Abstract
Individual members of the mammalian phospholipase D (PLD) superfamily undertake roles that extend from generating the second messenger signaling lipid, phosphatidic acid, through hydrolysis of the membrane phospholipid, phosphatidylcholine, to functioning as an endonuclease to generate small RNAs and facilitating membrane vesicle trafficking through seemingly nonenzymatic mechanisms. With recent advances in genome-wide association studies, RNA interference screens, next-generation sequencing approaches, and phenotypic analyses of knockout mice, roles for PLD family members are being uncovered in autoimmune, infectious neurodegenerative, and cardiovascular disease, as well as in cancer. Some of these disease settings pose opportunities for small molecule inhibitory therapeutics, which are currently in development.
Keywords: lipid signaling, phosphatidic acid, cancer
PHOSPHOLIPASE D OVERVIEW
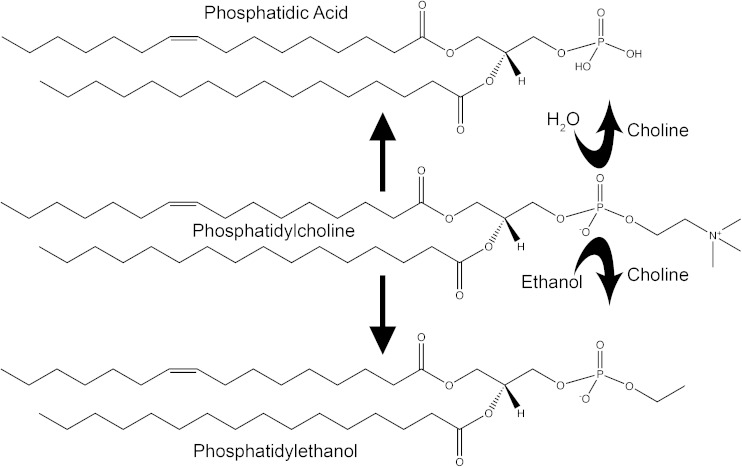
The mammalian phospholipase D (PLD) superfamily is best known for the catalytic action of its classical family members which hydrolyze phosphatidylcholine, the most abundant membrane phospholipid, to generate choline and the second messenger signaling lipid, phosphatidic acid (PA) (1). As transphosphatidylases, classical PLD enzymes more formally conduct headgroup exchange at the terminal phosphodiester bond on PA (2) (Fig. 1). In the most common cellular setting, water is used as the nucleophile to exchange an –OH group for the choline headgroup (3). However, because of a 1,000-fold higher preference for primary alcohols (2), production of phosphatidylalcohol (4) is the primary outcome when even relatively modest amounts (1–3%) of ethanol or 1-butanol are present (5). Phosphatidylethanol (PtdEtOH) has a long half-life relative to ethanol and can be detected in serum for up to a month subsequent to alcohol consumption (6), making it an increasingly popular biomarker for assessment of acute and even chronic drinking. PtdEtOH can be found at low levels even in nondrinking individuals though, because intestinal bacteria generate small amounts of ethanol through fermentation. PtdEtOH is generally thought to be physiologically inert; however, there is evidence to suggest that it may play beneficial or harmful roles in ethanol tolerance (7) and colon cancer (8), respectively. Primary alcohols have historically been used to block production of PA by the classical mammalian enzymes, PLD1 and PLD2, to assess their cellular signaling roles. However, alcohols have many other effects on cells, preventing definitive results from being obtainable with this approach (9, 10). The ability of PLD2 to also use glycerol as a nucleophile to generate phosphatidylglycerol has been proposed to play roles in wound healing (11, 12). Both PLD1 and PLD2 have been proposed to undertake roles in many cell biological and physiological settings, as will be described subsequently.
Fig. 1.
Schematic depiction of PLD generation of PA and PtdEtOH.
Enzymatic activities have not been discovered for the related proteins PLD3, PLD4, and PLD5; PLD5, in fact, has nonconservative substitutions in its putative catalytic site that make it very unlikely to be enzymatically active. PLD6 (MitoPLD) has been reported to hydrolyze cardiolipin on the outer surface of the mitochondria to generate PA (13), as well as to function as an endonuclease (via phosphodiesterase action) to generate specialized microRNAs (miRs) known as piwi-interacting RNAs (piRNAs) (14), which are critical during spermatogenesis (15). Despite the lack of evidence for PLD3 and PLD4 catalytic activity, they nonetheless have important functions, the loss of which creates pathology as discussed below. Definitive cellular and physiological roles for PLD5 have not yet been identified.
STRUCTURE AND REGULATION
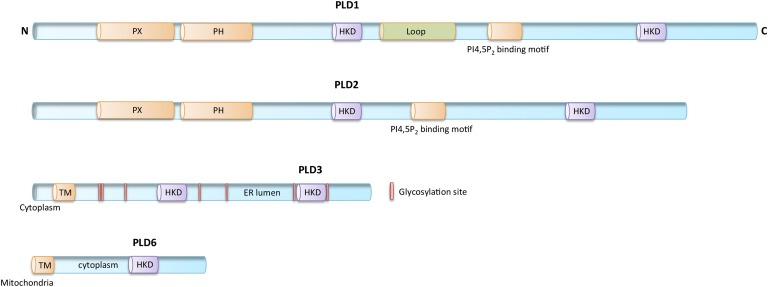
PLD1 (16) and PLD2 (17), which are ∼50% identical in protein sequence and have almost the same protein domain organization (Fig. 2), are widely expressed in different tissues and cell types and are activated by a variety of signaling molecules, including protein kinase C and the small GTPases, RhoA and ARF (1, 18–20). The PLD catalytic site is defined by the presence of two highly-conserved His-x-Lys-x-x-x-x-Asp sequences (x is any amino acid) termed the HKD motif (16), or more broadly, the PLD-c domain, each of which creates half of the split-catalytic site (21). The HKD motifs are essential for PLD enzymatic activity (2). A phox consensus sequence (PX), a pleckstrin homology (PH) domain, and an acidic PI(4,5)P2 binding motif are also found and are highly conserved in PLD1 and PLD2. These regions function in regulating subcellular localization (22, 23) through protein-protein interactions (24) and binding to phosphatidylinositol phosphates (23, 25, 26). PLD1 uniquely encodes an internal loop region that negatively regulates its activity (27) and, thus, may constitute the mechanism underlying the observation that the level of basal activity of PLD1 is lower than that of PLD2 (28).
Fig. 2.
Schematic depiction of PLD1, PLD2, PLD3, and PLD6. HKD, PLD superfamily catalytic motif; Loop, region found uniquely in PLD1. For PLD3: Cytoplasm, localization of the N-terminal region; TM, trans-membrane domain; ER lumen, localization of remainder of protein; pink vertical bars, glycosylation sites (29). For PLD6: TM, transmembrane domain that anchors PLD6 into the outer leaflet of the outer mitochondrial membrane; cytoplasm, localization of remainder of protein.
PLD3 (Hu-K4, SAM-9) encodes an abundant and widely expressed type 2 transmembrane protein that localizes to the endoplasmic reticulum (ER), where it is anchored by an N-terminal transmembrane domain and a short cytoplasmic sequence, with the putative catalytic domain localizing to the lumen of the ER (29) (Fig. 2). Similar to PLD1 and PLD2, PLD3 encodes two HKD motifs; however, it lacks both the PX and PH domains (29, 30). PLD3 has been linked to cellular differentiation and survival (29, 31–33). PLD4 is also an ER transmembrane glycoprotein that contains the canonical pair of HKD motifs and lacks PX and PH domains (34).
PLD6/MitoPLD (13) is most closely related to the bacterial protein Nuc (35), which is an endonuclease. PLD6, however, encodes an N-terminal extension that both localizes it to mitochondria and anchors it into the outer leaflet of the mitochondrial surface (13) (Fig. 2). PLD6 encodes only one HKD motif and dimerizes to exhibit catalytic activity. PLD6 promotes mitochondrial fusion, and through its ability to recruit lipin-1, a PA phosphatase (36), PLD6 indirectly facilitates mitochondrial fission (15). PLD6 also encodes an endonuclease activity that is required to generate piRNAs during spermatogenesis (14).
PHYSIOLOGICAL AND PATHOPHYSIOLOGICAL ROLES
PLD1
Roles for PLD1 in thrombotic disease (28, 37, 38), cancer (39), and auto-immunity (40), as revealed through animal model studies, have recently been summarized (5). Many other possible roles are suggested by cellular studies that have not yet been addressed in vivo, some of which will be reviewed here.
PLD1 and cancer.
PLD1 expression and activity are increased in many types of cancer (20, 41–43). However, the significance of this observation is uncertain because PLD1’s chromosomal location at 3q26 is adjacent to that of PI3Kinase-α, which is strongly amplified in numerous cancers. Most groups have studied the role of PLD1 in tumor cell viability, proliferation, and invasion, whereas our group has also shown a role for PLD1 in the tumor environment in the context of facilitating tumor neoangiogenesis and subsequent metastasis (39). As an example of the types of studies that have been reported, we will review the association of PLD1 with gliomas here.
Gliomas are the most common primary tumors of the CNS, diagnosed at a rate of 17,000 new cases per year in the United States (44). Despite clinical management, median time of survival after diagnosis is dismal, averaging between 12 and 15 months (45). Current treatment modalities consist of concomitant radiotherapy and chemotherapy, but are suboptimal in slowing disease progression (46). Patients incur frequent clinical complications including seizures, fluctuating neurological symptoms, and adverse effects of chemotherapy. PLD1 has been proposed to play important roles in the invasive migration of glioma cells (47), and in glioma cell proliferation (48), adhesion (48), and viability (49). The tumor signaling pathways and mechanisms relevant to PLD1 function are complex and have been proposed to include activation of AKT (49), upregulation of HIF1-α (50), and increased VEGF (50) and MMP-2 secretion (51). Overlapping roles have been proposed for PLD2 (52). Small molecule inhibitors of PLD1 and PLD2, such as FIPI (53) or isoform-selective analogs (20), have been shown to have dramatic effects on human glioma cell lines in tissue culture studies in the context of the PLD-driven roles above. How useful suppression of PLD activity will be for management of gliomas in vivo, though, remains to be determined.
PLD1 and fibrosis.
PLD1 has been speculated to participate in the process of fibrogenesis in multiple tissue types, including the liver (54, 55), lung (56), and heart (57, 58). PLD1 is known to be directly connected to autophagy (59, 60), the self-degradative process required for cellular homeostasis that is linked to several forms of liver disease (61–63). Based on recent reports, cardiac fibrosis is of particular interest. PLD mRNA, protein, and activity levels decrease during congestive heart failure subsequent to myocardial infarction in the scar tissue (57). The importance of this observation is suggested by a report that inhibition of PLD activity markedly attenuates left ventricular fibrosis, resulting in subsequent improvement in cardiac function (64). PLD would thus seem to be an attractive therapeutic target for scar remodeling and reducing left ventricular fibrosis. On the other hand, PLD1 deficiency, which blunts immune responses (65), hinders immune-driven elements of the repair process after myocardial infarction (38). Thus, there may be a balance between too little and too much PLD1 activity in this setting or specific sites at which PLD1 expression is harmful or beneficial. Similarly, there may be specific times during the repair process when PLD1 elevation is either helpful or harmful.
PLD2
Roles for PLD2 in thrombotic disease (37, 66), cancer (67, 68), Alzheimer’s disease (AD) (69), and immune function (65), based on animal model studies, have recently been summarized (5). Other potential functions have been raised by tissue culture studies, some of which will be reviewed here.
PLD2 and influenza.
Influenza epidemics and reoccurring pandemics continue to pose a great threat to public health worldwide, in part due to the viruses’ high mutation and replication rates (70, 71). As a consequence, treatment and prevention measures for influenza virus infections remain challenging. For example, in this current flu season, for which the immunization cocktail was largely ineffective through being directed at the incorrect strains, the anti-influenza therapeutic amantadine was also found to be of relatively little benefit due to extensive acquired viral resistance to it (72), suggesting the need for new therapeutic approaches. A genome-wide RNA interference screen identified 287 human host cell genes that influence the viruses’ ability to replicate, of which 29 were required for all of the viral strains tested (73). Among these, PLD2 was identified as a targetable candidate (73). A subsequent study using an isoform-selective PLD2 inhibitor further supported a critical role in the viral replication process; PLD2 was found to mediate rapid endocytosis of the virus, facilitating its escape from innate immune detection (74). As PLD2 knockout mice are grossly normal to inspection (69), PLD2 would appear to fit the category of a “temporarily dispensable host gene” that could be acutely targeted to suppress viral replication. One caution for this approach would entail potential effects of PLD2 inhibition on the immune system, which were previously reported to decrease macrophage phagocytosis and neutrophil migration (65). However, this might not be a substantive issue if the effects on the immune response to influenza were limited, whereas the effects on viral replication are profound.
PLD2 and cancer.
PLD2 polymorphisms, as well as upregulated protein activity levels, have been observed in several types of cancer, including gastric, colorectal, kidney, and breast (68, 75–77). In a particularly interesting recent report, it was observed that expression of miR-203 in high World Health Organization grade glioma tissues was significantly lower than in low World Health Organization grade gliomas and normal brain tissue. Transfection of a miR-203 mimic into human glioma cells strongly and directly downregulated PLD2 expression and, in parallel, suppressed proliferation and invasion of the glioma cells, whereas PLD2 overexpression rescued the effects induced by the miR-203 mimic. Taken together, these observations suggest important causal roles for PLD2 in glioma proliferation and invasive capacity (52). In a human breast cancer xenograft model, it was shown that increased PLD2 expression in tumor cells suppresses apoptosis, ultimately facilitating tumor growth and chemoresistance (68). PLD2 may also play roles in the tumor environment similar to those previously reported for PLD1 (39), because PLD2 ablation from endothelial cells suppresses their hypoxia-induced Hif1-α expression and VEGF secretion, reducing proximal tumor neovascularization and growth (67). Although the overall expression levels of PLD2 may vary in tumors, there is a significant correlation between PLD2 expression level and tumor size (P < 0.05), as well as with survival of patients with colorectal carcinoma (P < 0.05) (78). Immunohistochemical staining of 30 human colon cancer samples revealed a high level of correlation between Hif1-α and PLD2 (79). Moreover, Hif1-α and PLD2 expression levels are much higher in colon cancer tissues than in normal colon tissues (P < 0.01) (79), and under hypoxic conditions, Hif1-α upregulates PLD2 expression in colon cancer cells (79). Similar to PLD1, PLD2 should also be viewed as a major therapeutic target in the treatment of several forms of cancer.
PLD3
PLD3 and AD.
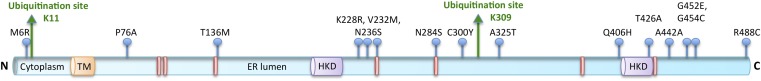
PLD1, PLD2, and PLD3 have all been implicated in AD (69, 80, 81). PLD3 is highly expressed in the brain, including in, but not limited to, mature neurons of the forebrain, hippocampus, and cortex (81–83). Rare coding variants in PLD3 have been associated with up to 9% of late-onset AD in 14 families of European ancestry (81) (Fig. 3). More specifically, Val232Met, a putative loss-of-function polymorphism, is proposed to increase pathogenic amyloid peptide (Aβ) secretion and, hence, increase the risk for late-onset AD (81). This increased risk is independent of the APOE genotype (81). Similarly, PLD3 putative loss-of-function polymorphisms have been reported to correlate with increased risk of AD in African Americans (81). Independent of the coding variants, PLD3 protein expression is downregulated in AD brains (84) and in cortical membrane lipid rafts prepared from the 3xTgAD murine model of AD (85).
Fig. 3.
Key features of PLD3 protein sequence. Blue lollipops, coding variants associated with late-onset AD (81); green arrows, ubiquitinated lysines (91, 93).
The mechanism of action of PLD3, as well as whether or not it encodes any type of catalytic activity, remains unknown; but its placement in the ER and secretory system suggests how it might suppress Aβ secretion. AβPP, the precursor protein to Aβ, is proteolytically processed to generate Aβ in early endosomes, and the extent of this processing depends on how rapidly it is trafficked from the early endosome to late endosomes and lysosomes. Key to this process is the phosphatidylinositol-3-phosphate effector Hrs, an early endosome-associated ubiquitin-interacting motif-containing protein that plays a central role in directing trafficking of membrane cargo proteins from early endosomes to luminal vesicles of multivesicular bodies (MVBs) for eventual degradation in the lysosome. Knockdown of Hrs or other proteins required for the transport of AβPP from early endosomes to luminal vesicles of MVBs results in increased amyloidogenic processing (86), supporting the general hypothesis that any defect that keeps AβPP and its processing enzyme, BACE1, in endosomes will increase Aβ production and drive pathology (87). Intriguingly, a recent screen for ubiquitinated proteins specifically recognized by Hrs identified 48 targets, among which were AβPP and PLD3 (88).
PLD3 has been reported in secretory granules in an insulin-producing pancreatic β-cell line (89) and in a pattern partially overlapping with lysosomes in HeLa cells (90), suggesting that PLD3 protein may traffic through endosomal pathways, even if the most abundantly observed steady-state location is in the ER in cultured cell lines (29, 32). PLD3 has been identified in multiple screens for proteins that become ubiquitinated (88, 91–93). One site for PLD3 ubiquitination is its short cytoplasmic N-terminal domain, K11 (Fig. 3). This key finding suggests that PLD3 undergoes cytoplasmic ubiquitination and could be recognized and sorted by Hrs to co-traffic with AβPP from endosomes to luminal vesicles of MVBs. Supporting this hypothesis, a PLD3 allele with significant association with late-onset AD, in which methionine 6 is substituted for by arginine (M6R), occurs in an amino acid residue close to K11 and could potentially affect ubiquitination, providing a basis for its disease linkage. These data, taken together, suggest that if ubiquitinated, Hrs-trafficked PLD3 plays a role in moving AβPP from early endosomes to luminal vesicles of MVBs for eventual lysosomal degradation; then a decrease in or a lack of ubiquitination, as well as nonfunctional PLD3, could cause AβPP retention in early endosomes and increased Aβ production to promote AD pathology.
Independently, a screen performed for targets of the FBOX6 ubiquitin ligase complex, which triggers ER-associated degradation (ERAD) by mediating glycoprotein ubiquitination, identified 29 targets including PLD3 (92). The ERAD system functions by recognizing improperly folded glycoproteins and poly-ubiquitinating and transferring them to the cytosol to be degraded by proteasomes. The second PLD3 site that becomes ubiquitinated is in the C-terminal ER-luminal portion of the protein (K309, Fig. 3) (91) and would be a candidate target site for this mechanism. A report on PLD3 in late-onset AD (81) identified six disease-associated alleles that are predicted by PolyPhen-2 (94) to be possibly or probably damaging, and are located in or near putative glycosylation sites (Fig. 3). If these missense mutations cause altered glycosylation or misfolding, then the ERAD system might target the PLD3 protein for degradation, causing a significant decrease in protein expression levels.
It is notable that none of the alleles identified encoded nonsense mutations (premature stop codons), suggesting that full or even heterozygous PLD3 loss might be deleterious. PLD3−/− mice have not been generated yet. A Drosophila line with a P element insertion into its PLD3 homolog does exist and is embryonic lethal when homozygous (unpublished observations). However, this line could have other genetic abnormalities or the P element could be affecting expression of other nearby genes, so additional studies would need to be performed to conclude that PLD3 loss creates lethality.
Finally, other groups have reported variable success in reproducing the genetic association of PLD3 polymorphisms with late-onset AD (95–97), suggesting that the linkage may be less robust than initially projected.
PLD4
PLD4 and autoimmune diseases.
As is the case for PLD3, it is not known whether PLD4 has a bona fide enzymatic function. Nonetheless, PLD4 clearly has important functional roles. Initial reports described PLD4 expression in microglia, the macrophage-like innate immune cells of the CNS, as well as in splenic cells, presumably macrophages. PLD4 expression increases with microglial activation, which is also characterized by increased phagocytic capacity (34, 98). siRNA knockdown of PLD4 suppressed phagocytosis, suggesting a role for PLD4 in the setting of CNS injury and infection (34, 98). A nonsense mutation in PLD4 (W215X) in Fleckvieh cattle causes severe skin lesions, generally poor health, and decreased survival (99). PLD4 deficiency in humans has been linked through genome-wide association studies to syndromes such as rheumatoid arthritis (100) and the autoimmune disease systemic sclerosis (101). Taken together, these findings suggest that PLD4 deficiency results in hyper-activation of the immune system, causing a variety of autoimmune-like syndromes.
PLD5
PLD5 and uterine fibroids.
Despite having no catalytic activity, PLD5 has been linked to a number of diseases, including a profibrotic uterine phenotype that occurs during childbearing years, and PLD5 polymorphisms may be associated with an increased risk of tumor progression in multiple cutaneous and uterine leiomyomatosis syndrome (102).
PLD5 is most widely known for its correlation with neuropsychiatric disorders. Autism, the neurological disorder associated with impaired social relationships and communication as well as repetitive behavior, is predominantly linked to de novo and inherited copy number variants of genes important for neuronal development (103–105). High-resolution genotyping of 1,558 families on the autism spectrum uncovered a PLD5 gene polymorphism as possibly being connected with autism physiopathology (106). Although the association signal of this SNP was borderline significant, further investigation is warranted, because autism has been proposed to be caused primarily by multigene interactions rather than solely by single rare mutations.
PLD6 (MitoPLD)
PLD6-deficient mice, which cannot generate piRNAs to suppress transposon mobilization during spermatogenesis, are completely sterile (15), but are otherwise grossly normal to inspection. PLD6 mutations do not appear to be a major cause of human infertility; sequencing of PLD6 in 400 azoospermic European men did not uncover any PLD6 polymorphisms (unpublished observation). Nonetheless, PLD6 may have other less obvious roles.
PLD6 and cervical cancer.
Even with the current advances in the diagnosis and characterization of cervical intraepithelial neoplasia (CIN), highly discriminating biomarkers are still needed (107, 108). Cervical cancer is the second most common cancer in women worldwide. In 2008, there were 529,800 cases of cervical cancer, with 14.5% occurring in developed countries and 85.5% occurring in developing countries, approximately 275,000 of which resulted in mortality (109). Cervical cancer is caused by infection with certain strains of the human papillomavirus (110, 111). Infection leads to the development of noninvasive neoplastic lesions, CIN (112). CIN is premalignant transformation and dysplasia of the cervix and is categorized into three major groups by the World Health Organization: CIN1, CIN2, and CIN3, where CIN1 is the least likely to progress into cervical cancer (113). Without proper diagnosis or medical intervention 5–30% of CIN2/CIN3 (collectively CIN2+) patients develop cervical cancer; however, 10–40% of women diagnosed with CIN2+ exhibit spontaneous regression of the lesion (114). This past year, PLD6 was identified as a predictive biomarker for regression of CIN2+ to CIN1 (108). PLD6 was expressed in 12 out of 20 cervical punch biopsy samples taken from women 25–40 years old who experienced spontaneous regression, whereas no PLD6 expression was found in any of the biopsy samples from women whose CIN2+ progressed to cervical cancer (108). piRNAs can be recovered from the human HeLa cervical cancer cell line, suggesting that the machinery to generate piRNAs is functional in cervical tissue (115). Adding PLD6 to the list of biomarkers for CIN2+ cervical lesions should further increase sensitivity in determining whether a patient’s CIN will spontaneously regress or persist and develop into cervical cancer.
CONCLUDING REMARKS
With many of the PLD-deficiency animal models recently generated, the field is in an explosive period of discovery for roles undertaken by this fascinating superfamily of enzymes. Some of the associated pathophysiological roles reflect undesirable PLD activity, whereas others occur as a consequence of inadequate activity (Table 1). With the on-going development of PLD small molecule inhibitors for several of the superfamily members, the former represent excellent therapeutic opportunities and it is likely that inhibitory strategies targeting PLD1 and PLD2 will find their application in several disease settings.
TABLE 1.
Physiological and pathophysiological processes affected by altered PLD activity
| Isoform | Pathophysiological Process | References |
| PLD1 | Thrombotic disease | (28, 37–40, 49, 50, 52, 55, 116) |
| Cancer: angiogenesis, metastasis, tumor invasion, and hypoxic response | ||
| Immune responses (autoimmunity) | ||
| Tissue fibrosis (cardiac, lung, and liver) | ||
| Muscle regeneration | ||
| PLD2 | Thrombotic disease | (37, 52, 65–69, 74) |
| Cancer: angiogenesis, metastasis, tumor invasion, and hypoxic response | ||
| Immune function | ||
| Influenza virus replication | ||
| AD | ||
| PLD3 | AD | (32, 81, 95–97) |
| Muscle development | ||
| PLD4 | Autoimmunity | (98, 100, 101) |
| PLD5 | Cancer | (102, 106) |
| Autism | ||
| PLD6 | Fertility | (15, 108) |
| Cancer |
Acknowledgments
The authors thank members of the Frohman laboratory for suggestions on the manuscript.
Footnotes
Abbreviations:
- AD
- Alzheimer’s disease
- CIN
- cervical intraepithelial neoplasia
- ER
- endoplasmic reticulum
- ERAD
- endoplasmic reticulum-associated degradation
- miR
- microRNA
- MVB
- multivesicular body
- PA
- phosphatidic acid
- PH
- pleckstrin homology
- piRNA
- piwi-interacting RNA
- PLD
- phospholipase D
- PtdEtOH
- phosphatidylethanol
- PX
- phox consensus sequence
This work was supported by National Institutes of Health Grants R01 GM084251 and GM100109 to M.A.F. and F31 NRSA DK097957 to R.K.N.
REFERENCES
- 1.Jenkins G. M., and Frohman M. A.. 2005. Phospholipase D: a lipid centric review. Cell. Mol. Life Sci. 62: 2305–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sung T. C., Roper R. L., Zhang Y., Rudge S. A., Temel R., Hammond S. M., Morris A. J., Moss B., Engebrecht J., and Frohman M. A.. 1997. Mutagenesis of phospholipase D defines a superfamily including a trans-Golgi viral protein required for poxvirus pathogenicity. EMBO J. 16: 4519–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selvy P. E., Lavieri R. R., Lindsley C. W., and Brown H. A.. 2011. Phospholipase D: enzymology, functionality, and chemical modulation. Chem. Rev. 111: 6064–6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alling C., Gustavsson L., and Anggård E.. 1983. An abnormal phospholipid in rat organs after ethanol treatment. FEBS Lett. 152: 24–28. [DOI] [PubMed] [Google Scholar]
- 5.Frohman M. A. 2015. The phospholipase D superfamily as therapeutic targets. Trends Pharmacol. Sci. 36: 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gnann H., Thierauf A., Hagenbuch F., Röhr B., and Weinmann W.. 2014. Time dependence of elimination of different PEth homologues in alcoholics in comparison with social drinkers. Alcohol. Clin. Exp. Res. 38: 322–326. [DOI] [PubMed] [Google Scholar]
- 7.Omodeo-Salé F., Lindi C., Palestini P., and Masserini M.. 1991. Role of phosphatidylethanol in membranes. Effects on membrane fluidity, tolerance to ethanol, and activity of membrane-bound enzymes. Biochemistry. 30: 2477–2482. [DOI] [PubMed] [Google Scholar]
- 8.Pannequin J., Delaunay N., Darido C., Maurice T., Crespy P., Frohman M. A., Balda M. S., Matter K., Joubert D., Bourgaux J. F., et al. 2007. Phosphatidylethanol accumulation promotes intestinal hyperplasia by inducing ZONAB-mediated cell density increase in response to chronic ethanol exposure. Mol. Cancer Res. 5: 1147–1157. [DOI] [PubMed] [Google Scholar]
- 9.Skippen A., Jones D. H., Morgan C. P., Li M., and Cockcroft S.. 2002. Mechanism of ADP ribosylation factor-stimulated phosphatidylinositol 4,5-bisphosphate synthesis in HL60 cells. J. Biol. Chem. 277: 5823–5831. [DOI] [PubMed] [Google Scholar]
- 10.Sato T., Hongu T., Sakamoto M., Funakoshi Y., and Kanaho Y.. 2013. Molecular mechanisms of N-formyl-methionyl-leucyl-phenylalanine-induced superoxide generation and degranulation in mouse neutrophils: phospholipase D is dispensable. Mol. Cell. Biol. 33: 136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arun S. N., Xie D., Howard A. C., Zhong Q., Zhong X., McNeil P. L., and Bollag W. B.. 2013. Cell wounding activates phospholipase D in primary mouse keratinocytes. J. Lipid Res. 54: 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bollag W. B., Xie D., Zheng X., and Zhong X.. 2007. A potential role for the phospholipase D2-aquaporin-3 signaling module in early keratinocyte differentiation: production of a phosphatidylglycerol signaling lipid. J. Invest. Dermatol. 127: 2823–2831. [DOI] [PubMed] [Google Scholar]
- 13.Choi S. Y., Huang P., Jenkins G. M., Chan D. C., Schiller J., and Frohman M. A.. 2006. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat. Cell Biol. 8: 1255–1262. [DOI] [PubMed] [Google Scholar]
- 14.Voigt F., Reuter M., Kasaruho A., Schulz E. C., Pillai R. S., and Barabas O.. 2012. Crystal structure of the primary piRNA biogenesis factor Zucchini reveals similarity to the bacterial PLD endonuclease Nuc. RNA. 18: 2128–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang H. Y., Gao Q., Peng X. X., Choi S. Y., Sarma K., Ren H. M., Morris A. J., and Frohman M. A.. 2011. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev. Cell. 20: 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammond S. M., Altshuller Y. M., Sung T. C., Rudge S. A., Rose K., Engebrecht J., Morris A. J., and Frohman M. A.. 1995. Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family. J. Biol. Chem. 270: 29640–29643. [DOI] [PubMed] [Google Scholar]
- 17.Colley W. C., Sung T. C., Roll R., Jenco J., Hammond S. M., Altshuller Y., Bar-Sagi D., Morris A. J., and Frohman M. A.. 1997. Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provokes cytoskeletal reorganization. Curr. Biol. 7: 191–201. [DOI] [PubMed] [Google Scholar]
- 18.Du G., Altshuller Y. M., Kim Y., Han J. M., Ryu S. H., Morris A. J., and Frohman M. A.. 2000. Dual requirement for rho and protein kinase C in direct activation of phospholipase D1 through G protein-coupled receptor signaling. Mol. Biol. Cell. 11: 4359–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammond S. M., Jenco J. M., Nakashima S., Cadwallader K., Gu Q., Cook S., Nozawa Y., Prestwich G. D., Frohman M. A., and Morris A. J.. 1997. Characterization of two alternately spliced forms of phospholipase D1. Activation of the purified enzymes by phosphatidylinositol 4,5-bisphosphate, ADP-ribosylation factor, and Rho family monomeric GTP-binding proteins and protein kinase C-alpha. J. Biol. Chem. 272: 3860–3868. [DOI] [PubMed] [Google Scholar]
- 20.Bruntz R. C., Lindsley C. W., and Brown H. A.. 2014. Phospholipase D signaling pathways and phosphatidic acid as therapeutic targets in cancer. Pharmacol. Rev. 66: 1033–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leiros I., Secundo F., Zambonelli C., Servi S., and Hough E.. 2000. The first crystal structure of a phospholipase D. Structure. 8: 655–667. [DOI] [PubMed] [Google Scholar]
- 22.Du G., Altshuller Y. M., Vitale N., Huang P., Chasserot-Golaz S., Morris A. J., Bader M. F., and Frohman M. A.. 2003. Regulation of phospholipase D1 subcellular cycling through coordination of multiple membrane association motifs. J. Cell Biol. 162: 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sciorra V. A., Rudge S. A., Wang J. Y., McLaughlin S., Engebrecht J., and Morris A. J.. 2002. Dual role for phosphoinositides in regulation of yeast and mammalian phospholipase D enzymes. J. Cell Biol. 159: 1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jang J. H., Lee C. S., Hwang D., and Ryu S. H.. 2012. Understanding of the roles of phospholipase D and phosphatidic acid through their binding partners. Prog. Lipid Res. 51: 71–81. [DOI] [PubMed] [Google Scholar]
- 25.Sugars J. M., Cellek S., Manifava M., Coadwell J., and Ktistakis N. T.. 2002. Hierarchy of membrane-targeting signals of phospholipase D1 involving lipid modification of a pleckstrin homology domain. J. Biol. Chem. 277: 29152–29161. [DOI] [PubMed] [Google Scholar]
- 26.Stahelin R. V., Ananthanarayanan B., Blatner N. R., Singh S., Bruzik K. S., Murray D., and Cho W. H.. 2004. Mechanism of membrane binding of the phospholipase D1 PX domain. J. Biol. Chem. 279: 54918–54926. [DOI] [PubMed] [Google Scholar]
- 27.Sung T. C., Zhang Y., Morris A. J., and Frohman M. A.. 1999. Structural analysis of human phospholipase D1. J. Biol. Chem. 274: 3659–3666. [DOI] [PubMed] [Google Scholar]
- 28.Elvers M., Stegner D., Hagedorn I., Kleinschnitz C., Braun A., Kuijpers M. E., Boesl M., Chen Q., Heemskerk J. W., Stoll G., et al. 2010. Impaired alpha(IIb)beta(3) integrin activation and shear-dependent thrombus formation in mice lacking phospholipase D1. Sci. Signal. 3: ra1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munck A., Bohm C., Seibel N. M., Hashemol Hosseini Z., and Hampe W.. 2005. Hu-K4 is a ubiquitously expressed type 2 transmembrane protein associated with the endoplasmic reticulum. FEBS J. 272: 1718–1726. [DOI] [PubMed] [Google Scholar]
- 30.Pedersen K. M., Finsen B., Celis J. E., and Jensen N. A.. 1998. Expression of a novel murine phospholipase D homolog coincides with late neuronal development in the forebrain. J. Biol. Chem. 273: 31494–31504. [DOI] [PubMed] [Google Scholar]
- 31.Kent D. G., Copley M. R., Benz C., Wohrer S., Dykstra B. J., Ma E., Cheyne J., Zhao Y. J., Bowie M. B., Zhao Y., et al. 2009. Prospective isolation and molecular characterization of hematopoietic stem cells with durable self-renewal potential. Blood. 113: 6342–6350. [DOI] [PubMed] [Google Scholar]
- 32.Osisami M., Ali W., and Frohman M. A.. 2012. A role for phospholipase D3 in myotube formation. PLoS ONE. 7: e33341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J., Chen S., Zhang S., Lu Z., Yang H., and Wang H.. 2009. Over-expression of phospholipase D3 inhibits Akt phosphorylation in C2C12 myoblasts [article in Chinese]. Sheng Wu Gong Cheng Xue Bao. 25: 1524–1531. [PubMed] [Google Scholar]
- 34.Yoshikawa F., Banno Y., Otani Y., Yamaguchi Y., Nagakura-Takagi Y., Morita N., Sato Y., Saruta C., Nishibe H., Sadakata T., et al. 2010. Phospholipase D family member 4, a transmembrane glycoprotein with no phospholipase D activity, expression in spleen and early postnatal microglia. PLoS ONE. 5: e13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Y., Stuckey J. A., Lohse D. L., and Dixon J. E.. 1997. Expression, characterization, and crystallization of a member of the novel phospholipase D family of phosphodiesterases. Protein Sci. 6: 2655–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reue K., and Dwyer J. R.. 2009. Lipin proteins and metabolic homeostasis. J. Lipid Res. 50: S109–S114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stegner D., Thielmann I., Kraft P., Frohman M. A., Stoll G., and Nieswandt B.. 2013. Pharmacological inhibition of phospholipase D protects mice from occlusive thrombus formation and ischemic stroke–brief report. Arterioscler. Thromb. Vasc. Biol. 33: 2212–2217. [DOI] [PubMed] [Google Scholar]
- 38.Schönberger T., Jürgens T., Müller J., Armbruster N., Niermann C., Gorressen S., Sommer J., Tian H., di Paolo G., Scheller J., et al. 2014. Pivotal role of phospholipase D1 in tumor necrosis factor-alpha-mediated inflammation and scar formation after myocardial ischemia and reperfusion in mice. Am. J. Pathol. 184: 2450–2464. [DOI] [PubMed] [Google Scholar]
- 39.Chen Q., Hongu T., Sato T., Zhang Y., Ali W., Cavallo J. A., van der Velden A., Tian H., Di Paolo G., Nieswandt B., et al. 2012. Key roles for the lipid signaling enzyme phospholipase d1 in the tumor microenvironment during tumor angiogenesis and metastasis. Sci. Signal. 5: ra79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gobel K., Schuhmann M. K., Pankratz S., Stegner D., Herrmann A. M., Braun A., Breuer J., Bittner S., Ruck T., Wiendl H., et al. 2014. Phospholipase D1 mediates lymphocyte adhesion and migration in experimental autoimmune encephalomyelitis. Eur. J. Immunol. 44: 2295–2305. [DOI] [PubMed] [Google Scholar]
- 41.Dhingra S., Rodriguez M. E., Shen Q., Duan X., Stanton M. L., Chen L., Zhang R., and Brown R. E.. 2011. Constitutive activation with overexpression of the mTORC2-phospholipase D1 pathway in uterine leiomyosarcoma and STUMP: morphoproteomic analysis with therapeutic implications. Int. J. Clin. Exp. Pathol. 4: 134–146. [PMC free article] [PubMed] [Google Scholar]
- 42.Gozgit J. M., Pentecost B. T., Marconi S. A., Ricketts-Loriaux R. S., Otis C. N., and Arcaro K. F.. 2007. PLD1 is overexpressed in an ER-negative MCF-7 cell line variant and a subset of phospho-Akt-negative breast carcinomas. Br. J. Cancer. 97: 809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen Q., Stanton M. L., Feng W., Rodriguez M. E., Ramondetta L., Chen L., Brown R. E., and Duan X.. 2010. Morphoproteomic analysis reveals an overexpressed and constitutively activated phospholipase D1-mTORC2 pathway in endometrial carcinoma. Int. J. Clin. Exp. Pathol. 4: 13–21. [PMC free article] [PubMed] [Google Scholar]
- 44.Omuro A., and DeAngelis L. M.. 2013. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 310: 1842–1850. [DOI] [PubMed] [Google Scholar]
- 45.Wen P. Y., and Kesari S.. 2008. Malignant gliomas in adults. N. Engl. J. Med. 359: 492–507. [DOI] [PubMed] [Google Scholar]
- 46.Oike T., Suzuki Y., Sugawara K., Shirai K., Noda S. E., Tamaki T., Nagaishi M., Yokoo H., Nakazato Y., and Nakano T.. 2013. Radiotherapy plus concomitant adjuvant temozolomide for glioblastoma: Japanese mono-institutional results. PLoS ONE. 8: e78943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Reilly M. C., Scott S. A., Brown K. A., Oguin T. H. 3rd, Thomas P. G., Daniels J. S., Morrison R., Brown H. A., and Lindsley C. W.. 2013. Development of dual PLD1/2 and PLD2 selective inhibitors from a common 1,3,8-triazaspiro[4.5]decane core: discovery of Ml298 and Ml299 that decrease invasive migration in U87-MG glioblastoma cells. J. Med. Chem. 56: 2695–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sayyah J., Bartakova A., Nogal N., Quilliam L. A., Stupack D. G., and Brown J. H.. 2014. The Ras-related protein, Rap1A, mediates thrombin-stimulated, integrin-dependent glioblastoma cell proliferation and tumor growth. J. Biol. Chem. 289: 17689–17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bruntz R. C., Taylor H. E., Lindsley C. W., and Brown H. A.. 2014. Phospholipase D2 mediates survival signaling through direct regulation of Akt in glioblastoma cells. J. Biol. Chem. 289: 600–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han S., Huh J., Kim W., Jeong S., Min do S., and Jung Y.. 2014. Phospholipase D activates HIF-1-VEGF pathway via phosphatidic acid. Exp. Mol. Med. 46: e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park M. H., Ahn B. H., Hong Y. K., and Min do S.. 2009. Overexpression of phospholipase D enhances matrix metalloproteinase-2 expression and glioma cell invasion via protein kinase C and protein kinase A/NF-kappaB/Sp1-mediated signaling pathways. Carcinogenesis. 30: 356–365. [DOI] [PubMed] [Google Scholar]
- 52.Chen Z., Li D., Cheng Q., Ma Z., Jiang B., Peng R., Chen R., Cao Y., and Wan X.. 2014. MicroRNA-203 inhibits the proliferation and invasion of U251 glioblastoma cells by directly targeting PLD2. Mol. Med. Rep. 9: 503–508. [DOI] [PubMed] [Google Scholar]
- 53.Su W., Yeku O., Olepu S., Genna A., Park J. S., Ren H., Du G., Gelb M. H., Morris A. J., and Frohman M. A.. 2009. 5-Fluoro-2-indolyl des-chlorohalopemide (FIPI), a phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis. Mol. Pharmacol. 75: 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seo H. Y., Jang B. K., Jung Y. A., Lee E. J., Kim H. S., Jeon J. H., Kim J. G., Lee I. K., Kim M. K., and Park K. G.. 2014. Phospholipase D1 decreases type I collagen levels in hepatic stellate cells via induction of autophagy. Biochem. Biophys. Res. Commun. 449: 38–43. [DOI] [PubMed] [Google Scholar]
- 55.Zhu X., Liu R., Kuang D., Liu J., Shi X., Zhang T., Zeng Y., Sun X., Zhang Y., and Yang W.. 2014. The role of phospholipase D1 in liver fibrosis induced by dimethylnitrosamine in vivo. Dig. Dis. Sci. 59: 1779–1788. [DOI] [PubMed] [Google Scholar]
- 56.Patel R. B., Kotha S. R., Sherwani S. I., Sliman S. M., Gurney T. O., Loar B., Butler S. O., Morris A. J., Marsh C. B., and Parinandi N. L.. 2011. Pulmonary fibrosis inducer, bleomycin, causes redox-sensitive activation of phospholipase D and cytotoxicity through formation of bioactive lipid signal mediator, phosphatidic acid, in lung microvascular endothelial cells. Int. J. Toxicol. 30: 69–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dent M. R., Singal T., Dhalla N. S., and Tappia P. S.. 2004. Expression of phospholipase D isozymes in scar and viable tissue in congestive heart failure due to myocardial infarction. J. Cell. Mol. Med. 8: 526–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tappia P. S., and Dhalla N. S.. 2014. Alterations in phospholipase D during the development of myocardial disease. In Phospholipases in Health and Disease. Vol. 10. P. S. Tappia and N. S. Dhalla, editors. Springer-Verlag, New York. 381–393. [Google Scholar]
- 59.Dall’Armi C., Hurtado-Lorenzo A., Tian H. S., Morel E., Nezu A., Chan R. B., Yu W. H., Robinson K. S., Yeku O., Small S. A., et al. 2010. The phospholipase D1 pathway modulates macroautophagy. Nat. Commun. 1: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bae E. J., Lee H. J., Jang Y. H., Michael S., Masliah E., Min D. S., and Lee S. J.. 2014. Phospholipase D1 regulates autophagic flux and clearance of alpha-synuclein aggregates. Cell Death Differ. 21: 1132–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rautou P. E., Mansouri A., Lebrec D., Durand F., Valla D., and Moreau R.. 2010. Autophagy in liver diseases. J. Hepatol. 53: 1123–1134. [DOI] [PubMed] [Google Scholar]
- 62.Levine B., and Kroemer G.. 2008. Autophagy in the pathogenesis of disease. Cell. 132: 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Domart M. C., Degli Esposti D., Sebagh M., Olaya N., Harper F., Pierron G., Franc B., Tanabe K. K., Debuire B., Azoulay D., et al. 2009. Concurrent induction of necrosis, apoptosis, and autophagy in ischemic preconditioned human livers formerly treated by chemotherapy. J. Hepatol. 51: 881–889. [DOI] [PubMed] [Google Scholar]
- 64.Yamamoto K., Takahashi Y., Mano T., Sakata Y., Nishikawa N., Yoshida J., Oishi Y., Hori M., Miwa T., Inoue S., et al. 2004. N-methylethanolamine attenuates cardiac fibrosis and improves diastolic function: inhibition of phospholipase D as a possible mechanism. Eur. Heart J. 25: 1221–1229. [DOI] [PubMed] [Google Scholar]
- 65.Ali W. H., Chen Q., Delgiorno K. E., Su W., Hall J. C., Hongu T., Tian H., Kanaho Y., Di Paolo G., Crawford H. C., et al. 2013. Deficiencies of the lipid-signaling enzymes phospholipase D1 and D2 alter cytoskeletal organization, macrophage phagocytosis, and cytokine-stimulated neutrophil recruitment. PLoS ONE. 8: e55325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hong K. W., Jin H. S., Lim J. E., Cho Y. S., Go M. J., Jung J., Lee J. E., Choi J., Shin C., Hwang S. Y., et al. 2010. Non-synonymous single-nucleotide polymorphisms associated with blood pressure and hypertension. J. Hum. Hypertens. 24: 763–774. [DOI] [PubMed] [Google Scholar]
- 67.Ghim J., Moon J. S., Lee C. S., Lee J., Song P., Lee A., Jang J. H., Kim D., Yoon J. H., Koh Y. J., et al. 2014. Endothelial deletion of phospholipase D2 reduces hypoxic response and pathological angiogenesis. Arterioscler. Thromb. Vasc. Biol. 34: 1697–1703. [DOI] [PubMed] [Google Scholar]
- 68.Henkels K. M., Boivin G. P., Dudley E. S., Berberich S. J., Gomez-Cambronero J., and Phospholipase D.. 2013. PLD) drives cell invasion, tumor growth and metastasis in a human breast cancer xenograph model. Oncogene. 32: 5551–5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oliveira T. G., Chan R. B., Tian H., Laredo M., Shui G., Staniszewski A., Zhang H., Wang L., Kim T. W., Duff K. E., et al. 2010. Phospholipase d2 ablation ameliorates Alzheimer’s disease-linked synaptic dysfunction and cognitive deficits. J. Neurosci. 30: 16419–16428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Horimoto T., and Kawaoka Y.. 2005. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Microbiol. 3: 591–600. [DOI] [PubMed] [Google Scholar]
- 71.Neumann G., Noda T., and Kawaoka Y.. 2009. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 459: 931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dong G., Peng C., Luo J., Wang C., Han L., Wu B., Ji G., and He H.. 2015. Adamantane-resistant influenza a viruses in the world (1902–2013): frequency and distribution of m2 gene mutations. PLoS ONE. 10: e0119115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karlas A., Machuy N., Shin Y., Pleissner K. P., Artarini A., Heuer D., Becker D., Khalil H., Ogilvie L. A., Hess S., et al. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 463: 818–822. [DOI] [PubMed] [Google Scholar]
- 74.Oguin T. H. 3rd, Sharma S., Stuart A. D., Duan S., Scott S. A., Jones C. K., Daniels J. S., Lindsley C. W., Thomas P. G., and Brown H. A.. 2014. Phospholipase D facilitates efficient entry of influenza virus, allowing escape from innate immune inhibition. J. Biol. Chem. 289: 25405–25417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Uchida N., Okamura S., and Kuwano H.. 1999. Phospholipase D activity in human gastric carcinoma. Anticancer Res. 19(1B): 671–675. [PubMed] [Google Scholar]
- 76.Yamada Y., Hamajima N., Kato T., Iwata H., Yamamura Y., Shinoda M., Suyama M., Mitsudomi T., Tajima K., Kusakabe S., et al. 2003. Association of a polymorphism of the phospholipase D2 gene with the prevalence of colorectal cancer. J. Mol. Med. (Berl). 81: 126–131. [DOI] [PubMed] [Google Scholar]
- 77.Toschi A., Edelstein J., Rockwell P., Ohh M., and Foster D. A.. 2008. HIF alpha expression in VHL-deficient renal cancer cells is dependent on phospholipase D. Oncogene. 27: 2746–2753. [DOI] [PubMed] [Google Scholar]
- 78.Saito M., Iwadate M., Higashimoto M., Ono K., Takebayashi Y., and Takenoshita S.. 2007. Expression of phospholipase D2 in human colorectal carcinoma. Oncol. Rep. 18: 1329–1334. [PubMed] [Google Scholar]
- 79.Liu M., Du K., Fu Z., Zhang S., and Wu X.. 2015. Hypoxia-inducible factor 1-alpha up-regulates the expression of phospholipase D2 in colon cancer cells under hypoxic conditions. Med. Oncol. 32: 394. [DOI] [PubMed] [Google Scholar]
- 80.Cai D., Netzer W. J., Zhong M., Lin Y., Du G., Frohman M., Foster D. A., Sisodia S. S., Xu H., Gorelick F. S., et al. 2006. Presenilin-1 uses phospholipase D1 as a negative regulator of beta-amyloid formation. Proc. Natl. Acad. Sci. USA. 103: 1941–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cruchaga C., Karch C. M., Jin S. C., Benitez B. A., Cai Y., Guerreiro R., Harari O., Norton J., Budde J., Bertelsen S., et al. 2014. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature. 505: 550–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lein E. S., Hawrylycz M. J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A. F., Boguski M. S., Brockway K. S., Byrnes E. J., et al. 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 445: 168–176. [DOI] [PubMed] [Google Scholar]
- 83.Hawrylycz M. J., Lein E. S., Guillozet-Bongaarts A. L., Shen E. H., Ng L., Miller J. A., van de Lagemaat L. N., Smith K. A., Ebbert A., Riley Z. L., et al. 2012. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 489: 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kong W., Mou X., Liu Q., Chen Z., Vanderburg C. R., Rogers J. T., and Huang X.. 2009. Independent component analysis of Alzheimer’s DNA microarray gene expression data. Mol. Neurodegener. 4: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chadwick W, Brenneman R, Martin B, Maudsley S.. Complex and multidimensional lipid raft alterations in a murine model of Alzheimer’s disease. Int J Alzheimers Dis. 2010;2010:604792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morel E., Chamoun Z., Lasiecka Z. M., Chan R. B., Williamson R. L., Vetanovetz C., Dall’Armi C., Simoes S., Point Du Jour K. S., McCabe B. D., et al. 2013. Phosphatidylinositol-3-phosphate regulates sorting and processing of amyloid precursor protein through the endosomal system. Nat. Commun. 4: 2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Small S. A., and Gandy S.. 2006. Sorting through the cell biology of Alzheimer’s disease: intracellular pathways to pathogenesis. Neuron. 52: 15–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pridgeon J. W., Webber E. A., Sha D., Li L., and Chin L. S.. 2009. Proteomic analysis reveals Hrs ubiquitin-interacting motif-mediated ubiquitin signaling in multiple cellular processes. FEBS J. 276: 118–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brunner Y., Coute Y., Iezzi M., Foti M., Fukuda M., Hochstrasser D. F., Wollheim C. B., and Sanchez J. C.. 2007. Proteomics analysis of insulin secretory granules. Mol. Cell. Proteomics. 6: 1007–1017. [DOI] [PubMed] [Google Scholar]
- 90.Palmieri M., Impey S., Kang H., di Ronza A., Pelz C., Sardiello M., and Ballabio A.. 2011. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 20: 3852–3866. [DOI] [PubMed] [Google Scholar]
- 91.Lee K. A., Hammerle L. P., Andrews P. S., Stokes M. P., Mustelin T., Silva J. C., Black R. A., and Doedens J. R.. 2011. Ubiquitin ligase substrate identification through quantitative proteomics at both the protein and peptide levels. J. Biol. Chem. 286: 41530–41538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu B., Zheng Y., Wang T. D., Xu H. Z., Xia L., Zhang J., Wu Y. L., Chen G. Q., and Wang L. S.. 2012. Proteomic identification of common SCF ubiquitin ligase FBXO6-interacting glycoproteins in three kinds of cells. J. Proteome Res. 11: 1773–1781. [DOI] [PubMed] [Google Scholar]
- 93.Shi Y., Chan D. W., Jung S. Y., Malovannaya A., Wang Y., and Qin J.. 2011. A data set of human endogenous protein ubiquitination sites. Mol. Cell. Proteomics. 10: 002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., Bork P., Kondrashov A. S., and Sunyaev S. R.. 2010. A method and server for predicting damaging missense mutations. Nat. Methods. 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Heilmann S., Drichel D., Clarimon J., Fernandez V., Lacour A., Wagner H., Thelen M., Hernandez I., Fortea J., Alegret M., et al. 2015. PLD3 in non-familial Alzheimer’s disease. Nature. 520: E3–E5. [DOI] [PubMed] [Google Scholar]
- 96.Hooli B. V., Lill C. M., Mullin K., Qiao D., Lange C., Bertram L., and Tanzi R. E.. 2015. PLD3 gene variants and Alzheimer’s disease. Nature. 520: E7–E8. [DOI] [PubMed] [Google Scholar]
- 97.Lambert J. C., Grenier-Boley B., Bellenguez C., Pasquier F., Campion D., Dartigues J. F., Berr C., Tzourio C., and Amouyel P.. 2015. PLD3 and sporadic Alzheimer’s disease risk. Nature. 520: E1. [DOI] [PubMed] [Google Scholar]
- 98.Otani Y., Yamaguchi Y., Sato Y., Furuichi T., Ikenaka K., Kitani H., and Baba H.. 2011. PLD4 is involved in phagocytosis of microglia: expression and localization changes of PLD4 are correlated with activation state of microglia. PLoS ONE. 6: e27544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jung S., Pausch H., Langenmayer M. C., Schwarzenbacher H., Majzoub-Altweck M., Gollnick N. S., and Fries R.. 2014. A nonsense mutation in PLD4 is associated with a zinc deficiency-like syndrome in Fleckvieh cattle. BMC Genomics. 15: 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Okada Y., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Kawaguchi T., Stahl E. A., Kurreeman F. A., Nishida N., et al. 2012. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat. Genet. 44: 511–516. [DOI] [PubMed] [Google Scholar]
- 101.Terao C., Ohmura K., Kawaguchi Y., Nishimoto T., Kawasaki A., Takehara K., Furukawa H., Kochi Y., Ota Y., Ikari K., et al. 2013. PLD4 as a novel susceptibility gene for systemic sclerosis in a Japanese population. Arthritis Rheum. 65: 472–480. [DOI] [PubMed] [Google Scholar]
- 102.Aissani B., Wiener H., and Zhang K.. 2013. Multiple hits for the association of uterine fibroids on human chromosome 1q43. PLoS ONE. 8: e58399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cook E. H. Jr., and Scherer S. W.. 2008. Copy-number variations associated with neuropsychiatric conditions. Nature. 455: 919–923. [DOI] [PubMed] [Google Scholar]
- 104.Lord C., Rutter M., and Le Couteur A.. 1994. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J. Autism Dev. Disord. 24: 659–685. [DOI] [PubMed] [Google Scholar]
- 105.Lord C., Risi S., Lambrecht L., Cook E. H. Jr., Leventhal B. L., DiLavore P. C., Pickles A., and Rutter M.. 2000. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J. Autism Dev. Disord. 30: 205–223. [PubMed] [Google Scholar]
- 106.Anney R., Klei L., Pinto D., Regan R., Conroy J., Magalhaes T. R., Correia C., Abrahams B. S., Sykes N., Pagnamenta A. T., et al. 2010. A genome-wide scan for common alleles affecting risk for autism. Hum. Mol. Genet. 19: 4072–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Uleberg K. E., Munk A. C., Skaland I., Furlan C., van Diermen B., Gudlaugsson E., Janssen E. A., Malpica A., Feng W., Hjelle A., et al. 2011. A protein profile study to discriminate CIN lesions from normal cervical epithelium. Cell. Oncol. (Dordr). 34: 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Uleberg K. E., Ovestad I. T., Munk A. C., Brede C., van Diermen B., Gudlaugsson E., Janssen E. A., Hjelle A., and Baak J. P.. 2014. Prediction of spontaneous regression of cervical intraepithelial neoplasia lesions grades 2 and 3 by proteomic analysis. Int. J. Proteomics. 2014: 129064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jemal A., Bray F., Center M. M., Ferlay J., Ward E., and Forman D.. 2011. Global cancer statistics. CA Cancer J. Clin. 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 110.zur Hausen H. 2009. Papillomaviruses in the causation of human cancers - a brief historical account. Virology. 384: 260–265. [DOI] [PubMed] [Google Scholar]
- 111.Walboomers J. M. M., Jacobs M. V., Manos M. M., Bosch F. X., Kummer J. A., Shah K. V., Snijders P. J. F., Peto J., Meijer C. J. L. M., and Munoz N.. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189: 12–19. [DOI] [PubMed] [Google Scholar]
- 112.Insinga R. P., Dasbach E. J., and Elbasha E. H.. 2009. Epidemiologic natural history and clinical management of human papillomavirus (HPV) disease: a critical and systematic review of the literature in the development of an HPV dynamic transmission model. BMC Infect. Dis. 9: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.World Health Organization. 2013. WHO Guidelines for Screening and Treatment of Precancerous Lesions for Cervical Cancer Prevention. World Health Organization, Geneva. PMID: 24716265. [PubMed] [Google Scholar]
- 114.Ostör A. G. 1993. Natural history of cervical intraepithelial neoplasia: a critical review. Int. J. Gynecol. Pathol. 12: 186–192. [PubMed] [Google Scholar]
- 115.Lu Y., Li C., Zhang K., Sun H., Tao D., Liu Y., Zhang S., and Ma Y.. 2010. Identification of piRNAs in Hela cells by massive parallel sequencing. BMB Rep. 43: 635–641. [DOI] [PubMed] [Google Scholar]
- 116.Teng S., Stegner D., Chen Q., Hongu T., Hasegawa H., Chen L., Kanaho Y., Nieswandt B., Frohman M. A., and Huang P.. 2015. Phospholipase D1 facilitates second-phase myoblast fusion and skeletal muscle regeneration. Mol. Biol. Cell. 26: 506–517. [DOI] [PMC free article] [PubMed] [Google Scholar]