Abstract
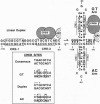
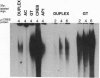
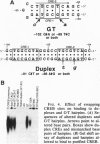
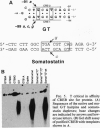
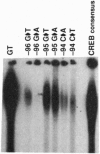
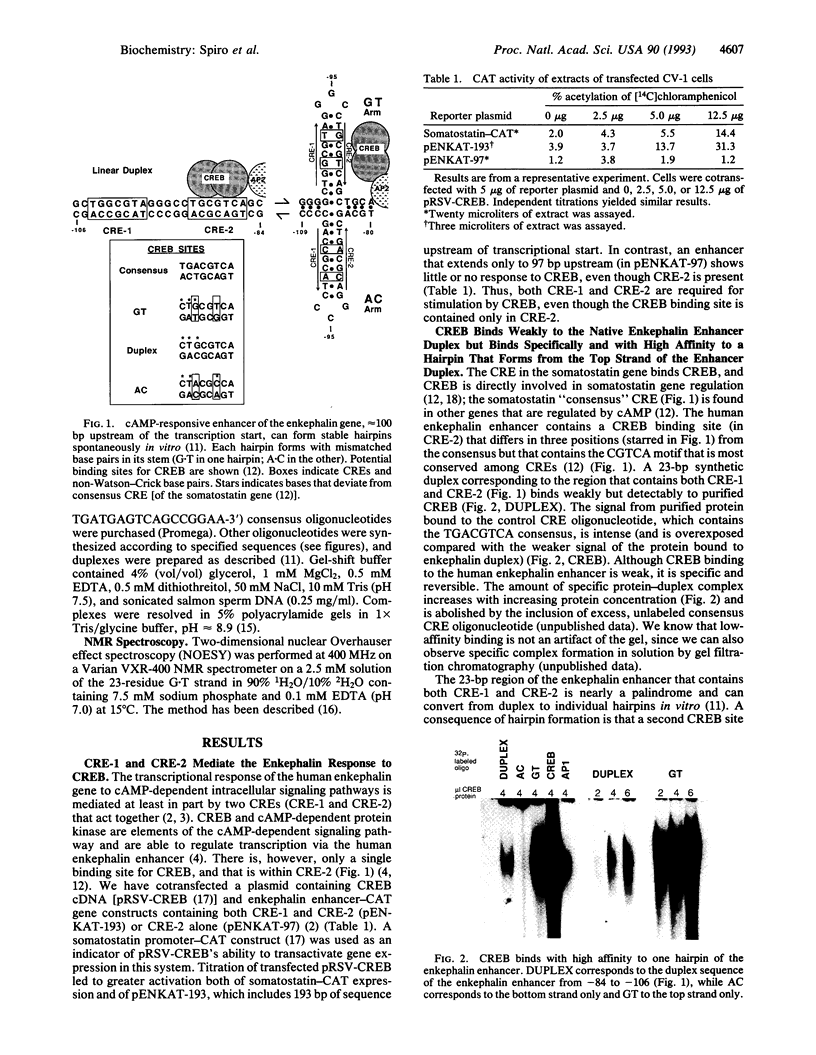
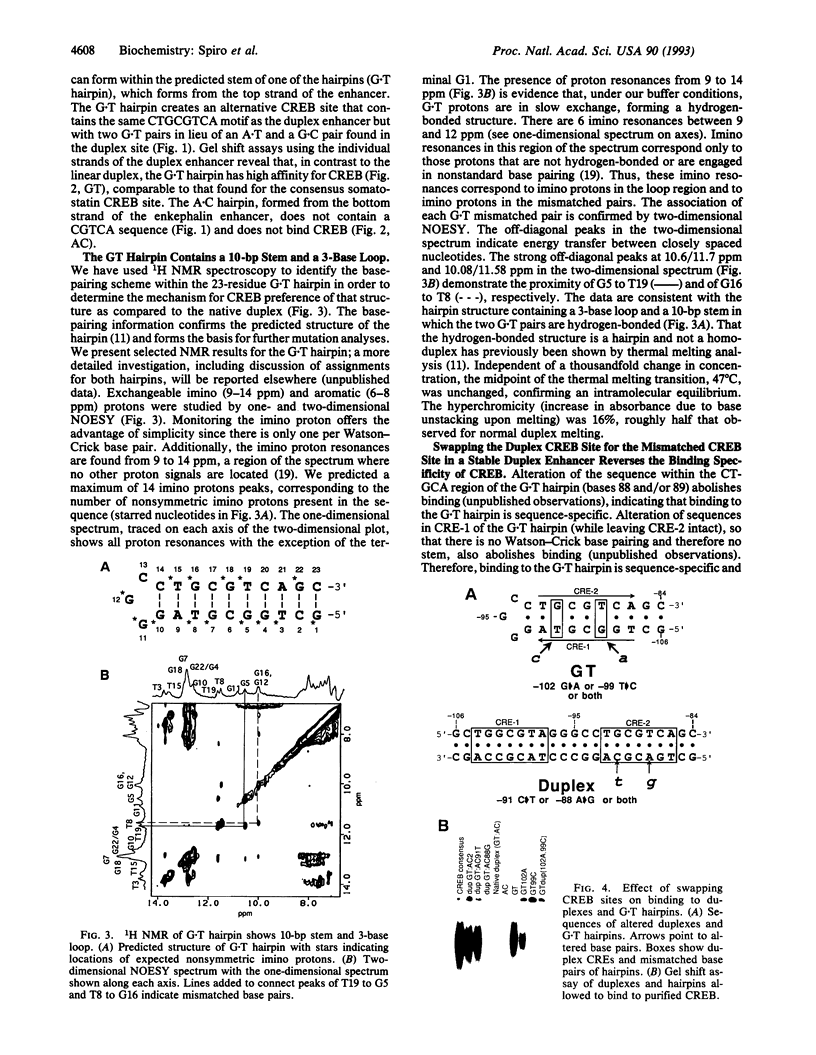
Transactivation studies of the enkephalin enhancer indicate that two cAMP response elements (CRE-1 and CRE-2) are needed to mediate the transcriptional response to cAMP and to the CRE-binding protein (CREB) transcription factor. CRE-1 and CRE-2 are contained within a nearly palindromic region that can form stable hairpin structures in vitro. CREB binds only weakly to the native duplex enhancer and only within CRE-2. In contrast, CREB binds with high affinity to the hairpin in which CRE-1 and CRE-2 come together to form a CREB site with two G.T base pairs. NMR and binding studies show that high-affinity binding to the G.T hairpin requires one of the mismatched G.T pairs. Insertion of that G.T pair into the duplex confers high-affinity binding. Parallel studies with the somatostatin CRE show that the T in one G.T pair is crucial for high-affinity binding. The existence within a short enhancer of alternative sites for a single factor suggests a mechanism for regulation of transcription by DNA structure.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bianchi M. E., Beltrame M., Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989 Feb 24;243(4894 Pt 1):1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- Comb M., Birnberg N. C., Seasholtz A., Herbert E., Goodman H. M. A cyclic AMP- and phorbol ester-inducible DNA element. 1986 Sep 25-Oct 1Nature. 323(6086):353–356. doi: 10.1038/323353a0. [DOI] [PubMed] [Google Scholar]
- Gartenberg M. R., Ampe C., Steitz T. A., Crothers D. M. Molecular characterization of the GCN4-DNA complex. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6034–6038. doi: 10.1073/pnas.87.16.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glucksmann M. A., Markiewicz P., Malone C., Rothman-Denes L. B. Specific sequences and a hairpin structure in the template strand are required for N4 virion RNA polymerase promoter recognition. Cell. 1992 Aug 7;70(3):491–500. doi: 10.1016/0092-8674(92)90173-a. [DOI] [PubMed] [Google Scholar]
- Gonzalez G. A., Montminy M. R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989 Nov 17;59(4):675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- He X., Rosenfeld M. G. Mechanisms of complex transcriptional regulation: implications for brain development. Neuron. 1991 Aug;7(2):183–196. doi: 10.1016/0896-6273(91)90257-z. [DOI] [PubMed] [Google Scholar]
- Hollenberg S. M., Evans R. M. Multiple and cooperative trans-activation domains of the human glucocorticoid receptor. Cell. 1988 Dec 2;55(5):899–906. doi: 10.1016/0092-8674(88)90145-6. [DOI] [PubMed] [Google Scholar]
- Horwitz M. S., Loeb L. A. An E. coli promoter that regulates transcription by DNA superhelix-induced cruciform extrusion. Science. 1988 Aug 5;241(4866):703–705. doi: 10.1126/science.2456617. [DOI] [PubMed] [Google Scholar]
- Huggenvik J. I., Collard M. W., Stofko R. E., Seasholtz A. F., Uhler M. D. Regulation of the human enkephalin promoter by two isoforms of the catalytic subunit of cyclic adenosine 3',5'-monophosphate-dependent protein kinase. Mol Endocrinol. 1991 Jul;5(7):921–930. doi: 10.1210/mend-5-7-921. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Curran T. DNA bending by Fos and Jun: the flexible hinge model. Science. 1991 Nov 22;254(5035):1210–1214. doi: 10.1126/science.1957173. [DOI] [PubMed] [Google Scholar]
- Kobierski L. A., Chu H. M., Tan Y., Comb M. J. cAMP-dependent regulation of proenkephalin by JunD and JunB: positive and negative effects of AP-1 proteins. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10222–10226. doi: 10.1073/pnas.88.22.10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray C. T., Devi L., Calavetta L., Douglass J. O. Regulated expression of the prodynorphin gene in the R2C Leydig tumor cell line. Endocrinology. 1989 Jan;124(1):49–59. doi: 10.1210/endo-124-1-49. [DOI] [PubMed] [Google Scholar]
- McMurray C. T., Wilson W. D., Douglass J. O. Hairpin formation within the enhancer region of the human enkephalin gene. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):666–670. doi: 10.1073/pnas.88.2.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plon S. E., Wang J. C. Transcription of the human beta-globin gene is stimulated by an SV40 enhancer to which it is physically linked but topologically uncoupled. Cell. 1986 May 23;45(4):575–580. doi: 10.1016/0092-8674(86)90289-8. [DOI] [PubMed] [Google Scholar]
- Rehfuss R. P., Walton K. M., Loriaux M. M., Goodman R. H. The cAMP-regulated enhancer-binding protein ATF-1 activates transcription in response to cAMP-dependent protein kinase A. J Biol Chem. 1991 Oct 5;266(28):18431–18434. [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Walton K. M., Rehfuss R. P. Molecular mechanisms of cAMP-regulated gene expression. Mol Neurobiol. 1990 Fall-Winter;4(3-4):197–210. doi: 10.1007/BF02780341. [DOI] [PubMed] [Google Scholar]