Abstract
HISAT (hierarchical indexing for spliced alignment of transcripts) is a highly efficient system for aligning reads from RNA sequencing experiments. HISAT uses an indexing scheme based on the Burrows-Wheeler transform and the Ferragina-Manzini (FM) index, employing two types of indexes for alignment: a whole-genome FM index to anchor each alignment and numerous local FM indexes for very rapid extensions of these alignments. HISAT’s hierarchical index for the human genome contains 48,000 local FM indexes, each representing a genomic region of ~64,000 bp. Tests on real and simulated data sets showed that HISAT is the fastest system currently available, with equal or better accuracy than any other method. Despite its large number of indexes, HISAT requires only 4.3 gigabytes of memory. HISAT supports genomes of any size, including those larger than 4 billion bases.
Since its introduction in 2008, RNA-seq1 has become ubiquitous as a tool for the study of gene expression, transcript structure and the identification of long noncoding RNAs and fusion transcripts2–5 As RNA-seq has matured, sequencing throughput and read lengths have increased dramatically to 100–500 million reads per run with lengths of 100 bp or longer. These large and ever-increasing data volumes necessitate fast and scalable computational analysis systems.
RNA-seq analysis begins by aligning reads against a reference genome to determine the location from which the reads originated6–8 a step that has become a time-consuming bottleneck; for example, widely used alignment programs such as TopHat2 (ref. 9) and GSNAP10 can take several days to process a single RNA-seq experiment. The recently introduced STAR program11 uses suffix arrays to provide substantially faster processing than most other methods, including TopHat2. However, the suffix-array method has very large memory requirements (28 gigabytes (GB) for the human genome) as compared to methods using the Burrows-Wheeler transform.
To create a fast spliced aligner that uses a modest amount of random access memory (RAM), we designed HISAT with a novel indexing strategy based on the Burrows-Wheeler transform12 and the FM index13.
As a result of HISAT’s greatly reduced memory requirements, users can shift these computations from special-purpose servers to a single conventional desktop computer, on which it is possible to run multiple samples at the same time. As developers of TopHat, we intend to make HISAT the core alignment engine for the next major version of that program, TopHat3. HISAT is open-source software freely available at http://www.ccb.jhu.edu/software/hisat/.
RESULTS
Design principles of HISAT
HISAT uses the Bowtie2 (ref. 14) implementation to handle many of the low-level operations required to construct and search an FM index. In contrast to most other aligners, our algorithm employs two different types of indexes: (i) a global FM index that represents the entire genome and (ii) numerous small FM indexes for regions that collectively cover the genome, where each index represents 64,000 bp. For the human genome, we create ~48,000 local FM indexes, each overlapping its neighbor by 1,024 bp, to cover the entire 3 billion bases. The overlapping boundaries make it easier to align reads that would otherwise span the regions covered by two indexes.
The program stores the large number of local indexes in a small set of files and implements other optimizations to minimize the memory requirements, allowing us to index the human genome in approximately 4 GB of space.
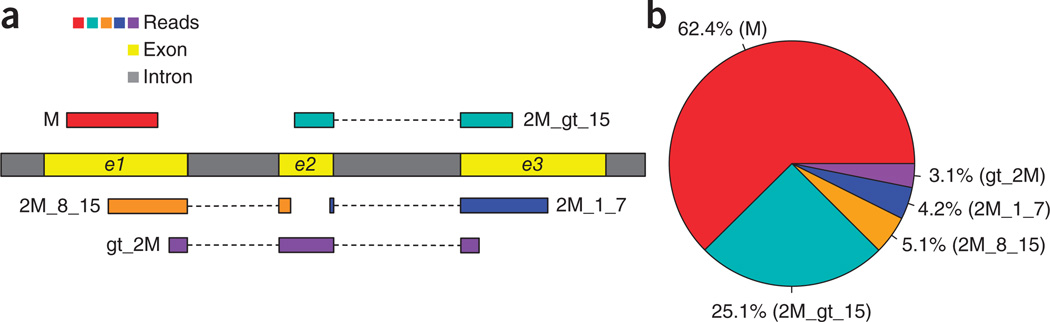
RNA-seq reads may span large gaps corresponding to introns, which in mammalian genomes can be over 1 Mbp in length. Exons are relatively short, and thus when 100-bp reads are used, a substantial proportion of reads (~34.5% in our simulated data set) will span two exons. For the purpose of alignment, we divide these exon-spanning reads into three categories: long-anchored reads, which have at least 16 bp in each of the two exons; intermediate-anchored reads, which have 8–15 bp in one exon; and short-anchored reads, with just 1–7 bp aligned to one of the exons (Fig. 1a).
Figure 1.
RNA-seq read types and their relative proportions from 20 million simulated 100-bp reads. (a) Five types of RNA-seq reads: (i) M, exonic read; (ii) 2M_gt_15, junction reads with long, >15-bp anchors in both exons; (iii) 2M_8_15, junction reads with intermediate, 8- to 15-bp anchors; (iv) 2M_1_7, junction reads with short, 1- to 7-bp, anchors; and (v) gt_2M, junction reads spanning more than two exons. (b) Relative proportions of different types of reads in the 20 million 100-bp simulated read data.
For a simulated human RNA-seq data set (100-bp reads) with realistic parameters (Supplementary Note), ~25.1% of the reads span two exons with long anchors (>15 bp) in both exons (Fig. 1b) which in most cases can be mapped to a unique location in the human genome. 5.1% of the reads spanned two exons with an intermediate-length anchor (8–15 bp) on one exon. Alignment programs that rely on a global index have great difficulty mapping these anchors uniquely (for example, an 8-bp sequence is expected to occur ~48,000 times in the human genome). This is where the use of a local index provides a substantial advantage. In HISAT, each local index covers 64,000 bp; thus, over 90% of annotated human introns are completely contained in one of these indexes. After mapping the longer part of a read to identify the relevant local index, HISAT can usually align the remaining small anchor within a single local index rather than searching across the whole genome. On average, an 8-bp sequence will occur just once in a local index of this size.
In our simulated data, 4.2% of the reads span two exons with a very short anchor (1–7 bp) in one exon. Because these anchors are so short, the best approach is, where possible, to align these reads by making use of splice site information found by aligning other reads in the same data or by using known splice sites. Note that ~3.1% of reads span more than two exons. In many mapping algorithms, the alignment of short- and intermediate-anchored reads and reads spanning more than two exons (12.4% of the total reads) takes up to 30–60% of the total run time, and many of those reads are ultimately aligned incorrectly or left unaligned.
HISAT solves these challenging spliced-alignment problems using hierarchical indexing and several alignment strategies specifically designed for handling different read types (Online Methods).
Comparison to other tools for accuracy and speed
We compared the accuracy and speed of HISAT to several of the leading spliced-alignment programs, including STAR11, GSNAP10, OLego15 and TopHat2 (ref. 9), using both simulated and real reads. We tested three versions of HISAT (HISATx1, HISATx2 and HISAT), which we ran with different parameters. HISATx1 uses a one-pass approach that aligns each pair of reads independently of others. HISATx2 is a two-pass version of HISAT to mimic the two-step approach used in TopHat2. In this version, the first run reports a list of splice sites supported by reads with long anchors. The second run makes use of that splice site information to align reads with short anchors (Online Methods). As expected, HISATx2 takes twice as long to run, but it discovers more alignments. The STAR program also has a two-pass mode, denoted here as STARx2, which we included in our evaluation. We found that STARx2 was more than twice as slow as STAR’s default one-pass mode because, before its second pass, STAR must build a new index for the splice junctions found in the first pass.
The third variant of HISAT (its default version) combines the first two ideas to gain sensitivity without the large performance cost incurred by running the program twice. In this algorithm, we allow HISAT to make use of splice sites found during the alignment of earlier reads when aligning later reads in the same run. This hybrid approach finds almost all the alignments found by HISATx2, with run time nearly as fast as that of HISATx1. To the best of our knowledge, this hybrid approach is the first such single-pass method that bypasses the time-consuming step of remapping reads but matches the sensitivity of two-pass methods. HISAT also includes an option to use known splice sites from gene annotations.
For our simulated data sets, we generated 20 million 100-bp reads with a mismatch rate of 0.5% and up to three mismatches per read from 17,647 randomly chosen transcripts from known protein-coding genes, based on the GRCh37 assembly of the human genome. Each transcript was assigned expression values according to a model provided by the Flux Simulator16 (Supplementary Note). Because we know the true alignments for the simulated reads, we can calculate alignment sensitivity as well as the sensitivity and precision of splice site detection for each program. We also ran all programs on an error-free simulated data set. These results are consistent with the results on data with mismatches (Supplementary Fig. 1 and Supplementary Table 1).
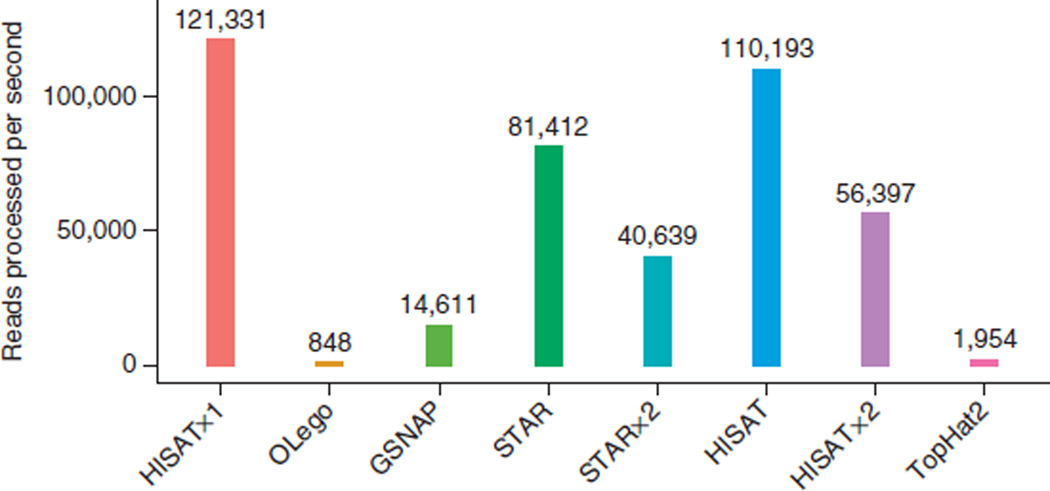
We plotted the alignment speed of the programs for all reads (Fig. 2). HISATx1 and HISAT were fastest, at 121,331 and 110,193 reads processed per second (r.p.s.), respectively, and STAR was third fastest at 81,412 r.p.s. As expected, HISATx2 (56,397 r.p.s.) and STARx2 (40,639 r.p.s.) took approximately twice as long as HISATx1 and STAR, respectively. Note that the speed reported for STARx2 did not include the index-building time. GSNAP was substantially slower at 14,611 r.p.s., and the slowest programs were TopHat2 (1,954 r.p.s.) and OLego (848 r.p.s.).
Figure 2.
Alignment speed of spliced alignment software for 20 million simulated 100-bp reads. Alignment speed for all read types (defined in Fig. 1) combined, measured as the number of reads processed per second by the indicated tools. supplementary Figure 2 provides the alignment speed for each type of read separately.
The current version of GSNAP uses a suffix array in addition to its use of a 15-mer hash table, which makes it several times faster than earlier versions that used only the hash table. OLego aligns reads using a global index based on an FM index, similarly to HISAT’s algorithm. However, OLego runs very slowly, presumably because it relies on a global index to handle all the different types of reads. Overall for this simulated data set, HISATx1 was 49% faster than STAR, eight times faster than GSNAP, 62 times faster than TopHat2 and 143 times faster than OLego. HISAT was more accurate and only 10% slower than HISATx1.
Comparison of sensitivity
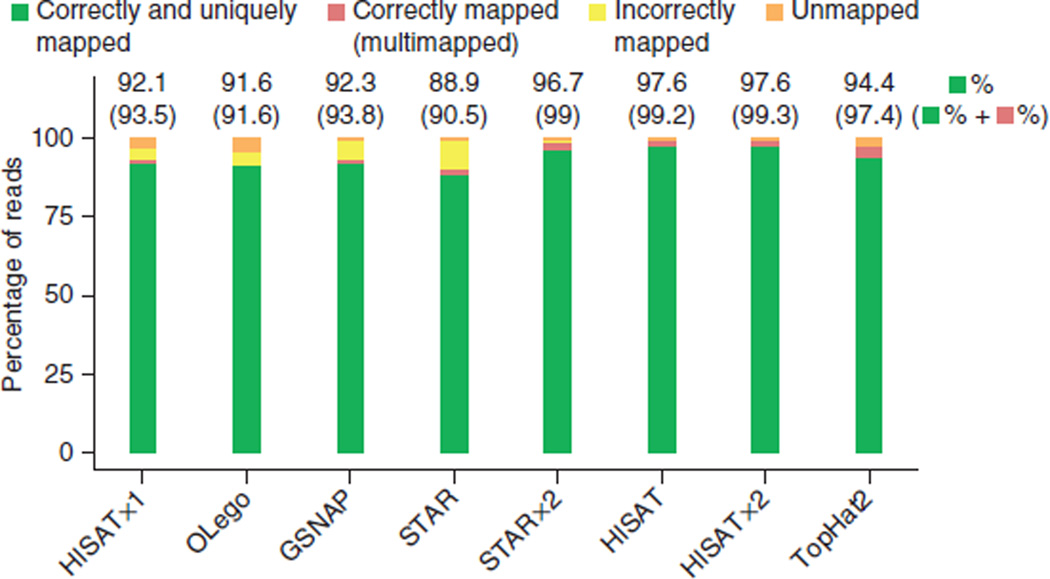
We calculated alignment sensitivity (reads that are aligned correctly, for which the beginning, end and all GT/AG splice sites within the alignment must match precisely) for all programs on the simulated reads (Fig. 3 and Supplementary Table 2). For non-GT/AG splice sites, an alignment was counted as correct if the intron boundaries matched within a 5-bp window. (Note that nonconsensus splice sites occur in just 0.6% of all reads. In Supplementary Table 3, we provided separate accuracies on this subset of splice sites when they were required to match precisely.) Among the one-pass algorithms (HISATx1, STAR, GSNAP and OLego), GSNAP and HISATx1 provided the highest alignment sensitivity at 93.8% and 93.5%, respectively. OLego and STAR yielded lower sensitivity, at 91.6% and 90.5%, respectively.
Figure 3.
Alignment accuracy of spliced alignment software for 20 million simulated 100-bp reads. Alignment results for all read types (defined in Fig. 1) on simulated data containing errors. Reads are categorized as indicated by the colors. For multimapped reads, an aligner was credited with a correct alignment if it mapped a read to multiple locations and one of those locations was correct. Note that the set of multimapped reads reported by the various aligners may be different, depending on each program’s alignment policy and default behavior. The upper numbers are the percentages corresponding to correctly and uniquely mapped reads. The numbers inside parentheses show percentages for cases correctly and uniquely mapped and correctly multimapped combined. In Supplementary Table 2, we provide detailed percentages on all four categories for each aligner.
Compared to the one-pass programs, two-pass approaches (HISAT, HISATx2, STARx2, TopHat2) obtained higher overall accuracies. These four methods had sensitivity from 97.4% to 99.3%, over 3% better than the one-pass methods.
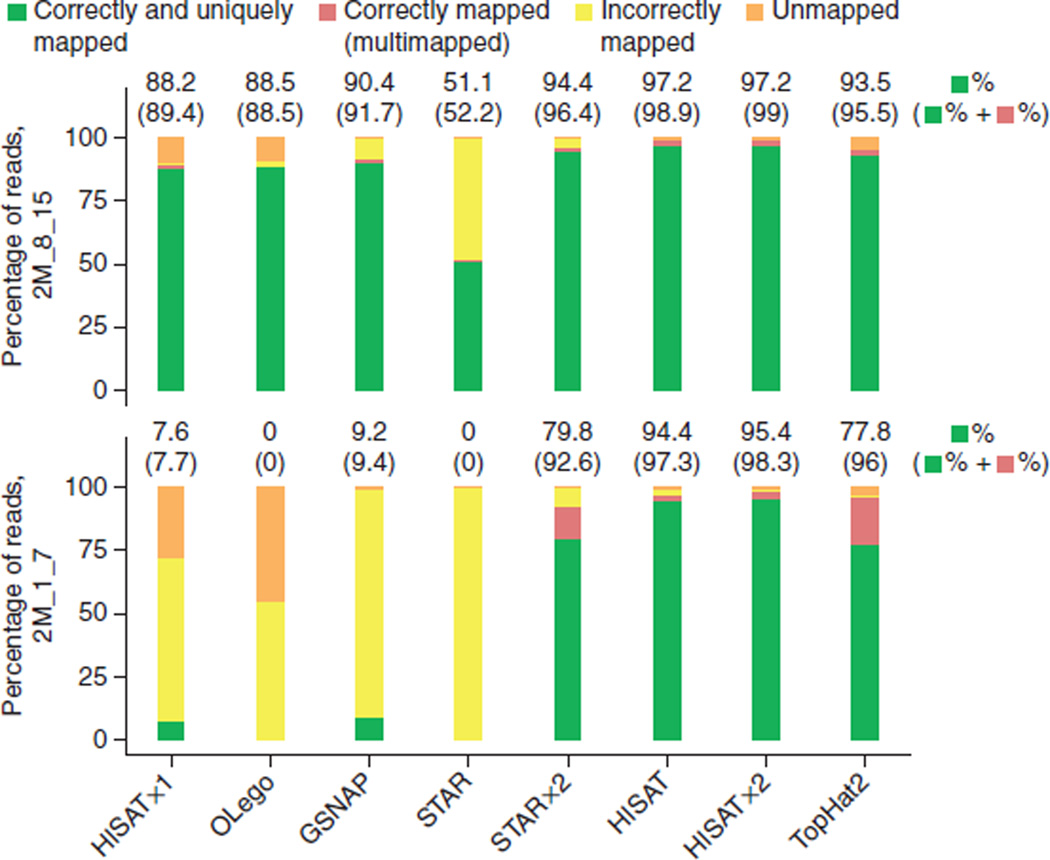
For reads with shorter anchors (1–7 bp), the two-pass algorithms (HISATx2, STARx2, TopHat2 and HISAT) generated much better alignment sensitivity, and we observed a similar result for reads spanning more than two exons (Fig. 4 and Supplementary Fig. 2). For reads with intermediate-length anchors, HISATx2, STARx2, HISAT and TopHat2 each correctly aligned >95.5% of the reads, whereas values for the one-pass methods ranged from 52.2% to 89.4%. For the reads with the shortest anchors, HISATx2, STARx2, HISAT and TopHat2 all provided sensitivity higher than 92%, whereas the other aligners correctly aligned fewer than 10% of these reads.
Figure 4.
Alignment accuracy of spliced-alignment software for reads with small anchors from 20 million simulated reads. This figure shows the alignment sensitivity for reads with small anchors (2M_8_15 and 2M_1_7). Reads are categorized as in Figure 3. The upper numbers on each bar show the percentages corresponding to correctly and uniquely mapped reads. The numbers inside parentheses represent the percentages for cases correctly and uniquely mapped and correctly multimapped combined. There were 1,022,348 and 843,420 reads in 2M_8_15 and 2M_1_7, respectively.
Accuracy of splice site detection
We separately calculated accuracy for detection of splice sites (Table 1). The simulated reads contained a total of 87,944 pairs of splice sites (acceptor and donor sites). We asked how many of these sites were correctly detected by each program, and we gave a program credit if at least one alignment supported a given splice site. We defined precision, or positive predictive value, as the percentage of predicted sites that matched a true splice site. By these measures, HISAT and GSNAP obtained the highest sensitivity (97.3%), and HISAT obtained the second highest precision (94.8%) among all the aligners. OLego yielded slightly higher precision (95.5%), but at the expense of lower sensitivity (94.2%).
Table 1.
Sensitivity and precision of leading spliced aligners
| Program | no. of splice sites reported |
no. of true splice sites reported |
sensitivity (%) |
Precision (%) |
|---|---|---|---|---|
| HISATx1 | 91,904 | 85,546 | 97.3 | 93.1 |
| HISATx2 | 90,331 | 85,603 | 97.3 | 94.8 |
| HISAT | 90,300 | 85,587 | 97.3 | 94.8 |
| STAR | 95,892 | 84,678 | 96.3 | 88.3 |
| STARx2 | 92,254 | 84,734 | 96.3 | 91.8 |
| GSNAP | 92,547 | 85,598 | 97.3 | 92.5 |
| OLego | 86,779 | 82,879 | 94.2 | 95.5 |
| TopHat2 | 96,474 | 79,705 | 90.6 | 82.6 |
Sensitivity and precision of leading spliced aligners for 87,944 true splice sites contained in 20 million simulated reads from the human genome, with a mismatch rate of 0.5%. Sensitivity is the percentage of true splice sites found out of the total that were present. Precision (or positive predictive value) is the percentage of reported splice sites that are correct.
Comparison on real data
We compared the aligners using 108,749,331 101-bp RNA-seq reads collected from fetal lung fibroblasts (GEO accession GSM981249; Supplementary Note). Because we do not know the true alignments for these reads, we evaluated alignment quality in two ways: (i) the cumulative number of alignments detected, up to an edit distance of 3, and (ii) the number of spliced alignments found that correspond to known human splice sites, based on the Ensembl GRCh37 gene annotation. At all distances, HISATx2, STARx2 and HISAT aligned the greatest number of reads, in a tight range from 95.9 million to 96 million (Supplementary Fig. 3). We then examined the cumulative number of spliced alignments that correspond to annotated human splice sites, also separated according to edit distance (Supplementary Fig. 4). At every distance and for the overall total, HISATx2, STARx2 and HISAT found the highest numbers of alignments, ranging from 34.6 million to 35.2 million. STAR and OLego found the lowest numbers of spliced alignments, at just 26.9 million and 26.2 million, respectively.
HISATx1 and HISAT took 23 and 27 min, respectively, and STAR took 25 min to process the 109 million reads. In contrast, TopHat2 took 1,170 min, OLego took 990 min and GSNAP took 292 min. In terms of memory usage, the suffix-array methods STAR and GSNAP used 28 and 20.2 GB of RAM. The Burrows-Wheeler transform–based programs (HISATx1, HISAT, HISATx2, OLego and TopHat2) required memory ranging from 3.7 to 4.3 GB of RAM (Table 2).
Table 2.
Run times and memory usage for HISAT and other spliced aligners
| Program | Run time (min) | Memory usage (GB) |
|---|---|---|
| HISATx1 | 22.7 | 4.3 |
| HISATx2 | 47.7 | 4.3 |
| HISAT | 26.7 | 4.3 |
| STAR | 25 | 28 |
| STARx2 | 50.5 | 28 |
| GSNAP | 291.9 | 20.2 |
| OLego | 989.5 | 3.7 |
| TopHat2 | 1,170 | 4.3 |
Run times and memory usage for HISAT and other spliced aligners to align 109 million 101-bp RNA-seq reads from a lung fibroblast data set. We used three CPU cores to run the programs on a Mac Pro with a 3.7 GHz Quad-Core Intel Xeon E5 processor and 64 GB of RAM.
We provide alignment results for additional sets of simulated reads and for an additional real data set from Chen et al.17 containing 217 million paired-end reads (Supplementary Figs. 5–7 and Supplementary Tables 4–6). In all cases, the relative performances of the alignment programs remained the same as described above. In Supplementary Table 7, we provide details of the input parameters and version numbers for all programs used in these evaluations.
DISCUSSION
Although HISAT is the first system to employ a hierarchical indexing strategy for spliced alignment, the strategy itself could be adopted by other methods if their data structures can be suitably redesigned. All the programs that were included in our study—GSNAP, STAR, OLego and TopHat2—could in principle use hierarchical indexing and thereby improve their alignment speed and quality. HISAT gains additional sensitivity from alignment algorithms specifically designed to handle different types of intron-spanning reads. The combination of these algorithms with hierarchical indexing enables dramatically faster alignment while matching or exceeding the accuracy of the best previous spliced aligners.
ONLINE METHODS
HISAT uses its hierarchical indexing algorithm along with several adaptive strategies, based on the position of a read with respect to splice sites, which we describe below. To begin processing each read, it first tries to find candidate locations across the target genome from which the read may have originated. It identifies these locations by first mapping part of each read using the global FM index, which in most cases identifies one or a small number of candidates (Supplementary Figs. 8–11). HISAT then selects one of the ~48,000 local indexes for each candidate and uses it to align the remainder of the read. For reads sequenced in pairs, each mate is separately aligned and the alignments of both mates are combined. If a read fails to align, then the alignments of its mate are used as anchors to map the unaligned mate. The extension of each alignment uses an efficient local index–based search as explained below.
Although searching the global FM index is much faster in principle than k-mer–based (or hashing) search, in practice it tends to be slower owing to properties of the low-level memory management strategy in a modern computer. The core memory includes both random access memory (RAM) and cache memory, with cache being much smaller but also much faster. When retrieving a block of data, the operating system searches cache first and looks in RAM only if the block is not found in cache.
Search through a global FM index of the human genome suffers from many cache ‘misses’ because the alignment algorithm proceeds one base at a time through a read and the corresponding locations in a very large FM index (about 913 MB for the human genome). As a match is extended base by base, the search jumps to completely different regions of the index, i.e., the FM index is not organized according to the sequence of the genome itself. Each time the search jumps, the computer has to search RAM and bring in a new piece of the FM index, which is rarely present in the cache already.
In contrast, the far smaller size of our local indexes, 24 KB, allows the entire index to fit in cache, which means that search through the local indexes generates considerably fewer cache misses and therefore runs much faster.
In addition to its two basic operations (global and local searches), HISAT uses an even faster operation for alignment extension. This operation, which performs direct comparisons of read sequences with genomic sequences, is used only when we know the genomic location to which a read is being mapped. The extension operation requires the entire genomic sequence to be loaded into memory for fast access; in the case of the human genome, this requires 682 MB. Strategically combining these three operations can dramatically reduce the use of relatively slow operations such as global search and even local search.
Here, we present three different alignment strategies based on three groupings of reads: (i) reads that map within an exon, across two exons with at least 15 bp in each exon, or across two exons with 8–15 bp mapping to one exon; (ii) reads that map to two exons with just 1–7 bp in one exon or that map across more than two exons; and (iii) reads that are likely to be incorrectly mapped to processed pseudogenes.
We illustrate each alignment strategy using examples (error-free reads) that for the purposes of illustration are relatively simple but that still provide insight into how hierarchical indexing enables fast and sensitive alignment. Although we use error-free reads in these examples, HISAT easily handles reads with both mismatches and indels (Supplementary Note and Supplementary Fig. 11). Note that HISAT is optimized for reads ranging from 75 to 150 bp, the most commonly used (and least expensive) type, but it will also handle the 250- to 300-bp reads generated by MiSeq instruments.
All the strategies that use local indexes initially retrieve just one index, based on the location of the current match. Among the 246,208 introns from the annotated protein-coding genes in the human genome, 222,503 (90.4%) are completely included in one local HISAT index, each of which spans 64,000 bp. One local index, therefore, is almost always sufficient to align a read. When reads involve long introns, HISAT uses two or more local indexes, up to a maximum intron length of 500,000 bp.
For the examples here, we search for matches in one direction, from right to left, in order to minimize HISAT’s memory foot-print, currently 4.3 GB. (Bidirectional search using our method would require 7.5 GB for the human genome.) This unidirectional search does not affect alignment sensitivity, though it might slightly reduce speed.
Alignment of exonic reads and long- and intermediate-anchored reads
Given two exons from a gene on human chromosome 22, separated by a 3,899-bp intron, suppose the genomic region is transcribed and spliced and we have three reads sequenced from the resulting transcript: (i) an exonic read, (ii) a read spanning two exons with an 8-bp anchor in one exon and (iii) a read spanning two exons with 50 bp in each exon (Supplementary Fig. 8). All the reads are assumed to be error free and 100 bp long. We can apply hierarchical indexing to align each of these reads rapidly and correctly. We align the first read using the global FM-index (Supplementary Fig. 8a). Because global search is relatively time consuming, we change strategies when the partial alignment meets two conditions: (i) it is at least 28 bp long and (ii) it maps onto exactly one location. For the read shown in the figure, the 28-bp sequence on its right end maps uniquely, allowing us to stop the global search operation at that point. From there, we extend the partial alignment by directly comparing the remaining sequence and the corresponding genomic sequence, which we can extract directly from the genome using the mapped location as an index. Because the read is error free and contained within one exon, the extension operation sweeps across the remaining 72 bp, completing the alignment for the read. Note that we could perform this alignment using the global FM index, as TopHat2 does, but the combination of global search and local extension is faster.
For the second read, which has a very short 8-bp anchor on the left side, we first try to map the read using global search, moving right to left (Supplementary Fig. 8b). The first 28 bp on the right end of the read maps uniquely, allowing us to anchor the alignment and halt the global search. We then extend the alignment until we encounter a mismatch at the 93rd base. This mismatch occurs when the alignment extension reaches the intron. At this point we pause the search, retrieve the local FM index that contains this location and align the remaining 8 bp using this index. Because the index covers only a small region, in this case we find just one match for the 8-bp segment. Finally, we check whether the two partial alignments (8 bp and 92 bp) are compatible with each other (for example, in the correct orientation) and then combine them to produce a spliced alignment of the original read.
Note that if we searched for an 8-bp sequence in the global index, we would expect to find an average of ~48,000 matching locations in the human genome (and sometimes many more). Instead of examining 48,000 possible locations, we use one of the local FM indexes, which is expected to contain just one copy of a given 8-bp sequence, on average. This two-stage hierarchical indexing allows us to avoid examining tens of thousands of candidate locations for short anchors, which in turn dramatically speeds up the overall alignment algorithm.
The third read has long anchors (50 bp each) in each exon. We first align the read beginning on the right, using global search as we did before. After the first 28 bp is uniquely mapped, we switch to the extension operation, which further aligns 22 bp and stops after a mismatch at the 51st base. We then choose a local FM index and perform a local search using the first 8 bp of the remaining part of the read. Once this 8 bp is found, HISAT again uses the extension operation to align the rest of the read (Supplementary Fig. 8c). Note that if the 8-bp prefix mapped to too many locations, HISAT would use a longer prefix to reduce the number of potential locations to 5 or fewer.
As we can see from these examples, we can combine global search, local search and directed read extension to achieve rapid yet sensitive alignment. Note that when a read has multiple spliced alignments, HISAT prefers to report alignments that use the canonical GT and AG dinucleotides on the ends of the intron. From any remaining alignments after this filter, it reports the one with the shortest intron length. HISAT provides several parameters with which users can customize its alignment strategy, including adjustable penalties for mismatches, indels and noncanonical splice sites (Supplementary Note).
Alignment of short-anchored reads and reads spanning three exons or more
Exon-spanning reads sometimes have very small anchors (defined here as 1–7 bp) in one of the exons. Correctly aligning these reads is extremely difficult because a 1- to 7-bp anchor will align to numerous locations, even in a local FM index. Arguably the most effective approach to align such short-anchored reads is to use splice site information to remove the introns computationally before alignment. We can identify and collect splice site locations when aligning reads with long anchors and then rerun HISAT for the short-anchored reads (Supplementary Fig. 9). This two-step approach is very similar to the two-step algorithm in TopHat2.
More specifically, in the two-step HISATx2 method, we use the first run of HISAT (HISATx1) to generate a list of splice sites supported by reads with long anchors. In the second run we then use the splice sites to align reads with small anchors. For example, consider the unmapped read spanning exons e2 and e3 (the upper portion of Supplementary Fig. 9). The right part of the read will be mapped to exon e3 using the global search and extension operations, leaving a short, 3-bp segment unmapped. We then check the splice sites found in the first run of HISAT to find any splice sites near this partial alignment. In this example, we find a splice site supported by a read spanning exons e2 and e3 with long anchors in each exon. On the basis of this information, we directly compare the 3 bp of the read and the corresponding genomic sequence in exon e2. If it matches, we combine the 3-bp alignment with the alignment of the rest of the read. This ‘junction extension’ procedure that makes use of previously identified splice sites is represented by brown arrows in the figure.
As we show in our experiments on simulated reads, this two-step strategy produces accurate alignment of reads with anchors as small as 1 bp (see Results). Although HISATx2 has considerably better sensitivity, it takes twice as long to run as HISATx1. As an alternative, we developed a hybrid method, HISAT, which has sensitivity almost equal to that of HISATx2 but with the speed of HISATx1. HISAT collects splice sites as it processes the reads, similarly to the first run of HISATx2. However, as it is processing, it uses the splice sites collected thus far to align short-anchored reads. In the vast majority of cases, it can align even the shortest anchors because it has seen the associated splice sites earlier. This result follows from the observation that most splice sites can be discovered within the first few million reads, and most RNA-seq data sets contain tens of millions of reads. As our results show, HISAT provides alignment sensitivity that very nearly matches the two-step HISATx2 algorithm, with a run time nearly as fast as the one-step HISAT method.
The hybrid approach is also effective in aligning reads spanning more than two exons, which are more likely to have small anchors. The alignment sensitivity for such reads increases from 53% using HISATx1 to 95% using HISAT (Supplementary Fig. 2).
Alignment of reads involving pseudogenes
Misalignments caused by pseudogenes present additional problems for spliced alignment. Processed pseudogenes are nonfunctional copies of genes that result when the original gene was transcribed, spliced to remove introns and reinserted at a different location in the genome. The most recently created pseudogenes are almost identical to the original genes, meaning that reads from these genes can map equally well to either version of the gene. Intron-spanning reads are a particular problem because they map end to end on the pseudogene but require a split (spliced) alignment to match the original, active gene. As we showed previously9, 2.7% of annotated human genes have pseudogene copies, and the corresponding genes can account for as much as 22.5% of an RNA-seq data set. Therefore, pseudogenes can introduce a strong mapping bias unless they are properly handled. In Supplementary Figure 10, we show a gene and its corresponding processed pseudogene, where the two exons shown on chromosome 1 have their nearly identical copies on chromosome 17 with only a single-base difference. Unlike the two exons on chromosome 1 that are separated by an intron, the two exons on chromosome 17 are adjacent. As a result, junction reads originally spanning the two exons on chromosome 1 are likely to be mismapped to chromosome 17, particularly if the alignment program prefers contiguous alignments.
In the presence of pseudogenes, HISAT correctly maps reads to their origin by considering several genomic locations (Supplementary Fig. 10). In this example, the rightmost portion of the read (48 bp) maps to chromosomes 1 and 17. The match continues on chromosome 17 (the pseudogene) but is interrupted on chromosome 1 at the 3′ end of the intron. Despite the mismatch, HISAT attempts to extend both partial alignments because both are sufficiently long (at least 22 bp by default). For the partial alignment on chromosome 1, we resume the search using a local FM index, which yields a spliced alignment with no mismatches. On chromosome 17, the extension of the alignment yields a nongapped alignment with one mismatch. Given the two candidate alignments, HISAT reports the spliced alignment because it has no mismatches, whereas the nonspliced alignment has one mismatch. If the two alignments were equally good, then HISAT would report both alignments. As shown in our results, this alignment strategy allows HISAT to detect more spliced alignments than any of the leading aligners.
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Pertea and L. Song for their invaluable contributions to our discussions on HISAT. We also thank C. Trapnell for the use of his TuxSim simulation program. This work was supported in part by the National Human Genome Research Institute (US National Institutes of Health) under grants R01-HG006102 and R01-HG006677 to S.L.S.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
D.K., B.L. and S.L.S. performed the analysis and discussed the results of HISAT. D.K. implemented HISAT. D.K., B.L. and S.L.S. wrote the manuscript. All authors read and approved the final manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 2.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12:R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garber M, Grabherr MG, Guttman M, Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods. 2011;8:469–477. doi: 10.1038/nmeth.1613. [DOI] [PubMed] [Google Scholar]
- 7.Grant GR, et al. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM) Bioinformatics. 2011;27:2518–2528. doi: 10.1093/bioinformatics/btr427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engström PG, et al. Systematic evaluation of spliced alignment programs for RNA-seq data. Nat. Methods. 2013;10:1185–1191. doi: 10.1038/nmeth.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burrows M, Wheeler DJA. Palo Alto: Digital Equipment Corp.; 1994. Block-sorting lossless data compression algorithm (Technical report 124) [Google Scholar]
- 13.Ferragina P, Manzini G. Proc. 41st Annual Symp. Found. Comput. Sci. IEEE; 2000. pp. 390–398. [Google Scholar]
- 14.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J, Anczuków O, Krainer AR, Zhang MQ, Zhang C. OLego: fast and sensitive mapping of spliced mRNA-Seq reads using small seeds. Nucleic Acids Res. 2013;41:5149–5163. doi: 10.1093/nar/gkt216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griebel T, et al. Modelling and simulating generic RNA-Seq experiments with the flux simulator. Nucleic Acids Res. 2012;40:10073–10083. doi: 10.1093/nar/gks666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen R, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.