Abstract
Hepatic steatosis is characterized by the accumulation of lipid droplets (LDs), which are composed of a neutral lipid core surrounded by a phospholipid monolayer embedded with many proteins. Although the LD-associated proteome has been investigated in multiple tissues and organisms, the dynamic changes in the murine LD-associated proteome in response to obesity and hepatic steatosis have not been studied. We characterized the hepatic LD-associated proteome of C57BL/6J male mouse livers following high-fat feeding using isobaric tagging for relative and absolute quantification. Of the 1,520 proteins identified with a 5% local false discovery rate, we report a total of 48 proteins that were increased and 52 proteins that were decreased on LDs in response to high-fat feeding. Most notably, ribosomal and endoplasmic reticulum proteins were increased and extracellular and cytosolic proteins were decreased in response to high-fat feeding. Additionally, many proteins involved in fatty acid catabolism or xenobiotic metabolism were enriched in the LD fraction following high-fat feeding. In contrast, proteins involved in glucose metabolism and liver X receptor or retinoid X receptor activation were decreased on LDs of high-fat-fed mice. This study provides insights into unique biological functions of hepatic LDs under normal and steatotic conditions.
Keywords: β-oxidation, liver, nutrition, obesity, proteomics
Non-alcoholic fatty liver disease (NAFLD), which is characterized by hepatic steatosis, is considered the hepatic component of metabolic syndrome. NAFLD is the most common chronic liver disease in Western nations and commonly occurs with comorbidities such as obesity and type 2 diabetes (1, 2). In support of a causative role for NAFLD, subjects with NAFLD have an increased risk of developing type 2 diabetes, cardiovascular disease, and liver cancer (3–5). Thus, understanding the pathobiology of NAFLD is critical for the prevention and treatment of this disease and its comorbidities.
During hepatic steatosis, excess lipids accumulate in lipid droplets (LDs), which are composed of a neutral lipid core of mainly TG and cholesterol esters surrounded by a phospholipid monolayer (6). Hepatic LDs are dynamic organelles that change in size and number in response to acute perturbations, such as fasting/feeding, and to chronic diseases, such as obesity. LDs play a role in protein quality control, protein storage, cell signaling, and viral replication, in addition to their major role as lipid metabolism mediators (7). The dynamic nature of protein association with LDs is not fully understood (8). Known LD proteins have diverse targeting mechanisms, from hydrophobic protein segments to protein-protein interactions to covalent lipid modifications (7) and some proteins dynamically shuttle to and from LDs (9–11). Additionally, LDs directly interact with numerous organelles and in some cases these interactions can result in protein transfer (12, 13). Recently, the human hepatic LD proteome has been described for the first time in NAFLD subjects (14). A few studies have examined the murine hepatic LD proteome (11, 15); however, changes in the LD proteome have not been quantified in a chronic animal model of steatosis. In the present study, we have quantified the hepatic LD-associated proteome and characterized its changes in response to diet-induced development of hepatic steatosis. The findings from this study further our understanding of LD biology and the dynamic nature of the LD-associated proteome.
MATERIALS AND METHODS
Animals and diets
Male C57BL/6J weaned mice (n = 3) were fed a 60% fat diet [high-fat diet (HFD), F3282; Bio-Serv] or a normal fat diet [normal diet (ND), F4031; Bio-Serv] for 9 weeks. The liver was excised from fed anesthetized mice at approximately 9:00 AM and a portion was reserved for microscopy prior to LD isolation. All procedures were approved by the University of Minnesota Institutional Animal Care and Use Committee.
LD isolation
LDs were isolated from freshly excised livers using the method of Zhang et al. (16) with the following modifications. The minced liver was resuspended in 4 ml buffer A [25 mM tricene (pH 7.6), 250 mM sucrose, and protease inhibitor cocktail] and dounce homogenized 30 times on ice. Cells were homogenized using a needle and syringe instead of a nitrogen bomb and then centrifuged for 10 min at 1,000 g. Following addition of 4 ml of buffer A, the postnuclear supernatant was divided into two ultracentrifuge tubes, overlaid with 3 ml buffer B [20 mM HEPES (pH 7.4), 100 mM KCl, 2 mM MgCl2, and protease inhibitor cocktail] and centrifuged at 300,000 g for 1 h at 4°C. The LD-containing band was transferred to a fresh tube and centrifuged at 20,000 g for 20 min at 4°C. The underlying liquid was carefully removed and the LD fraction was washed six times with 200 μl buffer B to remove copurifying membranes (16, 17) and acetone precipitated overnight.
Protein extraction
The LD precipitate was reconstituted with 65 μl of protein solubilization buffer [7 M urea, 2 M thiourea, 0.4 M triethylammonium bicarbonate (pH 8.5), 20% methanol, and 4 mM tris(2-carboxyethyl)phosphine]. The samples were bath sonicated for 2 min. The samples were then transferred to a pressure cycling technology tube with a 50 μl cap for the Barocycler NEP2320 (Pressure Biosciences, Inc.) and cycled between 35 kpsi for 30 s and 0 kpsi for 15 s for 40 cycles at 37°C. Two hundred millimoles of methyl methanethiosulfonate were added to a final concentration of 8 mM. Protein concentration was determined by Bradford assay.
In-solution proteolytic digestion and iTRAQ® labeling
A 28 μg aliquot of each sample was transferred to a new 1.5 ml microfuge tube and brought to the same volume with protein solubilization buffer plus 8 mM methyl methanethiosulfonate. All samples were diluted 4-fold with 80% ultra-pure water; 20% methanol and trypsin (Promega) were added in a 1:35 ratio of trypsin to total protein. Samples were incubated overnight for 16 h at 37°C after which they were frozen at −80°C for 0.5 h and dried in a vacuum centrifuge. Subsequently, samples were cleaned with a 4 ml Extract Clean™ C18 SPE cartridge (Grace-Davidson) and eluates were vacuum dried and resuspended in 0.5 M triethylammonium bicarbonate (pH 8.5) to a final 1 μg/μl concentration. Twenty-six micrograms of each sample were labeled with isobaric tagging for relative and absolute quantification (iTRAQ)® 8-plex reagent (AB Sciex, Foster City, CA). After labeling, the samples were multiplexed together and dried in vacuo. The multiplexed sample was cleaned with a 4 ml Extract Clean™ C18 SPE cartridge, and the eluate was dried in vacuo.
Peptide LC fractionation and MS
The iTRAQ®-labeled sample was resuspended in buffer A [10 mM ammonium formate (pH 10) in 98:2 water:acetonitrile] and fractionated offline by high pH C18 reversed-phase chromatography. A MAGIC 2002 HPLC system (Michrom BioResources, Inc.) was used with a C18 Gemini-NX column [150 mm × 2 mm internal diameter, 5 μm particle, 110 Å pore size (Phenomenex)]. The flow rate was 150 μl/min with a gradient from 0 to 35% buffer B [10 mM ammonium formate (pH 10) in 10:90 water:acetonitrile] over 60 min, followed by 35–60% over 5 min. Fractions were collected every 2 min and UV absorbances were monitored at 215 nm and 280 nm. Peptide-containing fractions were divided into two equal numbered groups, “early” and “late”. The first early fraction was concatenated with the first late fraction, and so on (18). Concatenated samples were dried in vacuo, resuspended in load solvent (98:2:0.01, water:acetonitrile:formic acid), and 1.5 μg aliquots were run on a Velos Orbitrap mass spectrometer (Thermo Fisher Scientific, Inc.) as described previously, with the exception that the activation energy was 40 ms (19).
Database searching, protein identification, and quantitation
The mass spectrometer RAW data (ProteoWizard files) were converted to mzXML using MSconvert software and to MGF files using TINT RAW-to-MGF converter (http://github.com/jmchilton/tint). ProteinPilot 4.5 (AB Sciex) searches were performed against the NCBI reference sequence Mus musculus (taxon 10090; April 21, 2012 version) protein FASTA database with canonical and isoform sequences (85,763 proteins), to which a contaminant database (http://www.thegpm.org/crap/) was appended. Search parameters were as follows: cysteine MMTS; iTRAQ 8plex (peptide labeled); trypsin; instrument Orbi MS (1–3 ppm) Orbi MS/MS; biological modifications ID focus; thorough search effort; and false discovery rate analysis (with reversed database). Proteins were accepted if they were identified by two or more unique peptides and fell within the 5% local false discovery rate cutoff in all of the biological replicates. Relative quantification of each protein was performed using ProteinPilot 4.5 (AB Sciex) with bias correction. Each ND sample (tag 113–115) was used as a denominator and protein summary results were exported to Microsoft Excel. The HFD/ND ratios and the corresponding P values from each report were combined, yielding nine total HFD/ND ratios. A subset of proteins was generated with the following criteria: Protein Pilot P values <0.05 for six out of nine protein ratios and a log fold change of ≥0.55 or less than or equal to −0.55, and two times the standard deviation of the nine ratios was not greater than the average log fold change.
Microscopy and immunoblotting
Formalin-fixed paraffin-embedded liver sections were processed for immunohistochemistry using standard methodologies. Slides were examined using a Nikon A1 spectral confocal microscope and NIS Elements imaging software. The nucleus was stained with DAPI and perilipin 2 (PLIN2) was immunolabeled with chicken polyclonal primary antibody (Abcam, Cambridge, MA; ab37516) and Rhodamine Red-X-conjugated donkey anti-chicken secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA). Acyl-CoA synthetase 1 (ACSL1) was immunolabeled with goat primary antibody (Santa Cruz Biotechnology) and Alexa-Fluor 647-conjugated donkey anti-goat secondary antibody (Jackson ImmunoResearch Laboratories). Similarly, carnitine palmitoyltransferase 2 (CPT2), glucose phosphate isomerase (GPI), malic enzyme (ME), and ribosomal protein large P0 (RPLP0) were labeled with specific primary antibodies (One World Lab, San Diego, CA) and Alexa-Fluor 488 anti-rabbit secondary antibody (Cell Signaling). Voltage-dependent anion channel was labeled with a specific primary antibody (Cell Signaling) and Alexa-Fluor 488 anti-rabbit secondary antibody. Images were obtained with a 60× magnification under oil immersion. For immunoblotting, equal amounts of protein, verified by Ponceau staining, were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membranes for Western blot analysis. Primary antibodies used were as follows: anti-ATP-citrate lyase (ATPCL), anti-binding immunoglobulin protein (BIP), anti-cyclooxygenase IV (COX IV) (Cell Signaling Technology), anti-β-actin (LI-COR Biosciences), anti- PLIN2 (kindly provided by Dr. Andrew Greenberg), anti-early endosomal antigen 1 (Bethel Laboratories), and anti-catalase and anti-anexin-2 (One World Lab).
For electron microscopy, livers of mice fed ND or HFD were perfused with 2.5% glutaraldehyde in 0.1 M sodium cacodylate. Tissues were subsequently washed, sectioned, and stained using standard methods (20). Sections were observed under a JEOL 1200 EX II transmission electron microscope (JEOL Ltd., Tokyo, Japan). Images were obtained using a Veleta 2K×2K camera with iTEM software (Olympus SIS, Munster, Germany). For quantitative analysis, images from 20 cells were randomly selected from each mouse (two mice per dietary group). LDs, mitochondria, and their physical interactions were subsequently quantified.
Bioinformatics analysis
Ingenuity Pathway Analysis (IPA) (QIAGEN) was used to assign enriched biological function and canonical pathway categories. The analysis settings were restricted to experimentally observed and hepatoma cell lines or liver. Suspension trapping (STRAP) was used for cellular component Gene Ontology (GO) term protein annotation (21). Other GO term annotation was retrieved using QuickGO from the UniProt Consortium GO annotation dataset (22, 23). Protein identifier conversion and retrieval of UniProt Reference Clusters 50 (UniRef50) were performed using UniProt (24). Ortholog identification was performed using the Drosophila RNAi Screening Center Integrative Ortholog Prediction Tool (25). KEGG pathway enrichment and protein identifier conversion were completed using Database for Annotation, Visualization, and Integrated Discovery (DAVID) (26–28). The network was constructed using the ClueGO plugin (29) and Cytoscape 3.0.2 (30).
RESULTS
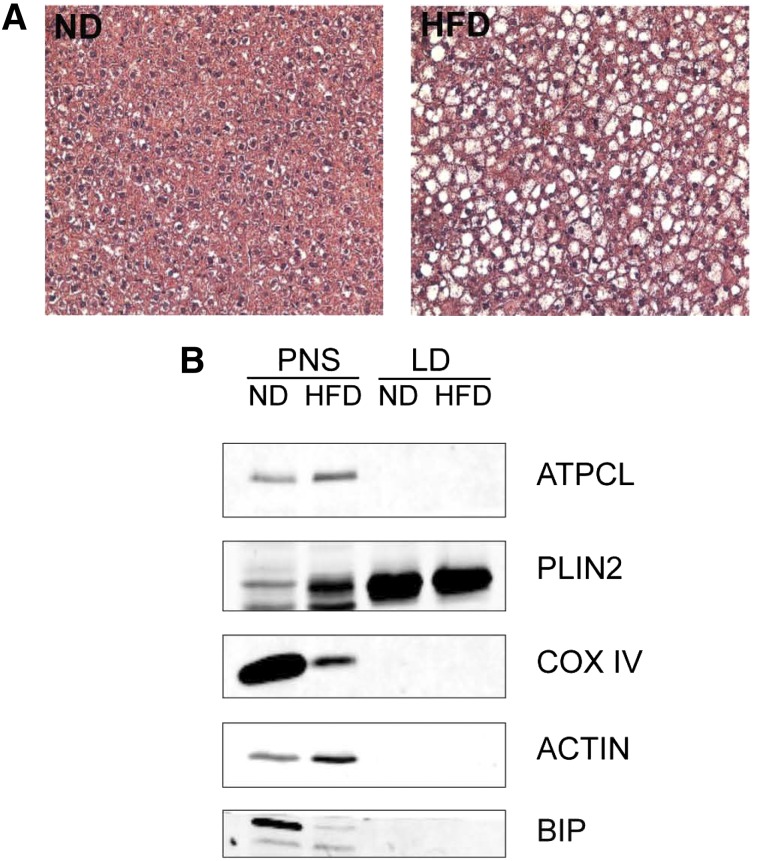
To identify the hepatic LD-associated proteome, LDs were isolated from freshly excised livers of C57BL/6J mice that had been fed either an ND or HFD for 9 weeks. Hematoxylin and eosin-stained liver sections (Fig. 1A) show that the HFD-fed mice accumulated more LDs than mice fed the ND, as expected. Postnuclear supernatant and LD fractions were evaluated for purity based on the presence of common copurifying organelles using Western blotting of marker proteins (Fig. 1B, supplementary Fig. 1). These data show that the LD fraction is highly enriched for the LD marker, PLIN2, while we were unable to detect common endoplasmic reticulum (ER) (BIP), mitochondrial (COX IV), cytosolic (ATPCL), plasma membrane (annexin-2), and endosome (early endosomal antigen 1) contaminating proteins. However, the peroxisomal marker protein, catalase, was not depleted in the LD fraction. Although the LD fraction is largely free of contaminant proteins, the sucrose centrifugation method to isolate LD does yield some contaminant proteins, as has been reported in other LD proteomic studies (14–16).
Fig. 1.
Liver morphology and purification of hepatic LDs. A: Hematoxylin and eosin-stained liver sections from mice fed the ND or HFD. B: Equal amounts of protein from the postnuclear supernatant (PNS) and LD fractions were analyzed by Western blotting with antibodies against protein markers of LD (PLIN2) or common contaminating organelles such as ER (BIP), mitochondria (COX IV), and cytosol (ATPCL).
Following LC-MS/MS analysis, we identified 1,520 LD-associated proteins (supplementary Table 1). The proteins identified through this method are proteins that are associated with the LD fraction and are not necessarily bona-fide LD proteins confirmed to reside primarily or exclusively on the LD and, thus, will be referred to as LD-associated proteins. For comparison, a list of the LD-associated proteins identified by previous proteomic studies of mammalian cells and tissues was compiled (10, 11, 15, 16, 31–42). To decrease the likelihood of similar isoforms preventing a protein match, we used the UniRef50, which merges proteins of similar sequence to one UniRef entry. We compared the UniRef50 and gene symbol identifiers between the previously identified LD proteins and our protein list (24). From this analysis, we determined that 671 out of 1,520 proteins identified have been reported in previous mammalian LD proteomics studies. In addition, 72 and 95% of the proteins found in previous studies on human (14) and mouse (15) hepatic LDs, respectively, were identified in our analysis (supplementary Table 1). Using confocal microscopy, we show that three of these proteins, RPLP0, GPI, and ME1, show partial colocalization with the LD marker protein, PLIN2, in mouse liver tissue sections (supplementary Fig. 2). It should be noted that these proteins do not primarily localize to LDs and the small amount of colocalization would suggest that these, and perhaps many other proteins identified, are not abundant LD-associated proteins.
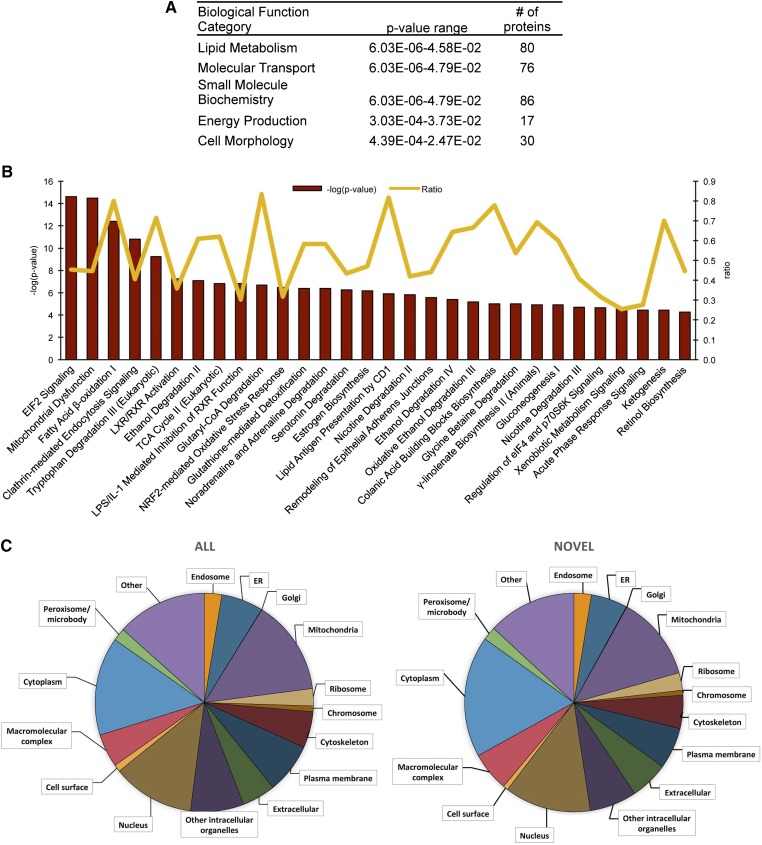
We next performed functional annotation of the hepatic LD-associated proteome. The top five enriched biological functions categories of LD-associated proteins, as determined by IPA, are listed in Fig. 2A. Within the functional categories of lipid metabolism, molecular transport, and small molecule biochemistry, the most highly enriched biological function annotations were related to lipid metabolism, as expected, and are as follows: 1) concentration of TG, acylglycerol, and lipid; 2) transport of molecules, fatty acid, and palmitic acid; 3) secretion of molecules and lipid; and 4) oxidation of lipid and fatty acid. Similar to the IPA analysis, the major enriched molecular functions by DAVID GO term annotation are oxidation-reduction, generation of precursor metabolites and energy, and protein localization (data not shown).
Fig. 2.
Enriched pathway analysis of hepatic LD-associated proteome. A: Top five enriched biological function categories as assigned by IPA. The P value range represents the lowest and highest enrichment within the subcategories. B: The top thirty enriched canonical pathways as assigned by IPA. The pathways are ranked left to right by – log (P value). The line graph represents the ratio of proteins from our dataset that map to the pathway divided by the total number of molecules in the same pathway. Pathways with fewer than three mapped proteins were discarded. C: The distribution of all 1,520 LD-associated proteins (ALL) versus the 849 novel mammalian LD-associated proteins (NOVEL) in a subset of GO terms as analyzed using STRAP.
We also performed a canonical pathway analysis using IPA and show the top 30 enriched canonical pathways (Fig. 2B). The most significantly enriched pathways are eIF2 signaling, mitochondrial dysfunction, fatty acid β-oxidation I, and clathrin-mediated endocytosis signaling. Because our dataset identified more LD-associated proteins than previous mammalian LD proteomic datasets, we were unsure whether the same pattern of organelle interactions would be observed. We used a GO slim assignment software to assign cellular component GO terms to both the novel and previously identified proteins from our dataset (21). The proportions of the cellular components are nearly identical to previous studies (Fig. 2C), suggesting that the increased number of proteins we identified was not due to new organelle associations.
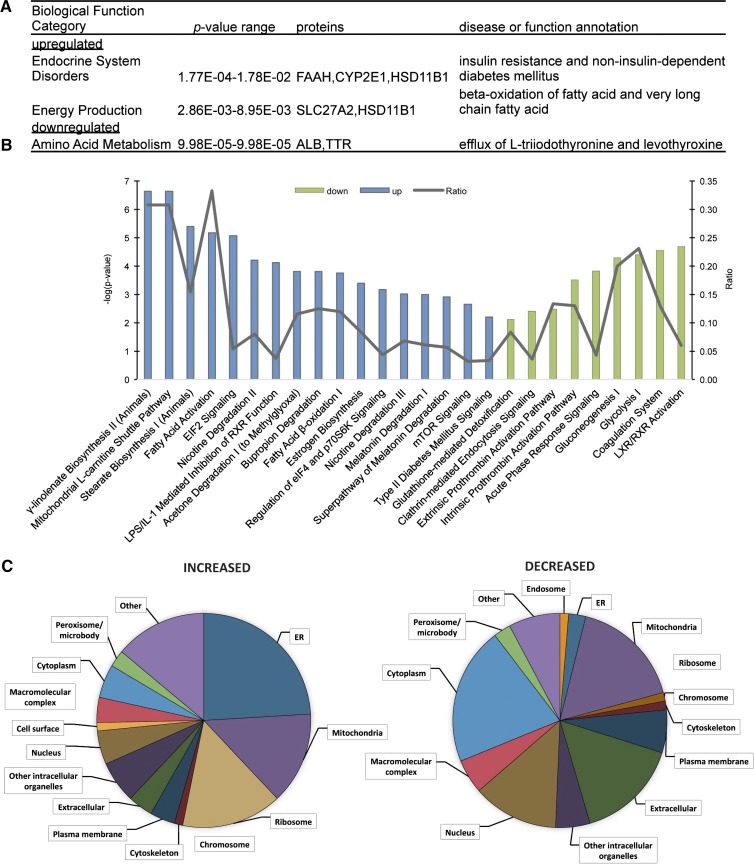
After filtering for statistical significance, 48 proteins were increased and 52 proteins decreased in response to high-fat feeding and development of hepatic steatosis (Table 1). Of the proteins that were increased, eight were also increased on LDs in humans with NAFLD (Table 1, supplementary Table 1) (14). IPA was used to identify biological function category enrichment (Fig. 3A). The increased functional categories of endocrine system disorders and energy production are consistent with the insulin resistance and altered lipid metabolism that characterize high-fat-fed mice. Canonical pathway analysis using IPA (Fig. 3B, Table 2) revealed that the most significantly enriched pathways for the increased proteins are those involved in fatty acid metabolism. Numerous LD-associated proteins were also decreased in pathways involving liver X and retinoic acid receptor activation, coagulation and prothrombin activation pathways, and glucose metabolism and clathrin-mediated endocytosis. As in the entire dataset, we used a GO slim assignment software to assign cellular component GO terms to the regulated proteins (Fig. 3C, Table 3) (21). A greater proportion of ER and ribosomal proteins are in the increased dataset; whereas, a greater proportion of extracellular and cytosolic proteins were decreased in response to high-fat feeding.
TABLE 1.
Hepatic LD-associated proteins regulated by the HFD
| Name | Accession | Symbol | Log Change | SD |
| Apolipoprotein A-IV | P06728 | Apoa4 | 1.7 | 0.1 |
| Apoc2 protein | Q3UJG0 | Apoc2 | 1.5 | 0.4 |
| Cytochrome P450 2D10 | P24456 | Cyp2d10 | 1.2 | 0.2 |
| Putative uncharacterized protein | Q3UNW2 | Cyp2d9 | 1.0 | 0.2 |
| Acyl-CoA dehydrogenase, very long chaina | B1AR27 | Acadvl | 1.0 | 0.3 |
| Cytochrome b-5 | Q544Z9 | Cyb5 | 1.0 | 0.2 |
| Long chain fatty acid-CoA ligase 1a | D3Z041 | Acsl1 | 0.9 | 0.4 |
| Cytochrome P450 2D26 | Q8CIM7 | Cyp2d26 | 0.9 | 0.4 |
| Putative uncharacterized protein | Q3TXJ7 | Bax | 0.9 | 0.2 |
| CYP3A13 | Q3UW87 | Cyp3a13 | 0.9 | 0.3 |
| Putative uncharacterized protein | Q9DBZ6 | Abcd3 | 0.8 | 0.3 |
| Monoglyceride lipasea | H7BXA7 | Mgll | 0.8 | 0.4 |
| Cytochrome c oxidase subunit 6B1 | P56391 | Cox6b1 | 0.8 | 0.2 |
| 40S ribosomal protein S19 | D3Z722 | Rps19 | 0.8 | 0.3 |
| Carnitine palmitoyltransferase 2, mitochondriala | P52825 | CPTase | 0.8 | 0.2 |
| MCG11809 | D3Z3K1 | Gm10288 | 0.8 | 0.3 |
| Calnexin | Q5SUC3 | Canx | 0.8 | 0.2 |
| Dimethylaniline monooxygenase (N-oxide-forming) 5a | P97872 | Fmo5 | 0.7 | 0.3 |
| MCG21688 | Q5M9K7 | Rps10 | 0.7 | 0.3 |
| Very long chain acyl-CoA synthetase | O35488 | Slc27a2 | 0.7 | 0.2 |
| Epoxide hydrolase 1a | Q9D379 | Ephx1 | 0.7 | 0.2 |
| Cytochrome P450 2E1a | Q05421 | Cyp2e1 | 0.7 | 0.3 |
| Cytochrome P450 2F2 | P33267 | Cyp2f2 | 0.7 | 0.2 |
| Erlin-2 | Q8BFZ9 | Erlin2 | 0.7 | 0.2 |
| Ribosomal protein, large, P0 | Q5M8R8 | Rplp0 | 0.7 | 0.3 |
| 60S ribosomal protein L27 | Q5BLJ9 | Rpl27 | 0.7 | 0.2 |
| Putative uncharacterized protein | Q3UAC2 | FTE1 | 0.7 | 0.2 |
| Putative uncharacterized protein | Q3TXS9 | Rps2 | 0.7 | 0.3 |
| Putative uncharacterized protein | Q8BT90 | Rps17 | 0.7 | 0.3 |
| 11B-hydroxysteroid dehydrogenase type 1 | Q4JHD9 | Hsd11b1 | 0.7 | 0.2 |
| Isoform 2 of pyridoxal-dependent decarboxylase domain-containing protein 1 | Q99K01-2 | Pdxdc1 | 0.6 | 0.1 |
| Ribosomal protein L23 | A2A6F9 | Rpl23 | 0.6 | 0.3 |
| MCG22987 | Q059I1 | Rps28 | 0.6 | 0.2 |
| Putative uncharacterized protein | Q3U944 | Lman1 | 0.6 | 0.3 |
| Bile acyl-CoA synthetasea | Q4LDG0 | Slc27a5 | 0.6 | 0.2 |
| ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | Q5DTI2 | Atp2a2 | 0.6 | 0.3 |
| Timm44 protein | Q8R3X4 | Timm44 | 0.6 | 0.2 |
| Cytoplasmic dynein 1 heavy chain 1 | Q9JHU4 | Dync1h1 | 0.6 | 0.2 |
| Uncharacterized protein | Q3U505 | Rpn2 | 0.6 | 0.2 |
| Putative uncharacterized protein | Q8BMR3 | Rpn1 | 0.6 | 0.2 |
| ADP-ribosylation factor GTPase-activating protein 1 | Q9EPJ9 | Arfgap1 | 0.6 | 0.3 |
| Ribosomal protein S7 | Q4FZE6 | Rps7 | 0.6 | 0.3 |
| Putative uncharacterized protein | Q3TWY6 | Cog1 | 0.6 | 0.3 |
| Coatomer subunit delta | Q5XJY5 | Arcn1 | 0.6 | 0.1 |
| Putative uncharacterized protein | Q8C541 | Faah | 0.6 | 0.3 |
| Isoform 2 of dynamin-like 120 kDa protein, mitochondrial | P58281-2 | Opa1 | 0.6 | 0.2 |
| Activator of 90 kDa heat shock protein ATPase homolog 1 | Q8BK64 | Ahsa1 | 0.5 | 0.2 |
| Gasdermin-D | Q9D8T2 | DF5L | 0.5 | 0.2 |
| Coagulation factor II | Q3TJ94 | F2 | −0.6 | 0.2 |
| Stress-induced-phosphoprotein 1 | Q60864 | Stip1 | −0.6 | 0.3 |
| Heterogeneous nuclear ribonucleoprotein F | Q9Z2X1 | Hnrnpf | −0.6 | 0.2 |
| Serotransferrin | Q921I1 | TFQTL1 | −0.6 | 0.2 |
| NSFL1 (P97) cofactor (P47) | Q3UVN5 | Nsfl1c | −0.6 | 0.3 |
| 1,4-α-Glucan-branching enzyme | Q9D6Y9 | Gbe1 | −0.6 | 0.3 |
| Alpha-1-antitrypsin 1-4 | Q00897 | Serpina1d | −0.7 | 0.1 |
| Antithrombin | Q543J5 | Serpinc1 | −0.7 | 0.2 |
| Hemopexin | Q91X72 | Hpx | −0.7 | 0.2 |
| Carboxylesterase 6 | Q8QZR3 | Ces2a | −0.7 | 0.1 |
| ATPCL | Q3V117 | Acly | −0.7 | 0.3 |
| Pyruvate kinase | Q3UEI4 | Pklr | −0.7 | 0.3 |
| 6-phosphogluconolactonase | Q9CQ60 | Pgls | −0.7 | 0.4 |
| Glutathione synthetase | Q541E2 | Gss | −0.7 | 0.4 |
| Putative uncharacterized protein | Q3U630 | Tars | −0.7 | 0.3 |
| Putative uncharacterized protein | Q3TW74 | Mthfd1 | −0.7 | 0.4 |
| Bifunctional purine biosynthesis protein PURH | Q9CWJ9 | Atic | −0.8 | 0.4 |
| Thyroid hormone responsive SP | Q543J4 | Lpgp | −0.8 | 0.4 |
| Dihydropyrimidinase | Q9EQF5 | Dpys | −0.8 | 0.4 |
| Ubiquitin carboxyl-terminal hydrolase | Q3U4W8 | Usp5 | −0.8 | 0.4 |
| Putative uncharacterized protein | Q3UGC0 | Eif4b | −0.8 | 0.4 |
| Hydroxyacyl glutathione hydrolase | G5E8T9 | Hagh | −0.8 | 0.3 |
| Mitochondrial peptide methionine sulfoxide reductase | Q9D6Y7 | Msra | −0.8 | 0.4 |
| Fatty acid binding protein 2, intestinal | Q53YP5 | Fabp2 | −0.8 | 0.3 |
| Carbohydrate kinase-like | B2KG58 | Shpk | −0.8 | 0.4 |
| Phytanoyl-CoA dioxygenase domain-containing protein 1 | D6RI61 | Phyhd1 | −0.9 | 0.4 |
| N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | Q9CWS0 | Ddah1 | −0.9 | 0.4 |
| Serine protease inhibitor A3K | P07759 | Serpina3k | −0.9 | 0.2 |
| Isoform LMW of kininogen-1 | O08677-2 | Kng1 | −0.9 | 0.2 |
| Transthyretin | Q5M9K1 | Ttr | −0.9 | 0.3 |
| Pyridoxine-5′-phosphate oxidase | Q91XF0 | Pnpo | −0.9 | 0.4 |
| Ester hydrolase C11orf54 homolog | Q91V76 | PTD012 | −0.9 | 0.4 |
| Phosphoglycerate mutase | Q3U7Z6 | Pgam1 | −1.0 | 0.4 |
| Selenium-binding protein 1 | P17563 | Selenbp1 | −1.0 | 0.5 |
| Acetoacetyl-CoA synthetase | Q9D2R0 | Aacs | −1.0 | 0.4 |
| Glucose-6-phosphate isomerase | P06745 | Gpi | −1.0 | 0.5 |
| Albumin 1 | Q546G4 | Alb | −1.0 | 0.3 |
| Malic enzyme | Q3TQP6 | Me1 | −1.0 | 0.5 |
| Dicarbonyl L-xylulose reductase | A2AC15 | Dcxr | −1.0 | 0.4 |
| Cytosolic 10-formyltetrahydrofolate dehydrogenase | Q8R0Y6 | Aldh1l1 | −1.0 | 0.5 |
| Cytoplasmic aconitase | Q8VDC3 | Aco1 | −1.0 | 0.5 |
| Carboxylesterase 1C | P23953 | Ces1c | −1.0 | 0.2 |
| MCG19655 | B1AXW7 | Prdx1 | −1.0 | 0.5 |
| Carboxymethylenebutenolidase homolog | Q8R1G2 | Cmbl | −1.0 | 0.3 |
| Peroxiredoxin-2 | Q61171 | Prdx2 | −1.0 | 0.5 |
| Glutathione S-transferase omega-1 | O09131 | Gsto1 | −1.1 | 0.6 |
| Phenazine biosynthesis-like domain-containing protein 1 | Q9DCG6 | Pbld1 | −1.1 | 0.6 |
| Glutathione S-transferase theta 3 | Q99L20 | Gstt3 | −1.1 | 0.6 |
| Farnesyl diphosphate synthetase | Q5M8R9 | Fdps | −1.2 | 0.6 |
| Superoxide dismutase (Cu-Zn) | P08228 | Sod1 | −1.2 | 0.6 |
| Carbonic anhydrase 3 | P16015 | Car3 | −1.3 | 0.5 |
| MCG12748 | B9EK13 | Urad | −1.4 | 0.3 |
| D-dopachrome tautomerase | Q3UNI8 | Ddt | −1.7 | 0.8 |
We identified 48 increased and 52 decreased LD-associated proteins that met our confidence criteria. Average log-fold change and SD are shown for each regulated protein.
Proteins that were also enriched on LDs in human subjects with NAFLD (10).
Fig. 3.
Enriched pathway analysis of hepatic LD-associated proteome regulated by diet. A: Enriched biological function categories for proteins increased and decreased on LDs in response to high-fat feeding as assigned by IPA. The P value range represents the lowest and highest enrichment within the subcategories. Categories containing fewer than two assigned proteins were discarded. B: The enriched canonical pathways as assigned by IPA. The blue bars represent increased pathways and the green bars represent decreased pathways. The line graph represents the ratio of proteins from our dataset that map to the pathway divided by the total number of molecules in the same pathway. Pathways with fewer than two proteins, P > 0.01, or with enrichment in both increased and decreased datasets were discarded. C: The distribution of increased or decreased proteins in a subset of cellular component GO terms as analyzed using STRAP.
TABLE 2.
Canonical pathway analysis of LD proteins regulated by diet and proteins within each component that were significantly altered
| Proteins | |
| Increased pathways | |
| γ-Linolenate biosynthesis II (animals) | SLC27A5, SLC27A2, CYB5A, ACSL1 |
| Mitochondrial L-carnitine shuttle pathway | SLC27A5, SLC27A2, CPT2, ACSL1 |
| Stearate biosynthesis I (animals) | SLC27A5, SLC27A2, CYP2E1, ACSL1 |
| Fatty acid activation | SLC27A5, SLC27A2, ACSL1 |
| EIF2 signaling | RPS7, RPS3A, RPS2, RPL27, RPS19, RPLP0 |
| Nicotine degradation II | CYP2D6, CYP2F1, CYP2E1, FMO5 |
| LPS/IL-1-mediated inhibition of RXR function | SLC27A5, SLC27A2, CPT2, APOC2, FMO5, ACSL1 |
| Bupropion degradation | CYP2D6, CYP2F1, CYP2E1 |
| Acetone degradation I (to methylglyoxal) | CYP2D6, CYP2F1, CYP2E1 |
| Fatty acid β-oxidation I | SLC27A5, SLC27A2, ACSL1 |
| Estrogen biosynthesis | CYP2D6, CYP2F1, CYP2E1 |
| Regulation of eIF4 and p70S6K signaling | RPS7, RPS3A, RPS2, RPS19 |
| Nicotine degradation III | CYP2D6, CYP2F1, CYP2E1 |
| Melatonin degradation I | CYP2D6, CYP2F1, CYP2E1 |
| Superpathway of melatonin degradation | CYP2D6, CYP2F1, CYP2E1 |
| mTOR signaling | RPS7, RPS3A, RPS2, RPS19 |
| Type II diabetes mellitus signaling | SLC27A5, SLC27A2, ACSL1 |
| Decreased pathways | |
| LXR/RXR activation | KNG1, TTR, ALB, HPX, TF, SERPINA1 |
| Coagulation system | KNG1, SERPINC1, SERPINA1, F2 |
| Glycolysis I | GPI,PKLR, PGAM1 |
| Gluconeogenesis I | GPI, PGAM1, ME1 |
| Acute phase response signaling | TTR, ALB, HPX, TF, SERPINA1, F2 |
| Intrinsic prothrombin activation pathway | KNG1, SERPINC1, F2 |
| Extrinsic prothrombin activation pathway | SERPINC1, F2 |
| Clathrin-mediated endocytosis signaling | ALB, TF, SERPINA1, F2 |
| Glutathione-mediated detoxification | Gstt3, GSTO1 |
TABLE 3.
Cellular component grouping of hepatic LD-associated proteins that were regulated by high-fat feeding and proteins within each component that were significantly altered
| Increased | Decreased | |
| Endosome | None | TF |
| Ribosome | RPS19, GM10288, CANX, RPS10, RPLP0, RPL27, RPS3A1, RPS17, RPS28, RPS7 | NONE |
| ER | CYP2D10, CYB5, CYP2D26, BAX, CANX, FMO5, SLC27A2, EPHX1, CYPE2E1, CYP2F2, ERLIN2, LMAN1, SLC27A5, ATP2A2, RPN1, RPN2, ARFGAP1, ARCN1, AHSA1 | ACO1, CES1C |
| Mitochondria | CYB5, ACSL1, BAX, ABCD3, COX6B1, CPT2, SLC27A2, CYP2E1, TIMM44, OPA1 | ACLY, MTHFD1, ATIC, HAGH, MSRA, DDAH1, ME1, ALDH1L1, ACO1, PRDX2, FDPS, SOD1 |
| Nucleus | CPT2, RPS3A1, ARFGAP1, RPS7 | STIP1, HNRNPF, ACLY, THRSP, MSRA, SELENBP1, SOD1 |
| Peroxisome/microbody | ABCD3, SLC27A2 | SOD1 |
| Cytoskeleton | DYNC1H1 | None |
| Plasma membrane | ERLIN2, SLC27A5, ARFGAP1 | ACLY, GPI, SOD1 |
| Cell surface | APOA4 | NONE |
| Extracellular | APOA4, APOC2, DYNCH1H1 | F2, TF, SERPINA1D, SPERINC1, HPX, SERPINA3K, KNG1, TTR, GPI, ALB, CES1C, SOD1 |
| Macromolecular complex | CANX, ERLIN2, SLC27A5, ARCN1 | TTR, ALB, SOD1 |
| Cytoplasm | APOA4, BAX, CANX, DYNC1H1, AHSA1 | STIP1, TF, PGLS, TARS, MTHFD1, DPYS, MSRA, SELENBP1, AACS, GPI, ALB, CMBL, GSTO1, SOD1, CA3 |
| Other | ARCN1, CYB5, CYP2E1, BAX, ABCD3, CANX FMO5, EPHX1, ERLIN2, LMAN1, ATP2A2, DYNC1H1, RPN2, RPN1, ARFGAP1, OPA1 | TF, ACO1, ACLY, MSRA, FABP2, SLENBP1, SOD1 |
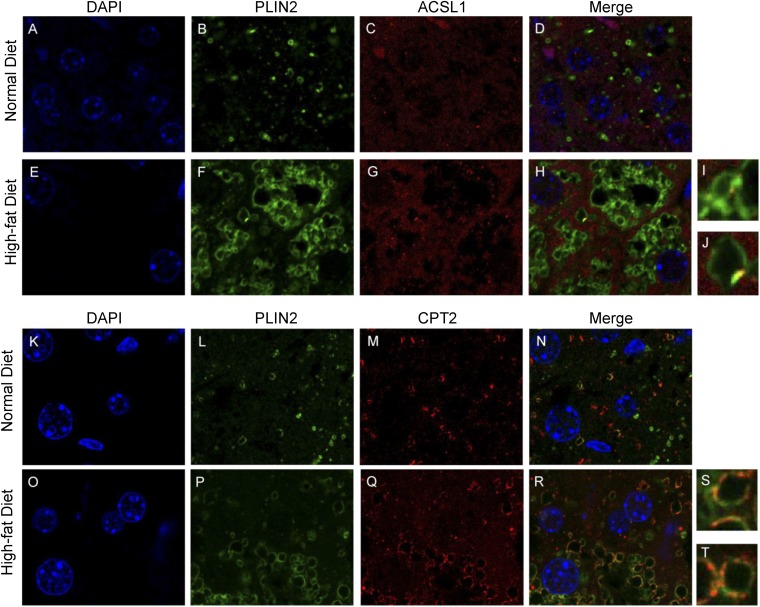
Two of the most robustly induced LD-associated proteins in response to the HFD were long chain ACSL1 and CPT2. ACSL1 has been identified to activate long chain fatty acids and selectively channel them to mitochondrial β-oxidation (43–45), and has also been shown to be present on LDs, in addition to its primary location on other organelles (13, 15). CPT2 is an inner mitochondrial membrane protein important for fatty acid transport and activation. Using confocal microscopy, we found both ACSL1 and CPT2 to have more colocalization with the LD marker protein, PLIN2, in response to high-fat feeding (Fig. 4). Despite increased colocalization on LDs in response to high-fat feeding, the staining patterns of ACSL1 and CPT2 were very different. Only a small amount of ACSL1 was found on LDs and it was generally confined to one region of a given LD. In contrast, much more CPT2 was found on LDs and it surrounded LDs similar to PLIN2 or other bona-fide LD proteins (Fig. 4). As a control, we stained mouse liver sections with a mitochondrial protein that was not identified in our proteomics screen. Voltage-dependent anion channel, an inner mitochondrial membrane protein, stained mitochondria in the cell but did not show colocalization with LD marker protein PLIN2 (supplementary Fig. 3). To further explore LD-mitochondria interactions, we examined electron microscopy micrographs of normal and high-fat-fed mouse livers. We quantified the number of mitochondria and LDs and calculated the percent of mitochondria interacting with LDs and the percent of LDs interacting with mitochondria. These data revealed that approximately 40% of LDs were in contact with mitochondria and 5% of mitochondria were in contact with LDs, but there was no change in either of these parameters between the livers of mice fed the ND or HFD (supplementary Fig. 4). Consistent with these data, we also did not observe an enrichment of mitochondria proteins in our GO analysis (Fig. 3C). Taken together, these data support the proteomic analysis showing that proteins involved in lipid catabolism are enriched on LDs in response to high-fat feeding and hepatic steatosis, and suggest that they are not the result of increased organelle interactions.
Fig. 4.
High-fat feeding results in an enriched of ACSL1 and CPT2 on LDs. Single plane image of a liver tissue section from a ND- and HFD-fed mouse stained for DAPI (A, E, K, O) or immunolabeled for PLIN2 (B, F, L, P), ACSL1 (C, G), or CPT2 (M, Q) or their respective merged images (D, H, N, R). I, J: Magnified from (H). S, T: Magnified from (R).
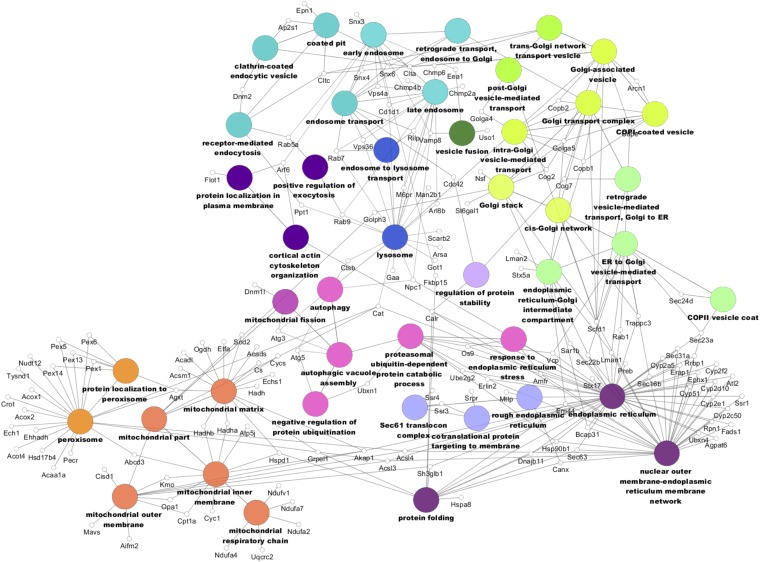
The nature of the interaction between primarily non-LD-localized proteins and the LD is unclear in most cases. The LD comes in contact with other organelles, so it is possible that other organelles, membrane fragments, and/or proteins copurify with the LD fraction. It is also possible that some of these proteins do indeed reside on the LD, but in a very transient or low stoichiometric basis. To represent the variety of proteins identified in the LD-associated protein dataset, we built a network using Cytoscape 3.0.1 with the ClueGO plugin (Fig. 5). Consistent with these LD-organelle interactions, many of the proteins in our dataset are annotated with multiple cellular components and/or biological functions, as shown by the many connecting edges. Nodes, which are GO terms, were selected to represent the variety of organelle interactions.
Fig. 5.
LD-associated proteome network. Only proteins found in the LD-associated proteome dataset are shown. Similar colors represent similar location and/or functional groups.
DISCUSSION
These data provide a quantitative analysis of changes in the hepatic LD-associated proteome in response to a model of diet-induced hepatic steatosis. Our analysis identified many proteins that have been previously found in LD proteomic analysis. We also found a greater number of proteins than previous hepatic LD proteomic analyses, which may in part be due to the higher level of detection of the mass spectrometer used in the current study or different LD isolation or analytical methodologies. A limitation of any proteomics study, including the current one, is determining whether proteins identified in our analysis are bona-fide LD proteins or contaminants. We did not detect contamination, via Western blotting, of any organelle except peroxisomes. However, Western blotting is much less sensitive than MS. As an example, we found the organelle markers ATP-CL and β-actin in our MS analysis of LD-associated proteins, but we were unable to detect them in the LD fraction by Western blotting. Thus, it is difficult to distinguish between bona-fide LD proteins and contaminant proteins, as is the case in any proteomic study. Additionally, many LD-associated proteins may be associated with LDs, but this only represents a small amount of the total intracellular distribution. Indeed, we found that several LD-associated proteins colocalized with PLIN2, but most of the proteins resided primarily in other intracellular locations. An additional caveat of these studies is that although PLIN2 is the most abundant hepatic LD protein, it is not present on all hepatic LDs and, therefore, is an imperfect LD marker (46).
Krahmer et al. (47) recently used a protein correlation profiles strategy to identify high confidence LD proteins from Drosophila SH2 cells. Given that this study identified a similar total number of proteins to our study (1,481 vs. 1,520), we determined the mammalian orthologs of the entire Drosophila LD protein dataset and compared them to our dataset (25). Roughly half of their proteins (694) matched to the mammalian orthologs of our list (supplementary Table 1). We also compared the bona-fide LD proteins from Krahmer et al. (47) to our dataset; seven of the Drosophila proteins had no mouse ortholog by our analysis. Of the 111 proteins they identified as bona-fide LD proteins, we found 60 homologs in our proteomic analysis.
The canonical pathway of fatty acid β-oxidation was enriched in our screen along with many proteins involved in lipid metabolism. We identified enzymes of both peroxisomal and mitochondrial fatty acid oxidation. Tight interaction between LDs and the peroxisome has been previously reported (48, 49). Consistent with these data, we detected the peroxisomal marker protein, catalase, in our LD fraction both by MS and Western blot (supplementary Fig. 1, supplementary Table 1). We identified the mouse homologs for seven of the eight yeast proteins involved in peroxisomal β-oxidation that were identified in a previous LD screen in yeast (48). It has also been demonstrated that the mitochondria directly interact with LDs (16, 50, 51). Consistent with this tight interaction, the most highly enriched GO annotation cluster by DAVID analysis was the mitochondrial part (data not shown).
In addition to mitochondria and peroxisomes, many organelles are in close proximity to LDs. ER proteins also commonly copurify with the LD fraction, which may be due to the fact that the ER, the reported origin of LDs, surrounds and/or protrudes into most, if not all, LDs (40, 52–54). Consistent with continuous membrane between LDs and the ER, membrane proteins have been observed to relocate between the LD and ER (13, 55). Welte (56) put forward the hypothesis that LDs are reservoirs for protein sequestration. The sequestration could be used to prevent highly abundant proteins from aggregating, to store unfolded proteins, to hold proteins that are targeted for degradation, or to inactivate proteins. It is possible that the abundance of vesicle trafficking proteins that localize to the LD bring other organelles into close contact with the LD. Proteins associated with vesicle trafficking, such as RAB GTPases and synaptosome-associated proteins, have been commonly reported on LDs (40), including in our analysis.
Pathways involving fatty acid catabolism were highly increased in the LD proteome in response to high-fat feeding. Isoform 2 of dynamin-like 120 kDa protein (OPA1), which was increased on LDs following high-fat feeding, has been shown to regulate protein kinase A-mediated lipolysis (57). Consistent with increased lipolysis, monoglyceride lipase (MGLL) was also enriched in LDs from mice fed the HFD. ACSL1 has been shown to promote the activation of fatty acids destined for mitochondrial oxidation in numerous tissues (43–45). The mitochondrial L-carnitine shuttle pathway is enriched due to the increase of CPT2, which converts medium and long chain acylcarnitines into acyl-CoAs that can be used for β-oxidation. Very long chain acyl-CoA dehydrogenase (ACADVL), which catalyzes the first step of β-oxidation in the mitochondria, was also enriched in LDs in response to the HFD. In support of this data, ACADVL, ACSL1, MGLL, and CPT2 were also found in higher abundance on hepatic LDs in human subjects with NAFLD, suggesting that these proteins are conserved in models of NAFLD (14). Taken together, these data are consistent with more recent studies showing that fatty acid oxidation is upregulated in models of hepatic steatosis (58–60). Recent studies in nonhepatocyte cell lines suggest that LD-mitochondria interactions are important for oxidation of hydrolyzed fatty acids (61). However, our studies do not show increased LD-mitochondria interactions in response to high-fat feeding. Clearly, much remains to be learned regarding the factors governing the oxidation of hydrolyzed fatty acids in the liver and how this changes during the development of steatosis.
We also observed an increase in many ribosomal and ER proteins on hepatic LDs in high-fat-fed mice. Ribosomal proteins have been previously observed on LDs in leukocytes via electron microscopy (40) and numerous ribosomal proteins, including eukaryotic initiation factor 1a, were found on LDs in Drosophila (62). Previous Drosophila RNAi screens have found that the knockdown of ribosomal proteins increases LD storage (63, 64). In obese mouse livers, ER-associated protein synthesis and the number of ribosome proteins bound to the ER membrane were found to be decreased, while lipid metabolism was increased (65, 66). It has been postulated that LDs provide a reservoir for unfolded proteins and toxic lipids from the ER, because during times of ER dysfunction, LDs accumulate (67). In addition, the increase in ER proteins may simply represent elevated LD biogenesis as discussed above.
Proteins involved in glycolysis, including pyruvate kinase, phosphoglycerate mutase, and glucose-6-phospate isomerase, were specifically depleted in hepatic LDs from mice fed the HFDs. Crunk et al. (15) also identified pyruvate kinase and phosphoglycerate mutase as hepatic LD-associated proteins and found that numerous proteins involved in carbohydrate metabolism were also decreased on LDs in response to acute high-fat feeding. Glycolytic enzymes are known to interact with caveolae (68), which are well-documented to be tightly linked to LD biogenesis and metabolism (69, 70). Additionally, the absence of caveolin-1 results in enhanced glycolysis (71), suggesting that alterations in the LD-associated proteome could directly influence carbohydrate metabolism.
It is not immediately clear why extracellular proteins would localize intracellularly to LDs or why their localization would be affected by hepatic steatosis; however, many of the identified extracellular proteins have links to hepatic steatosis, obesity, and diabetes (72–76). Additionally, it is not clear whether these extracellular proteins are in the secretory pathway or are being endocytosed. However, in support of the latter, proteins involved in endosomal trafficking have been previously identified on LDs (77). Additionally, multivesicular bodies/late endosomes interact with LDs in hepatocytes, suggesting that protein transfer could occur (78). The decreased presence of secreted proteins accounts for the reduction of numerous canonical pathways in response to high-fat feeding, including liver X and retinoic acid receptor activation, coagulation system acute phase response signaling, and prothrombin activation pathways.
In summary, our analysis has more comprehensively defined the hepatic LD-associated proteome and, for the first time, quantified the dynamic regulation of LD-associated proteins during chronic diet-induced hepatic steatosis. These data provide novel insights into the complexity of LD biology and point toward unique functions of LDs beyond simple lipid storage organelles.
Supplementary Material
Footnotes
Abbreviations:
- ACADVL
- very long chain acyl-CoA dehydrogenase
- ACSL1
- acyl-CoA synthetase 1
- ATPCL
- ATP-citrate lyase
- BIP
- binding immunoglobulin protein
- COX IV
- cyclooxygenase IV
- CPT2
- carnitine palmitoyltransferase 2
- DAVID
- Database for Annotation, Visualization, and Integrated Discovery
- ER
- endoplasmic reticulum
- GO
- Gene Ontology
- GPI
- glucose phosphate isomerase
- HFD
- high-fat diet
- IPA
- Ingenuity Pathway Analysis
- iTRAQ
- isobaric tagging for relative and absolute quantification
- LD
- lipid droplet
- ME
- malic enzyme
- MGLL
- monoglyceride lipase
- NAFLD
- non-alcoholic fatty liver disease
- ND
- normal diet
- OPA1
- isoform 2 of dynamin-like 120 kDa protein
- PLIN2
- perilipin 2
- RPLP0
- ribosomal protein large P0
- STRAP
- suspension trapping
- UniRef50
- UniProt Reference Clusters 50
This work was supported by National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases Grants DK085008 to D.G.M. and DK050456 to the Minnesota Obesity Center.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Milić S., and Štimac D.. 2012. Nonalcoholic fatty liver disease/steatohepatitis: epidemiology, pathogenesis, clinical presentation and treatment. Dig. Dis. 30: 158–162. [DOI] [PubMed] [Google Scholar]
- 2.Williams K. H., Shackel N. A., Gorrell M. D., McLennan S. V., and Twigg S. M.. 2013. Diabetes and nonalcoholic fatty liver disease: a pathogenic duo. Endocr. Rev. 34: 84–129. [DOI] [PubMed] [Google Scholar]
- 3.Duan X-Y., Zhang L., Fan J-G., and Qiao L.. 2014. NAFLD leads to liver cancer: do we have sufficient evidence? Cancer Lett. 345: 230–234. [DOI] [PubMed] [Google Scholar]
- 4.Adams L. A., Waters O. R., Knuiman M. W., Elliott R. R., and Olynyk J. K.. 2009. NAFLD as a risk factor for the development of diabetes and the metabolic syndrome: an eleven-year follow-up study. Am. J. Gastroenterol. 104: 861–867. [DOI] [PubMed] [Google Scholar]
- 5.Bhatia L. S., Curzen N. P., Calder P. C., and Byrne C. D.. 2012. Non-alcoholic fatty liver disease: a new and important cardiovascular risk factor? Eur. Heart J. 33: 1190–1200. [DOI] [PubMed] [Google Scholar]
- 6.Fujimoto T., and Parton R. G.. 2011. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb. Perspect. Biol. 3: a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walther T. C., and Farese R. V.. 2012. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81: 687–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodges B. D. M., and Wu C. C.. 2010. Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. J. Lipid Res. 51: 262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartz R., Zehmer J. K., Zhu M., Chen Y., Serrero G., Zhao Y., and Liu P.. 2007. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J. Proteome Res. 6: 3256–3265. [DOI] [PubMed] [Google Scholar]
- 10.Egan J. J., Greenberg A. S., Chang M. K., Wek S. A., Moos M. C. J., and Londos C.. 1992. Mechanism of hormone-stimulated lipolysis in adipocytes: translocation of hormone-sensitive lipase to the lipid storage droplet. Proc. Natl. Acad. Sci. USA. 89: 8537–8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turró S., Ingelmo-Torres M., Estanyol J. M., Tebar F., Fernández M. A., Albor C. V., Gaus K., Grewal T., Enrich C., and Pol A.. 2006. Identification and characterization of associated with lipid droplet protein 1: a novel membrane-associated protein that resides on hepatic lipid droplets. Traffic. 7: 1254–1269. [DOI] [PubMed] [Google Scholar]
- 12.Yang L., Ding Y., Chen Y., Zhang S., Huo C., Wang Y., Yu J., Zhang P., Na H., Zhang H., et al. 2012. The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J. Lipid Res. 53: 1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilfling F., Wang H., Haas J. T., Krahmer N., Gould T. J., Uchida A., Cheng J-X., Graham M., Christiano R., Fröhlich F., et al. 2013. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell. 24: 384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su W., Wang Y., Jia X., Wu W., Li L., Tian X., Li S., Wang C., Xu H., Cao J., et al. 2014. Comparative proteomic study reveals 17β-HSD13 as a pathogenic protein in nonalcoholic fatty liver disease. Proc. Natl. Acad. Sci. USA. 111: 11437–11442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crunk A. E., Monks J., Murakami A., Jackman M., MacLean P. S., Ladinsky M., Bales E. S., Cain S., Orlicky D. J., and McManaman J. L.. 2013. Dynamic regulation of hepatic lipid droplet properties by diet. PLoS One. 8: e67631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H., Wang Y., Li J., Yu J., Pu J., Li L., Zhang H., Zhang S., Peng G., Yang F., et al. 2011. Proteome of skeletal muscle lipid droplet reveals association with mitochondria and apolipoprotein A-I. J. Proteome Res. 10: 4757–4768. [DOI] [PubMed] [Google Scholar]
- 17.Ding Y., Zhang S., Yang L., Na H., Zhang P., Zhang H., Wang Y., Chen Y., Yu J., Huo C., et al. 2013. Isolating lipid droplets from multiple species. Nat. Protoc. 8: 43–51. [DOI] [PubMed] [Google Scholar]
- 18.Yang F., Shen Y., Camp D. G., and Smith R. D.. 2012. High pH reversed-phase chromatography with fraction concatenation as an alternative to strong-cation exchange chromatography for two-dimensional proteomic analysis. Expert Rev. Proteomics. 9: 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin-Moshier Y., Sebastian P. J., Higgins L., Sampson N. D., Hewitt J. E., and Marchant J. S.. 2013. Re-evaluation of the role of calcium homeostasis endoplasmic reticulum protein (CHERP) in cellular calcium signaling. J. Biol. Chem. 288: 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerbens F., Jansen A., van Erp A. J., Harders F., Meuwissen T. H., Rettenberger G., Veerkamp J. H., and te Pas M. F.. 1998. The adipocyte fatty acid-binding protein locus: characterization and association with intramuscular fat content in pigs. Mamm. Genome. 9: 1022–1026. [DOI] [PubMed] [Google Scholar]
- 21.Bhatia V. N., Perlman D. H., Costello C. E., and McComb M. E.. 2009. Software tool for researching annotations of proteins (STRAP): open-source protein annotation software with data visualization. Anal. Chem. 81: 9819–9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Binns D., Dimmer E., Huntley R., Barrell D., O’Donovan C., and Apweiler R.. 2009. QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics. 25: 3045–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dimmer E. C., Huntley R. P., Alam-Faruque Y., Sawford T., O’Donovan C., Martin M. J., Bely B., Browne P., Mun Chan W., Eberhardt R., et al. 2012. The UniProt-GO annotation database in 2011. Nucleic Acids Res. 40: D565–D570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.UniProt Consortium. 2014. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 42: D191–D198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., Perrimon N., and Mohr S. E.. 2011. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 12: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang da W., Sherman B. T., and Lempicki R. A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 27.Huang da W., Sherman B. T., and Lempicki R. A.. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M., and Goto S.. 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W-H., Pagès F., Trajanoski Z., and Galon J.. 2009. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 25: 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito R., Smoot M. E., Ono K., Ruscheinski J., Wang P-L., Lotia S., Pico A. R., Bader G. D., and Ideker T.. 2012. A travel guide to Cytoscape plugins. Nat. Methods. 9: 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beilstein F., Bouchoux J., Rousset M., and Demignot S.. 2013. Proteomic analysis of lipid droplets from Caco-2/TC7 enterocytes identifies novel modulators of lipid secretion. PLoS One. 8: e53017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brasaemle D. L., Dolios G., Shapiro L., and Wang R.. 2004. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J. Biol. Chem. 279: 46835–46842. [DOI] [PubMed] [Google Scholar]
- 33.Sato S. 2006. Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatitis C virus core protein. J. Biochem. 139: 921–930. [DOI] [PubMed] [Google Scholar]
- 34.Fujimoto Y., Itabe H., Sakai J., Makita M., Noda J., Mori M., Higashi Y., Kojima S., and Takano T.. 2004. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim. Biophys. Acta. 1644: 47–59. [DOI] [PubMed] [Google Scholar]
- 35.Kanshin E., Wang S., Ashmarina L., Fedjaev M., Nifant’ev I., Mitchell G. A., and Pshezhetsky A. V.. 2009. The stoichiometry of protein phosphorylation in adipocyte lipid droplets: analysis by N-terminal isotope tagging and enzymatic dephosphorylation. Proteomics. 9: 5067–5077. [DOI] [PubMed] [Google Scholar]
- 36.Kim S. C., Chen Y., Mirza S., Xu Y., Lee J., Liu P., and Zhao Y.. 2006. A clean, more efficient method for in-solution digestion of protein mixtures without detergent or urea. J. Proteome Res. 5: 3446–3452. [DOI] [PubMed] [Google Scholar]
- 37.Larsson S., Resjö S., Gomez M. F., James P., and Holm C.. 2012. Characterization of the lipid droplet proteome of a clonal insulin-producing β-cell line (INS-1 832/13). J. Proteome Res. 11: 1264–1273. [DOI] [PubMed] [Google Scholar]
- 38.Orban T., Palczewska G., and Palczewski K.. 2011. Retinyl ester storage particles (retinosomes) from the retinal pigmented epithelium resemble lipid droplets in other tissues. J. Biol. Chem. 286: 17248–17258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Umlauf E. 2004. Association of stomatin with lipid bodies. J. Biol. Chem. 279: 23699–23709. [DOI] [PubMed] [Google Scholar]
- 40.Wan H-C., Melo R. C. N., Jin Z., Dvorak A. M., and Weller P. F.. 2007. Roles and origins of leukocyte lipid bodies: proteomic and ultrastructural studies. FASEB J. 21: 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu C. C., Howell K. E., Neville M. C., Yates J. R. III, and McManaman J. L.. 2000. Proteomics reveal a link between the endoplasmic reticulum and lipid secretory mechanisms in mammary epithelial cells. Electrophoresis. 21: 3470–3482. [DOI] [PubMed] [Google Scholar]
- 42.Liu P., Ying Y., Zhao Y., Mundy D. I., Zhu M., and Anderson R. G. W.. 2004. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J. Biol. Chem. 279: 3787–3792. [DOI] [PubMed] [Google Scholar]
- 43.Ellis J. M., Mentock S. M., Depetrillo M. A., Koves T. R., Sen S., Watkins S. M., Muoio D. M., Cline G. W., Taegtmeyer H., Shulman G. I., et al. 2011. Mouse cardiac acyl coenzyme a synthetase 1 deficiency impairs Fatty Acid oxidation and induces cardiac hypertrophy. Mol. Cell. Biol. 31: 1252–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellis J. M., Li L. O., Wu P-C., Koves T. R., Ilkayeva O., Stevens R. D., Watkins S. M., Muoio D. M., and Coleman R. A.. 2010. Adipose acyl-CoA synthetase-1 directs fatty acids toward beta-oxidation and is required for cold thermogenesis. Cell Metab. 12: 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L. O., Ellis J. M., Paich H. A., Wang S., Gong N., Altshuller G., Thresher R. J., Koves T. R., Watkins S. M., Muoio D. M., et al. 2009. Liver-specific loss of long chain acyl-CoA synthetase-1 decreases triacylglycerol synthesis and beta-oxidation and alters phospholipid fatty acid composition. J. Biol. Chem. 284: 27816–27826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kassan A., Herms A., Fernández-Vidal A., Bosch M., Schieber N. L., Reddy B. J. N., Fajardo A., Gelabert-Baldrich M., Tebar F., Enrich C., et al. 2013. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J. Cell Biol. 203: 985–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krahmer N., Hilger M., Kory N., Wilfling F., Stoehr G., Mann M., Farese R. V. J., and Walther T. C.. 2013. Protein correlation profiles identify lipid droplet proteins with high confidence. Mol. Cell. Proteomics. 12: 1115–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Binns D. 2006. An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173: 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schrader M. 2001. Tubulo-reticular clusters of peroxisomes in living COS-7 cells: dynamic behavior and association with lipid droplets. J. Histochem. Cytochem. 49: 1421–1429. [DOI] [PubMed] [Google Scholar]
- 50.Jägerström S., Polesie S., Wickström Y., Johansson B. R., Schröder H. D., Højlund K., and Boström P.. 2009. Lipid droplets interact with mitochondria using SNAP23. Cell Biol. Int. 33: 934–940. [DOI] [PubMed] [Google Scholar]
- 51.Wang H., Sreenivasan U., Hu H., Saladino A., Polster B. M., Lund L. M., Gong D., Stanley W. C., and Sztalryd C.. 2011. Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J. Lipid Res. 52: 2159–2168. [Erratum. 2013. J. Lipid Res 54: 3539.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blanchette-Mackie E. J., Dwyer N. K., Barber T., Coxey R. A., Takeda T., Rondinone C. M., Theodorakis J. L., Greenberg A. S., and Londos C.. 1995. Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes. J. Lipid Res. 36: 1211–1226. [PubMed] [Google Scholar]
- 53.Hartman I. Z., Liu P., Zehmer J. K., Luby-Phelps K., Jo Y., Anderson R. G. W., and DeBose-Boyd R. A.. 2010. Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from endoplasmic reticulum membranes into the cytosol through a subcellular compartment resembling lipid droplets. J. Biol. Chem. 285: 19288–19298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohsaki Y., Cheng J., Fujita A., Tokumoto T., and Fujimoto T.. 2006. Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Mol. Biol. Cell. 17: 2674–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jacquier N., Choudhary V., Mari M., Toulmay A., Reggiori F., and Schneiter R.. 2011. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 124: 2424–2437. [DOI] [PubMed] [Google Scholar]
- 56.Welte M. A. 2007. Proteins under new management: lipid droplets deliver. Trends Cell Biol. 17: 363–369. [DOI] [PubMed] [Google Scholar]
- 57.Pidoux G., Witczak O., Jarnaess E., Myrvold L., Urlaub H., Stokka A. J., Kuntziger T., and Tasken K.. 2011. Optic atrophy 1 is an A-kinase anchoring protein on lipid droplets that mediates adrenergic control of lipolysis. EMBO J. 30: 4371–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iozzo P., Bucci M., Roivainen A., Någren K., Järvisalo M. J., Kiss J., Guiducci L., Fielding B., Naum A. G., Borra R., et al. 2010. Fatty acid metabolism in the liver, measured by positron emission tomography, is increased in obese individuals. Gastroenterology. 139: 846–856. [DOI] [PubMed] [Google Scholar]
- 59.Satapati S., Sunny N. E., Kucejova B., Fu X., He T. T., Méndez-Lucas A., Shelton J. M., Perales J. C., Browning J. D., and Burgess S. C.. 2012. Elevated TCA cycle function in the pathology of diet-induced hepatic insulin resistance and fatty liver. J. Lipid Res. 53: 1080–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sunny N. E., Parks E. J., Browning J. D., and Burgess S. C.. 2011. Excessive hepatic mitochondrial TCA cycle and gluconeogenesis in humans with nonalcoholic fatty liver disease. Cell Metab. 14: 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rambold A. S., Cohen S., and Lippincott-Schwartz J.. 2015. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell. 32: 678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cermelli S., Guo Y., Gross S. P., and Welte M. A.. 2006. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 16: 1783–1795. [DOI] [PubMed] [Google Scholar]
- 63.Beller M., Riedel D., Jänsch L., Dieterich G., Wehland J., Jäckle H., and Kühnlein R. P.. 2006. Characterization of the Drosophila lipid droplet subproteome. Mol. Cell. Proteomics. 5: 1082–1094. [DOI] [PubMed] [Google Scholar]
- 64.Guo Y., Walther T. C., Rao M., Stuurman N., Goshima G., Terayama K., Wong J. S., Vale R. D., Walter P., and Farese R. V.. 2008. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 453: 657–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu S., Yang L., Li P., Hofmann O., Dicker L., Hide W., Lin X., Watkins S. M., Ivanov A., and Hotamisligil G. S.. 2011. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 473: 528–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fu S., Fan J., Blanco J., Gimenez-Cassina A., Danial N. N., Watkins S. M., and Hotamisligil G. S.. 2012. Polysome profiling in liver identifies dynamic regulation of endoplasmic reticulum translatome by obesity and fasting. PLoS Genet. 8: e1002902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hapala I., Marza E., and Ferreira T.. 2011. Is fat so bad? Modulation of endoplasmic reticulum stress by lipid droplet formation. Biol. Cell. 103: 271–285. [DOI] [PubMed] [Google Scholar]
- 68.Lloyd P. G., and Hardin C. D.. 2001. Caveolae and the organization of carbohydrate metabolism in vascular smooth muscle. J. Cell. Biochem. 82: 399–408. [DOI] [PubMed] [Google Scholar]
- 69.Pol A., Martin S., Fernandez M. A., Ferguson C., Carozzi A., Luetterforst R., Enrich C., and Parton R. G.. 2004. Dynamic and regulated association of caveolin with lipid bodies: modulation of lipid body motility and function by a dominant negative mutant. Mol. Biol. Cell. 15: 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cohen A. W., Razani B., Schubert W., Williams T. M., Wang X. B., Iyengar P., Brasaemle D. L., Scherer P. E., and Lisanti M. P.. 2004. Role of caveolin-1 in the modulation of lipolysis and lipid droplet formation. Diabetes. 53: 1261–1270. [DOI] [PubMed] [Google Scholar]
- 71.Fernández-Rojo M. A., Restall C., Ferguson C., Martel N., Martin S., Bosch M., Kassan A., Leong G. M., Martin S. D., McGee S. L., et al. 2012. Caveolin-1 orchestrates the balance between glucose and lipid-dependent energy metabolism: Implications for liver regeneration. Hepatology. 55: 1574–1584. [DOI] [PubMed] [Google Scholar]
- 72.Kassel K. M., Sullivan B. P., Cui W., Copple B. L., and Luyendyk J. P.. 2012. Therapeutic administration of the direct thrombin inhibitor argatroban reduces hepatic inflammation in mice with established fatty liver disease. Am. J. Pathol. 181: 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang B., Zhou K. K., and Ma J.. 2010. Inhibition of connective tissue growth factor overexpression in diabetic retinopathy by SERPINA3K via blocking the WNT/beta-catenin pathway. Diabetes. 59: 1809–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takahashi E., Okumura A., Unoki-Kubota H., Hirano H., Kasuga M., and Kaburagi Y.. 2013. Differential proteome analysis of serum proteins associated with the development of type 2 diabetes mellitus in the KK-A(y) mouse model using the iTRAQ technique. J. Proteomics. 84: 40–51. [DOI] [PubMed] [Google Scholar]
- 75.Kaur P., Rizk N. M., Ibrahim S., Younes N., Uppal A., Dennis K., Karve T., Blakeslee K., Kwagyan J., Zirie M., et al. 2012. iTRAQ-based quantitative protein expression profiling and MRM verification of markers in type 2 diabetes. J. Proteome Res. 11: 5527–5539. [DOI] [PubMed] [Google Scholar]
- 76.Hernández-Espinosa D., Ordóñez A., Miñano A., Martínez-Martínez I., Vicente V., and Corral J.. 2009. Hyperglycaemia impairs antithrombin secretion: Possible contribution to the thrombotic risk of diabetes. Thromb. Res. 124: 483–489. [DOI] [PubMed] [Google Scholar]
- 77.Zehmer J. K., Huang Y., Peng G., Pu J., Anderson R. G. W., and Liu P.. 2009. A role for lipid droplets in inter-membrane lipid traffic. Proteomics. 9: 914–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schroeder B., Schulze R. J., Weller S. G., Sletten A. C., Casey C. A., and McNiven M. A.. 2015. The Small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology. 61: 1896–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.