Abstract
Global efforts are underway to combat antimicrobial resistance (AMR). A key target in this intervention is surveillance for local and national action. Data on AMR in Ghana are limited, and monitoring of AMR is nonexistent. We sought to generate baseline data on AMR, and to assess the readiness of Ghana in laboratory-based surveillance. Biomedical scientists in laboratories across Ghana with capacity to perform bacteriological culture were selected and trained. In-house standard operating protocols were used to perform microbiological investigations on clinical specimens. Additional microbiological tests and data analyses were performed at a centralized laboratory. Surveillance data were stored and analyzed using WHONET program files. A total of 24 laboratories participated in the training, and 1,598 data sets were included in the final analysis. A majority of the bacterial species were isolated from outpatients (963 isolates; 60.3%). Urine (617 isolates; 38.6%) was the most common clinical specimen cultured, compared to blood (100 isolates; 6.3%). Ten of 18 laboratories performed blood culture. Bacteria isolated included Escherichia coli (27.5%), Pseudomonas spp. (14.0%), Staphylococcus aureus (11.5%), Streptococcus spp. (2.3%), and Salmonella enterica serovar Typhi (0.6%). Most of the isolates were multidrug-resistant, and over 80% of them were extended-spectrum beta-lactamases-producing. Minimum inhibitory concentration levels at 50% and at 90% for ciprofloxacin, ceftriaxone, and amikacin on selected multidrug-resistant bacteria species ranged between 2 µg/mL and >256 µg/mL. A range of clinical bacterial isolates were resistant to important commonly used antimicrobials in the country, necessitating an effective surveillance to continuously monitor AMR in Ghana. With local and international support, Ghana can participate in global AMR surveillance.
Keywords: antimicrobial resistance, ESBL-producing, quinolone, multiple drug resistance
Introduction
Antimicrobial resistance (AMR) is emerging as a global health security threat.1,2 The World Health Organization’s Global Action Plan against AMR identified surveillance as one of the key pillars to combat this menace.3 In resource-rich settings, extensive national and regional programs have been developed to monitor AMR patterns over time.4 Examples include the Swedish Strategic Programme for the Rational Use of Antimicrobial Agents and Surveillance of Resistance,5 the European AMR Surveillance System,6 and the Antimicrobial Resistance Monitoring and Research programme.7 In contrast, the infrastructure and resources needed to implement such surveillance systems in resource-limited settings (RLS) are unavailable.4 Many RLS, including Ghana, have a high burden of infectious diseases that require antimicrobial therapy to save lives.8 In addition to the lack of infrastructure to investigate these infectious diseases in RLS, spurious, falsified, and counterfeit drugs often slip into the drug supply chain.9 These highlight the importance of AMR surveillance, especially in RLS. Such systems will feed into larger global platforms of AMR monitoring.
An AMR Working Group has been instituted in Ghana to create a policy platform, develop policy framework and implementation plan, and to help raise awareness on the menace of AMR.10 The Director of Pharmacy at the Ministry of Health (MOH) chairs this working group. Members include key stakeholders in health care, regulatory authorities, academia, research institutions, veterinary clinics, civil society organizations, and others. In addition, the MOH, in collaboration with local and international organizations, has developed Ghana’s Essential Medicines List and National Drug Policy.11 The first ever African Conference on Antibiotic Use and Resistance was also held in Ghana between March 18 and 20, 2015, to disseminate research information on AMR.12
Laboratory-based surveillance is required for local and national action in the monitoring of AMR and its spread.13,14 Individual scientists and researchers have been working on AMR in Ghana. However, their research findings are usually not well coordinated and they target either specific pathogens or infections.15–18 Between 2002 and 2003, Newman et al conducted the first ever surveillance of AMR in Ghana.19 Though data generated from this surveillance have been useful over the years, up-to-date information is needed to inform and direct policy issues in Ghana. The aim of the current study is to generate baseline data on AMR and to identify potential gaps that may affect future collaboration and data sharing in local and national surveillance efforts.
Methods
Laboratory selection and training workshop
Study laboratories were selected based on the recommendations from the AMR Working Group, Ghana.15 The categories of the laboratories included are as follows: three teaching hospitals – Korle-Bu (AcK), Komfo Anokye (AsK), and Tamale (NoT); seven regional hospitals – Ridge (AcR), Eastern (EaK), Central (CeT), Volta (VoV), Brong Ahafo (BaS), Upper East (UeB), and Upper West (UwR); three zonal public health reference laboratories (PHRLs) – National Public Health Reference Laboratory (NPHRL) (AcP), Kumasi (AsP), and Sekondi (WsE); four faith-based hospitals – St Patrick’s Holy Family Berekum (BaB), St Patrick’s Holy Family Nkawkaw (EaN), St Patrick, Offinso (AsO), and Presbyterian Hospital, Agogo (AsA); three district hospitals – LEKMA (AcL), Tema (AcT), and Tetteh Quarshie (EaT); two research laboratories – Kintampo (BaK) and War Memorial (UeW); and two quasi-government hospitals – 37 Military (Ac3) and Cape Coast University Hospital (CeU).
The AMR surveillance study spanned 6 months, from June to November 2014. Before commencement, a 3-day residential workshop was organized to harmonize susceptibility testing protocols. Issues related to logistics for the surveillance were also addressed. Geographically, Ghana was divided into two sectors (southern and northern) for the training workshop. The trainings were done 2 weeks apart at the University of Ghana Medical School, and the Kwame Nkrumah University of Science and Technology, for the southern and northern sectors, respectively. In total, 33 technologists from 24 laboratories participated in the two workshops. Training included classroom lectures, and practical sessions using the Clinical Laboratory Standard Institute20 guidelines for susceptibility testing. A predesigned data collection sheet was thoroughly discussed and adapted (Figure S1). The data collection sheet sought to capture basic information on patients, specimen types, bacteria isolated, antimicrobial agents tested and inhibitory zone sizes, as well as initials of the technologists submitting data. Study laboratories were requested to submit data on multidrug-resistant (MDR) isolates from all specimen types. Multidrug resistance was defined as resistance to two or more antibiotic classes.20
Antimicrobial susceptibility testing
Study laboratories performed routine microbiological investigations on all clinical specimens received using in-house standard operating procedures. Bacterial isolates were identified as far as possible using Gram morphology, routine biochemical tests, and in some instances the API 20E system (bioMérieux SA, Marcy l’Etoile, France). Susceptibility tests were performed by the disk diffusion method,21 and inhibition zones sizes were measured and reported in millimeters. Gram-negative and gram-positive antimicrobial disks were selected for gram-negative and gram-positive isolates, respectively. The disks tested and their concentrations in micrograms included: ampicillin (10), piperacillin (100), amoxicillin/clavulanic acid (20/10), cefuroxime (30), cefotaxime (30), meropenem (10), imipenem (10), amikacin (30), gentamicin (15), nalidixic acid (30), ciprofloxacin (5), ofloxacin (5), trimethoprim/sulfamethoxazole (1.25/23.75), erythromycin (15), nitrofurantoin (300), chloramphenicol (30), tetracycline (30), flucloxacillin, oxacillin (1), and cefoxitin (30). Pure isolates were stabbed on Mueller–Hinton agar slants and labeled appropriately using pathological codes of patients. Biomedical scientists additionally completed a surveillance data collection sheet (S1). Approximately a quarter of the study laboratories routinely participate in External Quality Assurance Systems.
Surveillance data and isolate submission
Biweekly, biomedical scientists sent completed data sheets together with bacterial isolates in cold boxes to the Medical Microbiology Department, School of Biomedical and Allied Health Sciences (MD-SBAHS). In-country courier systems were mostly used in the transportation of materials between the study laboratories and MD-SBAHS. A research assistant was employed for liaising with the study laboratories. The research assistant worked under the direct supervision of the research team. He was scheduled to conduct random quality checks, and additional tests, including minimum inhibitory concentration (MIC) and extended-spectrum beta-lactamase (ESBL) tests. Finally, he entered all data received into WHO-NET database files.22
Further microbiological tests
Klebsiella spp. and Escherichia coli isolates, found to be resistant to third-generation cephalosporins, were tested to detect the presence of ESBLs, using methods described elsewhere,23 and interpreted using the Clinical Laboratory Standards Institute guidelines.20 Cefotaxime–clavulanate combination versus cefotaxime or ceftazidime–clavulanate combination versus ceftazidime disk were used (MAST, Germany). ESBL-positive isolates were additionally tested to determine their susceptibility to meropenem by the disk diffusion method.21 MIC tests were carried out on randomly selected MDR isolates using E-test strips (bioMérieux SA, France). Antimicrobials tested included ceftriaxone, ciprofloxacin, and amikacin. The manufacturer’s instructions and recommendations were used to interpret results. E. coli ATCC 25922 and Staphylococcus aureus ATCC 25923 were included as control strains for susceptibility tests.
Data management and analysis
All surveillance data and isolates were cataloged and kept at MD-SBAHS. Soft copies of data were stored in the WHONET program file, which was also used to generate antibiogram profiles.22 For the purposes of analyses, the geographical border of Ghana was divided into southern, middle, and northern sectors (Figure 1). Data were summarized in tables and graphs. Chi-square test was used to analyze associations between the different sectors, after data was exported into Epi Info.24 Statistical significance was determined at P-value <0.05. Intermediate resistance of susceptibility test was considered resistant.
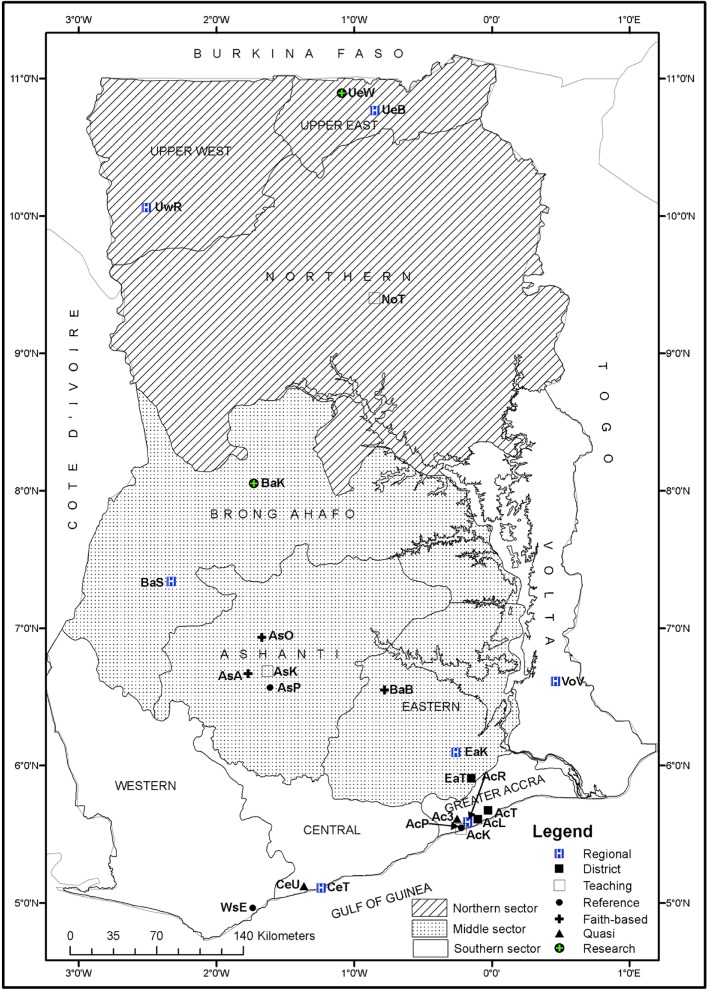
Figure 1.
Ten regional boundaries of Ghana showing study laboratories.
Notes: Regional hospitals: AcR – Ridge (25), EaK – Eastern (142), VoV – Volta (56), BaS – Brong Ahafo (209), UwR – Upper West (0), UeB – Upper East (0)*. District hospitals: AcT – Tema General (18), AcL – LEKMA (110), EaT – Tetteh Quarshie Memorial (13). Teaching hospitals: AcK – Korle-Bu (699), AsK – Komfo Anokye (7), NoT – Tamale (102), CeT – Cape Coast (6). Zonal public health reference laboratories: AcP – National (0)*, WsE – Sekondi (152), AsP – Kumasi (0)*. Faith-based hospitals: EaN – Holy Family, Nkawkaw (11), AsO – St Patrick, Offinso (14), AsA – Agogo Presbyterian (0)*, BaB – Holy Family, Brekum (19). Research laboratories: BaK – Kintampo (2), UeW – War Memorial (0)*. Quasi Hospitals: Ac3 – 37 Military Hospital (0)*, CeU – University, Cape Coast (3). () Data sets submitted. *Laboratories that did not submit any data during the surveillance period.
Ethical consideration
The MOH in conjunction with the Ghana Health Services (Institutional Care Division) granted permission to carry out the study. Additional permission was also sought from the medical directors of the respective health facilities where laboratories were located. To maintain confidentiality, patient information was codified. As this was a laboratory surveillance only, patient consent was not necessary.
Results
Participating laboratories and data received
Figure 1 shows all ten regional boundaries of Ghana with the exact geographical locations of the study laboratories. With the exception of the Upper West Region that did not submit any data, we received and analyzed data from the other nine regions of Ghana. A total of 1,606 data sets were received from 18 of 24 (75%) study laboratories. Out of this total, 1,598 data sets were included in the final analysis. The rest (eight) were excluded during data cleaning. Total data sets received from the southern, middle, and northern sectors were 1,069, 417, and 112, respectively. Six of the 24 laboratories did not submit any data within the surveillance period. Data received from teaching hospitals, PHRLs, and a district hospital were as follows: AcK 699 (43.7%), NoT 102 (6.4%), AsK 7 (0.4%); WsE 152 (9.5%), AcP 0 (0%); and AcL 110 (6.9%), respectively. Out of the three PHRLs that participated in the current surveillance, only one submitted data.
Bacteria species isolated
Bacterial species isolated included S. aureus (183), coagulase-negative Staphylococcus (45), Streptococcus spp. (37), Salmonella enterica serovar Typhi (9), nontyphoidal Salmonella (7), E. coli (440), Klebsiella spp. (114), Pseudomonas aeruginosa (118), Citrobacter spp. (163), and Vibrio cholerae (54) (Table 1). Bacteria were isolated from inpatients (428; 26.8%) and outpatients (963; 60.3%). The sources for the remaining isolates were not indicated (161; 10.1%). Specimen types that grew the bacterial isolates were diverse and varied. They included blood, urine, stool, swabs (ear, eye, wound, etc), and sputum (Table 2). In 2.8% (46/1,598) of the data sets, the specimen type was not indicated. More females (839; 53%) than males (660; 41%) were involved in the study; the sex data of the remaining patients were missing (99; 6%). The age distributions of the patients in years were as follows: <1,113 (7.1%); 1–40, 373 (23.3%); 41–60, 234 (14.6%); >61, 246 (15.4%). The ages of the rest were not indicated (396; 24.8%).
Table 1.
Bacterial species isolated during 6-month surveillance of antimicrobial resistance, Ghana, June–November 2014
| Organism | Number of isolates (%) | Inpatients | Outpatients | Source not indicated |
|---|---|---|---|---|
| Acinetobacter spp. | 25 (1.56) | 9 | 11 | 5 |
| Citrobacter koseri | 17 (1.06) | 1 | 15 | 1 |
| Citrobacter spp. | 146 (9.14) | 43 | 87 | 16 |
| Enterobacter spp. | 149 (9.32) | 42 | 83 | 24 |
| Enterococcus spp. | 3 (0.19) | 1 | 2 | N/A |
| Escherichia coli | 440 (27.53) | 94 | 309 | 37 |
| Klebsiella pneumoniae | 17 (1.06) | 3 | 12 | 2 |
| Klebsiella spp. | 97 (6.07) | 30 | 57 | 10 |
| Moraxella spp. | 3 (0.19) | N/A | 3 | N/A |
| Morganella spp. | 13 (0.81) | 4 | 9 | N/A |
| Proteus mirabilis | 60 (3.75) | 12 | 39 | 9 |
| Proteus spp. | 42 (2.63) | 10 | 30 | 2 |
| Providencia spp. | 5 (0.31) | 3 | 1 | 1 |
| Pseudomonas aeruginosa | 118 (7.38) | 17 | 83 | 18 |
| Pseudomonas spp. | 147 (9.20) | 46 | 90 | 11 |
| Salmonella enterica serovar Typhi | 9 (0.56) | 3 | 5 | 1 |
| Nontyphoidal Salmonella | 7 (0.44) | 5 | 1 | 1 |
| Serratia spp. | 15 (0.94) | 12 | 2 | 1 |
| Staphylococcus aureus | 183 (11.45) | 57 | 111 | 15 |
| Coagulase-negative Staphylococcus | 41 (2.57) | 22 | 15 | 4 |
| Stenotrophomonas maltophilia | 1 (0.06) | N/A | 1 | N/A |
| Streptococcus pneumoniae | 2 (0.13) | N/A | 2 | N/A |
| Other Streptococcus spp. | 35 (2.19) | 18 | 13 | 4 |
| Vibrio cholerae O1 Ogawa | 19 (1.19) | 9 | 9 | 1 |
| Total | 1,598 (100) | 428 | 963 | 161 |
Abbreviation: N/A, not applicable.
Table 2.
Specimen types cultured at laboratories during 6-month nationwide surveillance of antimicrobial resistance, Ghana, June–November 2014
| Specimen type | Number of isolates | Southern
|
Middle
|
Northern
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AcK | AcL | AcR | AcT | CeT | CeU | VoV | WsE | AsK | AsO | BaB | BaK | BaS | EaK | EaN | EaT | NoT | UeB | ||
| Abdominal fluid | 3 | 1 | N/A | N/A | N/A | N/A | N/A | 1 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Abscess | 4 | 1 | 1 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 1 | N/A | 1 | N/A | N/A | N/A | N/A | N/A |
| Aspirate | 33 | 20 | N/A | N/A | N/A | N/A | N/A | 1 | 3 | N/A | N/A | N/A | N/A | 3 | 2 | N/A | N/A | N/A | N/A |
| Blood | 100 | a | 16 | N/A | N/A | N/A | 1 | N/A | 14 | 7 | 1 | 1 | N/A | 45 | 1 | N/A | N/A | 13 | 1 |
| Burns | 1 | 1 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Catheter | 3 | 3 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Cerebrospinal fluid | 5 | 3 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 1 | N/A | N/A | 1 | N/A | N/A | N/A | N/A | N/A |
| Cervix | 2 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 2 | N/A | N/A | N/A | N/A |
| Cornea | 3 | 2 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 1 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Ear | 80 | 24 | 10 | N/A | 1 | N/A | N/A | 10 | 7 | N/A | 1 | N/A | N/A | N/A | 16 | 1 | 2 | N/A | 1 |
| Eyes | 25 | 18 | 1 | 1 | N/A | N/A | N/A | N/A | 1 | N/A | N/A | 2 | N/A | N/A | 1 | N/A | N/A | N/A | N/A |
| Gastric fluid | 1 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 1 | N/A | N/A | N/A | N/A | N/A |
| Heart valve | 1 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 1 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Joint | 1 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 1 | N/A | N/A | N/A | N/A | N/A |
| Nose | 2 | 1 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 1 | N/A | N/A | N/A | N/A | N/A |
| Pleural fluid | 3 | 1 | 1 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 1 | N/A | N/A | N/A | N/A | N/A |
| Prosthesis | 1 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 1 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Pus | 8 | 3 | N/A | N/A | 1 | N/A | N/A | N/A | 1 | N/A | N/A | N/A | N/A | 2 | N/A | N/A | N/A | N/A | N/A |
| Rectal | 2 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 2 | N/A | N/A | N/A | N/A |
| Sputum | 69 | 18 | 5 | 2 | N/A | 1 | N/A | N/A | 24 | N/A | N/A | 3 | N/A | 5 | N/A | N/A | N/A | 7 | N/A |
| Stool | 26 | 1 | 2 | N/A | N/A | N/A | N/A | 1 | N/A | N/A | N/A | N/A | N/A | 5 | 17 | N/A | N/A | N/A | N/A |
| Swab | 5 | 4 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Throat | 2 | 1 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 1 | N/A | N/A | N/A | N/A | N/A |
| Urethra | 26 | N/A | 3 | N/A | 2 | N/A | N/A | 5 | 2 | N/A | N/A | N/A | N/A | 11 | N/A | N/A | N/A | 1 | N/A |
| Urine | 617 | 293 | 40 | 9 | 2 | 1 | N/A | 23 | 29 | N/A | 4 | N/A | N/A | 66 | 58 | 2 | 4 | 63 | 3 |
| Vagina | 110 | 1 | 6 | 2 | 4 | 1 | 2 | 5 | 34 | N/A | 2 | N/A | N/A | 15 | 25 | N/A | 1 | 9 | 2 |
| Wound | 415 | 187 | 19 | 3 | 7 | 3 | N/A | 7 | 33 | N/A | 5 | 10 | 2 | 40 | 18 | 8 | 3 | 7 | 3 |
| Wound, surgical | 4 | 4 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Not indicated | 46 | 26 | 4 | 8 | 1 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 4 | N/A | N/A | N/A | N/A | N/A |
| Total | 1,598 | 613 | 108 | 25 | 18 | 6 | 3 | 53 | 150 | 7 | 14 | 18 | 2 | 199 | 142 | 11 | 10 | 100 | 10 |
Notes:
Korle-Bu Teaching Hospital was not performing blood culture during the surveillance period. Southern sector: AcT – Tema General Hospital; AcK – Korle-Bu Teaching Hospital; AcL – LEKMA Hospital; AcP – National Public Health Reference Laboratory; AcR – Ridge Hospital; Ac3 – 37 Military Hospital; CeT – Cape Coast Teaching Hospital; CeU – University Hospital, Cape Coast; VoV – Volta Regional Hospital; WsE – Public Health Laboratory, Sekondi. Middle sector: EaT – Tetteh Quarshie Memorial Hospital; EaK – Eastern Regional Hospital, Koforidua; EaN – Holy Family Hospital, Nkawkaw; AsK – Komfo Anokye Teaching Hospital; AsO – St Patrick’s Hospital, Offinso; AsA – Agogo Presbyterian Hospital; AsP – Public Health Laboratory, Kumasi; BaS – Brong Ahafo Regional Hospital, Sunyani; BaB – Holy Family Hospital, Berekum; BaK – Kintampo Health Research Centre. Northern sector: UeB – Upper East Regional Hospital, Bolgatanga; UeW – War Memorial Hospital, Navorongo; NoT – Tamale Teaching Hospital; UwR – Upper West Regional Hospital.
Abbreviation: N/A, not applicable.
Table 2 shows that though most of the laboratories have capacity for urine culture, many lacked capacity for blood culture. The highest number of blood culture specimens (45) was received from the Sunyani Regional Hospital, and none from the Korle-Bu Teaching Hospital.
Resistance profiles
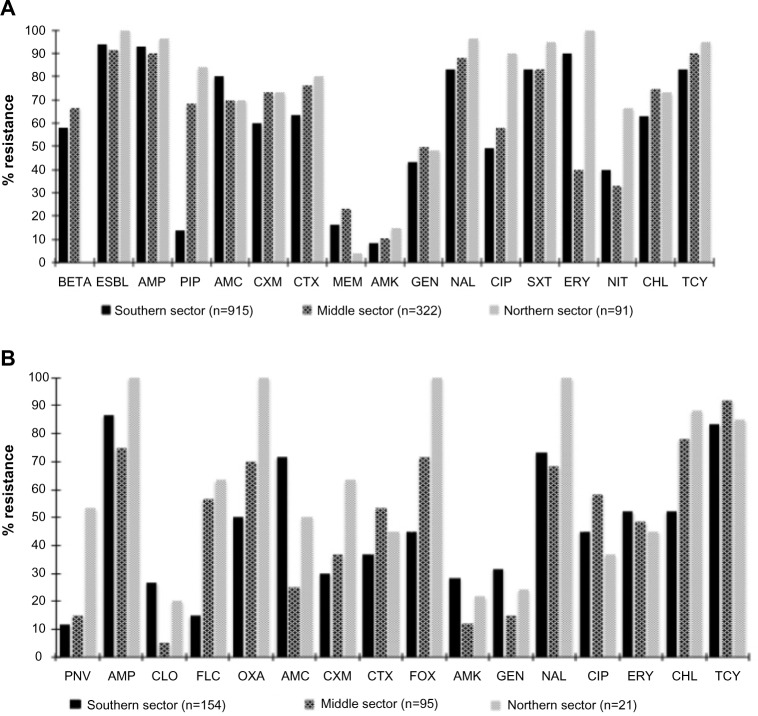
Figure 2 summarizes resistance profiles of gram-negative and gram-positive isolates that were tested against important antimicrobials. For both gram-negative and gram-positive isolates, the majority of the antimicrobial agents tested were ineffective, across the southern, middle, and northern sectors of Ghana. The WHONET expert rule indicated that over 50% of the gram-negative isolates from the southern and middle sectors were beta-lactamase-producing bacteria (Figure 2A). Additionally, nearly 90% of the gram-negative isolates were ESBL-producing (Figure 2A). Older drugs such as ampicillin, tetracycline, chloramphenicol, and trimethoprim sulfamethoxazole were ineffective (80%) against the isolates tested. However, the isolates showed resistance levels of <50% for injectables such as amikacin and gentamicin. Especially for gram-negative isolates, resistance profiles for the third-generation cephalosporins and quinolones such as nalidixic acid and ciprofloxacin were high – >50% across all the sectors of the country. For gram-positive isolates, cefoxitin resistance was <50% in the northern and southern sectors, but almost 100% for the middle sector (Figure 2B). Resistance profile for piperacillin was very high in the northern and middle sectors (~80% and 70%, respectively) compared to approximately 15% in the southern sector. Similarly, the nitrofurantoin resistance profile for the northern sector was high (~70%), compared to the middle and southern sectors (~30% and 40% respectively). Most of the gram-negative isolates were susceptible to meropenem across the country.
Figure 2.
Nationwide resistance profile of gram-negative isolates (A) and gram-positive isolates (B) from Ghana, June–November 2014.
Abbreviations: BETA, beta-lactamase; ESBL, extended-spectrum beta-lactamase; AMP, ampicillin; PIP, piperacillin; AMC, augmentin; CXM, cefuroxime; CTX, cefotaxime; MEM, meropenem; AMK, amikacin; NAL, nalidixic acid; CIP, ciprofloxacin; SXT, trimethoprim/sulfamethoxazole; ERY, erythromycin; NIT, nitrofurantoin; CHL, chloramphenicol; TCY, tetracycline; PNV, penicillin V; CLO, cloxacillin; FLC, flucloxacillin; OXA, oxacillin; FOX, cefoxitin; GEN, gentamicin.
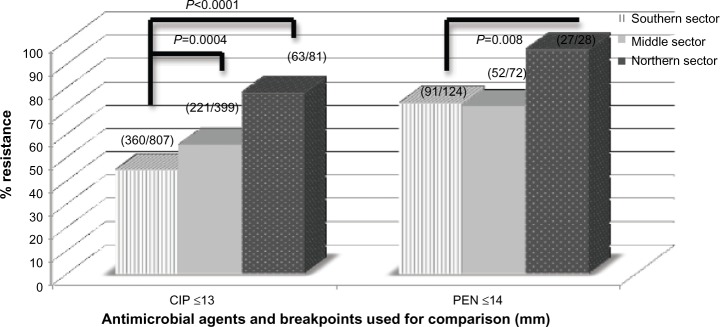
For several of the isolates tested, those from the northern sector had resistance profiles generally higher compared to that from the other sectors of the country. Especially for ciprofloxacin and penicillin, the differences observed in the resistance profiles between the southern and northern sectors were statistically significant (P<0.05; Figure 3).
Figure 3.
Comparison of resistance profile of selected antimicrobial between southern, middle, and northern sectors of Ghana.
Note: Differences observed in ciprofloxacin and penicillin is significant (P<0.05).
Abbreviations: CIP, ciprofloxacin; PEN, penicillin.
Phenotypic ESBL screening results
Seventy-four percent (86/117) of enterobacteria (E. coli, n=81 and Klebsiella spp., n=5) were observed to be ESBL-producing isolates. These positive ESBL-producing E. coli and Klebsiella spp. had percentage resistance ranges >85% to penicillins and third-generation cephalosporins (data not shown). As expected, these isolates were generally susceptible to cephamycins (cefoxitin) and carbapenemase (meropenem and imipenem).
MIC results
Table 3 shows the MIC50 and MIC90 results of randomly selected isolates that were MDR. MIC levels for gram-negative bacteria such as E. coli, Enterobacter spp., and Pseudomonas spp. were high (>256 µg/mL; Table 3). MIC50 and MIC90 of Pseudomonas spp. and Klebsiella spp. to ceftriaxone were <4 µg/mL and >256 µg/mL, respectively. In general, Pseudomonas had lower MIC50 and MIC90 to injectable antimicrobials such as amikacin (Table 3).
Table 3.
Minimum inhibitory concentration of randomly selected isolates to some antimicrobial agents
| Organism/antimicrobial agent | n | MIC range, μg/mL | MIC 50 | MIC 90 |
|---|---|---|---|---|
| Escherichia coli | ||||
| Ciprofloxacin | 23 | 0.019–256 | >256 | >256 |
| Ceftriaxone | 9 | 0.015–256 | >256 | >256 |
| Pseudomonas spp. | ||||
| Ciprofloxacin | 17 | 0.012–256 | >256 | >256 |
| Ceftriaxone | 8 | 0.38–256 | 4 | >256 |
| Amikacin | 17 | 0.75–128 | 3 | 48 |
| Klebsiella spp. | ||||
| Ciprofloxacin | 7 | 0.016–256 | 2 | >256 |
| Citrobacter spp. | ||||
| Ciprofloxacin | 8 | 0–256 | >256 | >256 |
| Enterobacter spp. | ||||
| Ciprofloxacin | 7 | >256 | >256 | >256 |
Abbreviations: n, number; MIC50, minimum inhibitory concentration at 50%; MIC90, minimum inhibitory concentration at 90%.
Discussion
Clinical specimen, isolates, and antibiogram
Clinical specimens submitted for culture were diverse in the current study. Urine, swabs, (especially wound) and blood formed >90% of all clinical specimens received by the study laboratories. Though this observation is consistent with studies conducted elsewhere,4,7 majority of the study laboratories did not perform blood cultures. For example, AcK, the largest tertiary referral hospital in Ghana, with over 2,500-bed capacity, did not process any blood culture during the surveillance period. Blood stream infections can be fatal.25 Apart from a few exceptions, including BaS (Regional), NoT (Teaching), WsE (PHRL), and AcL (District), the other study laboratories did not process enough blood cultures. This emphasizes the importance of surveillance in terms of promptly identifying possible gaps and taking necessary action in a timely and consistent manner to fix them.4
Both gram-positive and gram-negative bacteria were identified in the current study, as is typical with studies conducted elsewhere.4,7 In the last decade, there had been a divided opinion on whether to target specific pathogens or whether to consider specific clinical syndromes in AMR surveillance.3 The current opinion, however, is to include priority infections such as bloodstream, urinary tract, diarrhea, and gonorrhea infections.2,14 In the present study, we considered all clinical specimens submitted for culture at the study laboratories, and, therefore, captured both clinical syndromes as well as pathogens associated with those conditions. Gram-negative pathogens top the list of bacteria identified in the current surveillance. This finding is similar to both short- and long-duration surveillance conducted elsewhere.19,26–28 The few Vibrio cholerae isolates identified in the current study were from the last phases of the worst cholera outbreak that hit Ghana starting in January 2014 and lasting several months.29 Laboratories in Ghana lack capacity for anaerobic culture. Surveillance systems in well-endowed countries have cultured both aerobes and anaerobes.30,31
Across the southern, middle, and northern sectors of Ghana, both gram-negative and gram-positive isolates showed varied levels of susceptibilities to the antimicrobials tested. Commonly used antimicrobials such as ampicillin, tetracycline, chloramphenicol, and trimethoprim–sulfamethoxazole were ineffective (>70%) against gram-negative and gram-positive isolates. This is consistent with the studies conducted in Nigeria,32 Uganda,33 and Tanzania.34 These older antimicrobials are cheap, and their continued use (whether appropriate/inappropriate) in both humans and animals contribute to the high resistance levels.
In the present study, high prevalence levels (>50%) of resistance were also observed in third-generation cephalosporins, and fluoroquinolones. Syndromic infections are often treated with third-generation cephalosporins, and high levels of resistance to these antimicrobials are worrisome.35,36 MDR ESBL-producing bacteria were found to be generally resistant to ampicillin, third-generation cephalosporins, and fluoroquinolones. Several studies conducted in Ghana16,37 and elsewhere have reported this phenomenon.38
For treatment of urinary tract and blood stream infections, the Standard Treatment Guidelines, Ghana,39 recommends the use of ciprofloxacin. The high levels of ciprofloxacin resistance observed in the current study shows that we are gradually losing available treatment options. Globally, fluoroquinolones have been indiscriminately used for treating human infections40 and for controlling infections in farm animals.40 Most of the organisms studied in the current study had lower MIC levels, especially for ciprofloxacin. In an earlier study conducted in Ghana, MIC levels for ciprofloxacin were in the ranges of 0.004–32 µg/mL.19 MIC levels for ciprofloxacin in the current study ranged from 0.019 to >256 µg/mL. The seeming rise in resistance levels of ciprofloxacin over time in Ghana raises concerns about the need for action.
Preparedness of Ghana in local and national AMR surveillance
In the current laboratory-based surveillance of AMR, data sets were received and processed from >70% of the 24 laboratories that participated in the training workshop. However, six laboratories did not submit any data during the surveillance period. Our preliminary investigations revealed some lapses within these hospitals, including breakdown of culture facilities, clinicians not making request for culture, and some internal managerial issues. More than two-thirds of the data analyzed in the current surveillance were from the southern sector, with less than one-tenth from the northern sector. In 2003, a similar nationwide surveillance of AMR also received a relatively small number of isolates from the northern parts of Ghana.19 Such disparities may introduce some biases while interpreting the results to direct antibiotic policy in Ghana. Considerations such as the general lack of access to health care facilities in remote and rural parts in the northern parts of Ghana, economic and social reasons, and patronage of traditional medications (herbs) compared to orthodox medicine have to be factored into interpretation of surveillance data.41 The Korle-Bu Teaching Hospital alone submitted >50% of the total data sets. Generally, academic tertiary referral laboratories are known to be over-represented in national and multicenter surveillance systems.42 This overrepresentation of data introduces some biases in the overall AMR surveillance results. The Komfo Anokye Teaching Hospital did not submit enough data in the current surveillance compared to their output in the previous study.19
Ghana has four PHRLs, one each in Greater Accra, Ashanti, Brong Ahafo, and the northern regions. The Greater Accra PHRL doubles up as the NPHRL. Reference laboratories participated in the current surveillance but data were received from only one, Brong Ahafo. Some proposals have suggested that AMR surveillance systems should be coordinated by PHRLs.43 In the case of Ghana, perhaps PHRLs are not yet ready to spearhead AMR surveillance activities. The mandate, direction, and functions of PHRLs in Ghana must be critically aligned to address the global public health threat of AMR. Grundmann et al suggested that global AMR surveillance systems must have separate functions including reference work, quality assessment, and the actual surveillance.43 In Ghana, the NPHRL may play a vital role in future laboratory-based surveillance, by participating in quality assessments. In the present study, faith-based and district hospitals also submitted data. Since these hospitals contribute greatly to the health needs of Ghana, they should be included in future national AMR surveillance programs. In general, the current study did not observe disparities in susceptibility results compared to the results of the previous study in Ghana.19
Conclusion
This laboratory-based surveillance shows that important antimicrobial agents used in the country are not as highly effective against a range of clinical isolates as was previously believed. The study also highlights the need for con tinuous surveillance of AMR for local and national action. Additionally, the capacity and infrastructure for culture and susceptibility testing across Ghana needs improvement, especially in facilities in the northern parts of the country.
Supplementary material
Weekly data report form.
Acknowledgments
The following biomedical scientists were involved in the microbiological analysis and data collation at the laboratory levels and are gratefully acknowledged: Hodogbe P and Adade NE, Korle-Bu Teaching Hospital, Accra; Ampah EO, Ridge Hospital, Accra; Arthur F and Derban I, University of Cape Coast Hospital, Cape Coast; Mensah E, Holy Family Hospital, Nkawkaw; Twasam J and Opoku CN, LEKMA Hospital; Mensah E and Amedzro I, Sekondi Public Health Reference Laboratory; Agede C, Volta Regional Hospital; Tetteh F, Tema General Hospital, Accra; Kwakye R and Ehiem RC, St Patrick’s Hospital, Offinso; Tetteh-Ocloo G, Koforidua Regional Hospital, Koforidua; Ayivase J, Holy Family Hospital, Berekum; Kuma GK, Sunyani Regional Hospital; Tetteh I, Komfo Anokye Teaching Hospital, Kumasi; Asiedu B, Upper East Regional Hospital; and Bobzah BP, Tamale Teaching Hospital, Tamale. The authors also thank the staff of the Medical Microbiology Department, University of Ghana Medical School (now School of Biomedical and Allied Health Sciences), for technical support. The authors appreciate the invaluable role played by Mr Amos Akanwena.
Footnotes
Funding
ReAct through the Ministry of Health/Ghana National Drug Programme (GNDP) and the Antibiotic Drug use Monitoring and Evaluation of Resistance (ADMER) project jointly provided funding for this surveillance; grant numbers are MoH/DM-UGMS/2014 and SSI 09-099, respectively. GNDP and ADMER are funded by ReAct and DANIDA.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Cars O, Hedin A, Heddini A. The global need for effective antibiotics-moving towards concerted action. Drug Resist Updat. 2011;14(2):68–69. doi: 10.1016/j.drup.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Huttner A, Harbarth S, Carlet J, et al. Antimicrobial resistance: a global view from the 2013 world healthcare-associated infections forum. Antimicrob Resist Infect Control. 2013;2:31. doi: 10.1186/2047-2994-2-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simonsen GS, Tapsall JW, Allegranzi B, Talbot EA, Lazzari S. The antimicrobial resistance containment and surveillance approach – a public health tool. Bull World Health Organ. 2004;82(12):928–934. [PMC free article] [PubMed] [Google Scholar]
- 4.Holloway K, Mathai E, Gray A. Surveillance of community antimicrobial use in resource-constrained settings – experience from five pilot projects. Trop Med Int Health. 2011;16(2):152–161. doi: 10.1111/j.1365-3156.2010.02695.x. [DOI] [PubMed] [Google Scholar]
- 5.Mölstad S, Erntell M, Hanberger H, et al. Sustained reduction of antibiotic use and low bacterial resistance: 10-year follow-up of the Swedish Strama programme. Lancet Infect Dis. 2008;8(2):125–132. doi: 10.1016/S1473-3099(08)70017-3. [DOI] [PubMed] [Google Scholar]
- 6.Metz-Gercek S, Mittermayer H. The European surveillance activities EARSS and ESAC in the context of ABS International. Wien Klin Wochenschr. 2008;120(9–10):264–267. doi: 10.1007/s00508-008-0967-8. [DOI] [PubMed] [Google Scholar]
- 7.Lesho EP, Waterman PE, Chukwuma U, et al. The antimicrobial resistance monitoring and research (ARMoR) program: the US Department of Defense response to escalating antimicrobial resistance. Clin Infect Dis. 2014;59(3):390–397. doi: 10.1093/cid/ciu319. [DOI] [PubMed] [Google Scholar]
- 8.Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Almuzaini T, Choonara I, Sammons H. Substandard and counterfeit medicines: a systematic review of the literature. BMJ Open. 2013;3(8):e002923. doi: 10.1136/bmjopen-2013-002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.GNDP Ghana National Drug Programme. Ministry of Health. [Accessed March 3, 2015]. Available from: http://ghndp.org/antimicrobialresistance/
- 11.MOH. Ministry of Health. Republic of Ghana Ghana National Drugs Programme (GNDP) Ghana Essential Medicines List. 6th ed. 2010. [Accessed March 3, 2015]. Available from: http://apps.who.int/medicinedocs/documents/s18014en/s18014en.pdf.
- 12.AFCAR. African Conference on Antibiotic Use and Resistance Who is Winning the Antibiotic Resistance War – Bacteria or Man? 2015. [Accessed April 25, 2015]. Available from: http://admerproject.org/
- 13.WHO World Health Organization’s strategy to contain resistance to antimicrobial drugs. Rev Panam Salud Publica. 2001;10(4):284–294. [PubMed] [Google Scholar]
- 14.WHO. World Organization Health Antimicrobial Resistance: Global Report on Surveillance, Geneva. 2014. [Accessed April 27, 2014]. Available from: http://apps.who.int/iris/bitstream/10665/90975/1/WHO_HSE_PED_2013.10358_eng.pdf.
- 15.Acquah SE, Quaye L, Sagoe K, Ziem JB, Bromberger PI, Amponsem AA. Susceptibility of bacterial etiological agents to commonly-used antimicrobial agents in children with sepsis at the Tamale Teaching Hospital. BMC Infect Dis. 2013;13:89. doi: 10.1186/1471-2334-13-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feglo P, Adu-Sarkodie Y, Ayisi L, et al. Emergence of a novel extended-spectrum-beta-lactamase (ESBL)-producing, fluoroquinolone-resistant clone of extraintestinal pathogenic Escherichia coli in Kumasi, Ghana. J Clin Microbiol. 2013;51(2):728–730. doi: 10.1128/JCM.03006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labi AK, Obeng-Nkrumah N, Addison NO, Donkor ES. Salmonella blood stream infections in a tertiary care setting in Ghana. BMC Infect Dis. 2014;14:3857. doi: 10.1186/s12879-014-0697-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Opintan J, Newman MJ. Distribution of serogroups and serotypes of multiple drug resistant Shigella isolates. Ghana Med J. 2007;41(1):8–29. [PMC free article] [PubMed] [Google Scholar]
- 19.Newman MJ, Frimpong E, Donkor ES, Opintan JA, Asamoah-Adu A. Resistance to antimicrobial drugs in Ghana. Infect Drug Resist. 2011;4:215–220. doi: 10.2147/IDR.S21769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.CLSI. Clinical Standard Institute Laboratory . Performance Standards for Antimicrobial Susceptibility Testing: Eighteenth Informational Supplement Wayne: Clinical and Laboratory Standards Institute. Wayne: CLSI; 2010. [Google Scholar]
- 21.Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493–496. [PubMed] [Google Scholar]
- 22.Stelling JM, O’Brien TF. Surveillance of antimicrobial resistance: the WHONET program. Clin Infect Dis. 1997;24(Suppl 1):S157–S168. doi: 10.1093/clinids/24.supplement_1.s157. [DOI] [PubMed] [Google Scholar]
- 23.Ejaz H, Ul-Haq I, Mahmood S, Zafar A, Mohsin Javed M. Detection of extended-spectrum beta-lactamases in Klebsiella pneumoniae: comparison of phenotypic characterization methods. Pak J Med Sci. 2013;29(3):768–772. doi: 10.12669/pjms.293.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.CDC. Centers for Disease Control and Prevention . Epi-Info Version 7. Atlanta, GA: CDC; 2004. [Google Scholar]
- 25.Meremo A, Mshana SE, Kidenya BR, Kabangila R, Peck R, Kataraihya JB. High prevalence of Non-typhoid salmonella bacteraemia among febrile HIV adult patients admitted at a tertiary Hospital, North-Western Tanzania. Int Arch Med. 2012;5(1):28. doi: 10.1186/1755-7682-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Kraker ME, Jarlier V, Monen JC, Heuer OE, van de Sande N, Grundmann H. The changing epidemiology of bacteraemias in Europe: trends from the European Antimicrobial Resistance Surveillance System. Clin Microbiol Infect. 2013;19(9):860–868. doi: 10.1111/1469-0691.12028. [DOI] [PubMed] [Google Scholar]
- 27.Okeke IN, Lamikanra A, Steinrück H, Kaper JB. Characterization of Escherichia coli strains from Cases of Childhood Diarrhea in Provincial Southwestern Nigeria. J Clin Microbiol. 2000;38(1):7–12. doi: 10.1128/jcm.38.1.7-12.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Opintan JA, Newman MJ, Ayeh-Kumi PF, et al. Pediatric diarrhea in southern Ghana: etiology and association with intestinal inflammation and malnutrition. Am J Trop Med Hyg. 2010;83(4):936–943. doi: 10.4269/ajtmh.2010.09-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.UNICEF Cholera outbreak in the West and Central Africa: Regional Update, 2014-WEEK 52. 2014. [Accessed April 4, 2015]. Available from: http://www.unicef.org/cholera/files/Cholera_regional_update_W52_2014_West_and_Central_Africa.pdf.
- 30.Brazier J, Chmelar D, Dubreuil L, et al. ESCMID Study Group on Antimicrobial Resistance in Anaerobic Bacteria European surveillance study on antimicrobial susceptibility of Gram-positive anaerobic cocci. Int J Antimicrob Agents. 2008;31(4):316–320. doi: 10.1016/j.ijantimicag.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 31.Novak A, Rubic Z, Dogas V, Goic-Barisic I, Radic M, Tonkic M. Antimicrobial susceptibility of clinically isolated anaerobic bacteria in a University Hospital Centre Split, Croatia in 2013. Anaerobe. 2015;31:31–36. doi: 10.1016/j.anaerobe.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 32.Dada-Adegbola HO, Muili KA. Antibiotic susceptibility pattern of urinary tract pathogens in Ibadan, Nigeria. Afr J Med Med Sci. 2010;39(3):173–179. [PubMed] [Google Scholar]
- 33.Andabati G, Byamugisha J. Microbial aetiology and sensitivity of asymptomatic bacteriuria among ante-natal mothers in Mulago hospital, Uganda. Afr Health Sci. 2010;10(4):349–352. [PMC free article] [PubMed] [Google Scholar]
- 34.Moyo S, Aboud S, Kasubi M, Maselle SY. Bacteria isolated from bloodstream infections at a tertiary hospital in Dar es Salaam, Tanzania – antimicrobial resistance of isolates. S Afr Med J. 2010;100(12):835–838. doi: 10.7196/samj.4186. [DOI] [PubMed] [Google Scholar]
- 35.Marzouk M, Ferjani A, Haj Ali M, Boukadida J. Profile and susceptibility to antibiotics in urinary tract infections in children and newborns from 2012 to 2013: data from 1879 urine cultures. Arch Pediatr. 2015;22(5):505–509. doi: 10.1016/j.arcped.2015.02.015. [DOI] [PubMed] [Google Scholar]
- 36.Ntirenganya C, Manzi O, Muvunyi CM, Ogbuagu O. High prevalence of antimicrobial resistance among common bacterial isolates in a tertiary healthcare facility in rwanda. Am J Trop Med Hyg. 2015;92(4):865–870. doi: 10.4269/ajtmh.14-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Obeng-Nkrumah N, Twum-Danso K, Krogfelt KA, Newman MJ. High levels of extended-spectrum beta-lactamases in a major teaching hospital in Ghana: the need for regular monitoring and evaluation of antibiotic resistance. Am J Trop Med Hyg. 2013;89(5):960–964. doi: 10.4269/ajtmh.12-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Storberg V. ESBL-producing Enterobacteriaceae in Africa – a non-systematic literature review of research published 2008–2012. Infect Ecol Epidemiol. 2014;13(4) doi: 10.3402/iee.v4.20342. eCollection2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MOH Ministry of Health, Ghana. Standard Treatment Guidelines. 6th ed. 2010. [Accessed March 3, 2015]. Available from: who.int/medicinedocs/documents/s18015en/s18015en.pdf.
- 40.Poppe C, Ayroud M, Ollis G, et al. Trends in antimicrobial resistance of Salmonella isolated from animals, foods of animal origin, and the environment of animal production in Canada, 1994–1997. Microb Drug Resist. 2001;7(2):197–212. doi: 10.1089/10766290152045084. [DOI] [PubMed] [Google Scholar]
- 41.Radyowijati A, Haak H. Improving antibiotic use in low-income countries: an overview of evidence on determinants. Soc Sci Med. 2003;57(4):733–744. doi: 10.1016/s0277-9536(02)00422-7. [DOI] [PubMed] [Google Scholar]
- 42.Rempel O, Pitout JD, Laupland KB. Antimicrobial resistance surveillance systems: are potential biases taken into account? Can J Infect Dis Med Microbiol. 2011;22(4):e24–e28. doi: 10.1155/2011/276017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grundmann H, Klugman KP, Walsh T, et al. A framework for global surveillance of antibiotic resistance. Drug Resist Updat. 2011;14(2):79–87. doi: 10.1016/j.drup.2011.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Weekly data report form.