Summary
Eukaryotic transcription is pervasive, and many of the resulting RNAs are non-coding. It is unknown if ubiquitous transcription is functional or simply reflects stochastic transcriptional noise. By single-molecule visualization of the dynamic interplay between coding and non-coding transcription at the GAL locus in living yeast cells, we show that antisense GAL10 ncRNA transcription can switch between functional and spurious under different conditions. During galactose induction, GAL10 sense transcription occurs in short stochastic bursts which are unaffected by transcription of antisense GAL10 ncRNA, even when both are present simultaneously at the same locus. In contrast, when GAL10 is not induced, ncRNA transcription is critical to prevent transcriptional leakage of GAL1 and GAL10. Suppression of ncRNA transcription by strand-specific CRISPR/dCas9 results in transcriptional leakage of the inducer GAL1, leading to a more sensitive transcription activation threshold, an alteration of metabolic switching, and a fitness defect in competition experiments.
Introduction
Genome-wide transcriptome analyses have revealed that eukaryotic transcription is pervasive. For instance, the model organism baker’s yeast transcribes about 85% of its genome (David et al., 2006), including hundreds of non-coding transcripts. Functional roles for specific ncRNAs have been reported either on a case-by-case basis (Hongay et al., 2006; Camblong et al., 2007; Houseley et al., 2008), or by genome-wide correlation (Xu et al., 2011). However, given that transcription is also known to be stochastic, it has also been proposed that ncRNA represents transcriptional ‘noise’ that arises from leakiness of the transcription machinery (Struhl, 2007). For example, disruption of chromatin can result in cryptic initiation from regions where transcription is normally repressed (Kaplan et al., 2003). A general understanding of when ncRNA transcription is functional or spurious is still lacking.
One major difficulty in addressing the functional role of individual ncRNAs lies in the fact that these transcripts often originate from genomic regions that overlap coding genes. Therefore two primary tools for assessing function – evolutionary conservation and mutagenesis – are largely ineffective. In this article, we develop a new approach for dissecting the role of ncRNA and apply it to the ncRNA located within the GAL locus of S. cerevisiae. The locus consists of a cluster of adjacent genes (GAL1, GAL10, GAL7) involved in galactose metabolism. Among the different ncRNAs and antisense transcripts found in this cluster, at least two transcripts originate within GAL10 and extend through the coding region of GAL10 and into the promoter of GAL10 and GAL1 (Houseley et al., 2008). Initiation of these ncRNAs is regulated by the transcription factor Reb1, and requires different factors than GAL10 sense initiation (Houseley et al., 2008; Malik et al., 2013). How these ncRNAs are dynamically regulated in vivo is unknown. Moreover, there is conflicting data as to whether this ncRNA transcription has activating or repressive effects on transcription of the GAL coding genes (Houseley et al., 2008; Pinskaya et al., 2009; Geisler et al., 2012; Cloutier et al., 2013).
These disparate results may result from the complexity of gene regulation in this metabolic network. In raffinose, the locus is uninduced, because the transcriptional activator Gal4p is repressed by the inhibitor Gal80p. In glucose, the genes are actively repressed by additional mechanisms, including transcriptional down-regulation of the activator and upstream factors (Lamphier and Ptashne, 1992). Activation of the GAL network by galactose results in increased expression of the regulatory factors and the metabolic enzymes. One protein, Gal1p, plays a dual role in sensing and catalysis (Sellick et al., 2009). Because both activators and repressors are transcriptionally up-regulated by galactose (Fig. 1A), the system contains positive and negative feedback loops, shows a bimodal dose response, and displays hysteresis or ‘memory’ (Acar et al., 2005; Zacharioudakis et al., 2007; Venturelli et al., 2012). This bimodal behavior coupled with stochastic transcription results in heterogeneity in the galactose response among a population of cells (Gandhi et al., 2011). Therefore, any perturbations need to be considered in the context of this dynamic, inter-connected gene network.
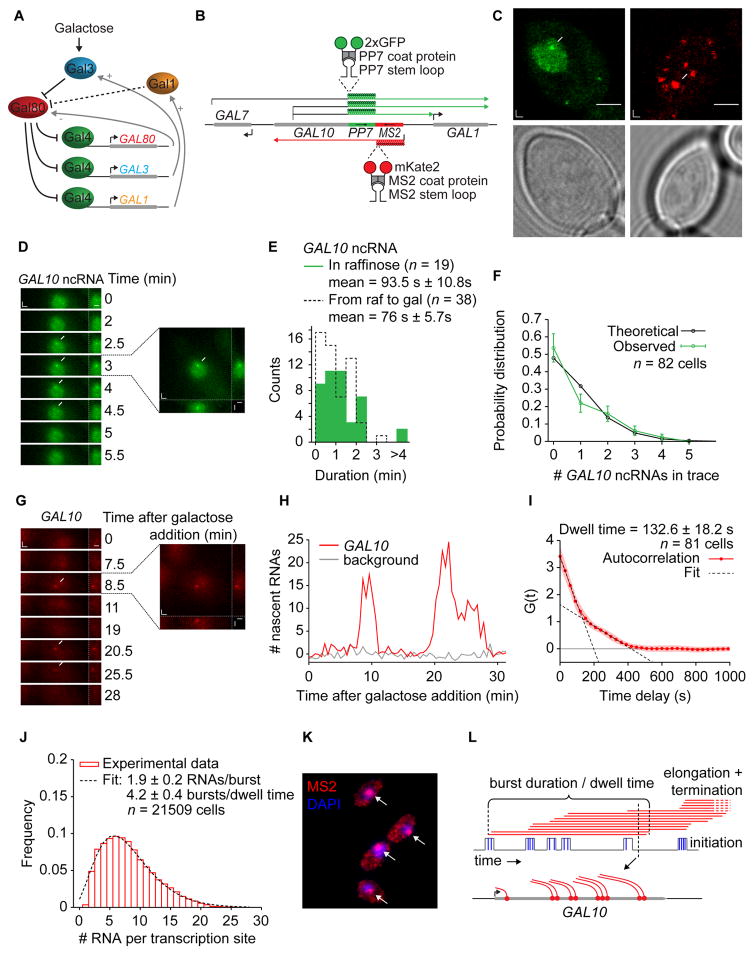
Figure 1. Visualizing GAL10 and GAL10 ncRNA transcription in live yeast cells.
(A) Schematic representation of the positive and negative feedback loops of the galactose response (B) Schematic representation of the PP7 and MS2 tagged GAL10 locus. (C) Scanning confocal microscope images of TSs (arrows) and individual transcripts (arrowheads) for GAL10 ncRNA in raffinose (left) and GAL10 in galactose (right) and transmitted light images (bottom). (D) Cropped frames from time-lapse movies of GAL10 ncRNA transcription in raffinose, imaged with 30s interval. The images show maximum intensity projections of 9 z-planes (left), and side-views (right). Arrows indicate the TS. See Movie S1. (E) Histogram of the signal duration of GAL10 ncRNA in raffinose (green) and after induction with galactose (dotted black line). (F) Probability distribution of the number of GAL10 ncRNA transcription events per trace in cells that are induced with galactose. The distribution fits a model of random initiation with exponentially distributed waiting times (black line). (G) GAL10 shows transcriptional bursting, with periods of transcriptional activity followed by periods of inactivity. See Movie S2. (H) Quantification of the TS intensity (red) and background (gray) of (G). The fluorescence intensity was normalized by smFISH and converted to number of RNA. (I) Autocorrelation of the GAL10 TS intensity traces corrected for non-steady-state effects (see Fig. S1C). The dwell time is 132.6 ± 18.2 s, which is given by the intersection of the fast and slow linear components (dotted lines). Shaded area indicates SEM. (J) Distribution of number of RNA per TS determined by smFISH after 30 min of galactose induction. The dotted line represents a fit using the random telegraph model. The average of 3 experimental fits gives 1.9 ± 0.2 RNAs/burst and 4.2 ± 0.4 bursts/dwell time. (K) Example cells of smFISH after 30 min of galactose induction, with MS2-Cy3 probes labeling the 5′ of GAL10. Arrows indicate TS. (L) Schematic of bursting at GAL10. Grey lines represent promoter states (“on” or “off”), blue lines represent initiation, red lines represent elongation and termination/release. The burst duration or dwell time, as measured by live cell microscopy, is indicated. See also Fig. S1.
Here, we use time-lapse single-molecule imaging in living cells in tandem with strand-specific transcriptional blocking of the ncRNA by CRISPR/dCas9 to interrogate the role of ncRNA transcription in the yeast galactose response. We devised a method based on the strand specific PP7 and MS2 RNA-labeling technique to visualize both GAL10 coding and non-coding transcription simultaneously in real-time (Coulon et al., 2014). We observed ncRNA synthesis before and even during sense transcription, suggesting that the template is highly permissive for transcription. By targeting an enzymatically-dead mutant of Cas9 to the locus, we were able to selectively block transcription of one strand without affecting expression levels of the opposite strand. Blocking ncRNA in this manner results in increased “transcriptional leakage” of both GAL1 and GAL10 under repressive conditions, revealing an important role of ncRNA transcription in gene repression beyond the existing mechanisms in the GAL network. Increased leakage of GAL1 alters the activation threshold and hysteresis of the network and resulted in a fitness defect in a direct competition assay. However, blocking ncRNA had no detectable effects on the actively transcribing gene. The appearance of the ncRNA is thus experimentally uncoupled from its function. Our results show that transcription of the same ncRNA is functional under repressive conditions but spurious under activating conditions, highlighting the nuanced roles that ncRNA can play in gene regulation.
Results
Real-time single-molecule visualization of GAL10 coding and non-coding transcription in live yeast cells
To visualize coding and non-coding transcription at the endogenous GAL10 locus in live yeast cells, RNAs were labeled using the orthogonal and strand-specific MS2 and PP7 systems. We introduced 14 MS2 repeats in the 5′ untranslated region of GAL10 in the sense orientation, and 14 PP7 repeats in the antisense orientation (Fig. 1B). When transcribed, each stem loop is selectively bound by a homodimer of the MS2 or PP7 bacteriophage coat protein fused to a nuclear localization signal and a fluorescent protein (mKate2 or 2xGFP, respectively). For GAL10 ncRNA, different start and termination sites have been reported (Houseley et al., 2008), but all were visualized here as soon as they passed the 5′ UTR of GAL10. All constructs were added to diploid cells and were minimally perturbative on transcript levels (Fig. S1).
GAL10 ncRNA is preferentially expressed under non-inducing conditions but is still produced in some cells before they switch to GAL10 transcription. In both repressive and activating conditions, GAL10 ncRNA is only present at the transcription site (TS) for 90 s or less (Fig. 1D,E, Movie S1). For an elongation rate of ~2 kb/min (Mason and Struhl, 2005), transcribing the different GAL10 ncRNA transcripts would take 50–100 s, suggesting that GAL10 ncRNA is only present at the TS while being synthesized. This finding does not support the proposed model that ncRNAs act through the formation of stable R-loops (Geisler et al., 2012).
We extended these results with single-molecule FISH (smFISH). The numbers of ncRNA per cell and at the TS follow a Poisson distribution (Fig. S1C,D). In addition, the distribution of the number of GAL10 ncRNA transcription events in live-cell traces is consistent with exponentially distributed waiting times, with a mean interval of 16 min (Fig. 1F). Both results are consistent with a simple model of uncorrelated initiation events and no transcriptional bursting.
When cells are induced with 2% galactose, GAL10 transcription displays transcriptional bursting, with periods of transcriptional activity followed by periods of inactivity (Fig. 1G,H, Movie S2). To our knowledge, this is the first time transcriptional bursting has been directly observed in yeast and not inferred through computational models (Zenklusen et al., 2008). Autocorrelation analysis of the fluorescence intensity at the TS revealed that the average RNA dwell time was 132.6 ± 18.2 s (Fig. 1I, Suppl. Exp. Proc.), which includes the duration of elongation, termination and release.
With smFISH, the intensity of the GAL10 TS is much brighter than the intensity of the individual RNAs, indicating that multiple RNAs are present at the active gene (Zenklusen et al., 2008; Larson et al., 2011). The normalized distribution of GAL10 RNA per TS after 30 min of galactose induction (Fig. 1J, K) did not fit a Poisson model, as expected (Fig. S1G), and was therefore fit to a model that assumes bursting behavior (Raj et al., 2006). The fit gave an average burst size of 1.9 ± 0.2 RNAs per burst and a frequency of 4.2 ± 0.4 bursts/dwell time. We therefore conclude that GAL10 shows transcriptional bursting with small burst sizes and a high burst frequency, resulting in many polymerases on the gene at the same time, as depicted schematically in Fig. 1L. In summary, single-molecule imaging indicates the same DNA template is transcribed in fundamentally different ways: the coding RNA is synthesized in transcriptional bursts, and the ncRNA is made in uncorrelated transcription events.
Dynamic switching between sense and antisense transcription
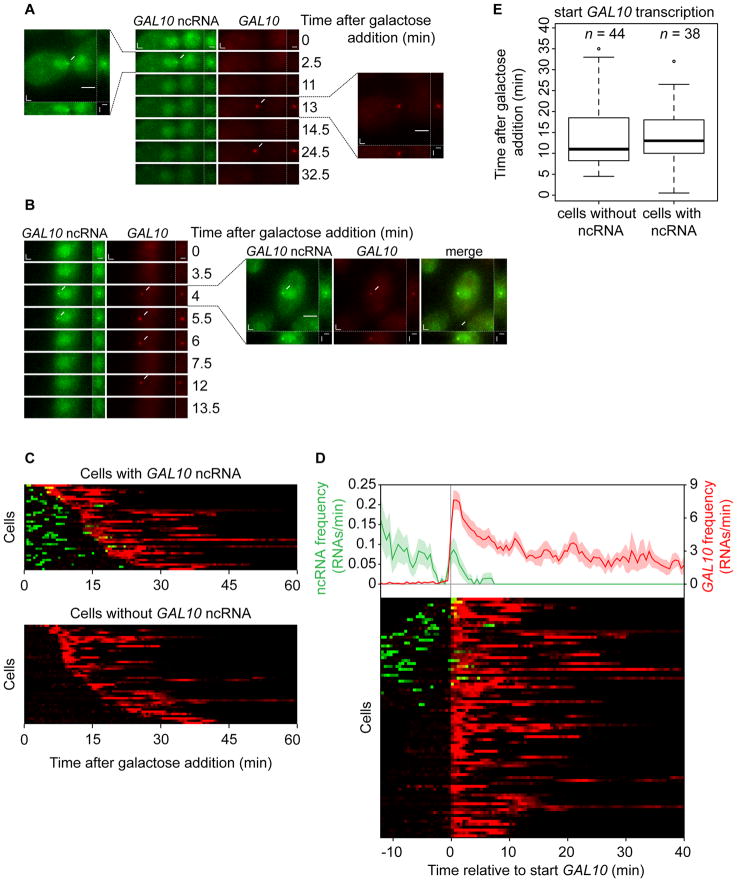
To investigate how individual cells switch from GAL10 ncRNA antisense transcription to GAL10 sense transcription, cells were tracked in time after galactose addition. Transcription of GAL10 ncRNA is seen either before or simultaneously with GAL10 transcription (Fig. 2A,B, Movies S3, S4) but mostly disappears after cells switch to GAL10 transcription (Fig. 2C). Stochastic differences in signaling and transcription factor binding result in heterogeneity in the onset of GAL10 transcription (Fig. 2C). When this variation is excluded by aligning all traces on the start of GAL10 transcription, it becomes clear that the frequency of GAL10 ncRNA transcription is not equal over time (Fig. 2D). Immediately before GAL10 activation, there is a 100 s period where GAL10 ncRNA is completely absent, suggesting that the chromatin template around the promoter may not be permissive for transcription in either direction. In addition to this depletion, there is a peak of GAL10 ncRNA frequency at the start of GAL10 transcription. Simultaneous transcription of GAL10 and GAL10 ncRNA likely reflect events where GAL10 transcription initiated immediately after GAL10 ncRNA passed the promoter of GAL10 in the antisense direction. In our assay, these diverging polymerases will be visible as a two color spot, because both RNAs remain visible at the TS during elongation. Almost half of the cells displayed GAL10 ncRNA transcription in the time period between galactose addition and GAL10 activation, which is expected based on random chance (Fig. 1F).
Figure 2. Switching from sense to antisense transcription.
(A) Example of a cell showing GAL10 ncRNA transcription before GAL10 transcription, similar to Fig. 1D and 1G. See Movie S3. (B) Example of a cell showing GAL10 ncRNA and GAL10 transcription simultaneously. See Movie S4. (C) Integrated view of the signal intensity traces of both GAL10 ncRNA (green) and GAL10 (red) transcription in individual cells (rows) after galactose addition. The disappearance of the red signal is due to nuclear depletion of coat proteins caused by the high transcriptional activity. (D) Alignment of individual traces based on the start of GAL10 transcription (bottom) shows the transcription frequency relative to the start of GAL10 (top). Shaded areas are SEM. (E) Boxplot of the start of GAL10 transcription for cells with and without GAL10 ncRNA transcription.
Previously, conflicting reports have suggested that GAL10 ncRNA acts either positively or negatively on GAL10 transcription (Houseley et al., 2008; Geisler et al., 2012; Cloutier et al., 2013). We therefore compared the onset of GAL10 transcription after galactose induction between cells with and without GAL10 ncRNA transcription. As is shown in Fig. 2E, the start of GAL10 transcription is indistinguishable for these two groups. These results indicate that GAL10 ncRNA transcription does not affect GAL10 activation kinetics and suggest that, in these conditions, transcription of GAL10 ncRNA is non-functional.
Strand-specific inhibition of GAL10 ncRNA with CRISPR/dCas9
Single-cell, single-molecule approaches allow one to examine correlations and timing between sense and antisense transcription in unperturbed cells. These data suggest that GAL10 ncRNA does not affect GAL10 activation, but a functional role for GAL10 ncRNA in other conditions could not be tested with this approach. We therefore sought to specifically repress GAL10 ncRNA without perturbing GAL10 sense transcription. Previous attempts to eliminate antisense transcription have relied on mutating transcription factor binding sites, inserting terminators or deleting the intergenic region (Houseley et al., 2008; Margaritis et al., 2012; Nguyen et al., 2014). However, genetic perturbations at the GAL locus have additional indirect effects (Murray et al., 2015).
A new strategy to control gene expression has recently emerged, using CRISPR and a catalytically dead version of Cas9, called CRISPR interference (CRISPRi) (Gilbert et al., 2013; Qi et al., 2013). In E. coli, CRISPRi can interfere with recruitment of polymerase and transcription factors when targeted to a DNA sequence in the gene promoter, or it can strand-specifically repress transcriptional elongation when targeted to the non-template (NT) strand of the gene body (Qi et al., 2013). In eukaryotes, however, the same level of inhibition has only been reported by fusing dCas9 with a repressor domain, hence ablating its strand-specificity (Gilbert et al., 2013).
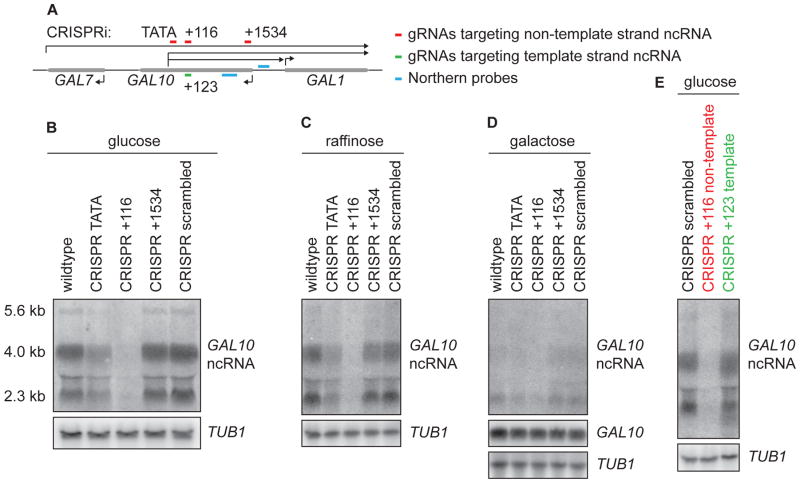
To investigate whether strand-specific repression with CRISPRi could be achieved in eukaryotes, guide RNAs (gRNA) were designed targeting the NT strand at the TATA box and at two positions in the body of GAL10 ncRNA (+116 nt and +1534 nt downstream of the transcription start site (TSS), Fig. 3A). dCas9 was used without a repressor domain. Both in glucose and in raffinose conditions, wt cells transcribe three GAL10 ncRNA transcripts that have been detected previously (Fig. 3B,C) (Houseley et al., 2008). Surprisingly, targeting of the CRISPR complex to the TATA box decreases the GAL10 ncRNA levels by ~50%, whereas targeting to +116 nt downstream of the TSS completely abolishes all GAL10 ncRNA transcription (Fig. 3B,C). A gRNA designed against +1534 nt downstream of the TSS or a random control gRNA (scrambled) did not have any effect on ncRNA levels. When induced with galactose, the levels of GAL10 transcript appeared similar for all CRISPRi strains after 30 minutes of induction (Fig. 3D), suggesting that binding of CRISPR to the NT strand of GAL10 ncRNA (template strand of GAL10) does not interfere with GAL10 transcription. To test whether our approach is really strand-specific, another gRNA was designed close to gRNA +116, but on the opposite strand (+123, Fig. 3A). In contrast to gRNA +116, gRNA +123 did not repress transcription of GAL10 ncRNA (Fig. 3E). In summary, by targeting dCas9 to the NT strand of GAL10 antisense, we report strand specific repression of the ncRNA without affecting GAL10 transcription. Because the approach does not rely on genetic modification, it is a powerful and versatile method to specifically inhibit antisense transcription.
Figure 3. Strand-specific inhibition of GAL10 ncRNA with CRISPRi.
(A) Schematic representation of the GAL locus with the positions of the gRNAs targeting GAL10 ncRNA. Numbers are relative to the TSS of GAL10 ncRNA. (B) Northern blot of the GAL10 ncRNA in glucose in strains without and with CRISPRi using gRNAs shown in (A). TUB1 is the loading control (C) Same as (B), but for cells grown in raffinose. (D) Northern blot of GAL10 ncRNA and GAL10 after 30 minutes of galactose induction. (E) Northern blot of GAL10 ncRNA in strains with CRISPRi using gRNAs scrambled, + 116, and a gRNA against the template strand of GAL10 ncRNA (+123) at the same genomic position as gRNA +116.
GAL10 ncRNA regulates gene activation kinetics and threshold
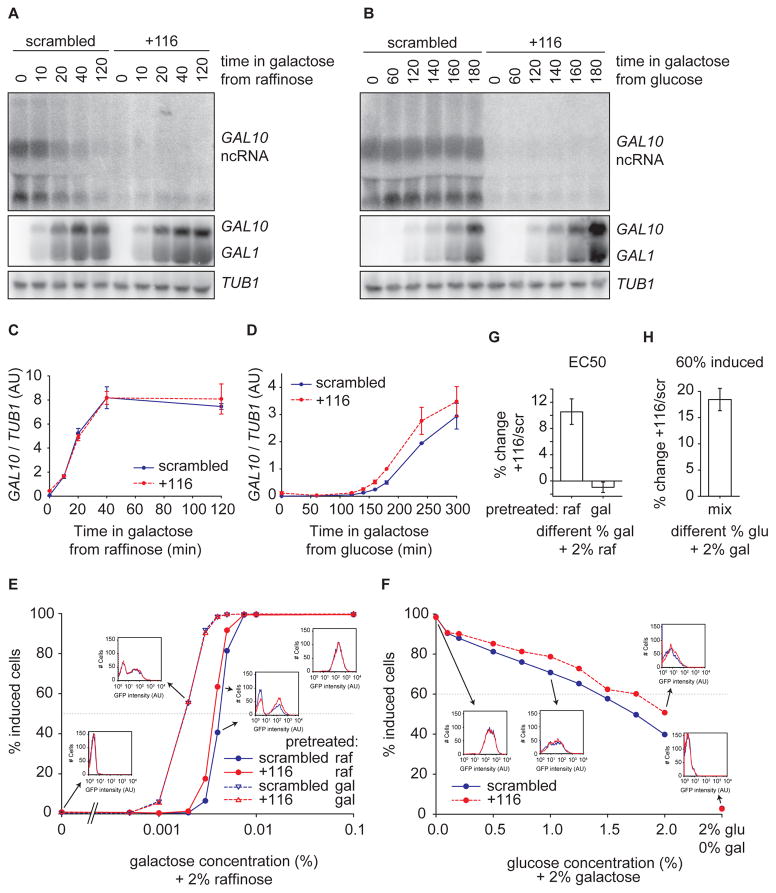
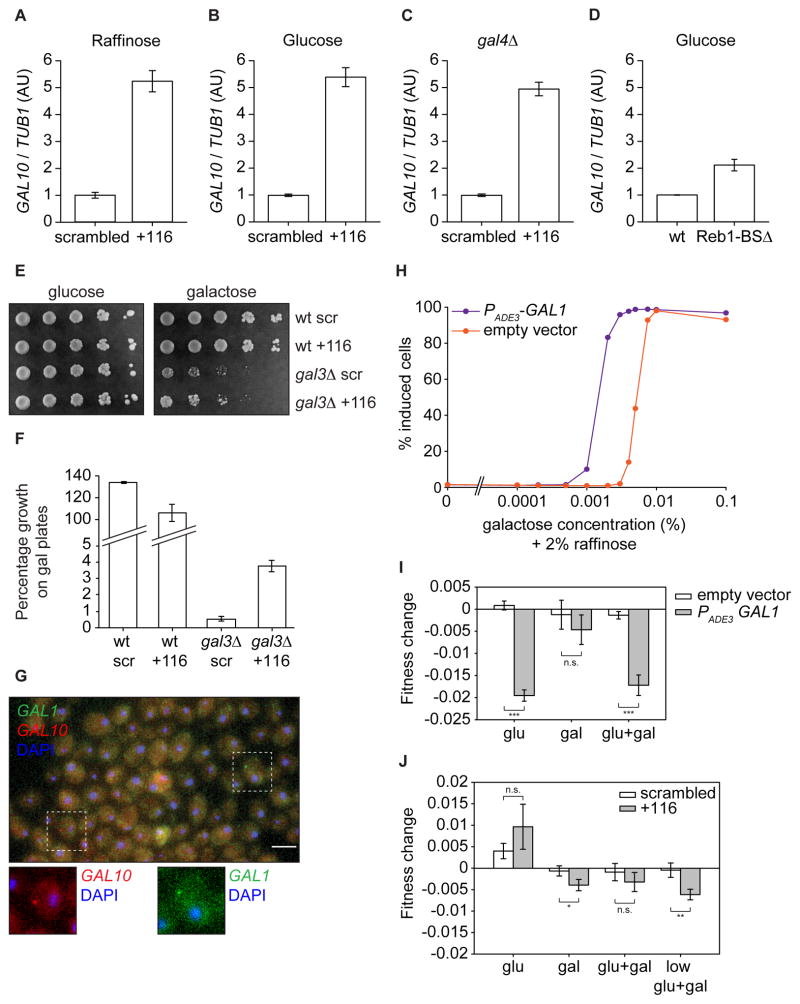
To investigate whether GAL10 ncRNA inhibition has any effect on GAL10 activation kinetics, cells were grown in raffinose or glucose to mid-log, induced with galactose, and analyzed by Northern blot at various times. Inhibition of GAL10 ncRNA resulted in faster induction from glucose but not from raffinose conditions (Fig. 4A–D). Since GAL10 shares an upstream activating sequence with GAL1, their expression has been shown to be highly correlated at the single cell level (Gandhi et al., 2011). Consistently, the effect of CRISPRi on GAL1 expression showed the same behavior as GAL10 in both experiments (Fig. 4A–D). Importantly, these results indicate that GAL10 ncRNA can repress in both antisense (GAL10) and sense (GAL1), but not under all conditions.
Figure 4. GAL10 ncRNA inhibition results in a higher percentage of GAL10 expressing cells.
(A) Northern blot of GAL10 ncRNA, GAL10 and GAL1 in strains with CRISPRi scrambled and +116 pre-grown in raffinose at indicated time points after galactose addition. (B) Same as (A), but for cells pre-grown in glucose. (C) Quantification of GAL10 expression relative to TUB1 expression of 3 Northern blot experiments as in (A). (D) Same as (C), but after galactose addition from glucose. At least 2 measurements were used for quantification. (E) Percentage of induced cells in steady state as determined by GAL10-2xGFP expression with flow cytometry. See Fig. S2 (F) Percentage of induced cells in steady state populations in media with 2% galactose and different glucose concentrations. (G) Quantification of fold change in EC50 of +116/scrambled, normalized to scrambled, of 3 experiments as in (A). (H) Quantification of the fold change at 60% induced of +116/scrambled of 3 experiments as in (B). The dotted lines in (E) and (F) indicate the values that were used to determine the fold change in (C) and (D), respectively. Error bars are SEM.
More rapid activation of GAL1 and GAL10 transcription by population-based Northern blot measurements can arise either from a faster uniform up-regulation of these genes in every cell, or a faster switch from an inactive to an active state in a subset of cells (Castelnuovo et al., 2013). In order to distinguish between these two possibilities, GAL10 was fused to 2xGFP (Fig. S2) and analyzed by flow-cytometry. The number of induced cells expressing fluorescent GAL10 was measured as a function of galactose concentration (Fig. 4E). As observed previously, GAL10 induction displays both a bimodal response and hysteresis (Acar et al., 2005; Venturelli et al., 2012). This hysteresis or ‘memory’ is caused by elevated cytoplasmic concentrations of GAL1 when cells are pretreated with galactose, resulting in a more sensitive activation than when cells are pretreated in raffinose (Zacharioudakis et al., 2007) (see below).
We observe that GAL10 ncRNA has differential effects depending on the history of the cells. GAL10 ncRNA inhibition with CRISPRi +116 results in a 10.6% (± 1.9%) shift in EC50 for cells pre-grown in raffinose but not for cells pre-grown in galactose, meaning the dose response is more sensitive (Figs. 4E,G). The EC50 of the galactose response presumably reflects the evolutionary competition between utilization of glucose as the preferred carbon source and switching metabolic programs when glucose is absent (Lamphier and Ptashne, 1992). To test the role of GAL10 ncRNA in this metabolic switching, we grew cells under mixed glucose/galactose conditions (Fig. 4F,H). When GAL10 ncRNA is inhibited, more cells switch to galactose metabolism over a wide range of mixed media conditions (Fig. 4F), similar to previous observations (Houseley et al., 2008). Thus, the ncRNA is involved in modulating the relative preference for galactose over glucose by controlling the activation threshold of the network.
Inhibition of GAL10 ncRNA results in transcriptional leakage of GAL1 and GAL10
Because GAL10 ncRNA inhibition affects only the naïve state (raffinose history) and not the exposed state (galactose history), it is necessary to consider the underlying causes of hysteresis in the galactose response. The galactokinase Gal1p and the galactose inducer Gal3p are paralogs that arose by whole genome duplication (Hittinger and Carroll, 2007). In organisms such as K. lactis Gal1p has both sensing and galactokinase ability, whereas in S. cerevisiae both proteins have specialized: Gal3p has lost its kinase activity, but the kinase Gal1p still possesses weak activation activity (Sellick et al., 2009). In non-inducing conditions, GAL3 is basally expressed, whereas GAL1 transcription is repressed, with the result that Gal3p is the main galactose sensor. Importantly, in cells where Gal3p is absent or in cells with previous exposure to galactose, Gal1p becomes the primary sensor of galactose (Abramczyk et al., 2012; Kar et al., 2014).
Since GAL10 ncRNA regulates expression of both GAL1 and GAL10 (Fig. 4A,B), we hypothesized that removal of GAL10 ncRNA increases the basal levels of GAL1 and GAL10 expression. Elevated expression of Gal1p could then result in higher sensitivity to galactose. When GAL10 levels were compared by qRT-PCR in cells grown in raffinose, the strain with CRISPRi +116 displays 5-fold higher levels of GAL10 than the control strain (Fig. 5A). The same increase was observed in glucose (Fig. 5B), or cells where GAL4 is deleted (Fig. 5C), indicating that this transcriptional leakage is not affected by glucose repression or Gal4p activity. Repression by GAL10 ncRNA thus forms an additional layer of repression that is not described in existing models. Notably, this result indicates that transcriptional leakage is independent of the activator Gal4p.
Figure 5. GAL10 ncRNA inhibition increases transcriptional leakage of GAL10 and GAL1.
(A) Quantification of GAL10 levels relative to TUB1 using qRT-PCR in cells with CRISPRi scrambled and +116, grown in raffinose. GAL10 levels were measured using primers against the 3′ of GAL10, to prevent interfering effects of GAL10 ncRNA removal. (B),(C) and (D) Same as (A) but for cells grown in glucose (B) or for gal4Δ cells (C) or for a Reb1-BSΔ mutant grown in glucose (D). (E) Growth assay of wt and gal3Δ cells with CRISPRi scrambled and +116. Serial dilution (5 fold) are shown after 3 days of growth on glucose and galactose plates. (F) Percentage of colonies that grow on galactose + EtBr containing plates, normalized to glucose plates, for wt and gal3Δ cells with CRISPRi scrambled and +116. (G) smFISH with GAL1-cy3 (green) and GAL10-cy5 (red) probes on cells with CRISPRi +116 grown in glucose. Higher magnifications are shown at the bottom. (H) Percentage of induced cells, as measured in Fig. 4E, for cells with a plasmid expressing GAL1 from the ADE3 promoter. (I) Fitness change measured by a competition assay of cells expressing GAL1 from a plasmid versus empty vector (J) Fitness change of cells with CRISPRi scrambled versus +116. Glu = 2% glucose, gal = 2% galactose, glu+gal = 2% glucose + 2% galactose, low glu+gal = 0.02% glucose + 0.01% galactose + 2% raffinose. * p<0.05, ** p<0.01 *** p<10−4. Error bars are SEM.
To test whether the increased leakage could be an indirect effect of the CRISPR targeting, we measured GAL10 leakage in a strain in which GAL10 ncRNA was prevented by mutating of the Reb1 binding sites. Similarly to CRISPRi ncRNA inhibition, the Reb1-BSΔ mutant showed increased transcriptional leakage (Fig. 5D), arguing for a direct role of the ncRNA in repression of transcriptional leakage. The change in transcriptional leakage in this mutant is lower than after CRISPRi, possibly because the Reb1-BSΔ mutations were shown to result in additional transcripts in the locus (Murray et al., 2015).
We next utilized a functional measure of transcriptional leakage. Stains with a GAL3 deletion exhibit a partial penetrance phenotype in which growth on galactose is dependent on GAL1 leakage (Kar et al., 2014). We find that after GAL10 ncRNA inhibition, gal3Δ strains grow faster on galactose containing plates (Fig. 5E) due to increased GAL1 leakage. To quantify this observation, cells were grown on plates containing glucose and galactose + ethidium bromide, where growth on galactose reflects activation of the GAL pathway (Kar et al., 2014). In wt cells, CRISPRi of GAL10 ncRNA has no effect on the percentage of colonies formed on galactose plates, but in gal3Δ cells the percentage increased from 0.56% ± 0.13 to 3.73% ± 0.34 colonies, a 6.7 fold increase (Fig. 5F). Since Gal1p is the only inducer in gal3Δ cells, this phenotypic result shows that CRISPRi inhibition of GAL10 ncRNA results in higher transcriptional leakage of GAL1.
smFISH reveals that transcriptional leakage of GAL10 and GAL1 is a rare event with few cells expressing and at the most only one transcript per cell (Fig. 5G). Surprisingly, cells no longer exhibit correlated expression between GAL1 and GAL10, suggesting that transcriptional leakage arises from independent initiation events for GAL10 and GAL1.
Finally, to test the effect of increased GAL1 levels, a plasmid with an additional copy of GAL1 under the ADE3 promoter was introduced in the GAL10-2xGFP strain. As predicted, increased expression of GAL1 results in a more sensitive dose response (Fig. 5H). The shift in this gain-of-function experiment is much more extreme than after GAL10 ncRNA inhibition, because the expression of GAL1 from a plasmid is much higher than the increased transcriptional leakage of 5–7 fold.
Taken together, we conclude that GAL10 ncRNA prevents transcriptional leakage of GAL10 and GAL1, and elevated levels of Gal1p result in a more sensitive activation threshold. The kinetic induction experiments (Fig. 4) also agree with this model. In raffinose, galactose induction would mostly rely on Gal3p, with little contribution of Gal1p and thus no effect of GAL10 ncRNA knockdown. However, in glucose GAL3 transcription is repressed and elevated GAL1 levels now result in faster galactose sensing and more rapid activation.
Increased transcriptional leakage results in fitness defect
Our data suggest a model where a 5–7 fold change in GAL1 mRNA levels results in a modest 11% change in the EC50 of the galactose dose response. It seems counterintuitive that cells have evolved a mechanism to repress leakage of GAL1 considering that low levels of Gal1p lead to more sensitive activation: one might argue that more sensitive or faster activation would be beneficial (Venturelli et al., 2015). Conversely, full repression of GAL1 might give cells an evolutionary advantage by preventing inappropriate activation of the GAL genes in glucose-galactose mixtures where glucose is the preferred carbon source (Wang et al., 2015). To distinguish between these models, we measured the fitness effect of GAL1 overexpression and ncRNA inhibition in a competition assay in multiple conditions (Hittinger and Carroll, 2007). Expression of GAL1 from a plasmid resulted in a 2% fitness defect in both glucose and mixed glucose/galactose conditions (Fig. 5I). ncRNA inhibition resulted in a fitness defect of 0.3–0.6% in several conditions, with the most pronounced effect in mixtures of low glucose and galactose (Fig. 5J). To put these fitness defects in context, a population with equal numbers of yeast with and without ncRNA will converge after ~750 generations to a population where more than 99% of the culture expresses GAL10 ncRNA. In evolutionary timescales, yeast without ncRNA is thus quickly outcompeted. Surprisingly, in both experiments, having increased GAL1 expression also resulted in a slight fitness defect in full induction conditions (2% galactose). Even though GAL1 and GAL10 steady-state levels did not measurably change in galactose after ncRNA inhibition, it is possible that targeting CRISPR to an actively transcribed gene may change elongation kinetics without affecting total RNA levels, which might have residual effects on growth. In summary, the primary selective advantage of this ncRNA appears to be in metabolic decision making in mixed carbon sources. The corollary to this observation is that the dose response is evolutionarily optimized at the expense of the activation time.
Transcriptional leakage repression is the only detectable role of GAL10 ncRNA
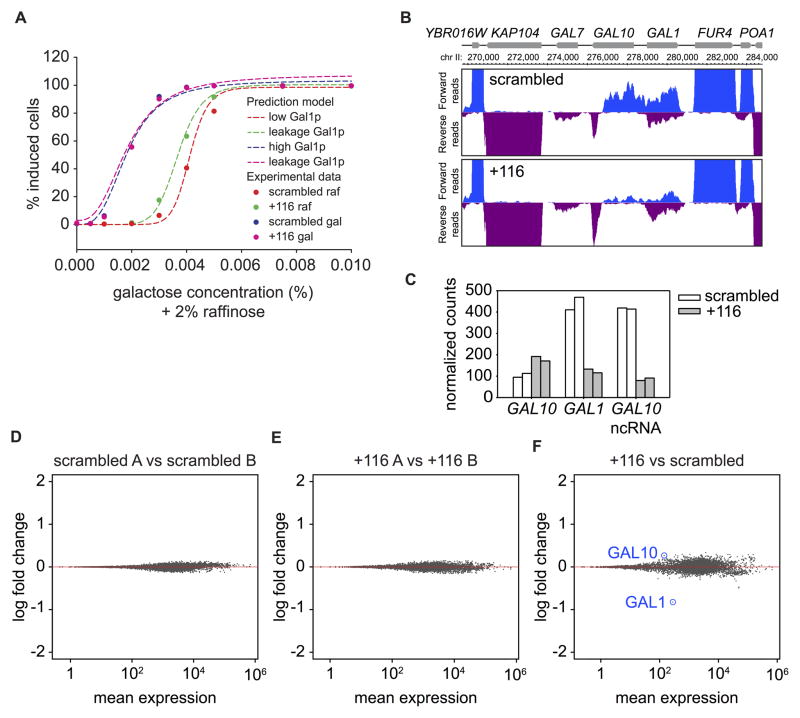
Since the galactose response contains multiple positive and negative feedback loops resulting in bistability and hysteresis, it seems possible that the ncRNA could be acting at multiple points in the network. To assess the quantitative role of GAL10 ncRNA, we adapted a previously reported model to quantitatively fit our dose response data (Venturelli et al., 2012). This ordinary differential equation model, which recapitulates the behavior of many mutants, is based on the dynamic interaction of the four main components of the network (Gal1p, Gal3p, Gal4p, Gal80p, Fig. S3A). Using the previously determined parameters with minor modification, this model gave bistability and hysteresis that were in quantitative agreement with the data (Fig. 6A).
Figure 6. Transcriptional leakage repression is the only detectable role of GAL10 ncRNA.
(A) Computational model of the dose response. The dotted lines represent the Venturelli model prediction; data points are from Fig. 4E. The model is fit using the scrambled data with Gal4 synthesis heterogeneity as a free parameter (red and blue). The predicted dose responses with a 5-fold leakage of GAL1 (green and pink) agree with the measured dose response shift when GAL10 ncRNA is inhibited (+116 raf and gal). See also Fig. S3. (B) Normalized coverage of forward and reverse reads at the GAL locus from RNA-seq. (C) Normalized counts at GAL10, GAL1 and GAL10 ncRNA (antisense of GAL10) for both replicates. (D) MA plot of scrambled A vs B. (E) MA plot of +116 A vs B. (F). MA plot of +116 vs scrambled.
We then asked whether the model could account for the +116 dose response shift. We found that increasing the Gal1p transcriptional leakage rate automatically leads to a shift in the EC50 of raffinose pretreated cells without having a major impact on galactose pretreated cells (Fig. S3). By allowing a 5-fold increase in Gal1p leakage, as observed experimentally, the computed dose response is in excellent quantitative agreement with the measured response upon ncRNA inhibition (Fig. 6A) and is sufficient to explain an 11% shift in the dose response from the naïve, but not from the exposed state.
To further examine whether GAL10 ncRNA could have additional functions, genome-wide expression was analyzed in glucose by RNA-seq in the presence and absence of the ncRNA. As expected, in strains with CRISPR +116, GAL10 ncRNA levels were reduced, and GAL10 levels were increased, although with a lower fold-change than observed by qRT-PCR (Figs. 6B,C). Since GAL1 is on the same strand as GAL10 ncRNA, the number of reads in the GAL1 locus decreased, but when normalized to GAL10 ncRNA, its levels increased. Replicates were highly reproducible (Figs. 6D,E). Apart from these changes at the GAL1-10 locus, no other genes changed more than 1.5 fold upon ncRNA inhibition (Fig. 6F). This analysis highlights the specificity of the CRISPRi approach. In summary, both quantitative modeling and RNA-seq suggest a singular role for GAL10 ncRNA in leakage suppression under repressive conditions.
GAL10 ncRNA knockdown has no effect on GAL10 activation
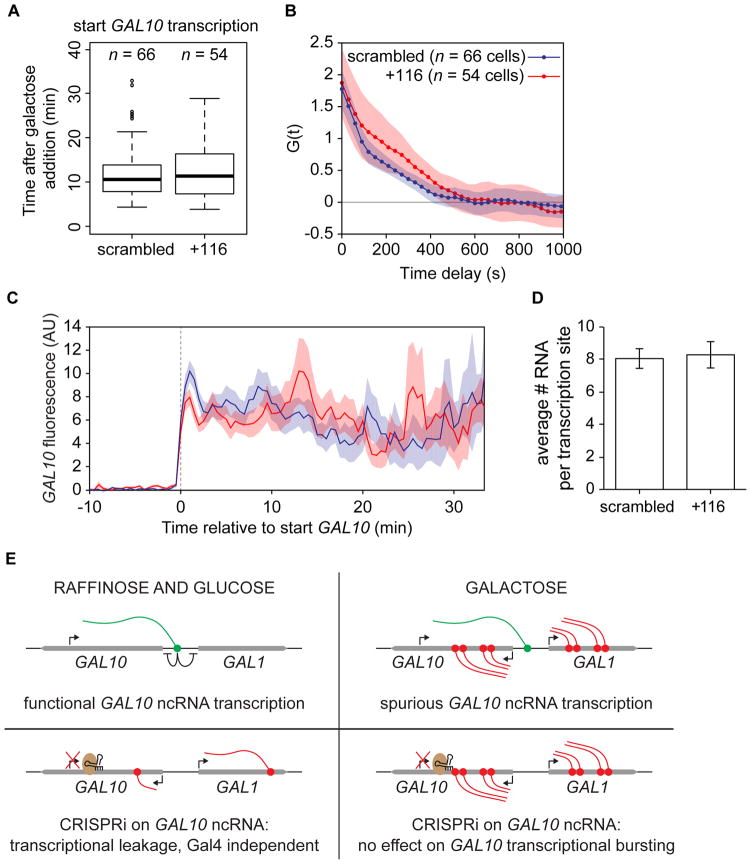
GAL10 ncRNA has a functional role in repression of GAL1 and GAL10 transcriptional leakage, but the initial imaging experiments and our computational model suggest that there is no function for GAL10 ncRNA during activation, i.e. after addition of galactose. To confirm this conjecture, CRISPRi scrambled and +116 were introduced into the PP7 and MS2 tagged GAL10 strain (Fig. S4). Consistent with our earlier findings, live-cell imaging revealed no difference in the start of GAL10 transcription (Fig. 7A), the burst duration (Fig. 7B) or GAL10 transcription frequency (Fig. 7C) when GAL10 ncRNA was removed. Additionally, the average number of RNAs per TS in the two strains is identical (Fig. 7D). Upon galactose activation, Gal4p activity thus seems to override all effects of GAL10 ncRNA, whose transcription switches from functional to spurious. In addition, binding of the CRISPRi complex does not appear to interfere with GAL10 sense transcription, underscoring its strand-specific nature.
Figure 7. GAL10 ncRNA transcription does not affect GAL10 activation.
(A). Boxplot of the start of GAL10 transcription determined by imaging after galactose addition, in cells with CRISPRi scrambled or +116. (B) Autocorrelations of TS intensity traces for CRISPRi scrambled (blue) and +116 (red), respectively, corrected for non-steady-state effects. Shaded areas are SEM. (C) GAL10 frequency of CRISPRi scrambled and +116, measured as the average fluorescent intensity of all traces, after aligning the traces on the start of GAL10 transcription. Color code and cell numbers are the same as (B). (D) Average number of RNA per TS, as measured by smFISH on MS2 in cells with CRISPRi scrambled and +116. Each bar represents 4 experiments. See Fig. S4. (E) Model for switch between functional and spurious GAL10 ncRNA transcription.
Discussion
Our results indicate that ncRNA transcription can be both functional and spurious, but under different conditions (Fig. 7E). In non-inducing conditions, such as raffinose or glucose, GAL10 ncRNA transcription is repressive to genes located both in sense and antisense to the ncRNA. Specifically, this repression manifests itself in the galactose network by suppression of transcriptional leakage of the gene encoding the metabolic enzyme and sensor Gal1p. However, ncRNA transcription is also pervasive under activating conditions but seems to have no inhibitory effect on GAL10 transcription. In fact, there is no detectable role for this ncRNA when galactose is present, leading us to conclude that it results from spurious transcription and is indeed ‘transcriptional noise.’
Repression by ncRNA transcription
The primary indication that this ncRNA is repressive comes from strand-specific blocking of ncRNA synthesis by CRISPR/dCas9. Previous attempts at inhibition with CRISPR in eukaryotes have required the addition of a repressor domain to dCas9 for full repression, resulting in a loss of strand-specificity (Gilbert et al., 2013). In our case, dCas9 alone was sufficient for repression, perhaps because of the low-level expression of GAL10 ncRNA. Partial repression could be achieved using a gRNA targeted to the TATA box, indicating that CRISPRi is a versatile tool to tune ncRNA expression. The advantage of this approach is that this perturbation does not rely on genetic modifications, which might disrupt other cis-acting sequences. By blocking GAL10 ncRNA synthesis with CRISPR, we show that GAL1 and GAL10 basal levels are 5–7 fold increased (Fig. 5). Importantly, the ncRNA-based repression was independent of Gal4p, suggesting a mechanism of repression that is outside the existing galactose regulatory network.
Several models have been proposed for ncRNA-mediated gene repression. One model suggests that antisense transcripts form R-loops, or RNA-DNA hybrids, which can repress transcription (Cloutier et al., 2013; Geisler et al., 2012). However, the formation of stable R-loops is inconsistent with our live-cell imaging results, which indicate that GAL10 ncRNA is only present at the locus while being transcribed. In addition, our live-cell data does not support the occurrence of long-lived stalled polymerases from head-to-head collisions (Hobson et al., 2012), suggesting that collisions are rare or that stalled polymerases are quickly removed.
The short dwell time of ncRNA suggest that repression occurs by the act of ncRNA transcription and not by the RNA itself, in agreement with a previous observation that the GAL10 ncRNA cannot act in trans (Houseley et al., 2008) and does not change expression of genes outside the GAL locus (Fig. 6). More specifically, the ncRNA likely exerts its physiological effects through transcriptional interference of the GAL1-10 promoter. Non-coding transcription in promoter regions has been proposed to be activating, for example by removal of nucleosomes (Uhler et al., 2007) or repressive by removal of transcription factors (Bumgarner et al., 2012). However, we find that repression is independent of Gal4p, arguing against the latter explanation. Another mechanism of repression is the formation of a repressive chromatin environment. Factors such as the methyltransferases Set1 and Set2 and the deacetylases Set3 and Rpd3S are recruited to chromatin by the act of transcription, after which deacetylation in the promoter region prevents transcription initiation of the sense transcripts (Houseley et al., 2008; Kim et al., 2012; Margaritis et al., 2012; Pinskaya et al., 2009; van Werven et al., 2012). Removal of several of these chromatin modifications in the GAL1-10 promoter have been associated with elimination of ncRNA-mediated repression (Houseley et al., 2008; Pinskaya et al., 2009). In summary, the mechanism of transcriptional interference by ncRNAs appears to be highly gene-specific.
The role of stochastic transcription in the GAL network
Single-molecule imaging reveals multiple, hitherto unappreciated, roles for stochastic transcription in this eukaryotic transcriptional paradigm. Under repressed conditions, 0.6% of cells show transcriptional leakage of GAL1 (Fig. 5F), which likely reflects a stochastic transcription event resulting in the production of single GAL1 mRNAs. Such events are in stark contrast to induced GAL10 transcriptional bursting, because they are Gal4p independent, uncorrelated between GAL10 and GAL1, and cannot be repressed by glucose repression mechanisms.
Transcription of the ncRNA is also stochastic and plays an essential role in carbon source selection. Upon GAL10 ncRNA inhibition, an increase in the number of cells with GAL1 leakage results in a cell population that is overall more sensitive to galactose, with several measurable phenotypes, including faster activation from glucose, a shift in the dose response, and a higher percentage of induced cells in glucose-galactose mixtures. In fact, we can quantitatively account for the degree of repression and the shift in the EC50 by using a previously-reported model of the GAL network (Venturelli et al., 2012). The fact that the shift in the dose response can be explained only by changing the leakage rate of Gal1p indicates that the singular role of the ncRNA in the network is to prevent transcriptional leakage of a signaling molecule and supports the supposition that the ncRNA affects neither the rate constants nor the topology of the network. These data are further supported by RNA-seq which indicates the high specificity of action for this ncRNA. The view engendered by these results is that the cell uses stochastic transcription of one RNA to suppress the stochastic transcription of another RNA in cis.
Under repressive conditions, stochastic transcription plays a discernible functional role in the galactose response, but we also identify conditions where the ncRNA is present but has no detectable function. The existence of a spurious phase of ncRNA transcription once galactose is present suggests that the Gal4p activator is able to completely override the effects of ncRNA transcription. Although it is nearly impossible to establish the absence of function, several observations support this interpretation of spurious transcription. First, we see no difference in activation times between cells that do or do not show ncRNA transcription (Fig. 2). Second, the kinetics of GAL10 transcription do not change when ncRNA synthesis is blocked (Fig. 7). Finally, a genome-wide study reported that antisense transcription represses genes in the lower expression range, but has no effect in the higher expression range (Xu et al., 2011). Thus, the mechanism we propose here may be generally applicable to other genes. Moreover, our competition experiments suggest the cell is willing to tolerate spurious, energy-consuming transcription of the ncRNA in order to achieve repression of a metabolic gene. We speculate that ncRNA transcription has evolved as a mechanism of deeper repression in an organism that possesses little heterochromatin and no RNA interference machinery.
Experimental procedures
Yeast strains and plasmids
Diploid yeast (BY4743) was transformed with a PCR product containing the MS2+PP7 loop cassette and loxP-kanMX-loxP. The kanMX marker was removed with CRE recombinase (pSH47). Coat protein were expressed from two plasmids: pHIS PMET17 MCP-NLS-mKate2, and pURA PADE3 PCP-NLS-2xGFP. Strains, plasmids and oligos used to construct the strains are listed in Tables S1, S2 and S3, respectively.
Microscopy and image processing
Cells were imaged at mid-log (OD 0.2–0.4) on concanavalin A-coated MatTek dishes at 30°C. Imaging was performed on a confocal laser scanning microscope (Zeiss 780) with 488/594 nm excitation or on a custom build wide-field microscope, consisting of an AxioObserver inverted microscope (Zeiss), a 150x NA 1.35 objective, two Evolve 512 EMCCD cameras (Photometrics), a Tokai Hit stage incubator (INUB-LPS) and laser excitation at 488 nm and 594 nm (Excelsior, Spectra Physics). Wide-field images were recorded in 2 colors simultaneously every 30s as 9 z-stacks (Δz 0.5 μm) with 150 ms exposure using micromanager software (Edelstein et al., 2010). Image analysis was done as described previously (Coulon et al., 2014) with minor modifications (Suppl. Exp. Procedures)
Single-molecule FISH
Yeast cultures were grown to early mid-log, fixed with PFA and permeabilized with lyticase as described in (Trcek et al., 2012). Coverslips were hybridized for 4h at 37°C with hybridization buffer containing 10% dextran sulfate, 10% formamide, 2xSSC and 7.5 pmol probe. For FISH targeting the repeats, two MS2 or four PP7 probes labeled with Cy3 were targeted to the loops (Integrated DNA Technologies). For FISH on GAL1 and GAL10, 48 probes were targeted to the coding regions and labeled with Cy3 and Cy5, respectively, (Biosearch Technologies). Coverslips were washed 2x for 30 min with 10% formamide, 2xSSC at 37°C, 1x with 2xSSC, and 1x for 5 min with 1xPBS at room temperature. Coverslips were mounted on microscope slides using mounting media with DAPI (ProLong Gold, Life Technologies).
Flow-cytometry
Cells were pre-grown for 20h in media with 2% raffinose or 2% galactose. Cultures were grown to steady state for 30h in 2% raffinose with different galactose concentration or 2% galactose with different glucose concentrations, Cultures were maintained at low cell density (below OD600 0.1). Flow-cytometry measurements were taken on a DB FACSCalibur, using 488 nm excitation and a 530/30 nm emission filter. For each measurement, 10,000 cells were collected. Competition experiments we performed as in (Hittinger and Carroll, 2007), with minor modifications (Suppl. Exp. Procedures).
Northern blot and quantitative RT-PCR
RNA from mid-log cultures was isolated by phenol extraction. Northern blot analysis was performed using standard protocols. Strand-specific detection was ensured by hybridizing with a hot ssDNA PCR product generated by linear PCR from a cold PCR template. Quantification of the signals was carried out using a Storm 860 phosphoimager. For qPCR, RNA was cleaned with Qiagen RNAeasy columns with DNAse treatment. cDNA was made with poly-dT primers and protoScript II and quantified by qPCR. Primer pairs are in Table S3.
Colony assay
Diluted cultures were plated on selective plates with2% glucose or 2% galactose with 20 μg/ml Ethidium Bromide. The percentage of colonies on galactose plates after 4 days of growth was normalized to the number of colonies on glucose plates.
Modeling of the GAL network
Computational modeling was based on numerical analysis of the solution of coupled nonlinear differential equations described in (Venturelli et al., 2012) with an added leakage rate α0G1 = 0.13 nM/min and a reduced Gal4p to Gal1p feedback parameter αG1 = 7 nM/min. We implement heterogeneity in Gal4p by assuming that the baseline synthesis rate αG4 is normally distributed with mean 0.2 nM/min (as specified by Venturelli) and σαG4 = 0.07. See Suppl. Exp. Procedures.
RNA-seq
PolyA selected mRNA was sequenced on one HiSeq2500 lane using Illumina TruSeq v4 chemistry. Paired-end reads were aligned with Tophat and analyzed with the R packages GenomicAlignments and DESEq2 (Suppl. Exp Procedures). Data is deposited in GEO with accession number: GSE72032
Supplementary Material
Acknowledgments
We thank M.A. Basrai, M. Vogelauer, C. Hittinger, V. Verkhusha, T.S. Karpova, G.L. Hager, M.H. Larson, J.S. Weissman, R.H. Singer and D.R. Zenklusen for strains and plasmids. We thank T.S. Karpova for assistance with confocal imaging, K.M. McKinnon for assistance with flow cytometry, the NCI/CCR Sequencing Facility for RNA-seq, D.R. van Dijk and J. Rodriguez for help with RNA-seq analysis, and members of the Larson lab and of the Janelia Research Campus/HHMI Transcription Imaging Consortium for helpful discussions. Supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, National Institute of Diabetes and Digestive and Kidney Diseases, the European Molecular Biology Organization and the KWF Dutch Cancer Society, grants ALTF 37-2012, KWF 2012-5394 (TLL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abramczyk D, Holden S, Page CJ, Reece RJ. Interplay of a ligand sensor and an enzyme in controlling expression of the Saccharomyces cerevisiae GAL genes. Eukaryot Cell. 2012;11:334–342. doi: 10.1128/EC.05294-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acar M, Becskei A, van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–232. doi: 10.1038/nature03524. [DOI] [PubMed] [Google Scholar]
- Bumgarner SL, Neuert G, Voight BF, Symbor-Nagrabska A, Grisafi P, van Oudenaarden A, Fink GR. Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Mol Cell. 2012;45:470–482. doi: 10.1016/j.molcel.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Castelnuovo M, Rahman S, Guffanti E, Infantino V, Stutz F, Zenklusen D. Bimodal expression of PHO84 is modulated by early termination of antisense transcription. Nat Struct Mol Biol. 2013;20:851–858. doi: 10.1038/nsmb.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier SC, Wang S, Ma WK, Petell CJ, Tran EJ. Long noncoding RNAs promote transcriptional poising of inducible genes. PLoS Biol. 2013;11:e1001715. doi: 10.1371/journal.pbio.1001715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon A, Ferguson ML, de Turris V, Palangat M, Chow CC, Larson DR. Kinetic competition during the transcription cycle results in stochastic RNA processing. eLife. 2014;3 doi: 10.7554/eLife.03939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci U A. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using μManager. In: Frederick M, Ausubel Al, editors. Curr Protoc Mol Biol. Unit14.20. Chapter 14. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi SJ, Zenklusen D, Lionnet T, Singer RH. Transcription of functionally related constitutive genes is not coordinated. Nat Struct Mol Biol. 2011;18:27–34. doi: 10.1038/nsmb.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Lojek L, Khalil AM, Baker KE, Coller J. Decapping of long noncoding RNAs regulates inducible genes. Mol Cell. 2012;45:279–291. doi: 10.1016/j.molcel.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger CT, Carroll SB. Gene duplication and the adaptive evolution of a classic genetic switch. Nature. 2007;449:677–681. doi: 10.1038/nature06151. [DOI] [PubMed] [Google Scholar]
- Hobson DJ, Wei W, Steinmetz LM, Svejstrup JQ. RNA polymerase II collision interrupts convergent transcription. Mol Cell. 2012;48:365–374. doi: 10.1016/j.molcel.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- Kar RK, Qureshi MT, DasAdhikari AK, Zahir T, Venkatesh KV, Bhat PJ. Stochastic galactokinase expression underlies GAL gene induction in a GAL3 mutant of Saccharomyces cerevisiae. FEBS J. 2014;281:1798–1817. doi: 10.1111/febs.12741. [DOI] [PubMed] [Google Scholar]
- Kim T, Xu Z, Clauder-Münster S, Steinmetz LM, Buratowski S. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 2012;150:1158–1169. doi: 10.1016/j.cell.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphier MS, Ptashne M. Multiple mechanisms mediate glucose repression of the yeast GAL1 gene. Proc Natl Acad Sci U S A. 1992;89:5922–5926. doi: 10.1073/pnas.89.13.5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011;332:475–478. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Durairaj G, Bhaumik SR. Mechanisms of antisense transcription initiation from the 3′ end of the GAL10 coding sequence in vivo. Mol Cell Biol. 2013;33:3549–3567. doi: 10.1128/MCB.01715-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaritis T, Oreal V, Brabers N, Maestroni L, Vitaliano-Prunier A, Benschop JJ, van Hooff S, van Leenen D, Dargemont C, Géli V, et al. Two Distinct Repressive Mechanisms for Histone 3 Lysine 4 Methylation through Promoting 3′-End Antisense Transcription. PLoS Genet. 2012;8:e1002952. doi: 10.1371/journal.pgen.1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Murray SC, Haenni S, Howe FS, Fischl H, Chocian K, Nair A, Mellor J. Sense and antisense transcription are associated with distinct chromatin architectures across genes. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Fischl H, Howe FS, Woloszczuk R, Serra Barros A, Xu Z, Brown D, Murray SC, Haenni S, Halstead JM, et al. Transcription mediated insulation and interference direct gene cluster expression switches. eLife. 2014;3 doi: 10.7554/eLife.03635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinskaya M, Gourvennec S, Morillon A. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 2009;28:1697–1707. doi: 10.1038/emboj.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick CA, Jowitt TA, Reece RJ. The Effect of Ligand Binding on the Galactokinase Activity of Yeast Gal1p and Its Ability to Activate Transcription. J Biol Chem. 2009;284:229–236. doi: 10.1074/jbc.M807878200. [DOI] [PubMed] [Google Scholar]
- Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- Trcek T, Chao JA, Larson DR, Park HY, Zenklusen D, Shenoy SM, Singer RH. Single-mRNA counting using fluorescent in situ hybridization in budding yeast. Nat Protoc. 2012;7:408–419. doi: 10.1038/nprot.2011.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhler JP, Hertel C, Svejstrup JQ. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc Natl Acad Sci U A. 2007;104:8011–8016. doi: 10.1073/pnas.0702431104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturelli OS, El-Samad H, Murray RM. Synergistic dual positive feedback loops established by molecular sequestration generate robust bimodal response. Proc Natl Acad Sci U S A. 2012;109:E3324–E3333. doi: 10.1073/pnas.1211902109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturelli OS, Zuleta I, Murray RM, El-Samad H. Population Diversification in a Yeast Metabolic Program Promotes Anticipation of Environmental Shifts. PLoS Biol. 2015;13:e1002042. doi: 10.1371/journal.pbio.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Atolia E, Hua B, Savir Y, Escalante-Chong R, Springer M. Natural Variation in Preparation for Nutrient Depletion Reveals a Cost–Benefit Tradeoff. PLoS Biol. 2015;13:e1002041. doi: 10.1371/journal.pbio.1002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven FJ, Neuert G, Hendrick N, Lardenois A, Buratowski S, van Oudenaarden A, Primig M, Amon A. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell. 2012;150:1170–1181. doi: 10.1016/j.cell.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Clauder-Münster S, Smolik M, Huber W, Steinmetz LM. Antisense expression increases gene expression variability and locus interdependency. Mol Syst Biol. 2011;7:468. doi: 10.1038/msb.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharioudakis I, Gligoris T, Tzamarias D. A yeast catabolic enzyme controls transcriptional memory. Curr Biol CB. 2007;17:2041–2046. doi: 10.1016/j.cub.2007.10.044. [DOI] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol. 2008;15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.