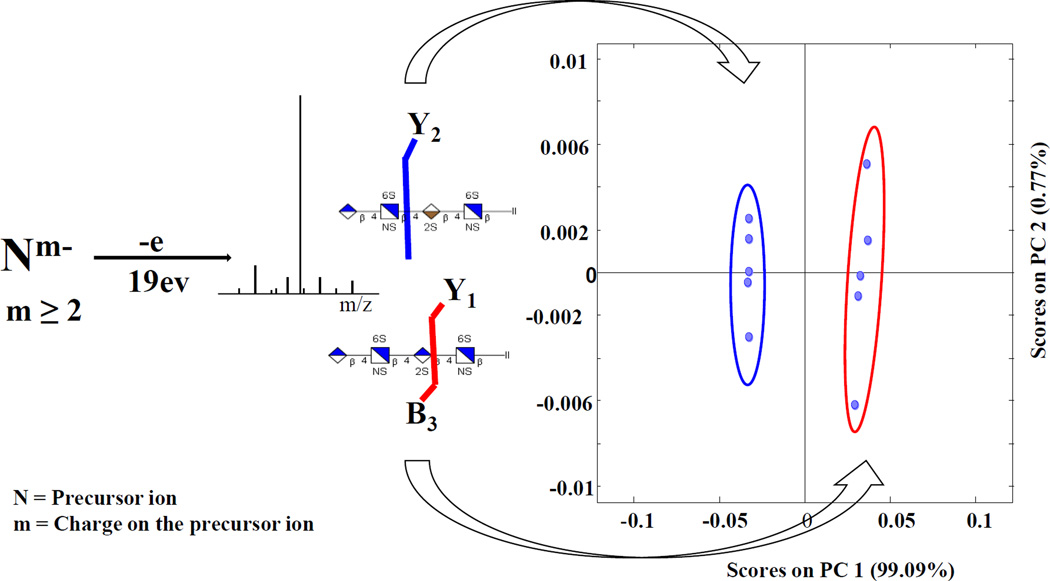
Abstract
The present work focuses on the assignment of uronic acid stereochemistry in heparan sulfate (HS) oligomers. The structural elucidation of HS glycosaminoglycans is the subject of considerable importance due to the biological and biomedical significance of this class of carbohydrates. They are highly heterogeneous due to their non-template biosynthesis. Advances in tandem mass spectrometry activation methods, particularly electron detachment dissociation (EDD), has led to the development of methods to assign sites of sulfo modification in glycosaminoglycan oligomers, but there are few reports of the assignment of uronic acid stereochemistry, necessary to distinguish glucuronic acid (GlcA) from iduronic acid (IdoA). Whereas preceding studies focused on uronic acid epimers with no sulfo modification, the current work extends the assignment of the hexuronic acid stereochemistry to 2-O-sulfo uronic acid epimeric tetrasaccharides. The presence of a 2-O-sulfo group on the central uronic acid was found to greatly influence the formation of B3, C2, Z2, and Y1 ions in glucuronic acid and Y2 and 1,5X2 for iduronic acid. The intensity of these peaks can be combined to yield a diagnostic ratios (DR), which can be used to confidently assign the uronic acid stereochemistry.
Graphical abstract
INTRODUCTION
Heparan sulfate (HS) glycosaminoglycans (GAGs) are a unique class of proteoglycans present on the surface of mammalian cells and in the extracellular matrix [1, 2]. These compounds are known to bind to a variety of proteins, and are implicated in several biological activities including angiogenesis, embryonic development, inflammation, and microbial infections [3, 4]. Structurally, they are synthesized as alternating disaccharide units of uronic acid [D-glucuronic acid (GlcA) or L-iduronic acid (IdoA)] linked to a glucosamine in the Golgi apparatus [5]. Selective sulfo modification of the disaccharide may occur at the N, 3-O and 6-O positions of the glucosamine while 2-O-sulfo modification can be observed on the acidic sugar [1, 4, 6]. The enzyme, 2-O-sulfotransferase, responsible for the transfer of the sulfo group to the 2-O position of a uronic acid, preferentially modifies IdoA, but can also modify GlcA to a limited extent, forming IdoA2S and GlcA2S [4]. These structural modifications of the HS chain are known to contribute to its various biological functions [1]. A well-documented example is the role of IdoA2S in binding several proteins including antithrombin, the fibroblast growth factors [7], and hepatocyte growth factors [8]. Even though the function of its epimeric isomer, GlcA2S, has yet to be elucidated, substantial amounts have been found in the human cerebral cortex [9], implicating an important role for this less commonly observed modification. Robust analytical methods for obtaining all structural details of GAG oligomers, including uronic acid stereochemistry, are required to aid researchers in their quest to gain a deeper understanding of the biological role GAGs.
Tandem mass spectrometry is well known as a tool for the analysis of biomolecules, offering high sensitivity, throughput, and accuracy [10]. However, MS/MS analysis of HS oligosaccharides is challenging due to their heterogeneity in oligomer length, degree and sites of sulfo modifications, and hexuronic acid C-5 stereochemistry. The labile nature of a sulfate half-ester hinders structural characterization, especially during ion activation, due to the facile decomposition of the sulfo group. To address these challenges, researchers have focused their efforts on the development and refinement of ion activation methods to minimize sulfo decomposition while maximizing the fragmentation that provides useful structural information.
Chemical derivatization methods such as permethylation and acetylation followed by MS/MS have been successfully employed to locate sites of sulfation in HS oligomers [11]. Another approach to reducing sulfo decomposition is via metal cation exchange, which has been used with positive ionization to facilitate fragmentation by electron capture dissociation [12–14]. An alternative method involves efficient deprotonation of the acidic groups using supercharging agents like sulfolane [15]. This approach works well with less sulfated GAGs, but is limited in its applicability to highly sulfated GAG oligomers due to charge repulsion. Recent work showed that by adding sodium hydroxide to the spray solution, one can ionize sulfo groups in highly sulfated GAGs, while reducing the overall charge of the precursor ion. With this approach, one can obtain very rich and structurally informative MS/MS spectra [16, 17].
Electron based activation methods such electron detachment dissociation (EDD) [16–21], electron induced dissociation (EID) [18–20], and negative electron transfer dissociation (NETD) [22, 23] have been particularly useful for the tandem mass spectrometry analysis of GAGs. Compared to threshold activation methods like collision induced dissociation (CID) and infrared multi-photon dissociation (IRMPD), EDD produces more structurally relevant ions with less SO3 loss [19]. Prior studies have shown the capability of EDD for sequencing dp4-dp10 chondroitin sulfate (CS) oligomers, and assigning the hexuronic acid stereochemistry of HS tetrasaccharides [18, 20]. With the aid of sodium cation pairing of ionizable acidic protons, EDD has been established as an excellent tool for characterizing highly sulfated heparin tetrasaccharides [24]. Recent EDD applications to the highly sulfated pentasaccharide, Arixtra, showed remarkable product ion coverage for assigning sites of modifications [25]. The density of peaks in EDD mass spectra has driven the application of multivariate statistical methods like principal component analysis (PCA) to maximize the amount of information retrieved in a data set. PCA was used to differentiate the EDD spectra of four diastereomeric HS tetrasaccharides with very similar spectral features [26], and also used for the analysis of mixtures of epimer pairs that were resolved by field asymmetric ion mobility spectrometry (FAIMs) [27].
Most of the previous efforts to develop tandem mass spectrometry methods for analyzing GAG oligomers have focused on assigning sites of sulfo modification. However, a few examples of the assignment of C-5 stereochemistry in the uronic acids of glycosaminoglycans using tandem mass spectrometry can be found in the literature. Zaia and coworkers have been able to assign the global ratio of chondroitin sulfate (CS) versus dermatan sulfate (DS) in mammalian extracellular matrix by using collision induced dissociation [28, 29]. The ability to distinguish the C-5 stereochemistry in uronic acid residues of 4-O-sulfo chondroitin sulfate epimers, based on diagnostic ion intensities in EDD mass spectra has been reported by Leach et al [18]. The results showed doubly deprotonated 0,2X and Y3 were indicative of CS-A containing GlcA, in agreement with the prior CID results for CS oligomers examined by Zaia and co-workers (vide infra). Wolff and Amster have reported diagnostic ions that can be used for assigning IdoA versus GlcA in unsulfated and monosulfated heparan sulfate (HS) tetramers by using EDD [20].
Despite these promising results, there is still a need for a more general approach to identifying uronic acid C-5 stereochemistry in GAG oligomers, particularly in HS and heparin (Hp) oligomers, where 2-O-sulfo modification of the uronic acid can occur. In prior work using EDD to examine HS tetramers, stereospecific product ions B3’-CO2 and 0,2A3 indicated the presence of glucuronic acid in enzymatically produced HS tetrasaccharides (0–0.25 sulfates per disaccharide) [20]. For IdoA residues, the relative intensities of C3’’ is higher compared to C3. Later work on more highly sulfated HS tetrasaccharides found that these same ions were not as useful for assigning uronic acid stereochemistry [26]. This has motivated us to examine in more detail the capability of EDD mass spectrometry to assign uronic acid stereochemistry in HS GAGs.
In the present work, we study the significance of 2-O-sulfo modification of the uronic acid residue on product ion formation during EDD activation. Synthetically produced epimeric 2-O-sulfo modified HS tetrasaccharides, with varying degrees of sulfation (0.5–2.5 sulfates per disaccharide subunit) serve as standards to examine differences in the EDD mass spectra. Principal component analysis identifies statistically significant peaks that are diagnostic of the C-5 stereochemistry in a uronic acid. With these data, we can assign the stereochemistry of the reducing end uronic acid in a tetrasaccharide with a single stage of tandem mass spectrometry, based on diagnostic ratios of selected product ions.
EXPERIMENTAL METHODS
Sample Preparation
Epimeric heparan sulfate tetrasaccharides; IA[GlcA-GlcNAc-IdoA2S-GlcNAc-(CH2)5NH2], IB [GlcA-GlcNAc-GlcA2S-GlcNAc-(CH2)5NH2], IIA [GlcA-GlcNAc-IdoA2S-GlcNAc6S-(CH2)5NH2], IIB [GlcA-GlcNAc-GlcA2S-GlcNAc6S-(CH2)5NH2], IIIA [GlcA-GlcNS-IdoA2S-GlcNS-(CH2)5NH2], IIIB [GlcA-GlcNS-GlcA2S-GlcNS-(CH2)5NH2, IVA [GlcA-GlcNS-IdoA2S-GlcNS6S- (CH2)5NH2], IVB [GlcA-GlcNSGlcA2S-GlcNS6S- (CH2)5NH2], VA [GlcA-GlcNS6S-Ido2SA-GlcNS6S- (CH2)5NH2], and VIA [IdoA-GlcNS6S-Ido2SA-GlcNS6S- (CH2)5NH2], were synthesized by the modular approach [30]. FT-ICR MS accurate mass measurements and 1H NMR were used to confirm all structures.
Mass Spectrometry Analysis
All tandem mass spectrometry experiments were performed on a 9.4T Bruker Apex Ultra QeFTMS (Billerica, MA) fitted with an indirectly heated hollow cathode (HeatWave, Watsonville, CA) to generate electrons for EDD. 0.1 mg/ml sample solutions in 50:50 methanol/H2O were infused at a rate of 120 µL/h and ionized by electrospray using a metal capillary (Agilent Technologies, Santa Clara, CA, #G2427A) [19]. For the EDD experiments, precursor ions were isolated in the external quadrupole and accumulated for 1 to 3s before injection into the FT-MS cell. Two isolation cell fills were applied per scan, and the selection of the precursor ion was further refined by using in-cell isolation with a coherent excitation frequency (CHEF) event. Selected precursor ions were then irradiated with electrons for 1s. For electron irradiation, the cathode bias was set to 19 and extraction lens set to −18.5 ± 0.5V with the cathode heater set to 1.5A. 36 acquisitions were signal averaged per spectrum. 512 K points were acquired for each spectrum, padded with one zero fill, and apodized using a sine bell window. Internal calibration was also performed using confidently assigned glycosidic bond cleavage products as internal calibrants, providing mass accuracy of <1 ppm. Due to the large number of low intensity products formed by EDD, only peaks with S/N> 10 are reported. Product ions were assigned using accurate mass measurement and GlycoWorkbench. All products are reported using the annotation proposed by Wolff and Amster, derived from the Domon and Costello nomenclature [31].
Principal Component Analysis
Principal component analysis (PCA) was performed using PLS Toolbox (Eigenvector Research, Inc., Wenatchee, WA), as reported earlier [26]. The abundances of 20 assigned product ions were normalized with respect to the total ion current in each EDD spectrum. An input matrix with each row containing the mass spectrum of a single tetrasaccharide epimer and each column the normalized abundance of an assigned fragment ion (variables) is constructed. For each sample quintuplicate EDD spectra were acquired in the same day. Each data set was mean-centered and cross-validated prior to PCA analysis. Principal component scores plot shows the relationship between the samples, whereas the loading plot reveals the variables that contribute to the sample differences.
RESULTS AND DISCUSSIONS
The compounds selected for this study provide the means to examine differences between HS tetramers that differ only in C-5 stereochemistry of the uronic acid closest to the reducing end of the oligomer. The uronic acid at the nonreducing end is of little consequence, as this residue is often converted to a Δ-uronic acid during the enzymatic processing of proteoglycans to produce oligomers of a size that can be sequenced by tandem mass spectrometry. Such enzymatic processing eliminates stereochemical differences between IdoA and GlcA in the nonreducing end sugar. In the following discussion, references to the uronic acid are to the residue closest to the reducing end. Several EDD mass spectra were recorded for each compound, so that statistical significant differences between compounds could be differentiated from variations that occur spectrum-to-spectrum for the same compound. The variations in product ion abundance for quintuplicate EDD spectra for each compound are shown in Fig. S1–4. Previous studies of differences in the EDD mass spectra of epimeric GAGs focused on uronic acids lacking 2-O-sulfo, and have little or no sulfo modifications elsewhere in the oligomer chain. Here, all the samples have 2-O-sulfo modifications in the uronic acid closest to the reducing end. Furthermore, we examine HS tetramers with up to five sulfo modifications, to find product ions that are diagnostic of the uronic acid stereochemistry, yet insensitive to the degree of sulfation.
EDD and PCA results for mono-sulfated HS tetrasaccharides epimers IA and IB
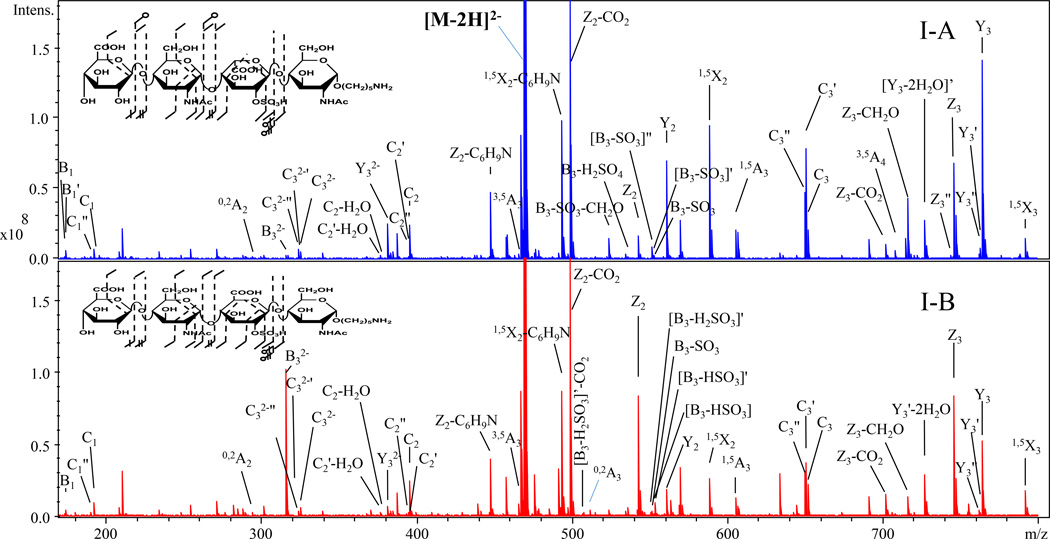
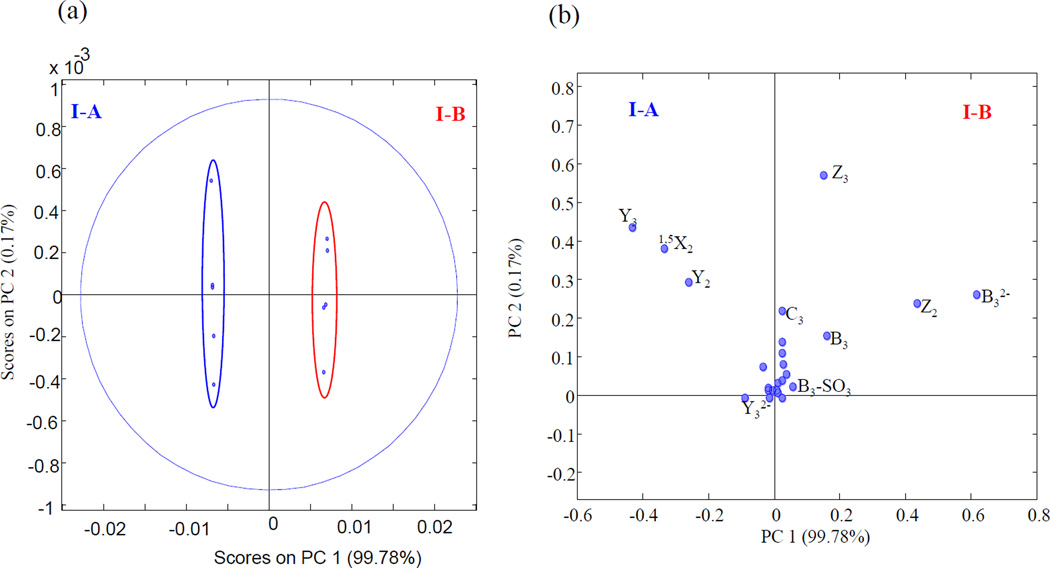
EDD mass spectra and annotated structures for [M-2H]2− precursor ions for I-A (contains IdoA2S) and I-B (contains GlcA2S) are shown in Fig. 1. Abundant cross-ring cleavages occur on the hexuronic acid near the reducing end, consistent with the prior observations that electron detachment is favored on a carboxyl group, particularly if it is ionized [19, 20]. Glycosidic cleavage products, B3 and C2, locate a sulfo modification on the uronic acid; definitive assignment of the 2-O-sulfo modification in compound IB can be made with the 0,2A3 ion, which is absent in IA as shown in S5. This ion has been shown to indicate the presence of GlcA in previous EDD studies [20]. B3' and B3'-CO2, indicative of GlcA in prior studies of oligomers lacking 2-O-sulfo modification, were not observed in I-B. We attribute this difference to the presence of 2-O-sulfo, which introduces extra charge on the uronic acid residue. We find instead the products [B3-HSO3], [B3-H2SO3] and [B3-H2SO3-CO2] in the EDD spectrum of I-B. To better identify diagnostic ions that distinguish the epimer pair, the data was subjected to PCA, and the scores and loadings plots are shown in Fig. 2. 99.78% of the differences between the EDD spectra of the epimers are found in a single principal component. Ions contributing to the clear distinction of these isomers are B3, Y1, Z2, and Z3, which correlate with GlcA2S, while Y2, Y3, and 1,5X2 correlate with IdoA2S.
Figure 1.
EDD spectra and annotated structures for [M-2H]2− precursor ion for I-A and I-B.
Figure 2.
PCA projection of EDD results for I-A and I-B. Score Plot (a) PC1 and PC2 (b) loadings plot showing PC1 and PC2
EDD and PCA results for di-sulfated HS tetrasaccharides epimers IIA and IIB
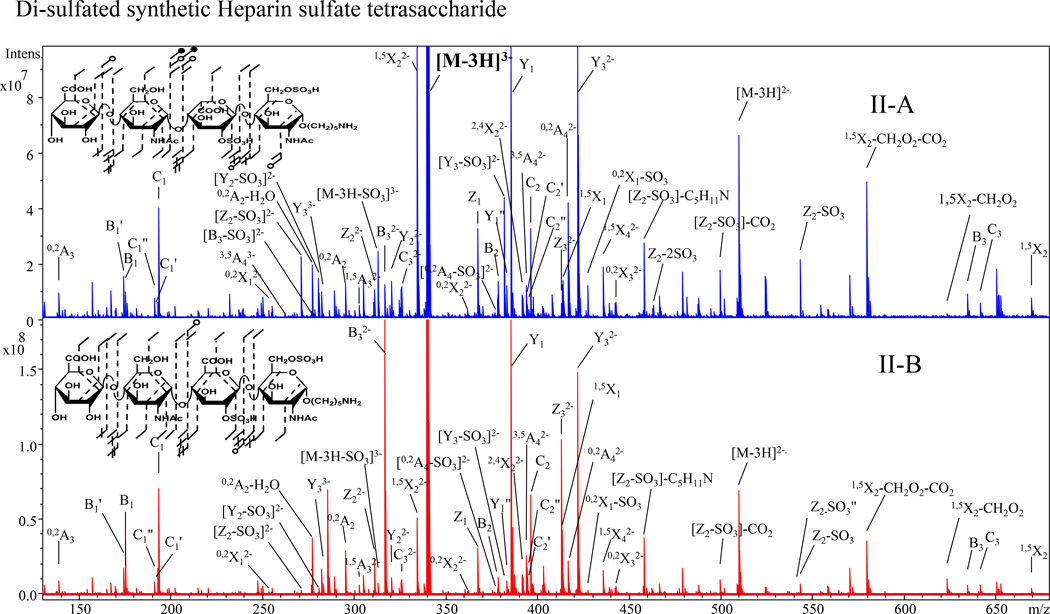
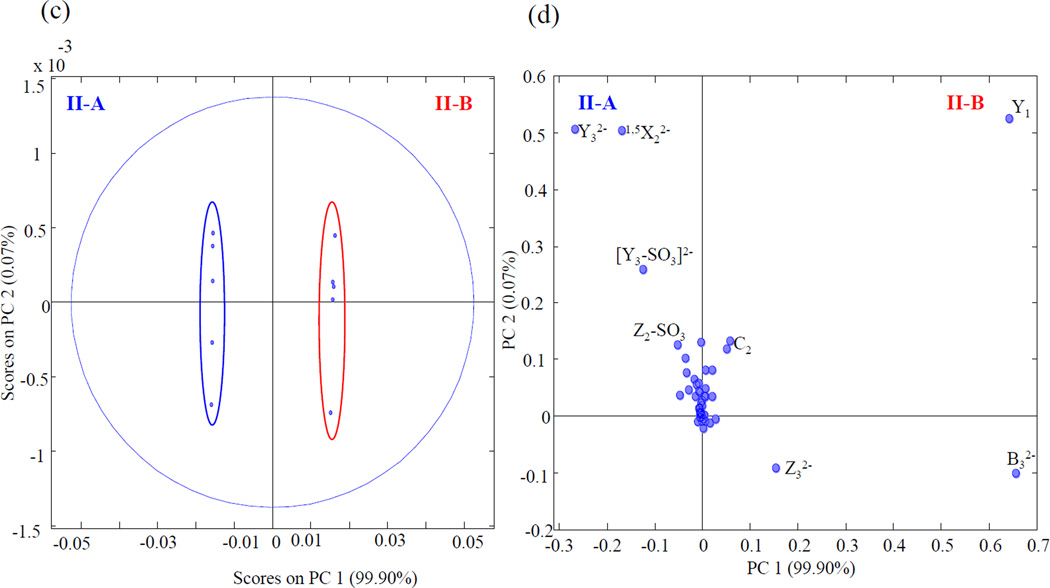
The introduction of a 6-O-sulfo modification on the reducing end amino sugar distinguishes epimer pair II-A and II-B from the first set of standards. The EDD mass spectra and annotated structure of the [M-3H]3− precursor ion for both for epimers are shown in Fig. 3. The EDD data unambiguously determine the locations of the two sulfo modifications. The mass difference between 0,2X1 and B3 assigns 2-O-sulfo on the central uronic acid, while 3,5A4 and Z1 locates the 6-O-sulfo on the reducing terminus amino sugar. PCA of the EDD mass spectra for II-A and II-B established a high variance of 99.9% in a single principal component. Fig. 4 shows the scores and loadings plot for IIA and IIB. From the loadings plot, B32−, Y1, C2 and Z3, are observed to be diagnostic for IIB (containing GlcA2S) and Y32−, 1,5X22−, [Y3-SO3]2− and Z2-SO3 for II-A (containing IdoA2S).
Figure 3.
EDD spectra with structural annotations of [M-3H]3− precursor ion for II-A and II-B.
Figure 4.
PCA scores plot (c) and loadings plot (d) for II-A and II-B
EDD and PCA results for tri-sulfated HS tetrasaccharides epimers IIIA and IIIB
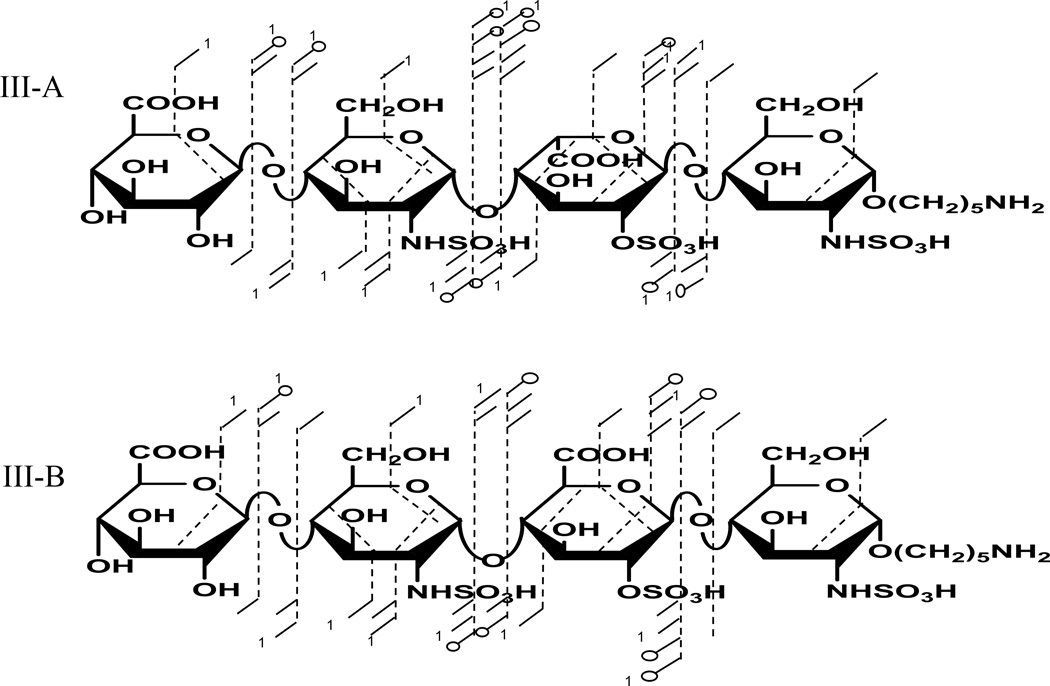
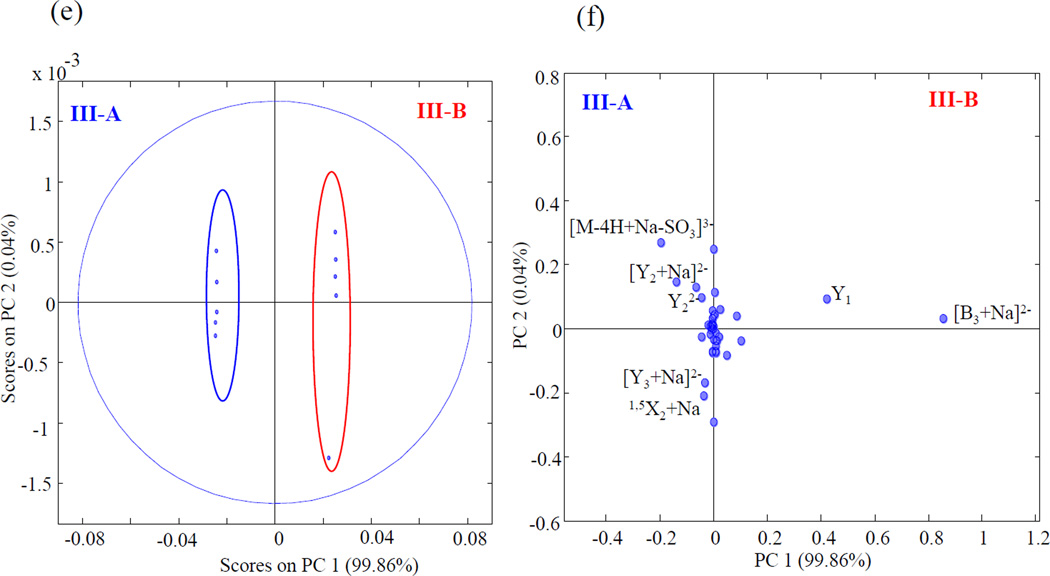
Product ion assignments for the EDD mass spectra of epimers with three sulfo modifications are shown in Fig. 5. The sulfo modifications are distributed on the three reducing end saccharides. Prior work has shown that all sulfo groups must be ionized for such high sulfated HS oligomers to fragment by pathways other than SO3 loss [16]. The [M-4H+Na]3− precursor ion for IIIA and IIIB was thus selected for EDD fragmentation. Supplemental Fig. S6, shows the tandem mass spectrum for IIIA and IIIB .Complete glycosidic product ions with cross ring product ions observed for these epimers were enough to locate all the sites of sulfation. EDD-PCA scores and loadings plot shown in Fig. 6. show that 99.9% of variance between the epimers occurs in one principal component. The loadings plot shows very high positive loading values for complementary ions [B3+Na]2− and Y1 for IIIB, and negative loadings value for [Y2+Na]2−, Y22−, [Y3+Na]2− and 1,5X2+Na for IIIA.
Figure 5.
Annotated structures for EDD fragmentation of [M-4H+Na]3− for III-A and III-B
Figure 6.
PCA scores plot (e) and loadings plot (f) for III-A and III-B
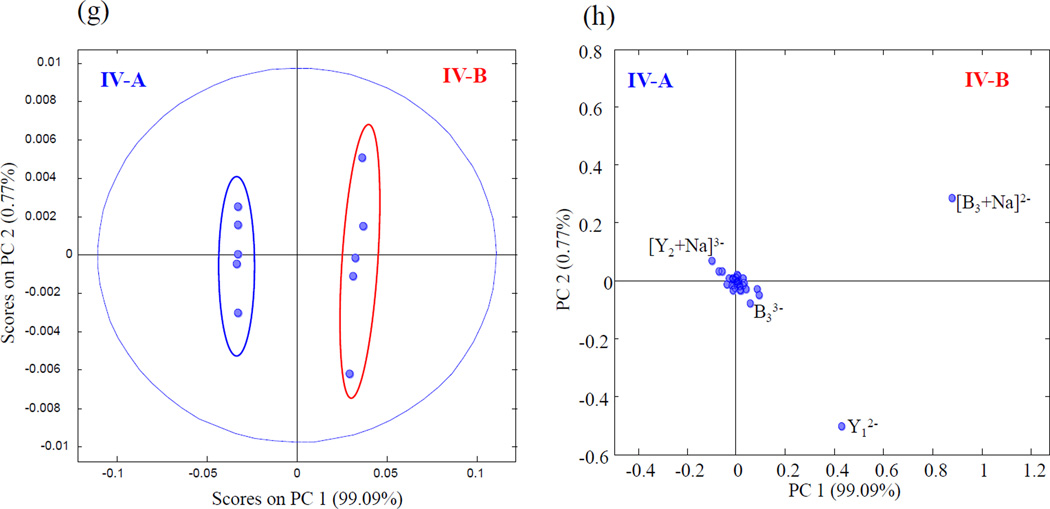
EDD and PCA results for tetra-sulfated HS tetrasaccharides epimers IVA and IVB
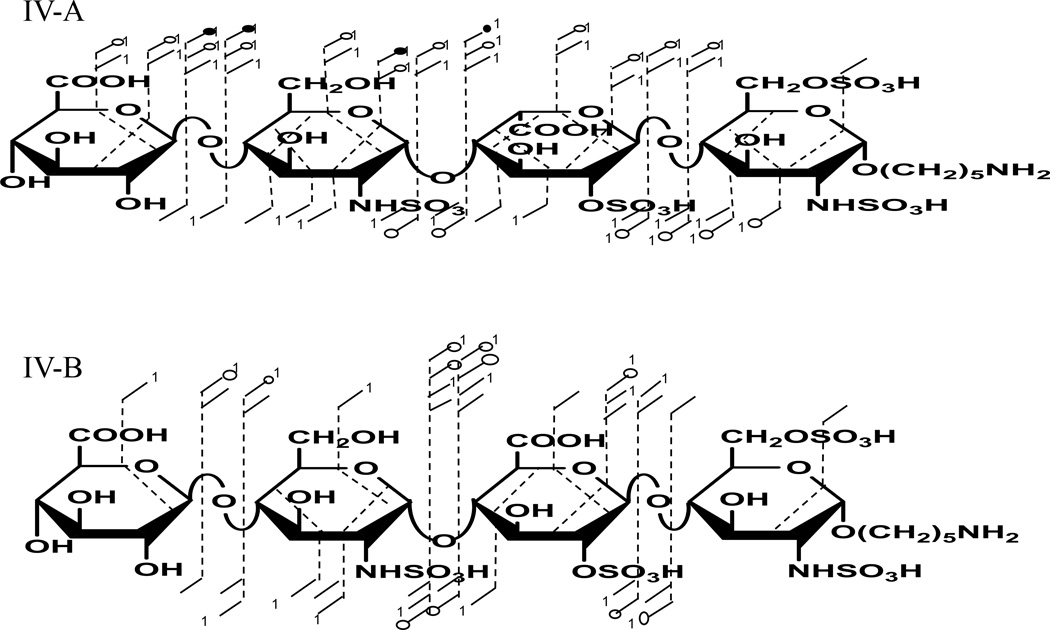
Sulfo modification at the 6-O position on the reducing end amino-sugar of IV-A and IVB distinguish these compounds from the tri-sulfated HS tetrasaccharides described above. This modification increases the number of sulfates per disaccharides from 1.5 to 2, making it even more prone to SO3 loss. To prevent excessive sulfo decomposition from occurring, the [M-5H+Na]4− precursor ion was selected for the EDD experiment. EDD product ions spanning the range m/z 120–600 as shown in the supplemental Fig. S7, were sufficient to locate all four sites of sulfo modification. Fig. 7 shows the structural annotations for IV-A and IV-B. PCA projection of the EDD results shown in Fig. 8 revealed these two epimeric tetrasaccharides could be differentiated with a single principal component (99.1% of the variance is in PC1). High positive loadings values again for [B3+Na]2− and Y1 were observed for IV-B and [Y2+Na]3− showed negative loading values for IVA.
Figure 7.
Annotated structures for EDD fragmentation of [M-5H+Na]4− for IV-A and IV-B
Figure 8.
PCA scores plot (g) and loadings plot (h) for IV-A and IV-B
Careful analysis of EDD-PCA results for all eight synthetic 2-O-sulfo tetrasaccharides revealed a pattern of glycosidic and cross-ring product ions that correlate with hexuronic acid stereochemistry in their EDD spectra, specifically, the glycosidic cleavages surrounding the uronic residue, B3, Y1, Z2, and C2 were observed to be diagnostic for GlcA2S. The more remote glycosidic cleavage, Y2, as well as the cross ring fragment in the adjacent amino sugar, 1,5X2, are found to be diagnostic for IdoA2S.
Diagnostic ratio (DR) analysis of 2-O-sulfo HS tetrasaccharides
A tandem mass spectrometric method for assigning uronic acid stereochemistry in HS oligomers, and that is applicable to compounds with a wide variation in sulfo modification, has not previously been reported. Contrary to initial tandem mass spectrometric assignments of uronic acid stereochemistry in chondroitin sulfate oligosaccharides [28] which has a defined sulfation pattern, HS oligosaccharides presents a more complex challenge due to the variability in sulfation pattern and hexuronic acid stereochemistry. Earlier attempt using EDD to assign the hexuronic acid stereochemistry in HS tetrasaccharides yielded promising results. However, the sulfate density was very low (0–0.25 sulfates per disaccharide) for the compounds that were tested, and 2-O-sulfo was not present [19]. To provide a more robust analysis for assigning the hexuronic acid stereochemistry in a single mass spectrum using EDD, we have considered 4 epimers pairs with varying degree of sulfation (0.25–2.0). From the PCA loadings plots, we observe strong positive loading values for Glc2AS diagnostic ions and negative loadings values for IdoA2S. The strong variations between these epimer pairs from the EDD PCA loadings plots suggest the possibility of combining diagnostic ions could to assign the stereochemistry in a mass spectrum. Combining the results from all 4 PCA plots, we have selected diagnostic ions indicative of Glc2AS (B3, Y1, C2 and Z2) and Ido2AS (Y2 and 1,5X2). The ratio of these summed peaks could predict the hexuronic acid stereochemistry for all the 2-O-sulfated HS tetrasaccharides examined. Equation 1 below compares the intensity ratios of Glc2AS ions to Ido2AS.
| (1) |
In equation 1, DR is the diagnostic ratio calculated from the abundance of the designated fragment ions in the EDD spectra. I(GlcA2S), I(IdoA2S) are the intensities for all diagnostic ions, including those accompanied by sulfo decomposition, for the fragment ions that are diagnostic of Glc2AS and Ido2AS. DR results for the four epimer pairs are found to segregate in to low ratios (less than 1.5) for IdoA2S containing tetrasaccharides, and high ratios (8.9 or higher) for tetrasaccharides with GlcA2S, as shown in Table 1, with up to four sulfo modifications. The large gap between the DR values of the GlcA2S and the IdoA2S tetrasaccharides allows the confident assignment of stereochemistry for the uronic acid residue closest to the reducing end. This was tested on even more highly sulfo modified oligomers, two penta-sulfo modified tetramers that contain IdoA2S, GlcAGlcNS6S-IdoA2S-GlcNS6S-(CH2)5NH2 (DR = 1.4) and IdoA-GlcNS6S-IdoA2S-GlcNS6S-(CH2)5NH2 (DR=0.6), shown Table 1. EDD spectra for the pentasulfated tetrasaccharides are shown in supplementary material as Fig. S8 and Fig. S9. Also included in the supplementary material are mass (m/z) - intensity table for the ions used for the DR calculations. These data suggest that the DR calculation from assigned peaks in EDD mass spectra of HS tetramers can be used to accurately and confidently assign uronic acid C-5 stereochemistry.
Table 1.
Diagnostic ratio (DR) values for the 10 synthetic 2-O-sulfo HS tetrasaccharides investigated in this study.
| IdoA2S | DR | GlcA2S | DR | ||
|---|---|---|---|---|---|
| IA | GlcA-GlcNAc-IdoA2S-GlcNAc-(CH2)5NH2 | 0.5 | IB | GlcA-GlcNAc-GlcA2S-GlcNAc-(CH2)5NH2 | 12.5 |
| IIA | GlcA-GlcNAc-IdoA2S-GlcNAc6S-(CH2)5NH2 | 1.0 | IIB | GlcA-GlcNAc-GlcA2S-GlcNAc6S-(CH2)5NH2 | 13.5 |
| IIIA | GlcA-GlcNS-IdoA2S-GlcNS-(CH2)5NH2 | 1.6 | IIIB | GlcA-GlcNS-GlcA2S-GlcNS-(CH2)5NH2 | 21.6 |
| IVA | GlcA-GlcNS-IdoA2S-GlcNS6S-(CH2)5NH2 | 1.2 | IVB | GlcA-GlcNS-GlcA2S-GlcNS6S-(CH2)5NH2 | 9.3 |
| VA | GlcA-GlcNS6S-IdoA2S-GlcNS6S-(CH2)5NH2 | 1.3 | |||
| VIA | IdoA-GlcNS6S-IdoA2S-GlcNS6S-(CH2)5NH2 | 0.6 | |||
CONCLUSIONS
EDD fragmentation is known to provide useful data for the assignment of sulfo locations in HS oligomers, but the assignment of uronic acid stereochemistry has remained an open issue. This work demonstrates for the first time the assignment of C-5 stereochemistry in 2-O-sulfated uronic acid epimers using EDD data. The relative abundance of glycosidic products B3, Y1, Z2 and C2 as projected onto the PCA loadings plots are found to be diagnostic of GlcA2S while Y2, Y3 and 1,5X2 are diagnostic of IdoA2S near the reducing end of HS or Hp tetramers. By summing the intensities of the GlcA2S diagnostic fragments, and dividing by the sum of the IdoA2S diagnostic fragments, a ratio is obtained that allows one to assign the hexuronic acid stereochemistry for a number of HS/Hp tetramers with a broad range of sulfo modifications. Since there are several structural variations of HS oligomers, future work will test the value of the diagnostic ratio (DR) for a full range of HS tetrasaccharides. We also plan to test this approach on hexasaccharides and longer oligomers, to see if it extends to the assignment of more than one uronic acid residue in an oligomer.
Supplementary Material
Highlights of paper.
The assignment of uronic acid stereochemistry in glycosaminoglycans using tandem mass spectrometry presents a major analytical challenge.
Ten synthetic 2-O-sulfo heparan sulfates tetrasaccharide oligomers, varying in degree of sulfation, were subjected to tandem mass spectrometry analysis using electron detachment dissociation (EDD).
B3 and it complementary Y1 ions are diagnostic for glucuronic acid, independent of the degree of sulfation.
The ratios of the sum of diagnostic ions for glucuronic acid (Z2, C2, B3 and Y1) and iduronic acid (Y2 and 1,5X2) predict the uronic acid stereochemistry for all ten 2-O-sulfo heparan sulfate tetramers examined.
ACNOLWEDGEMENTS
The authors acknowledge generous financial support from the National Institutes of Health, grant P41 GM103390.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Chi L, Amster J, Linhardt RJ. Current Analytical Chemistry. 2005;1:223–240. doi: 10.2174/157341105774573929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rabenstein DL. Natural product reports. 2002;19:312–331. doi: 10.1039/b100916h. [DOI] [PubMed] [Google Scholar]
- 3.Desai UR, Wang H, Ampofo SA, Linhardt RJ. Anal. Biochem. 1993;213:120–127. doi: 10.1006/abio.1993.1394. [DOI] [PubMed] [Google Scholar]
- 4.Liu C, Sheng J, Krahn JM, Perera L, Xu Y, Hsieh P-H, Dou W, Liu J, Pedersen LC. J. Biol. Chem. 2014;289:13407–13418. doi: 10.1074/jbc.M113.530535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warda M, Toida T, Zhang F, Sun P, Munoz E, Xie J, Linhardt RJ. Glycoconjugate J. 2006;23:555–563. doi: 10.1007/s10719-006-7668-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ai X, Do A-T, Lozynska O, Kusche-Gullberg M, Lindahl U, Emerson CP. J. Cell Biol. 2003;162:341–351. doi: 10.1083/jcb.200212083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pellegrini L. Curr. Opin. Struct. Biol. 2001;11:629–634. doi: 10.1016/s0959-440x(00)00258-x. [DOI] [PubMed] [Google Scholar]
- 8.Fuster MM, Wang L. Progress in Molecular Biology and Translational Science. 2010;93:179–212. doi: 10.1016/S1877-1173(10)93009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindahl B, Eriksson L, Lindahl U. Biochem. J. 1995;306:177–184. doi: 10.1042/bj3060177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaia J, Costello CE. Anal. Chem. 2003;75:2445–2455. doi: 10.1021/ac0263418. [DOI] [PubMed] [Google Scholar]
- 11.Huang R, Pomin VH, Sharp JS. J. Am. Soc. Mass Spectrom. 2011;22:1577–1587. doi: 10.1007/s13361-011-0174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou W, Hakansson K. J. Am. Soc. Mass Spectrom. 2013;24:1798–1806. doi: 10.1007/s13361-013-0700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaia J. Mass Spectrom. Rev. 2004;23:161–227. doi: 10.1002/mas.10073. [DOI] [PubMed] [Google Scholar]
- 14.Liu HC, Hakansson K. Int. J. Mass spectrom. 2011;305:170–177. [Google Scholar]
- 15.Shi X, Huang Y, Mao Y, Naimy H, Zaia J. J. Am. Soc. Mass Spectrom. 2012;23:1498–1511. doi: 10.1007/s13361-012-0429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kailemia MJ, Li L, Ly M, Linhardt RJ, Amster IJ. Anal. Chem. 2012;84:5475–5478. doi: 10.1021/ac3015824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kailemia MJ, Li L, Xu Y, Liu J, Linhardt RJ, Amster IJ. Mol. Cell. Proteomics. 2013;12:979–990. doi: 10.1074/mcp.M112.026880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leach FE, III, Ly M, Laremore TN, Wolff JJ, Perlow J, Linhardt RJ, Amster IJ. J. Am. Soc. Mass Spectrom. 2012;23:1488–1497. doi: 10.1007/s13361-012-0428-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolff JJ, Amster IJ, Chi L, Linhardt RJ. J. Am. Soc. Mass Spectrom. 2007;18:234–244. doi: 10.1016/j.jasms.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolff JJ, Chi L, Linhardt RJ, Amster IJ. Anal. Chem. 2007;79:2015–2022. doi: 10.1021/ac061636x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolff JJ, Laremore TN, Busch AM, Linhardt RJ, Amster IJ. J. Am. Soc. Mass Spectrom. 2008;19:294–304. doi: 10.1016/j.jasms.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leach FE, III, Wolff JJ, Xiao Z, Ly M, Laremore TN, Arungundram S, Al-Mafraji K, Venot A, Boons G-J, Linhardt RJ. Eur. J. Mass Spectrom. 2011;17:167. doi: 10.1255/ejms.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolff JJ, Leach FE, III, Laremore TN, Kaplan DA, Easterling ML, Linhardt RJ, Amster IJ. Anal. Chem. 2010;82:3460–3466. doi: 10.1021/ac100554a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leach FE, Xiao Z, Laremore TN, Linhardt RJ, Amster IJ. Int. J. Mass spectrom. 2011;308:253–259. doi: 10.1016/j.ijms.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y, Yu X, Mao Y, Costello CE, Zaia J, Lin C. Anal. Chem. 2013;85:11979–11986. doi: 10.1021/ac402931j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh HB, Leach FE, III, Arungundram S, Al-Mafraji K, Venot A, Boons G-J, Amster IJ. J. Am. Soc. Mass Spectrom. 2011;22:582–590. doi: 10.1007/s13361-010-0047-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kailemia MJ, Park M, Kaplan DA, Venot A, Boons G-J, Li L, Linhardt RJ, Amster IJ. J. Am. Soc. Mass Spectrom. 2014;25:258–268. doi: 10.1007/s13361-013-0771-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaia J, Li X-Q, Chan S-Y, Costello CE. J. Am. Soc. Mass Spectrom. 2003;14:1270–1281. doi: 10.1016/S1044-0305(03)00541-5. [DOI] [PubMed] [Google Scholar]
- 29.Miller MJC, Costello CE, Malmstrom A, Zaia J. Glycobiology. 2006;16:502–513. doi: 10.1093/glycob/cwj093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arungundram S, Al-Mafraji K, Asong J, Leach FE, Amster IJ, Venot A, Turnbull JE, Boons GJ. J. Am. Chem. Soc. 2009;131:17394–17405. doi: 10.1021/ja907358k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Domon B, Costello CE. Glycoconjugate J. 1988;5:397–409. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.