.1038/bjc.1995.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRENNER W, GROSS S, STEINBACH F, HORN S, HOHENFELLNER R, THUROFF JW. Differential inhibition of renal cancer cell invasion mediated by fibronectin, collagen IV and laminin. Cancer Lett. 2000;155:199–205. doi: 10.1016/s0304-3835(00)00429-8. [DOI] [PubMed] [Google Scholar]
- BRONICH TK, NGUYEN HK, EISENBERG A, KABANOV AV. Recognition of DNA Topology in Reactions between Plasmid DNA and Cationic Copolymers. 2000.
- BURAK Y, ARIEL G, ANDELMAN D. Onset of DNA aggregation in presence of monovalent and multivalent counterions. Biophys J. 2003;85:2100–10. doi: 10.1016/s0006-3495(03)74638-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLEMENTS BA, BAI J, KUCHARSKI C, FARRELL LL, LAVASANIFAR A, RITCHIE B, GHAHARY A, ULUDAG H. RGD conjugation to polyethyleneimine does not improve DNA delivery to bone marrow stromal cells. Biomacromolecules. 2006;7:1481–8. doi: 10.1021/bm060073w. [DOI] [PubMed] [Google Scholar]
- COLIN M, HARBOTTLE RP, KNIGHT A, KORNPROBST M, COOPER RG, MILLER AD, TRUGNAN G, CAPEAU J, COUTELLE C, BRAHIMI-HORN MC. Liposomes enhance delivery and expression of an RGD-oligolysine gene transfer vector in human tracheal cells. Gene Ther. 1998;5:1488–98. doi: 10.1038/sj.gt.3300760. [DOI] [PubMed] [Google Scholar]
- DASH PR, READ ML, FISHER KD, HOWARD KA, WOLFERT M, OUPICKY D, SUBR V, STROHALM J, ULBRICH K, SEYMOUR LW. Decreased binding to proteins and cells of polymeric gene delivery vectors surface modified with a multivalent hydrophilic polymer and retargeting through attachment of transferrin. J Biol Chem. 2000;275:3793–802. doi: 10.1074/jbc.275.6.3793. [DOI] [PubMed] [Google Scholar]
- ENNS A, KORB T, SCHLUTER K, GASSMANN P, SPIEGEL HU, SENNINGER N, MITJANS F, HAIER J. Alphavbeta5-integrins mediate early steps of metastasis formation. Eur J Cancer. 2005;41:1065–72. doi: 10.1016/j.ejca.2004.12.031. [DOI] [PubMed] [Google Scholar]
- HARVIE P, DUTZAR B, GALBRAITH T, CUDMORE S, O'MAHONY D, ANKLESARIA P, PAUL R. Targeting of lipid-protamine-DNA (LPD) lipopolyplexes using RGD motifs. J Liposome Res. 2003;13:231–47. doi: 10.1081/lpr-120026389. [DOI] [PubMed] [Google Scholar]
- HAUBNER R, WESTER HJ, BURKHART F, SENEKOWITSCH-SCHMIDTKE R, WEBER W, GOODMAN SL, KESSLER H, SCHWAIGER M. Glycosylated RGD-containing peptides: tracer for tumor targeting and angiogenesis imaging with improved biokinetics. J Nucl Med. 2001;42:326–36. [PubMed] [Google Scholar]
- HOUK BE, HOCHHAUS G, HUGHES JA. Kinetic modeling of plasmid DNA degradation in rat plasma. AAPS PharmSci. 1999;1:E9. doi: 10.1208/ps010309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOUK BE, MARTIN R, HOCHHAUS G, HUGHES JA. Pharmacokinetics of plasmid DNA in the rat. Pharm Res. 2001;18:67–74. doi: 10.1023/a:1011078711008. [DOI] [PubMed] [Google Scholar]
- HYNES RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–87. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- KIM S, BELL K, MOUSA SA, VARNER JA. Regulation of angiogenesis in vivo by ligation of integrin alpha5beta1 with the central cell-binding domain of fibronectin. Am J Pathol. 2000;156:1345–62. doi: 10.1016/s0002-9440(10)65005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM WJ, YOCKMAN JW, LEE M, JEONG JH, KIM YH, KIM SW. Soluble Flt-1 gene delivery using PEI-g-PEG-RGD conjugate for anti-angiogenesis. J Control Release. 2005;106:224–34. doi: 10.1016/j.jconrel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- KUNATH K, MERDAN T, HEGENER O, HABERLEIN H, KISSEL T. Integrin targeting using RGD-PEI conjugates for in vitro gene transfer. J Gene Med. 2003;5:588–99. doi: 10.1002/jgm.382. [DOI] [PubMed] [Google Scholar]
- LAI E, VAN ZANTEN JH. Monitoring DNA/poly-L-lysine polyplex formation with time-resolved multiangle laser light scattering. Biophys J. 2001;80:864–73. doi: 10.1016/S0006-3495(01)76065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANG SH, CLARKE NW, GEORGE NJ, TESTA NG. Primary prostatic epithelial cell binding to human bone marrow stroma and the role of alpha2beta1 integrin. Clin Exp Metastasis. 1997;15:218–27. doi: 10.1023/a:1018465213641. [DOI] [PubMed] [Google Scholar]
- MAYORAL R, FERNANDEZ-MARTINEZ A, BOSCA L, MARTIN-SANZ P. Prostaglandin E2 promotes migration and adhesion in hepatocellular carcinoma cells. Carcinogenesis. 2005;26:753–61. doi: 10.1093/carcin/bgi022. [DOI] [PubMed] [Google Scholar]
- MEEROVITCH K, BERGERON F, LEBLOND L, GROUIX B, POIRIER C, BUBENIK M, CHAN L, GOURDEAU H, BOWLIN T, ATTARDO G. A novel RGD antagonist that targets both alphavbeta3 and alpha5beta1 induces apoptosis of angiogenic endothelial cells on type I collagen. Vascul Pharmacol. 2003;40:77–89. doi: 10.1016/s1537-1891(02)00339-7. [DOI] [PubMed] [Google Scholar]
- MITRA A, MULHOLLAND J, NAN A, MCNEILL E, GHANDEHARI H, LINE BR. Targeting tumor angiogenic vasculature using polymer-RGD conjugates. J Control Release. 2005;102:191–201. doi: 10.1016/j.jconrel.2004.09.023. [DOI] [PubMed] [Google Scholar]
- NIVEN R, PEARLMAN R, WEDEKING T, MACKEIGAN J, NOKER P, SIMPSON-HERREN L, SMITH JG. Biodistribution of radiolabeled lipid-DNA complexes and DNA in mice. J Pharm Sci. 1998;87:1292–9. doi: 10.1021/js980087a. [DOI] [PubMed] [Google Scholar]
- OSAKA G, CAREY K, CUTHBERTSON A, GODOWSKI P, PATAPOFF T, RYAN A, GADEK T, MORDENTI J. Pharmacokinetics, tissue distribution, and expression efficiency of plasmid [33P]DNA following intravenous administration of DNA/cationic lipid complexes in mice: use of a novel radionuclide approach. J Pharm Sci. 1996;85:612–8. doi: 10.1021/js9504494. [DOI] [PubMed] [Google Scholar]
- OUPICKY D, HOWARD KA, KONAK C, DASH PR, ULBRICH K, SEYMOUR LW. Steric stabilization of poly-L-Lysine/DNA complexes by the covalent attachment of semitelechelic poly[N-(2-hydroxypropyl)methacrylamide] Bioconjug Chem. 2000;11:492–501. doi: 10.1021/bc990143e. [DOI] [PubMed] [Google Scholar]
- OUPICKY D, OGRIS M, HOWARD KA, DASH PR, ULBRICH K, SEYMOUR LW. Importance of lateral and steric stabilization of polyelectrolyte gene delivery vectors for extended systemic circulation. Mol Ther. 2002a;5:463–72. doi: 10.1006/mthe.2002.0568. [DOI] [PubMed] [Google Scholar]
- OUPICKY D, PARKER AL, SEYMOUR LW. Laterally stabilized complexes of DNA with linear reducible polycations: strategy for triggered intracellular activation of DNA delivery vectors. J Am Chem Soc. 2002b;124:8–9. doi: 10.1021/ja016440n. [DOI] [PubMed] [Google Scholar]
- PAN H, ZHENG Q, GUO X, LIU Y, LI C, SONG Y. A RGD-containing oligopeptide (K)16GRGDSPC: a novel vector for integrin-mediated targeted gene delivery. J Huazhong Univ Sci Technolog Med Sci. 2006;26:513–6. doi: 10.1007/s11596-006-0506-7. [DOI] [PubMed] [Google Scholar]
- PATRYCJA BARANSKA HJ, PAWLOWSKA ZOFIA, KOZIOLKIEWICZ WIKTOR, CIERNIEWSKI CZESLAWS. Expression of Integrins and Adhesive Properties of Human Endothelial Cell Line EA.hy 926. CANCER GENOMICS & PROTEOMICS. 2005;2:265–270. [PubMed] [Google Scholar]
- RAY S, CHATTOPADHYAY N, BISWAS N, CHATTERJEE A. Regulatory molecules in tumor metastasis. J Environ Pathol Toxicol Oncol. 1999;18:251–9. [PubMed] [Google Scholar]
- ROKHLIN OW, COHEN MB. Expression of cellular adhesion molecules on human prostate tumor cell lines. Prostate. 1995;26:205–12. doi: 10.1002/pros.2990260406. [DOI] [PubMed] [Google Scholar]
- ROMANOV VI, GOLIGORSKY MS. RGD-recognizing integrins mediate interactions of human prostate carcinoma cells with endothelial cells in vitro. Prostate. 1999;39:108–18. doi: 10.1002/(sici)1097-0045(19990501)39:2<108::aid-pros5>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- RUOSLAHTI E. Specialization of tumour vasculature. Nat Rev Cancer. 2002;2:83–90. doi: 10.1038/nrc724. [DOI] [PubMed] [Google Scholar]
- RUPONEN M, YLA-HERTTUALA S, URTTI A. Interactions of polymeric and liposomal gene delivery systems with extracellular glycosaminoglycans: physicochemical and transfection studies. Biochim Biophys Acta. 1999;1415:331–41. doi: 10.1016/s0005-2736(98)00199-0. [DOI] [PubMed] [Google Scholar]
- SCHIFFELERS RM, ANSARI A, XU J, ZHOU Q, TANG Q, STORM G, MOLEMA G, LU PY, SCARIA PV, WOODLE MC. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 2004;32:e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHIRAS A, BHOSALE A, PATEKAR A, SHEPAL V, SHASTRY P. Differential expression of CD44(S) and variant isoforms v3, v10 in three-dimensional cultures of mouse melanoma cell lines. Clin Exp Metastasis. 2002;19:445–55. doi: 10.1023/a:1016305611858. [DOI] [PubMed] [Google Scholar]
- SMOLARCZYK R, CICHON T, GRAJA K, HUCZ J, SOCHANIK A, SZALA S. Antitumor effect of RGD-4C-GG-D(KLAKLAK)2 peptide in mouse B16(F10) melanoma model. Acta Biochim Pol. 2006;53:801–5. [PubMed] [Google Scholar]
- SOUNDARA MANICKAM D, BISHT HS, WAN L, MAO G, OUPICKY D. Influence of TAT-peptide polymerization on properties and transfection activity of TAT/DNA polyplexes. J Control Release. 2005;102:293–306. doi: 10.1016/j.jconrel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- SUH W, HAN SO, YU L, KIM SW. An angiogenic, endothelial-cell-targeted polymeric gene carrier. Mol Ther. 2002;6:664–72. [PubMed] [Google Scholar]
- USTACH CV, TAUBE ME, HURST NJ, JR., BHAGAT S, BONFIL RD, CHER ML, SCHUGER L, KIM HR. A potential oncogenic activity of platelet-derived growth factor d in prostate cancer progression. Cancer Res. 2004;64:1722–9. doi: 10.1158/0008-5472.can-03-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VARGA CM, HONG K, LAUFFENBURGER DA. Quantitative analysis of synthetic gene delivery vector design properties. Mol Ther. 2001;4:438–46. doi: 10.1006/mthe.2001.0475. [DOI] [PubMed] [Google Scholar]
- WIETHOFF CM, SMITH JG, KOE GS, MIDDAUGH CR. The potential role of proteoglycans in cationic lipid-mediated gene delivery. Studies of the interaction of cationic lipid-DNA complexes with model glycosaminoglycans. J Biol Chem. 2001;276:32806–13. doi: 10.1074/jbc.M007940200. [DOI] [PubMed] [Google Scholar]
- ZHOU QH, MILLER DL, CARLISLE RC, SEYMOUR LW, OUPICKY D. Ultrasound-enhanced transfection activity of HPMA-stabilized DNA polyplexes with prolonged plasma circulation. J Control Release. 2005;106:416–27. doi: 10.1016/j.jconrel.2005.05.002. [DOI] [PubMed] [Google Scholar]