Abstract
Purpose
Breast cancer patients’ response to treatment is highly dependent on the primary tumor molecular features, with triple-negative breast tumors having the worst prognosis of all subtypes. According to the molecular features, tumors stimulate the microenvironment to induce distinct immune responses, baseline immune activation being associated with higher likelihood of pathologic response. In this study, we investigated the deconvolution of the immunological status of triple-negative tumors in comparison with luminal tumors and the association with patients’ clinicopathological characteristics.
Patients and methods
Gene expression of 84 inflammatory molecules and their receptors were analyzed in 40 peripheral blood samples from patients with Her2− primary breast cancer tumors. We studied the association of triple-negative phenotype with age, clinical stage, tumor size, lymph nodes, and menopausal status.
Results
We observed that more patients with estrogen (ER)/progesterone (PR)-negative tumors had grade III, while more patients with ER/PR-positive tumors had grade II tumors. Gene expression analysis revealed a panel of 14 genes to have differential expression between the two groups: several interleukins: IL13, IL16, IL17C and IL17F, IL1A, IL3; interleukin receptors: IL10RB, IL5RA; chemokines: CXCL13 and CCL26; and cytokines: CSF2, IFNA2, OSM, TNSF13.
Conclusion
The expression levels of these genes have been previously shown to be associated with reduced immunological status; indeed, the triple-negative breast cancer patients presented with lower counts of lymphocytes and eosinophils than the ER/PR-positive ones. These results contribute to a better understanding of the possible role of antitumor immune responses in mediating the clinical outcome.
Keywords: triple-negative breast cancer, gene expression, molecular mechanism, immunological status, clinical outcome
Introduction
Breast cancer is the most prevalent cancer in women, in both developed and developing countries.1 Being a heterogeneous disease, breast tumors have been classified into four main molecular subtypes: luminal A and B, Her2+, and basal-like.2 The luminal subtype is characterized by a high expression of receptors for estrogen (ER) and progesterone (PR), while Her2+ tumors are negative for ER and PR, but express high levels of Her2. The basal-like class of breast tumors comprises the triple-negative breast cancers (TNBCs) and is defined by lack of ER, PR, and Her2 expression. Breast cancer treatment is designed on the basis of this classification, luminal tumors having a more favorable prognosis, while TNBC patients are refractory to common targeted therapies, leading to the worst overall and disease-free survival of all subtypes.3
In order to achieve the most effective treatment response, it is imperative to understand the pathogenic mechanisms of breast cancer. There is a clear link between cancer and inflammation,4 the local microenvironment been known to aid in tumor growth and progression.5 Cancer microenvironment and cross talk (tumor cells, fibroblasts, endothelial cells, and infiltrating leukocytes) are tightly regulated through cytokines, key signaling molecules5 that can determine stimulatory or suppressive tumor responses.6 Due to their pleiotropic actions, cytokines are assembled in complex signaling networks that regulate tumor proliferation, progression, and host immune responses,7 thereby affecting prognosis and response to therapy.8
Studies have shown that immune activation at baseline is associated with a higher likelihood of pathological response (reviewed in Andre et al9); therefore, in this study, we compared the systemic cytokine expression of two molecular subtypes of breast cancer patients. The patients were grouped according to the ER/PR expression on the primary tumors, ER/PR+, Her2− patients being considered as treatment responsive, and TNBC patients as nonresponsive. With the view to better characterizing the baseline immune status, we evaluated the expression levels of a panel of 84 inflammatory molecules in the peripheral blood of our patients in correlation with histological and clinical data. These results could aid not only in the prediction of chemotherapeutic efficacy or treatment response monitoring, but also in the development of strategies to specifically modulate and reverse immune defects in breast cancer patients.
Patients and methods
Patients and blood samples
For this study, we collected 40 peripheral blood samples from patients with Her2− primary breast cancer tumors diagnosed at The Oncology Institute “I. Chiricuta”, Cluj-Napoca, Romania, between 2010 and 2012. Informed consent was obtained from all participants included in the study. This study is in accordance with the ethical standards of the institute’s research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. Blood samples were collected in ethylenediaminetetraacetic acid (EDTA) anticoagulant tubes within a 4-hour interval (8 am–12 noon) before patients underwent any type of treatment: steroids, chemotherapy, or surgery. The plasma was removed and stored for protein analysis, while red blood cells were lysed and white cells were processed for RNA isolation with TriReagent (Sigma-Aldrich, St Louis, MO, USA). RNA samples were purified with RNeasy Mini Kit (Qiagen, Valencia, CA, USA), only the RNA samples with RNA integrity number (RIN) >7 were used for further analysis. At the same time, for clinical purposes, blood was collected in EDTA tubes for full blood count with Sysmex XT-4000i (Sysmex, Kobe, Japan). AJCC criteria were used for histopathological analysis and staging of the patients. The status confirmation of ER, PR, and Her2 receptors was done by immunohistochemical analysis. Table 1 comprises the patients’ clinicopathological characteristics.
Table 1.
Patients’ clinicopathological characteristics
| Number | Age (years) | Allred score
|
Clinical stage | TNM staging | Nottingham staging | Age at menopause (years) | ||
|---|---|---|---|---|---|---|---|---|
| ER | PR | Her2 | ||||||
| 1 | 58 | 0 | 0 | 0 | III | T4bN3M0 | II | 50 |
| 2 | 58 | 0 | 0 | 0 | II | T2N1M0 | II | 0 |
| 3 | 59 | 0 | 0 | 0 | III | T3N2M0 | III | N/A |
| 4 | 53 | 0 | 0 | 0 | III | T2N2M0 | II | 50 |
| 5 | 52 | 0 | 0 | 0 | II | T2N0M0 | III | 48 |
| 6 | 45 | 0 | 0 | 0 | III | T3N1M0 | III | 0 |
| 7a | 48 | 0 | 0 | 0 | II (rb) + I (lb) | T2N1M0 (rb) + T1N0M0 (lb) | III | 32 |
| 8 | 51 | 0 | 0 | 0 | III | T4bN2M0 | I | 0 |
| 9 | 49 | 0 | 0 | 0 | II | T2N1M0 | III | 0 |
| 10 | 50 | 0 | 0 | 0 | III | T4bN1M0 | II | 0 |
| 11a | 55 | 0 | 0 | 0 | III (rb) + I (lb) | T4bN2M0 (rb) + T1N0M0 (lb) | III | 51 |
| 12 | 56 | 0 | 0 | 0 | II | T2N1M0 | III | N/A |
| 13 | 59 | 0 | 0 | 0 | III | T4cN2M0 | III | 52 |
| 14 | 60 | 0 | 0 | 0 | II | T1N1MO | III | 45 |
| 15 | 35 | 0 | 0 | 0 | II | T2N1M0 | III | 0 |
| 16 | 53 | 0 | 0 | 0 | III | T2N2M0 | III | 50 |
| 17 | 59 | 0 | 0 | 0 | II | T2N1M0 | III | 46 |
| 18 | 42 | 0 | 0 | 0 | N/A | T4bN2Mx | III | 38 |
| 19 | 40 | 0 | 0 | 0 | II | T2N1M0 | III | 0 |
| 20 | 74 | 0 | 0 | 0 | III | T4bN2M0 | III | 48 |
| 21 | 56 | 0 | 0 | 0 | III | T4bN1M0 | III | 46 |
| 22 | 59 | 0 | 0 | 0 | I | T1cN0Mx | I | 49 |
| 23 | 70 | 0 | 0 | 0 | III | T4bNxMx | II | N/A |
| 24 | 50 | 6 | 4 | 0 | II | T2N1M0 | II | 50 |
| 25 | 45 | 7 | 7 | 0 | II | T2N1M0 | I | 45 |
| 26 | 52 | 8 | 8 | 0 | III | T2N2M0 | III | N/A |
| 27 | 54 | 7 | 7 | 0 | III | T2N2M0 | II | 0 |
| 28 | 65 | 8 | 7 | 0 | III | T4bN2M0 | II | 47 |
| 29 | 50 | 8 | 7 | 0 | I | T1cN0Mx | III | 45 |
| 30 | 62 | 7 | 6 | 0 | III | T4bN2M0 | II | 54 |
| 31 | 52 | 8 | 7 | 0 | III | T4bN2M0 | III | 50 |
| 32 | 62 | 4 | 6 | 0 | III | T3N1M0 | II | 52 |
| 33 | 55 | 8 | 6 | 0 | III | T3N1M0 | II | 52 |
| 34 | 68 | 3 | 7 | 0 | II | T3N0M0 | I | 50 |
| 35 | 49 | 7 | 7 | 0 | III | T3N1M0 | I | 0 |
| 36 | 43 | 7 | 8 | 0 | III | T3N1M0 | I | N/A |
| 37 | 63 | 8 | 8 | 0 | III | T4aN0Mx | II | 40 |
| 38 | 62 | 7 | 7 | 0 | N/A | N/A | II | 54 |
| 39 | 52 | 8 | 7 | 0 | II | T2N1M0 | II | 52 |
| 40 | 48 | 6 | 8 | 0 | II | T2N0M0 | II | 0 |
Note:
Patients with bilateral breast cancer.
Abbreviations: ER, estrogen; PR, progesterone; rb, right breast tumor; lb, left breast tumor; N/A, not available; TNM, tumor, lymph nodes, metastasis.
PCR array
Approximately 300 ng of the total RNA of each sample was used for cDNA synthesis using RT2 First Strand kit (SABiosciences by Qiagen, Valencia, CA, USA). The cDNA was amplified using LightCycler 480 II termocycler (Hoffmann-La Roche, Basel, Switzerland) in 96-well Human Inflammatory Cytokines & Receptors PCR Array plates (SABiosciences by Qiagen, Valencia, CA, USA). Amplification and SYBR Green detection were performed according to the manufacturer’s recommendations. The array consists of 84 primers for chemokines, cytokines, interleukins and their receptors, five housekeeping genes for sample-to-sample normalization, and several controls for reverse transcription, genomic DNA contamination, and polymerase chain reactions (PCRs). Threshold cycle (Ct) calculations were done using the automated second derivative analysis method.
Data analysis
Correlations between clinical data were calculated using the Fisher’s exact test or the chi-square test. Cell count differences were assessed by the Mann–Whitney test. The PCR array Ct raw results were imported into the Web-based software for Cataloged and Custom Arrays (SABiosciences by Qiagen, Valencia, CA, USA) for expression analysis. Of interest to us were genes with a fold regulation cutoff ±1.5 and a P-value <0.05 after the Benjamini and Hochberg correction method (*P-value <0.05, **P-value <0.01, ***P-value <0.001). The patients were grouped according to ER/PR expression on the primary tumor at diagnosis. Genes of interest and the corresponding fold regulation values were uploaded into Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood City, CA, USA) for functional analysis. Using Ingenuity Knowledge Base (IKB), the genes were assigned into networks and biological functions on the basis of hierarchical organization of right-tailed Fisher’s exact test calculations (P<0.05). Patients’ Ct raw expression levels and blood count are available upon request.
Results
ER/PR expression and clinicopathological characteristics
All the patients included in this study were diagnosed with invasive ductal breast carcinomas. Twenty-three patients did not express ER or PR on the primary tumors, classifying the patients into the TNBC phenotype. The average age at the time of diagnosis for each group was 54 years; 34% of the patients were under 50 years of age with a mean age of 45.7 years, while patients over 50 years of age had a mean age of 58.4 years. Twenty-five of the patients had reached menopause at the time of diagnosis. Approximately 46% of the patients already had grade III tumors, and almost all had positive lymph nodes, but with no detectable secondary distant tumors. The clinicopathological characteristics of the patients are presented in Table 1.
The correlations between the clinical data were done with respect to ER/PR expression in primary tumors (Table 2). No association was observed for age, clinical stage, tumor size, lymph nodes, or menopausal status. Statistical significance was observed between the tumor grading and the ER/PR status (P<0.005); more patients with ER/PR-negative tumors had grade III, while more patients with ER/PR-positive tumors had grade II.
Table 2.
Correlations between ER/PR status and patients’ clinicopathological features
| Patient characteristics | Number of patients | % | ER/PR+ Her2− | ER/PR− Her2− | P-value |
|---|---|---|---|---|---|
| All patients | 40 | 100 | 17 | 23 | |
| Age (years) | |||||
| ≤50 | 13 | 34.15 | 6 | 7 | 1.000 |
| >50 | 27 | 65.85 | 11 | 16 | |
| Clinical stage | |||||
| I | 2 | 4.87 | 1 | 1 | 0.731 |
| II | 14 | 34.15 | 5 | 9 | |
| III | 22 | 56.10 | 10 | 12 | |
| Tumor size | |||||
| T1 and T2 | 19 | 43.35 | 7 | 12 | 0.747 |
| T3 and T4 | 20 | 51.22 | 9 | 11 | |
| Lymph nodes | |||||
| N0 | 6 | 14.64 | 4 | 2 | 0.450 |
| N1 | 18 | 43.90 | 7 | 11 | |
| N2 | 13 | 34.15 | 5 | 8 | |
| N3 | 1 | 2.44 | 0 | 1 | |
| Nottingham grading | |||||
| I | 6 | 14.63 | 4 | 2 | 0.005** |
| II | 15 | 39 | 10 | 5 | |
| III | 19 | 46.34 | 3 | 16 | |
| Premenopause | 5 | 12.20 | 2 | 3 | 1.000 |
| Postmenopause | 14 | 34.15 | 6 | 7 |
Note:
Percentage <100% is attributed to missing information.
P<0.01.
Abbreviations: ER, estrogen; PR, progesterone.
ER/PR expression and blood gene profile
We profiled the expression of 84 key genes involved in the regulation of signal transduction in the inflammatory cascades as a response to immune reaction. We observed a change in expression for a panel of 14 genes between the ER/PR-positive/negative group (Table 3). All but three genes were downregulated in the blood of TNBC patients. We identified several interleukins as being of interest: IL13, IL16, IL17C and F, IL1A, IL3; interleukin receptors: IL10RB, IL5RA; chemokines: CXCL13 and CCL26; and cytokines: CSF2, IFNA2, OSM, TNSF13.
Table 3.
Gene differentially expressed in the blood of TNBC patients versus that of ER/PR+ patients
| Gene | Name | Fold regulation | 95% CI | Adj P-value |
|---|---|---|---|---|
| IL10RB | Interleukin 10 receptor, beta | 1.83 | (0.24, 3.42) | 0.012 |
| IFNA2 | Interferon, alpha 2 | 1.58 | (1.15, 2.01) | 0.038 |
| CXCL13 | Chemokine (C-X-C motif) ligand 13 | 1.52 | (1.24, 1.79) | 0.002 |
| IL17C | Interleukin 17C | −3.91 | (−12.5, −2.32) | 0.000 |
| IL17F | Interleukin 17F | −3.38 | (−12.5, −1.96) | 0.004 |
| IL13 | Interleukin 13 | −2.92 | (−12.5, −1.64) | 0.038 |
| CCL26 | Chemokine (C-C motif) ligand 26 | −2.55 | (−12.5, −1.4) | 0.023 |
| CSF2 | Colony-stimulating factor 2 (granulocyte-macrophage) | −2.49 | (0.21, 0.59) | 0.008 |
| IL3 | Interleukin 3 (colony-stimulating factor, multiple) | −2.49 | (−4.76, −1.7) | 0.011 |
| OSM | Oncostatin M | −2.42 | (−7.14, −1.45) | 0.021 |
| IL1A | Interleukin 1, alpha | −1.96 | (−3.57, −1.35) | 0.036 |
| IL16 | Interleukin 16 | −1.68 | (−3.84, −1.07) | 0.008 |
| IL5RA | Interleukin 5 receptor, alpha | −1.62 | (−2.85, −1.13) | 0.041 |
| TNFSF13 | Tumor necrosis factor (ligand) superfamily, member 13 | −1.54 | (−2.32, −1.14) | 0.025 |
Abbreviations: TNBC, triple-negative breast cancer; ER, estrogen; PR, progesterone; CI, confidence interval; Adj, adjusted.
The data set was imported into IPA software to predict the chemical, molecular, and cellular interactions between these genes in the context of cellular phenotype and disease. Based on gene expression, the software predicted that these genes mediate the inflammatory response (P=2.57×10−12–1.8×10−3) through cell-to-cell signaling and interaction (P=1.4×10−11–1.8×10−3) in the hematological system development and function (P=1.4×10−11–1.8×10−4). The main identified canonical pathway was, as expected, the cytokines’ mediated communication between immune cells (P=4.39×10−12).
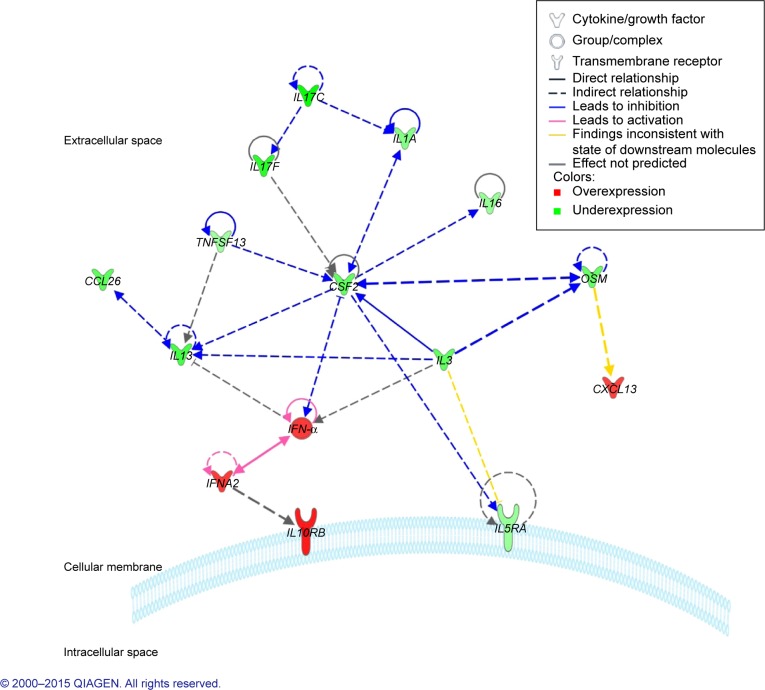
The software also generated two plausible regulatory networks, which we merged in order to get a more comprehensive view on how these genes are regulated and how they might interact with each other (Figure 1). Most of the identified molecules interact in the extracellular space and regulate each other’s expression through direct (solid lines) or indirect (dashed lines) interactions. Furthermore, they seem to have the ability to self-regulate their expression (circular arrows that originate from one molecule and pointing back at that same molecule). To ascertain whether our findings are in accordance with the previous reports in the field, we ran our gene expression results through a Molecule Activity Predictor (MAP) algorithm. We observed that most of the identified gene expression levels corresponded to predicted inhibition (blue arrows) or activation (pink arrows) by the upstream/downstream molecules. However, there were exceptions with the literature findings of OSM regulation of CXCL13 expression and IL3 regulation of IL5RA expression, which in our study seem to be the opposite of what was predicted. CSF2 seems to be a nodal molecule in our network, its expression being regulated by OSM, IL3, TNFSF13, IL1A, IL17, and IFN-α, and in turn regulates the expression of other molecules through positive feedback loops as in the case of IFN-α, OSM, and IL1A.
Figure 1.
IPA prediction of the interactive network between the differentially expressed genes in the blood of triple negative compared to ER/PR+ breast cancer patients.
Notes: Red – genes overexpressed; Green – genes underexpressed. Data were analyzed through the use of QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity).
Abbreviations: IPA, Ingenuity Pathway Analysis; ER, estrogen; PR, progesterone.
According to our results, upregulation of IFNA2 and IFN-α suggests an activation of downstream signaling cascade through IL10RB receptors, while downregulation of CSF2 expression suggests signaling repression through IL5RA receptors.
ER/PR expression and white cells count
On the basis of gene expression results, we retrospectively examined each patient’s chart for the white blood cells count at the time of blood collection. Most of the patients had cell counts within the normal limits; however, we did observe differences in the individual cell population distribution (Table 4). The TNBC group had almost two times less eosinophils than the ER/PR+ group, but the difference was not significant, although the trend is clearly evident (P=0.06). Significant differences (P<0.03) were observed for the lymphocyte counts, which were approximately 30% lower in the TNBC group compared with ER/PR+. The differences between the other cell types were negligible.
Table 4.
Patients’ differential blood count according to ER/PR expression on the primary tumors
| Cell type | Reference values | ER/PR− | ER/PR+ | P-value |
|---|---|---|---|---|
| White blood cells | 4,000–10,000/μL | 7,340±321 | 7,780±555 | 0.72 |
| Neutrophils | 2,000–8,000/μL | 4,864±273 | 4,689±345 | 0.51 |
| 45%–80% | 65.6±1.2 | 60.6±1.8 | 0.05* | |
| Eosinophils | 50–700/μL | 101±19 | 195±60 | 0.06 |
| ≤1.5% | 1.4±0.2 | 22±0.4 | 0.11 | |
| Basophils | ≤200/μL | 21±3 | 27±4 | 0.21 |
| ≤0.2% | 0.3±0 | 0.3±0 | 0.33 | |
| Lymphocytes | 1,000–4,000/μL | 1,805±71 | 2,304±230 | 0.03* |
| 20%–55% | 25.2±1.1 | 29.5±1.9 | 0.05* | |
| Monocytes | 300–1,000/μL | 545±33 | 565±48 | 0.83 |
| ≤15% | 7.5±0.3 | 7.4±0.5 | 0.78 | |
| Thrombocytes | 150,000–450,000/μL | 268,000±19,696 | 264,765±18,055 | 0.93 |
Notes:
Data presented as mean ± standard error.
P<0.05.
Abbreviations: ER, estrogen; PR, progesterone.
Discussion
Increasing evidence suggests that the immune system can influence breast tumors’ initiation, progression, and response to therapy; furthermore, the chemotherapeutic effect of some conventional drugs such as doxorubicin seems to be mediated through immunological signaling pathways.9 In a previous study,10 we observed that primary tumor molecular features induce distinct gene expression levels in the peripheral blood of breast cancer patients, genes that are known to be involved in immune signaling. Therefore, in this study, we wanted to gain a more comprehensive view of the baseline immunological status of our patients by investigating the expression profile of several cytokines, chemokines, interleukins and their receptors, as these are the direct mediators of the immune response. Expression of ER, PR, and Her2 has been previously associated with distinct patterns of cytokines stimuli;11,12 however, these studies evaluated a variable series of breast cancer cases and a limited panel of cytokines. Our study does not suffer from these limitations; we evaluated a broad panel of molecules, and our analysis was restricted to a homogeneous group of Her2− patients (ER+, PR+ vs ER, PR−).
TNBC patients have the worst outcome of all breast cancer patients, regardless of the stage at diagnosis.13 Specific clinical features include early age at onset, premenopausal status, and prevalence among racial/ethnicity population. Although no relation was observed with tumor size and node positivity,14 these patients are more likely to develop visceral metastases,15,16 suggesting that the metastatic spread occurs predominantly through hematogenous mechanisms (reviewed in Rastelli et al17). The analysis of the clinicopathological data of our patients showed that the TNBC patients enrolled in this study present clinical factors associated with poor prognosis, such as advanced tumor grade at time of diagnosis18 and a reduced number of eosinophils19 and lymphocytes.19–21 Although we did not reach statistical significance in blood eosinophils count, the difference between the two cohorts of patients is clear (P<0.06). Eosinophils can have both pro- and anti-inflammatory and immunoregulatory activities, and therefore, the role of these cells in cancer is still unclear; however, lower counts of eosinophils in breast patients have been associated with worse prognosis.19 Eosinophils production and activation requires IL-5, IL-3, and CSF2, and they also display high-affinity receptors of IL-5.22 We did find significant lower levels for IL-3, CSF2, and IL5RA gene expressions in the peripheral blood of TNBC patients, which could be responsible for the lower count of eosinophils.
Pretreatment lymphopenia has been associated not only with poor cancer survival, but also with poor response to chemotherapy and radiotherapy, thereby also suggesting predictive potential value.20,23 Lymphocytopenia was previously reported as an important predictive, prognostic, and survival parameter in metastatic breast cancer;24 however, this is the first report that specifically associates lower counts of lymphocytes with TNBC phenotype. We hypothesize that this could be one of the factors that influence the disease-free and overall survival of these patients. The lymphocyte levels indicate the cell-mediated immunity status, which is important in the host’s defense against tumors. Low counts of lymphocytes could reveal a previous immunosuppressed state, and therefore an inappropriate host response.21 Sup-pressions of lymphocyte levels might be a tumor progression mechanism,25 therefore indicating a therapeutic potential for lymphocytopenia correctors such as anti-CTLA4 antibody, IL-7 LGF, etc.24
The most downregulated cytokines in the TNBC group were IL-17C and IL-17F, which belong to the IL-17 family of cytokines and cooperate to induce tissue inflammation through T helper 17 cells (Th17). IL-17C, better known as IL-21, is produced by the lymphoid population and shows pleiotropic actions on both lymphoid and nonlymphoid cells, indicating broad functions with implications in innate and adaptive immune response, inflammation, allergies, and cancer (reviewed in Spolski and Leonard26). C > T polymorphism in the promoter region of IL-21 has been shown to control the transcriptional levels, the CT/TT genotypes having a higher transcriptional activity27 with better prognosis and higher survival probability than the CC genotype.27 Whether the incidence of CT/TT genotypes is lower in TNBC compared to ER/PR+ patients remains to be investigated, but could explain the reduced levels of IL-21 in TNBC patients. Recent reports have indicated that IL-21 is a negative regulator of Treg development,28 prevailing the Treg-mediated immunosuppression.29,30 Downregulation of IL-21 in TNBC patients’ blood might lead to increased Tregs, which have been associated with a poor prognosis in patients with early breast cancer.31 Also, IL-21 has been reported to have strong antitumor effects by increasing cytotoxicity toward cancer cells,32 including breast cancer cells,33 by directing CD8+ T-cells to infiltrate tumors.34 Reduced blood IL-21 levels in TNBC patients might be an indicator of the tumor-infiltrating lymphocytes (TILs) status (especially of those of the CD8+), which has been shown to be predictive of therapy outcome.35
Triple-negative breast tumors present high vascular endothelial growth factor (VEGF) concentrations36 and increased micrometastases that seem to be angiogenesis dependent,37 and IL-21 has been shown to inhibit tumor angiogenesis.38 Breast cancer angiogenesis has also been shown to be regulated by CSF2, which stimulates monocytes39 and macrophages40 to produce VEGFR1 that binds and inactivates VEGF. Reduced IL-21 and CSF2 levels could suggest increased angiogenesis in these patients. CSF2 also enhances macrophage antigen presentation and immune response.41 Breast cancer patients who received CSF2 showed substantial activity in controlling the disease,42 and patients treated after transplant lived longer and had fewer relapses.43
OSM is secreted in low amounts by resting circulating cells such as macrophages, neutrophils, and T lymphocytes;44,45 however, when these cells get activated, they quickly express OSM. This molecule has been shown to have pleiotropic effects: whereas it can inhibit breast cancer cells proliferation in vitro, recent studies have also shown that they effectively mediate macrophage-assisted45 cancer progression.44 OSM secretion is conditioned by cell priming with tumor cells;44,45 furthermore, in neutrophils, OSM release seems to be CSF2 dependent.44 Both cohorts of our patients have shown similar neutrophil counts concomitantly with reduced blood expression of CSF2 and OSM, suggesting that neutrophils in TNBC patients are rather in a “resting” state.
We identified several other molecules to be differentially expressed between the two groups of patients; however, the available literature regarding their role in breast cancer immune status is limited, warranting further investigations into their association with treatment response. IL-1A46 is secreted by macrophages and monocytes and has been shown to stimulate breast cancer proliferation. TNFSF13 is important for B-cell development, and IL-16 has been identified as a critical chemotactic factor that contributes to monocytes’ and macrophages’ migration toward tumor cells.47
Conclusion
Overall, our findings suggest that the evaluation of the immunologic blood profile in TNBC patients may yield important clinical information to guide breast cancer prognosis and therapy. We showed a distinct systemic cytokine profile associated with the ER/PR expression on breast tumors, results that could also explain the singular behavior of each of the tumor subtypes, allowing future immunotherapeutic interventions. Furthermore, these results could contribute to a better understanding of the possible role of antitumor immune responses in mediating the clinical outcome. The recent initiation of the first clinical trials regarding targeted T-cells immunotherapy of TNBC patients (ClinicalTrials.gov Identifier: NCT01147016, NCT02316457) attests to the relevance of the results presented.
Acknowledgments
This work was financed by the POSCCE 709/2010 grant with the title: “Clinical and economical impact of proteome and transcriptome molecular profiling in neoadjuvant therapy of triple negative breast cancer (BREASTIMPACT)”, and published under the aegis of the European Social Fund, Human Resources Development Operational Programme 2007–2013, project number: POSDRU/159/1.5/S/138776.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.American Cancer Society . Global Cancer Facts & Figures 2nd Edition. Atlanta, GA: American Cancer Society; 2011. [Google Scholar]
- 2.Guiu S, Michiels S, Andre F, et al. Molecular subclasses of breast cancer: how do we define them? The IMPAKT 2012 Working Group Statement. Ann Oncol. 2012;23(12):2997–3006. doi: 10.1093/annonc/mds586. [DOI] [PubMed] [Google Scholar]
- 3.Onitilo AA, Engel JM, Greenlee RT, Mukesh BN. Breast cancer subtypes based on ER/PR and Her2 expression: comparison of clinicopathologic features and survival. Clin Med Res. 2009;7(1–2):4–13. doi: 10.3121/cmr.2009.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357(9255):539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Wilson J, Balkwill F. The role of cytokines in the epithelial cancer microenvironment. Semin Cancer Biol. 2002;12(2):113–120. doi: 10.1006/scbi.2001.0419. [DOI] [PubMed] [Google Scholar]
- 7.Korkaya H, Liu S, Wicha MS. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Invest. 2011;121(10):3804–3809. doi: 10.1172/JCI57099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levano KS, Jung EH, Kenny PA. Breast cancer subtypes express distinct receptor repertoires for tumor-associated macrophage derived cytokines. Biochem Biophys Res Commun. 2011;411(1):107–110. doi: 10.1016/j.bbrc.2011.06.102. [DOI] [PubMed] [Google Scholar]
- 9.Andre F, Dieci MV, Dubsky P, et al. Molecular pathways: involvement of immune pathways in the therapeutic response and outcome in breast cancer. Clin Cancer Res. 2013;19(1):28–33. doi: 10.1158/1078-0432.CCR-11-2701. [DOI] [PubMed] [Google Scholar]
- 10.Tudoran O, Virtic O, Balacescu L, et al. Differential peripheral blood gene expression profile based on Her2 expression on primary tumors of breast cancer patients. PLoS One. 2014;9(7):e102764. doi: 10.1371/journal.pone.0102764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrera AC, Panis C, Victorino VJ, et al. Molecular subtype is determinant on inflammatory status and immunological profile from invasive breast cancer patients. Cancer Immunol Immunother. 2012;61(11):2193–2201. doi: 10.1007/s00262-012-1283-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muraro E, Martorelli D, Turchet E, et al. A different immunologic profile characterizes patients with HER-2-overexpressing and HER-2-negative locally advanced breast cancer: implications for immune-based therapies. Breast Cancer Res. 2011;13(6):R117. doi: 10.1186/bcr3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dent R, Trudeau M, Pritchard KI, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13(15 Pt 1):4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan HG, Malmgren JA, Atwood M. T1N0 triple negative breast cancer: risk of recurrence and adjuvant chemotherapy. Breast J. 2009;15(5):454–460. doi: 10.1111/j.1524-4741.2009.00789.x. [DOI] [PubMed] [Google Scholar]
- 15.Ihemelandu CU, Naab TJ, Mezghebe HM, et al. Basal cell-like (triple-negative) breast cancer, a predictor of distant metastasis in African American women. Am J Surg. 2008;195(2):153–158. doi: 10.1016/j.amjsurg.2007.09.033. [DOI] [PubMed] [Google Scholar]
- 16.Haffty BG, Yang Q, Reiss M, et al. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J Clin Oncol. 2006;24(36):5652–5657. doi: 10.1200/JCO.2006.06.5664. [DOI] [PubMed] [Google Scholar]
- 17.Rastelli F, Biancanelli S, Falzetta A, et al. Triple-negative breast cancer: current state of the art. Tumori. 2010;96(6):875–888. [PubMed] [Google Scholar]
- 18.Howlader N, Noone AM, Krapcho M, Garshell J, et al., editors. SEER Cancer Statistics Review, 1975–2010. Bethesda, MD: National Cancer Institute; 2013. [Google Scholar]
- 19.Ownby HE, Roi LD, Isenberg RR, Brennan MJ. Peripheral lymphocyte and eosinophil counts as indicators of prognosis in primary breast cancer. Cancer. 1983;52(1):126–130. doi: 10.1002/1097-0142(19830701)52:1<126::aid-cncr2820520123>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 20.Vicente Conesa MA, Garcia-Martinez E, Gonzalez Billalabeitia E, et al. Predictive value of peripheral blood lymphocyte count in breast cancer patients treated with primary chemotherapy. Breast. 2012;21(4):468–474. doi: 10.1016/j.breast.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Ray-Coquard I, Cropet C, Van Glabbeke M, et al. Lymphopenia as a prognostic factor for overall survival in advanced carcinomas, sarcomas, and lymphomas. Cancer Res. 2009;69(13):5383–5391. doi: 10.1158/0008-5472.CAN-08-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothenberg ME, Hogan SP. The eosinophil. Annu Rev Immunol. 2006;24:147–174. doi: 10.1146/annurev.immunol.24.021605.090720. [DOI] [PubMed] [Google Scholar]
- 23.Huang JJ, Jiang WQ, Lin TY, et al. Absolute lymphocyte count is a novel prognostic indicator in extranodal natural killer/T-cell lymphoma, nasal type. Ann Oncol. 2011;22(1):149–155. doi: 10.1093/annonc/mdq314. [DOI] [PubMed] [Google Scholar]
- 24.De Giorgi U, Mego M, Scarpi E, et al. Relationship between lymphocytopenia and circulating tumor cells as prognostic factors for overall survival in metastatic breast cancer. Clin Breast Cancer. 2012;12(4):264–269. doi: 10.1016/j.clbc.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Fogar P, Sperti C, Basso D, et al. Decreased total lymphocyte counts in pancreatic cancer: an index of adverse outcome. Pancreas. 2006;32(1):22–28. doi: 10.1097/01.mpa.0000188305.90290.50. [DOI] [PubMed] [Google Scholar]
- 26.Spolski R, Leonard WJ. The Yin and Yang of interleukin-21 in allergy, autoimmunity and cancer. Curr Opin Immunol. 2008;20(3):295–301. doi: 10.1016/j.coi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.You Y, Deng J, Zheng J, et al. IL-21 gene polymorphism is associated with the prognosis of breast cancer in Chinese populations. Breast Cancer Res Treat. 2013;137(3):893–901. doi: 10.1007/s10549-012-2401-1. [DOI] [PubMed] [Google Scholar]
- 28.Korn T, Bettelli E, Gao W, et al. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 2007;448(7152):484–487. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peluso I, Fantini MC, Fina D, et al. IL-21 counteracts the regulatory T cell-mediated suppression of human CD4+ T lymphocytes. J Immunol. 2007;178(2):732–739. doi: 10.4049/jimmunol.178.2.732. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Yee C. IL-21 mediated Foxp3 suppression leads to enhanced generation of antigen-specific CD8+ cytotoxic T lymphocytes. Blood. 2008;111(1):229–235. doi: 10.1182/blood-2007-05-089375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menetrier-Caux C, Gobert M, Caux C. Differences in tumor regulatory T-cell localization and activation status impact patient outcome. Cancer Res. 2009;69(20):7895–7898. doi: 10.1158/0008-5472.CAN-09-1642. [DOI] [PubMed] [Google Scholar]
- 32.Tian Y, Yuan C, Ma D, et al. IL-21 and IL-12 inhibit differentiation of Treg and TH17 cells and enhance cytotoxicity of peripheral blood mononuclear cells in patients with cervical cancer. Int J Gynecol Cancer. 2011;21(9):1672–1678. doi: 10.1097/IGC.0b013e3182358955. [DOI] [PubMed] [Google Scholar]
- 33.Park YK, Shin DJ, Cho D, et al. Interleukin-21 increases direct cytotoxicity and IFN-gamma production of ex vivo expanded NK cells towards breast cancer cells. Anticancer Res. 2012;32(3):839–846. [PubMed] [Google Scholar]
- 34.Sondergaard H, Frederiksen KS, Thygesen P, et al. Interleukin 21 therapy increases the density of tumor infiltrating CD8+ T cells and inhibits the growth of syngeneic tumors. Cancer Immunol Immunother. 2007;56(9):1417–1428. doi: 10.1007/s00262-007-0285-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.West NR, Milne K, Truong PT, Macpherson N, Nelson BH, Watson PH. Tumor-infiltrating lymphocytes predict response to anthracycline-based chemotherapy in estrogen receptor-negative breast cancer. Breast Cancer Res. 2011;13(6):R126. doi: 10.1186/bcr3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foekens JA, Peters HA, Grebenchtchikov N, et al. High tumor levels of vascular endothelial growth factor predict poor response to systemic therapy in advanced breast cancer. Cancer Res. 2001;61(14):5407–5414. [PubMed] [Google Scholar]
- 37.Holmgren L, O’Reilly MS, Folkman J. Dormancy of micrometastases: balanced proliferation and apoptosis in the presence of angiogenesis suppression. Nat Med. 1995;1(2):149–153. doi: 10.1038/nm0295-149. [DOI] [PubMed] [Google Scholar]
- 38.Castermans K, Tabruyn SP, Zeng R, et al. Angiostatic activity of the antitumor cytokine interleukin-21. Blood. 2008;112(13):4940–4947. doi: 10.1182/blood-2007-09-113878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eubank TD, Roberts R, Galloway M, Wang Y, Cohn DE, Marsh CB. GM-CSF induces expression of soluble VEGF receptor-1 from human monocytes and inhibits angiogenesis in mice. Immunity. 2004;21(6):831–842. doi: 10.1016/j.immuni.2004.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eubank TD, Roberts RD, Khan M, et al. Granulocyte macrophage colony-stimulating factor inhibits breast cancer growth and metastasis by invoking an anti-angiogenic program in tumor-educated macrophages. Cancer Res. 2009;69(5):2133–2140. doi: 10.1158/0008-5472.CAN-08-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Armstrong CA, Botella R, Galloway TH, et al. Antitumor effects of granulocyte-macrophage colony-stimulating factor production by melanoma cells. Cancer Res. 1996;56(9):2191–2198. [PubMed] [Google Scholar]
- 42.Kurbacher CM, Kurbacher JA, Cramer EM, et al. Continuous low-dose GM-CSF as salvage therapy in refractory recurrent breast or female genital tract carcinoma. Oncology (Williston Park) 2005;19(4 Suppl 2):23–26. [PubMed] [Google Scholar]
- 43.Kuzhan O, Ozet A, Ulutin C, et al. Survival benefit with GM-CSF use after high-dose chemotherapy in high-risk breast cancer. Tumori. 2007;93(6):550–556. doi: 10.1177/030089160709300606. [DOI] [PubMed] [Google Scholar]
- 44.Queen MM, Ryan RE, Holzer RG, Keller-Peck CR, Jorcyk CL. Breast cancer cells stimulate neutrophils to produce oncostatin M: potential implications for tumor progression. Cancer Res. 2005;65(19):8896–8904. doi: 10.1158/0008-5472.CAN-05-1734. [DOI] [PubMed] [Google Scholar]
- 45.Vlaicu P, Mertins P, Mayr T, et al. Monocytes/macrophages support mammary tumor invasivity by co-secreting lineage-specific EGFR ligands and a STAT3 activator. BMC Cancer. 2013;13:197. doi: 10.1186/1471-2407-13-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicolini A, Carpi A, Rossi G. Cytokines in breast cancer. Cytokine Growth Factor Rev. 2006;17(5):325–337. doi: 10.1016/j.cytogfr.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Milke L, Schulz K, Weigert A, Sha W, Schmid T, Brune B. Depletion of tristetraprolin in breast cancer cells increases interleukin-16 expression and promotes tumor infiltration with monocytes/macrophages. Carcinogenesis. 2013;34(4):850–857. doi: 10.1093/carcin/bgs387. [DOI] [PubMed] [Google Scholar]