Summary
A hallmark of bacterial biofilms is a self-produced extracellular matrix of exopolysaccharide, extracellular DNA (eDNA) and proteins that hold bacterial cells together in the community. However, interactions among matrix components and how the interactions contribute to the formation of matrix remain unclear. Here, we show the physical interaction between exopolysaccharide Psl and eDNA, the two key biofilm matrix components of the opportunistic pathogen Pseudomonas aeruginosa. The interaction allows the two components to combine to form a web of eDNA–Psl fibres, which resembles a biofilm skeleton in the centre of pellicles to give bacteria structural support and capability against agents targeted on one matrix component. The web of eDNA–Psl fibres was also found in flow-cell biofilms at microcolonies initiation stage. The colocalization of eDNA or Psl fibres with bacterial cell membrane stain suggests that fibre-like eDNA is likely derived from the lysis of dead bacteria in biofilms. Psl can interact with DNA from diverse sources, suggesting that P. aeruginosa has the ability to use DNA of other organisms (such as human neutrophils and other bacterial species) to form its own communities, which might increase the survival of P. aeruginosa in multispecies biofilms or within a human host.
Introduction
The complex multicellular communities of microorganisms known as biofilms are of high significance to industrial, environmental and clinical settings. Biofilms, for example, are a source of persistent infections (Costerton et al., 1999). The biofilm matrix is a poorly defined mixture of extracellular DNA (eDNA), exopolysaccharides and proteins that enmesh microbial cells together into a community and provide protection for resident cells in the biofilm. To date, the interactions among the components of the biofilm matrix, especially between two critical components, eDNA and exopolysachcharide, remain unclear.
Extracellular DNA is an important and abundant matrix component of many single- and multispecies cultured biofilms (Whitchurch et al., 2002; Flemming and Wingender, 2010). Extracellular DNA strengthens biofilms, confers antibiotic resistance, and acts as a nutrient source during starvation and a gene pool for the horizontal gene transfer (Molin and Tolker-Nielsen, 2003; Dominiak et al., 2011; Chiang et al., 2013). Extracellular DNA can also facilitate twitching motility-mediated biofilm expansion (Gloag et al., 2013). That eDNA functions as a matrix component was first reported in Pseudomonas aeruginosa, an important opportunistic pathogen and a paradigm organism for biofilm research (Whitchurch et al., 2002). DNase I can disrupt ‘young’ but not established, ‘aged’ biofilms of P. aeruginosa (Whitchurch et al., 2002; Parks et al., 2009), suggesting the complexity and dynamic of biofilm matrix. As a matrix component, eDNA is mostly found in microbial communities rather than in the tissues of animals and plants.
Unlike eDNA, exopolysaccharide is a common matrix component in plant and animal. It is also a critical biofilm matrix component of many Gram-positive and Gram-negative bacteria (Ma et al., 2009; Kolodkin-Gal et al., 2012; Xiao et al., 2012). Despite highly aggressive antimicrobial therapy, P. aeruginosa (a Gram-negative bacterium) causes life-threatening, persistent infections in cystic fibrosis (CF) patients. Persistence is due to the ability of these bacteria to form biofilms (Singh et al., 2000). Thus, many studies have focused on biofilm formation of P. aeruginosa. Exopolysaccharide Psl is a key scaffolding component in P. aeruginosa that promotes bacterial cell–cell and cell–surface interactions by acting as ‘molecular glue’; Psl forms a fibre-like matrix to maintain the biomass of flow-cell biofilms as well as floating biofilms known as pellicles at the air–liquid interface of standing cultures (Ma et al., 2009; Wang et al., 2013). Psl can also function as a signal to stimulate biofilm formation by P. aeruginosa (Irie et al., 2012). Researchers have proposed that the formation of Psl fibres or tracks is a result of Psl release from the bacterial cell surface during bacterial migration mediated by type IV pili (Wang et al., 2013). Zhao and colleagues (2013) also showed that P. aeruginosa could deposit Psl trails during migration on a surface and that such trails guided subsequent bacterial exploration, leading to the formation of microcolonies (Zhao et al., 2013).
In this study, we demonstrate the existence of the physical interaction between eDNA and Psl, which enables the formation of an eDNA–Psl–membrane combined fibres web in the centre of biofilms, resembling a skeleton of biofilms. Psl can interact not only with DNA of P. aeruginosa, but also the genomic DNA from human neutrophils and Gram-positive bacterium Staphylococcus aureus, implying that P. aeruginosa has the ability to use DNA of other organisms to form its own communities.
Results and discussion
A skeleton-like web of eDNA–Psl fibres located in the centre of air–liquid interface biofilms (pellicles) of P. aeruginosa PAO1
We previously showed that the exopolysaccharide Psl can form a fibre-like matrix in pellicles and flow-cell biofilms of P. aeruginosa (Wang et al., 2013). While detecting DNA in P. aeruginosa pellicles with SYTO9, a green fluorescent DNA dye that stains genomic DNA within bacteria (concentrated fluorescent dot) as well as eDNA (diffused fluorescence or fibre-like structure) in biofilms, we found that eDNA can form a fibre-like structure similar to the Psl matrix. The DNA fibre-like structure was frequently observed in an established pellicle that was several micrometres thick (Fig. 1A and B). Analysis of 45 image stacks indicated that the frequency of DNA fibres observed in 2 day old pellicles was over 90%. Like the Psl fibre matrix in pellicles (Wang et al., 2013), the DNA fibre web has a radial pattern and is located in the middle of pellicles (Fig. 1B). By using the DNA dye SYTO9 along with the Psl-staining Hippeastrum hybrid lectin from amaryllis (HHA), we found that most of the fibre-like DNA was colocalized with Psl fibres in P. aeruginosa pellicles (Fig. 1C; Fig. S1). The analysis of five image stacks by three different methods showed that the DNA–Psl colocalization coefficients were all above 0.5 (Fig. 2). The visible DNA–Psl colocalization was mostly associated with a fibre-like/rope-like matrix structure (Fig. 1C and E; Fig. S1). Such eDNA–Psl fibres were mostly found in the middle to the air face of pellicles (Fig. 2; Fig. S1). The DNA–Psl fibre structure resembled a ‘skeleton’ of biofilms or backbones that was located at the centre of pellicles and that was surrounded by bacteria (Fig. 1A–F). The eDNA–Psl fibres that had strong fluorescent signals (indicated by a big arrow in Fig. 1F) or had weak fluorescent signals (indicated by a small arrow in Fig. 1F) were both found in a pellicle.
Fig. 1.
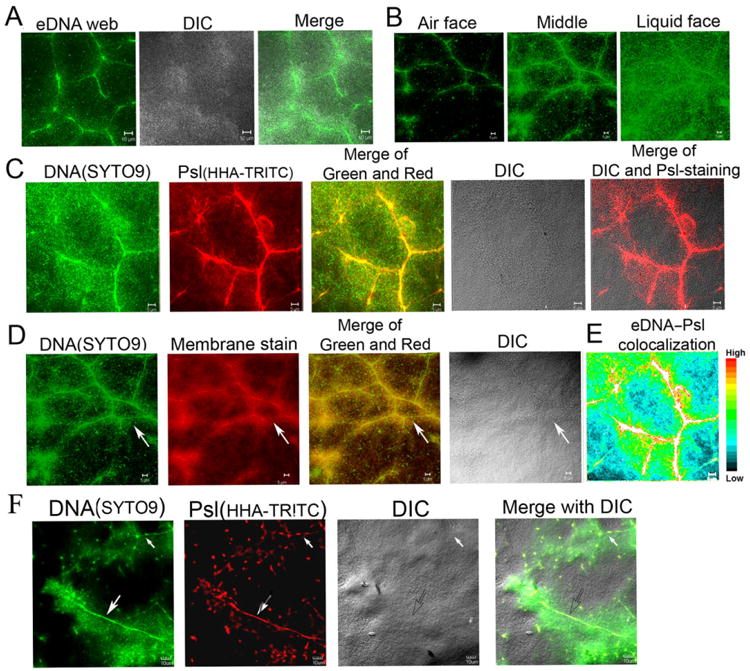
The web of DNA fibres and its association with Psl polysaccharide and bacterial cell membrane in the air–liquid interface biofilms (pellicles) of P. aeruginosa.
A. A web of fibre-like DNA (stained in green by SYTO9) was clearly visualized at the middle of pellicles grown at air–liquid interface of standing culture.
B. The optical sectioned images showed the location of eDNA fibres web (green) in a pellicle.
C. The eDNA fibres web (green) was associated with the fibres of Psl polysaccharide (stained in red by lectin HHA-TRITC) in pellicles.
D. The eDNA fibres were colocalized with bacterial membrane in a 46 h pellicle stained by SYTO9 and the cell membrane stained by FM6-64 (the arrow indicated the eDNA-membrane fibres web).
E. The eDNA–Psl colocalization was mostly associated with the fibre-like matrix structure depicted by the colour map, which was made by the ImageJ software according to the colour image series of (C).
F. The eDNA–Psl fibres with strong (indicated by a big arrow) or weak (indicated by a small arrow) fluorescent signal both found in the middle of a pellicle. Scale bar in all panels: 5 μm.
Fig. 2.
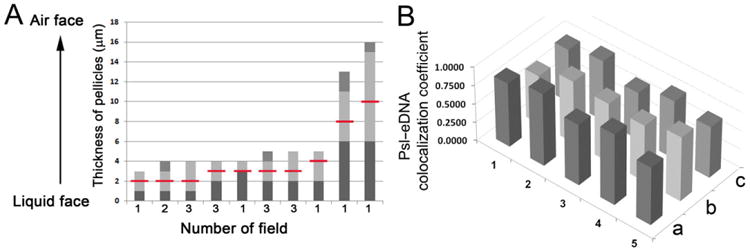
The area in pellicles found eDNA–Psl fibres and the analysis of eDNA–Psl colocalization coefficients.
A. Analysis of 19 CLSM image stacks from 2-day old pellicles showed the thickness of pellicles, the area (light grey columns) with eDNA–Psl fibres, and the location found the eDNA–Psl fibres with highest fluorescent intensity (indicated by the red lines).
B. Columns showed the eDNA–Psl colocalization coefficients analysed from five images (No #1–5) by three methods. a, Pearson's correlation coefficient; b, Manders' colocalization coefficient M1; c, Manders' colocalization coefficient M2.
To further confirm that the DNA fibres were eDNA, we used propidium iodide (PI), a red fluorescent dye that is not permeable to cell membranes. PI is often used for staining eDNA in biofilms and it can also stain genomic DNA within bacteria that had compromised cell membranes. Thus bacterial cells stained by PI is considered to be dead or dying. The results of PI and SYTO9 double stainings showed that SYTO9-stained DNA fibres in pellicles were overlapped with eDNA fibres stained by PI (Fig. 3A). In contrast, little overlapping was found between SYTO9-stained genomic DNA within live bacteria and PI-stained genomic DNA in dead bacteria (Fig. 3A). This data further confirmed that DNA fibres in pellicles were eDNA. In addition, PI and Psl double stainings also indicated the colocalization of eDNA fibres and Psl (Fig. 3B).
Fig. 3.
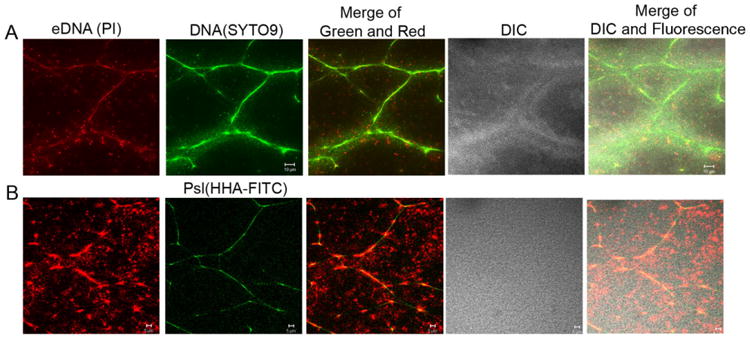
The eDNA and dead bacteria in pellicles of P. aeruginosa PAO1 were stained by Propidium iodide (PI) to confirm that fibre-like DNA was eDNA. Shown were the optical sectioned images in the middle of pellicles. (A) PI (red) and SYTO9 (green) double-stained images of a 2-day-old pellicle. (B) PI (red) and HHA-FITC (a lectin that stains Psl in green) double-stained images of a 2-day-old pellicle. Scale bar: 10 μm for (A); 5 μm for (B).
The eDNA–Psl fibres are also present in flow-cell biofilms
Double staining with SYTO9/HHA-TRITC (Fig. 4A) or PI/HHA-FITC (Fig. 4B) indicated that eDNA colocalized with Psl and formed fibre-like structures in flow-cell biofilms at microcolonies initiation stage. Most of the eDNA–Psl combined fibres were either surrounding multiple bacterial cell aggregates or in areas with a few bacteria (Fig. 4). The distribution pattern was similar to that of Psl fibre matrices in flow-cell biofilms reported previously (Wang et al., 2013). Some eDNA–Psl fibres (indicated by the white arrows in Fig. 4) were also visualized in differential interference contrast (DIC) images. Many dead cells that were stained by PI and had a localized staining pattern (indicated by the black arrow in Fig. 4B) were often associated with eDNA–Psl fibres. The web of eDNA–Psl fibres in flow-cell biofilms did not appear identical to the web of eDNA–Psl fibres in pellicles. The differences in the pattern of eDNA–Psl fibres in pellicles versus flow-cell biofilms were most likely due to differences in growth conditions and flow stress. The flow-cell biofilms were attached to a solid glass surface and grew with continuously flowing media, while the pellicles were floating on liquid media with limited flow. Taken together, our data suggested that the Psl–eDNA association and the formation of eDNA–Psl fibres were not specific to air–liquid interface biofilms but occurred in both types of biofilms.
Fig. 4.
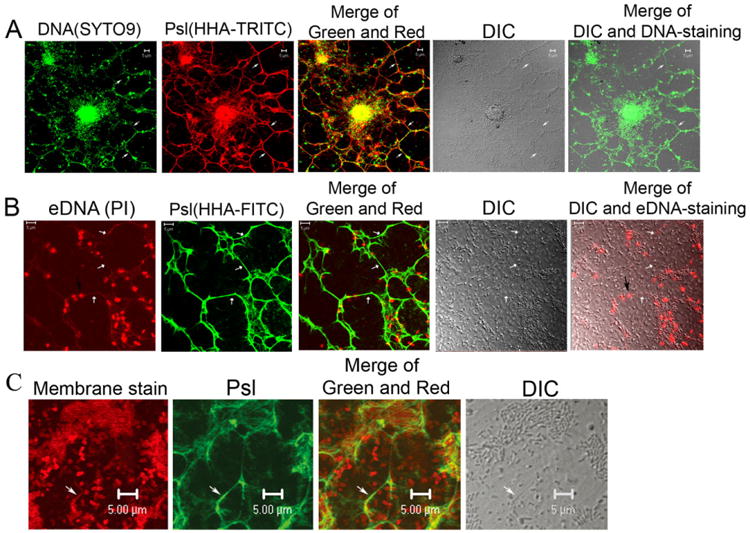
The eDNA–Psl fibres in flow-cell biofilms of P. aeruginosa.
A. SYTO9 and Psl staining (HHA-TRITC) showed the colocalization of Psl fibres with eDNA in a flow-cell biofilm.
B. eDNA fibres (stained in red by PI) were colocalized with Psl fibres (stained in green by lectin HHA-FITC) in a flow-cell biofilm. The black arrow indicated the dead bacteria associated with the eDNA–Psl fibres. The white arrows indicated the visible eDNA–Psl fibres on DIC images.
C. A track of bacterial membrane (stain in red by FM4-64) was colocalized with Psl (green, stained by HHA-FITC) in a flow-cell biofilm. The white arrow indicated a Psl-membrane track and its corresponding DIC image. Scale bar in all panels: 5 μm.
eDNA fibres were likely derived from dead bacteria in biofilms
While a living bacterial cell is doubled-stained by SYTO9 and FM4-64, the genomic DNA appears usually as a concentrate fluorescent dot within bacterial cell membrane. The DNA fibres visualized in pellicles were overlapped with bacterial membrane stained by FM4-64 (Fig. 1D) and DNA stained by PI (Fig. 3), which suggested that eDNA fibres were likely derived from dead or dying bacteria (membrane-compromised bacteria). In support of this suggestion, a track of bacterial cell membrane staining was observed in a flow-cell biofilm that appeared following a bacterial cell and was colocalized with Psl staining (Fig. 4C). This membrane track was also clearly visible in the DIC image (indicated by big white arrows in Fig. 4C), which was similar to the eDNA–Psl fibres depicted in Fig. 4A and B. The membrane of a live bacterium would not be likely peeled off to leave a membrane track; thus, these results suggested that eDNA fibres were likely derived from dead bacteria in biofilms, and flow stress might peel off the membrane of membrane-compromised bacteria and stretch out their chromosome DNA, leading to the formation of eDNA–Psl-membrane fibres.
Previous reports have not found detectable eDNA associated with Psl trails/tracks from living bacteria (Wang et al., 2013; Zhao et al., 2013). Psl released from a living bacterium may be associated with limited eDNA (≈2.5 × 10−8 μg per bacterial cell) (Allesen-Holm et al., 2006), which may be under the fluorescence-detecting level. The formation of Psl–eDNA fibres with strong fluorescent signals (Fig. 1F, the stronger fluorescent signal indicates that the fibre has more Psl and DNA material) is likely a result of multiple events (such as lysis of multiple bacteria). Initially, the eDNA or Psl–eDNA tracks may have a weak fluorescent signal (such as the one indicated by the small arrow in Fig. 1F). However, bacteria attached to eDNA or Psl–eDNA fibres (or tracks) may die, and their DNA and Psl may merge with the original fibre and thereby increase their volume.
Psl interacts with DNA from diverse sources
The association of eDNA and Psl in biofilms suggested that there may be physical interaction between the two components. To determine whether there is physical interaction between the two main biofilm matrix components, Psl and eDNA, we used a modified competitive binding assay. Psl can be coated on microtiter plates and detected by ELISA with anti-Psl serum (Byrd et al., 2010); thus, the blocking of Psl coated on wells of ELISA plates by DNA represents a form of competitive binding assay that tests the interaction between Psl and DNA. To perform the assay, we used P. aeruginosa genomic DNA (eDNA in P. aeruginosa biofilm is derived from random chromosomal DNA) from an exopolysaccharide-non-producing strain (algC mutant) (Ma et al., 2012), which eliminated potential interference from exopolysaccharides associated with purified DNA. The results showed that P. aeruginosa genomic DNA inhibited the detection of Psl by the anti-Psl antibody (Fig. 5A). P. aeruginosa DNA 120 μg μl−1 (black column in Fig. 5A) reduced detection of Ps1 by 50% (compared with the white column in Fig. 5A). When a lower concentration of DNA was used, the inhibition was reduced (grey column in Fig. 5A). These results indicated that DNA can bind to Psl coated on ELISA wells and can thereby interfere with the binding of anti-Psl antibody to Psl, suggesting that Psl physically interacts with P. aeruginosa DNA.
Fig. 5.
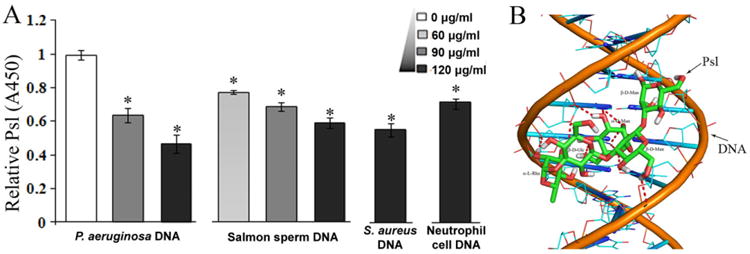
The interaction between DNA and Psl polysaccharide.
A. The detection of Psl can be interfered by DNA isolated from P. aeruginosa, salmon sperm, S. aureus and neutrophil cells. Psl was detected by anti-Psl antibody in ELISA assay. The samples without DNA blocking were normalized to 1 (A450 = 0.635 ± 0.012). *P< 0.01.
B. A plausible model for DNA–Psl interaction. The docking result suggested that hydrogen bonds (indicated by dash lines) can be formed between a Psl polysaccharide repeat unit (carbon, green; oxygen, red; hydrogen, white) and the standard B-DNA duplex (phosphorus, orange; carbon, cyan; oxygen, red; nitrogen, blue).
To examine whether the interaction between Psl and DNA is specific, we used salmon sperm DNA for the competitive-binding assay. Like P. aeruginosa DNA, salmon sperm DNA interfered with the binding of anti-Psl antibody to Psl, and the degree of interference promoted with an increase of salmon sperm DNA concentration (Fig. 5A). This result suggested that Psl polysaccharide can bind to both prokaryotic and eukaryotic DNA.
P. aeruginosa and the Gram-positive bacterium S. aureus are two prevalent species that often infect CF patients (Harrison, 2007). P. aeruginosa can lyse S. aureus and other Gram-positive bacteria (Mashburn et al., 2005; Harrison, 2007). Although bacterial DNA can activate neutrophils (Fuxman Bass et al., 2010), the presence of human neutrophils promotes biofilm formation by P. aeruginosa (Walker et al., 2005). In addition, human neutrophils can release granule proteins and chromatin that together form extracellular fibres to trap bacteria (Brinkmann et al., 2004), yet most of the trapped bacteria on neutrophils extracellular traps (NETs) remain alive (Menegazzi et al., 2012). P. aeruginosa was recently showed to induce NET formation (Yoo et al., 2014). Since the Psl–eDNA interaction is not specific, we tested the hypothesis that P. aeruginosa Psl is able to bind the genomic DNA from human neutrophils and S. aureus by the competitive binding assay. The results showed that both S. aureus and neutrophils DNA inhibited the ability of anti-Psl serum to detect Psl in the ELISA assay, suggesting that Psl can interact with DNA from S. aureus and human neutrophils (Fig. 5A). This result also implies that the eDNA–Psl interaction might increase the survival of P. aeruginosa in the lungs of CF patients by allowing the bacterial cells of P. aeruginosa to use NETs or eDNA from other microbial species as a scaffold on which to grow its own communities. In consistent with our perspective, recent reports also suggested the non-mucoid P. aeruginosa strains (produce Psl, but little alginate exopolysaccharide) induces NET formation better than the mucoid strains (produce predominant alginate, but little Psl), and in the case of CF, NET release may actually support initial bacterial sequestration and drive mutability (Dwyer et al., 2014; Rahman and Gadjeva, 2014).
Modelling of Psl–DNA interaction
To know how Psl may interact with DNA, we used the Glycam Biomolecule Builder to generate an energy-minimized spatial structure model of the Psl repeat unit (Appendix S1). AutoDock software was used to mimic the physical interaction between a single repeat unit of Psl and the DNA double helix. The results suggested that Psl was able to fit into the minor groove of the DNA double helix, allowing hydrogen bonds to form between DNA and Psl (Fig. 5B). The estimated interaction energy for the Psl–DNA complex was −2.18 kcal mol−1.
Psl–DNA interaction determined by isothermal titration calorimetry (ITC)
Isothermal titration calorimetry is an important technique to study the thermodynamics of molecular interactions. This technique was recently used to study the binding of protein to lipid polysaccharide (LPS) (Gries et al., 2014). We used ITC to further confirm the interaction between Psl and DNA. Salmon sperm DNA was used to titrate Psl dispersion. Data showed in Fig. 6 indicate an exclusively exothermic reaction, which however does not show a sigmoidal saturation profile. A similar profile was reported for the LPS–protein interaction. The observed enthalpy change of DNA–Psl interaction is in the range of −8 to −4.5 kJ mol−1 (Fig. 6A, lower panel), which indicates a binding of salmon sperm DNA to Psl. A variety of Psl/DNA ratios gave similar results (data not shown). When genomic DNA of P. aeruginosa was used to titrate Psl, it showed a stronger DNA–Psl interaction with the enthalpy change in range of −23 to −6.0 kJ mol−1 (Fig. 6B, lower panel). These results were consistent with the data shown in Fig. 5A, in which P. aeruginosa gave better inhibition than salmon sperm DNA.
Fig. 6.
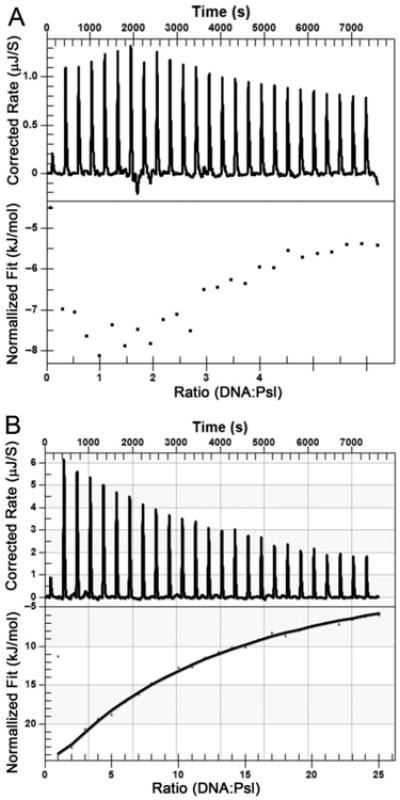
Isothermal titration calorimetry measurement to determine the interaction of Psl and salmon sperm DNA.
A. Salmon sperm DNA to titrate Psl dispersion.
B. The genomic DNA of P. aeruginosa to titrate Psl dispersion. The upper panel shows raw titration data, and the bottom panel is integrated and dilution-corrected peak area plotting of titration data. To a Psl dispersion (2.25 mg ml−1), DNA (200 μg ml−1) is titrated in 10 μl portion.
Significance of the eDNA–Psl interaction
The Psl–eDNA interaction may benefit biofilms in many ways. First, the interaction allows Psl to associate with eDNA or fusion of multiple DNA strands together to form a super DNA–Psl fibre, which functions as a skeleton of biofilm or gives a backbone support, allowing bacteria to attach with and grow. Second, as a signal molecular, Psl can stimulate bacteria adhered on matrix to synthesize cyclic-di-GMP (Irie et al., 2012), which enhances the production of exopolysaccharide and retains the bacteria in the biofilm communities. Third, the association of eDNA with Psl polysaccharide may limit access of agents that target only one component of biofilm matrix. This was supported by our data that the biofilm of a Psl-deficient strain was more sensitive than its isogenic wild-type strain to DNase I treatment (Fig. S2), and by the previously published finding that DNase I cannot disrupt established, ‘aged’ biofilms (Whitchurch et al., 2002). Fourth, a high concentration of eDNA was suggested to cause chelating cations to lyse bacterial cells (Mulcahy et al., 2008), and the covering of eDNA by Psl might reduce this cationchelating of eDNA in biofilm and protect bacteria. Finally, the eDNA–Psl interaction might increase P. aeruginosa survival in multispecies biofilms or within a human host because Psl of P. aeruginosa could interact with DNA from other bacteria as well as DNA from human neutrophils, which might enable P. aeruginosa to attach on NET or eDNA from other bacteria to grow its biofilms.
Pseudomonas aeruginosa can also produce an exopolysaccharide named Pel, which is required for pellicles formation. Reports have shown that P. aeruginosa strain, such as PAO1, primarily uses Psl as matrix component (Ma et al., 2009; Colvin et al., 2012). We have previously demonstrated that the formation of Psl fibres does not require Pel. The purified Psl used in this study was prepared from PAO1-derived strain WFPA801 (Psl-overproduced strain), which has reported to produce little Pel (Wang et al., 2013). Therefore, it is unlikely that Pel could be involved in the eDNA–Psl association or interaction in pellicles.
Conclusion
Our data suggest that the two main biofilm matrix components of P. aeruginosa, eDNA and Psl, cooperate by physically interacting in a biofilm to form the web of Psl–eDNA fibres, which functions as a skeleton to allow bacteria to adhere and grow. Bacterial membrane is overlapped with eDNA or Psl fibres, suggesting the plausible formation of eDNA–Psl-membrane fibres in biofilms. Once the eDNA–Psl-membrane combined matrix forms, the biofilm and matrix are protected against DNase I, or agents targeted one kind of biofilm matrix components. Psl can interact with DNA from diverse sources, suggesting that P. aeruginosa has the ability to use DNA of other organisms (such as human neutrophils and other bacterial species) as a scaffold to form its own communities. DNA was reported to interact with glucan (Sakurai and Shinkai, 2001). In this study, we showed that DNA interacted with Psl, a repeating pentasaccharide containing d-mannose, d-glucose and l-rhamnose. This suggests that DNA may interact with different kinds of polysaccharide, and the strategy used in P. aeruginosa biofilm formation might be common for bacteria in natural and clinical settings. Taken together, the formation of biofilm matrix is a complex, dynamic process with contribution of multiple factors, including bacterial migration, cell death, the release of polysaccharide and eDNA, and the interaction between the matrix components.
Experimental procedures
Strains and growth conditions
The P. aeruginosa strains used in this study are listed in Table S1. Unless otherwise indicated, P. aeruginosa was grown at 37°C in Luria–Bertani medium lacking sodium chloride (LBNS) or in Jensen's, a chemically defined medium (Ma et al., 2006). Biofilms of P. aeruginosa were cultured in Jensen's medium at room temperature (RT). To induce the transcription of the psl operon, 0.2–2.0% arabinose was added to Jensen's medium.
Biofilm and matrix staining
The air–liquid interface biofilms were grown in glass chambers with individual chamber dimensions of 1 × 1 × 4 cm (Chambered #1.5 German Coverglass System, Nunc) as described previously (Wang et al., 2013). DNase I (Sigma, 100 U/chamber) was added to glass chambers 1 h post-inoculation. For Confocal Laser Scanning Microscopy (CLSM) observation, buffer was gently removed from glass chambers to allow the pellicles to drop onto coverslips. The flow-cell biofilms were grown at RT in three-channel flow cell with individual channel dimensions of 1 × 4 × 40 mm (Stovall Life Science) as previously described (Ma et al., 2006). The mid-log phase culture was used for inoculation. The biofilms were stained with membrane stain FM4-64 (1 μm ml−1 final concentration, Molecular Probes, Invitrogen) or DNA stain SYTO9 (Molecular Probes, Invitrogen). Extracellular DNA in biofilms was stained with PI and SYTOX Green (Molecular Probes, Invitrogen). The Psl matrix was stained with fluorescence-labelled lectin HHA at 100 μg ml−1 (EY Lab) as we described elsewhere (Ma et al., 2009).
Image acquisition and analysis
All fluorescent images were acquired with a Zeiss 510 CLS microscope (Carl Zeiss, Jena, Germany). Images were obtained using a 63×/1.3 objective. An LSM image browser generated the 3D images and optical Z-sections. CLSM-captured images were subjected to quantitative image analysis using comstat software to determine biofilm biomass, thickness and other properties as previously described (Heydorn et al., 2000). The biofilm biomass was quantified from SYTO9-stained images or by crystal violet staining of pellicles. ImageJ software was used to analyse the colocalization of eDNA and Psl and to make the corresponding colour map (Hartig, 2013; Zinchuk et al., 2011). The colocalization coefficient and correlation coefficient of eDNA and Psl in biofilm images were analysed and calculated as previously described (Zinchuk et al., 2013).
DNA purification
Salmon sperm DNA was obtained from Sigma-Aldrich. Neutrophils were isolated from the whole blood of healthy volunteers (25–35 years old) by dextral sedimentation, followed by Ficoll-Hypaque density-gradient centrifugation (400 g, 30 min) as described previously. Erythrocytes were removed by hypotonic lysis (Ottonello et al., 1999). Genomic DNAs of neutrophils, P. aeruginosa and S. aureus ATCC 6538 were purified with Wizard Genomic DNA Purification Kits (Promega).
Preparation of Psl extract
Psl polysaccharide extract was prepared from overnight culture of WFPA801 as previously described (Byrd et al., 2009) with modification. To remove the DNA, crude Psl polysaccharide was precipitated with three volumes of ethanol and dissolved in pyrogen-free distilled water with 5 mM MgCl2, followed by DNase I (Sigma) treatment (0.1 mg ml−1 final concentration) for 5 h at 37°C and treatment with proteinase K (0.1 mg ml−1 final concentration) for 1 h at 60°C. Psl samples without DNase I treatment were only treated with proteinase K. Enzymes were inactivated at 80°C for 30 min, and Psl samples were quantified by ELISA as described previously (Byrd et al., 2009).
Examination of the Psl–DNA interaction by ELISA
To determine Psl and DNA interact, Psl post-treatment of DNase and proteinase was diluted with phosphate buffer (PBS, 0.01 M phosphate, 0.15 M NaCl; pH 7.2), and Psl at 0.25 μg ml−1 was used to coat 96-well MaxiSorp plates (Jet Biofil, 100 μl/well) for overnight at 4°C. Psl-coated plates were washed five times with PBST buffer (PBS containing 0.05% Tween 20) and were blocked for 2 h at RT with 10% newborn calf serum (MP chromato Pur) in PBS. DNA from different origins was diluted in PBS to 60, 90 or 120 μg ml−1. An aliquot of DNA (100 μl per well) was transferred into the Psl-coated plates and incubated overnight at 4°C. After five washes with PBST, Psl was detected with anti-Psl serum as previously described. The absorbance at 450 nm (A450) was determined by the Multiskan Ascent version 2.6 plate reader.
Isothermal titration calorimetry
Microcalorimetric measurements of the binding of salmon sperm DNA to Psl were performed on a NANO ITC 2G at 25°C(TA Instruments, USA) as described previously with modification (Ni et al., 2013). Psl and salmon sperm DNA were dissolved into 0.01 M PBS (pH 7.4). Psl (2.25 mg ml−1) was dispensed into the microcalorimetric cell (volume 1.3 ml), and the DNA solution (200 μg ml−1) was filled into the syringe compartment (volume 250 μl). DNA was titrated in 10 μl portions (3.14 μl for the first injection) into the Psl-containing cell under constant stirring, and the heat of reaction was plotted versus time. Data analysis was executed by the NanoAnalyze software.
Modelling of the DNA–Psl interaction by docking
The spatial structure of the DNA duplex in an ideal B-DNA conformation for docking was obtained by using the w3dna software package (Zheng et al., 2009). Docking calculations were carried out using AutoDock version 4.2 with general protocols (Morris et al., 2008). The averaged conformations of the Psl polysaccharide repeat unit (generated from molecular dynamics (MD) simulation, see Appendix S1) and the DNA duplex were converted into the proper file formats for AutoDock using AutoDock Tools version 1.5.4 (Morris et al., 2009). The DNA was enclosed in a box with 70 × 70 × 70 grid points in x × y × z directions and a grid spacing of 0.375 Å. The maximum number of energy evaluation was set to 2.5 × 106, and the number of Lamarckian genetic algorithm runs was set to 100. Lamarckian genetic algorithms, as implemented in AutoDock, were used to perform docking calculations. During the docking, Psl was kept in a flexible state while the DNA was kept in fixed state. All other parameters were given default settings. One hundred solutions were generated for the docking run. The docking results were then subjected to cluster analysis (RMS tolerance of 2.0 Å), and the lowest energy conformation was selected. The docked structure suggested that the Psl polysaccharide repeat unit fitted the minor groove of DNA and was almost parallel with the DNA helix. The estimated interaction energy for the polysaccharide-DNA complex was −2.18 kcal mol−1. The complex was stabilized by hydrogen bonds.
Supplementary Material
Fig. S1. A set of images to show where eDNA–Psl fibres were observed in a pellicle of PAO1. Serial horizontal optical section images of a 5 μm thick pellicle were shown. Psl was stained by HHA-TRITC in red. eDNA and bacterial chromosomal DNA were stained by SYTO9 in green. The chromosomal DNA in bacteria were concentrated dots, yet eDNA appeared diffused and had fibre-look.
Fig. S2. The comparison of DNase I treatment on the biofilm biomass of Psl-negative strain WFPA800 and wild-type strains PAO1. Shown are microtiter dish biofilm assay of PAO1 and WFPA800 with/without DNase I treatment. DNase I treatment caused 50% reduction of biofilm biomass in Psl-negative strain compared with the non-treatment control. The same treatment only resulted in a 10% reduction of biofilm biomass in wild-type strain.
Table S1. The P. aeruginosa strains used in this study and their motility phenotype.
Appendix S1. Supplemental methods and results.
Acknowledgments
The authors thank Dr. Alan K Chang at Liaoning University, Dr. Joseph Lam at the University of Guelph, and Dr. Di Wang and Dr. Qing Wei at the Institute of Microbiology, Chinese Academy of Sciences, for their contribution to the revision of the manuscript; Dr. Tong Li at Peking University for assistance with isolation of neutrophils; Mr. Likai Hao at Eberhard Karls University Tuebingen for assistance with image analysis. This work was supported by the National Basic Research Program of China (973 Program, Grant 2014CB846002) (L.Z.M.) and the National Natural Science Foundation of China Grant 31270177 (L.Z.M.).
Footnotes
Supporting information: Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
References
- Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, et al. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- Byrd MS, Sadovskaya I, Vinogradov E, Lu HP, Sprinkle AB, Richardson SH, et al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol. 2009;73:622–638. doi: 10.1111/j.1365-2958.2009.06795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd MS, Pang B, Mishra M, Swords WE, Wozniak DJ. The Pseudomonas aeruginosa exopolysaccharide Psl facilitates surface adherence and NF-kappaB activation in A549 cells. MBio. 2010;1:e00140–10. doi: 10.1128/mBio.00140-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang WC, Nilsson M, Jensen PO, Hoiby N, Nielsen TE, Givskov M, Tolker-Nielsen T. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2013;57:2352–2361. doi: 10.1128/AAC.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, et al. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol. 2012;14:1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Dominiak DM, Nielsen JL, Nielsen PH. Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ Microbiol. 2011;13:710–721. doi: 10.1111/j.1462-2920.2010.02375.x. [DOI] [PubMed] [Google Scholar]
- Dwyer M, Shan Q, D'Ortona S, Maurer R, Mitchell R, Olesen H, et al. Cystic fibrosis sputum DNA has NETosis characteristics and neutrophil extracellular trap release is regulated by macrophage migration-inhibitory factor. J Innate Immun. 2014;6:765–779. doi: 10.1159/000363242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Fuxman Bass JI, Russo DM, Gabelloni ML, Geffner JR, Giordano M, Catalano M, et al. Extracellular DNA: a major proinflammatory component of Pseudomonas aeruginosa biofilms. J Immunol. 2010;184:6386–6395. doi: 10.4049/jimmunol.0901640. [DOI] [PubMed] [Google Scholar]
- Gloag ES, Turnbull L, Huang A, Vallotton P, Wang H, Nolan LM, et al. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc Natl Acad Sci USA. 2013;110:11541–11546. doi: 10.1073/pnas.1218898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gries A, Prassl R, Fukuoka S, Rossle M, Kaconis Y, Heinbockel L, et al. Biophysical analysis of the interaction of the serum protein human beta2GPI with bacterial lipopolysaccharide. FEBS Open Bio. 2014;4:432–440. doi: 10.1016/j.fob.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison F. Microbial ecology of the cystic fibrosis lung. Microbiology. 2007;153:917–923. doi: 10.1099/mic.0.2006/004077-0. [DOI] [PubMed] [Google Scholar]
- Hartig SM. Basic image analysis and manipulation in ImageJ. Curr Protoc Mol Biol. 2013;102:14.15.1–14.15.12. doi: 10.1002/0471142727.mb1415s102. [DOI] [PubMed] [Google Scholar]
- Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- Irie Y, Borlee BR, O'Connor JR, Hill PJ, Harwood CS, Wozniak DJ, Parsek MR. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2012;109:20632–20636. doi: 10.1073/pnas.1217993109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Cao S, Chai L, Bottcher T, Kolter R, Clardy J, Losick R. A self-produced trigger for biofilm disassembly that targets exopolysaccharide. Cell. 2012;149:684–692. doi: 10.1016/j.cell.2012.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol. 2006;188:8213–8221. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 2009;5:e1000354. doi: 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Wang J, Wang S, Anderson EM, Lam JS, Parsek MR, Wozniak DJ. Synthesis of multiple Pseudomonas aeruginosa biofilm matrix exopolysaccharides is post-transcriptionally regulated. Environ Microbiol. 2012;14:1995–2005. doi: 10.1111/j.1462-2920.2012.02753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn LM, Jett AM, Akins DR, Whiteley M. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol. 2005;187:554–566. doi: 10.1128/JB.187.2.554-566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegazzi R, Decleva E, Dri P. Killing by neutrophil extracellular traps: fact or folklore? Blood. 2012;119:1214–1216. doi: 10.1182/blood-2011-07-364604. [DOI] [PubMed] [Google Scholar]
- Molin S, Tolker-Nielsen T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr Opin Biotechnol. 2003;14:255–261. doi: 10.1016/s0958-1669(03)00036-3. [DOI] [PubMed] [Google Scholar]
- Morris GM, Huey R, Olson AJ. Using AutoDock for ligand-receptor docking. Curr Protoc Bioinformatics. 2008;8:8.14.1–8.14.40. doi: 10.1002/0471250953.bi0814s24. [DOI] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy H, Charron-Mazenod L, Lewenza S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008;4:e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni B, Huang Z, Fan Z, Jiang CY, Liu SJ. Comamonas testosteroni uses a chemoreceptor for tricarboxylic acid cycle intermediates to trigger chemotactic responses towards aromatic compounds. Mol Microbiol. 2013;90:813–823. doi: 10.1111/mmi.12400. [DOI] [PubMed] [Google Scholar]
- Ottonello L, Tortolina G, Amelotti M, Dallegri F. Soluble Fas ligand is chemotactic for human neutrophilic polymorphonuclear leukocytes. J Immunol. 1999;162:3601–3606. [PubMed] [Google Scholar]
- Parks QM, Young RL, Poch KR, Malcolm KC, Vasil ML, Nick JA. Neutrophil enhancement of Pseudomonas aeruginosa biofilm development: human F-actin and DNAas targets for therapy. J Med Microbiol. 2009;58:492–502. doi: 10.1099/jmm.0.005728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Gadjeva M. Does NETosis contribute to the bacterial pathoadaptation in cystic fibrosis? Front Immunol. 2014;5:378. doi: 10.3389/fimmu.2014.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K, Shinkai S. Novel DNA-polysaccharide triple helixes and their application to a gene carrier. J Incl Phenom Macrocycl Chem. 2001;41:173–178. [Google Scholar]
- Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- Walker TS, Tomlin KL, Worthen GS, Poch KR, Lieber JG, Saavedra MT, et al. Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infect Immun. 2005;73:3693–3701. doi: 10.1128/IAI.73.6.3693-3701.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Parsek MR, Wozniak DJ, Ma LZ. A spider web strategy of type IV pili-mediated migration to build a fibre-like Psl polysaccharide matrix in Pseudomonas aeruginosa biofilms. Environ Microbiol. 2013;15:2238–2253. doi: 10.1111/1462-2920.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- Xiao J, Klein MI, Falsetta ML, Lu B, Delahunty CM, Yates JR, et al. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 2012;8:e1002623. doi: 10.1371/journal.ppat.1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo DG, Floyd M, Winn M, Moskowitz SM, Rada B. NET formation induced by Pseudomonas aeruginosa cystic fibrosis isolates measured as release of myeloperoxidase-DNA and neutrophil elastase-DNA complexes. Immunol Lett. 2014;160:186–194. doi: 10.1016/j.imlet.2014.03.003. [DOI] [PubMed] [Google Scholar]
- Zhao K, Tseng BS, Beckerman B, Jin F, Gibiansky ML, Harrison JJ, et al. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature. 2013;497:388–391. doi: 10.1038/nature12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Lu XJ, Olson WK. Web 3DNA – a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res. 2009;37:W240–W246. doi: 10.1093/nar/gkp358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinchuk V, Wu Y, Grossenbacher-Zinchuk O, Stefani E. Quantifying spatial correlations of fluorescent markers using enhanced background reduction with protein proximity index and correlation coefficient estimations. Nat Protoc. 2011;6:1554–1567. doi: 10.1038/nprot.2011.384. [DOI] [PubMed] [Google Scholar]
- Zinchuk V, Wu Y, Grossenbacher-Zinchuk O. Bridging the gap between qualitative and quantitative colocalization results in fluorescence microscopy studies. Sci Rep. 2013;3:1365. doi: 10.1038/srep01365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. A set of images to show where eDNA–Psl fibres were observed in a pellicle of PAO1. Serial horizontal optical section images of a 5 μm thick pellicle were shown. Psl was stained by HHA-TRITC in red. eDNA and bacterial chromosomal DNA were stained by SYTO9 in green. The chromosomal DNA in bacteria were concentrated dots, yet eDNA appeared diffused and had fibre-look.
Fig. S2. The comparison of DNase I treatment on the biofilm biomass of Psl-negative strain WFPA800 and wild-type strains PAO1. Shown are microtiter dish biofilm assay of PAO1 and WFPA800 with/without DNase I treatment. DNase I treatment caused 50% reduction of biofilm biomass in Psl-negative strain compared with the non-treatment control. The same treatment only resulted in a 10% reduction of biofilm biomass in wild-type strain.
Table S1. The P. aeruginosa strains used in this study and their motility phenotype.
Appendix S1. Supplemental methods and results.


