SUMMARY
Despite minimal disparity at the sequence level, mammalian H3 variants bind to distinct sets of polypeptides. Though histone H3.1 predominates in cycling cells, our knowledge of the soluble complexes that it forms en route to deposition or following eviction from chromatin remains limited. Here, we provide a comprehensive analysis of the H3.1-binding proteome, with emphasis on its interactions with histone chaperones and components of the replication fork. Quantitative mass spectrometry revealed 170 protein interactions, whereas a large-scale biochemical fractionation of H3.1 and associated enzymatic activities uncovered over twenty stable protein complexes in dividing human cells. The sNASP and ASF1 chaperones play pivotal roles in the processing of soluble histones, but do not associate with the active CDC45/MCM2-7/GINS (CMG) replicative helicase. We also find TONSL-MMS22L to function as a H3-H4 histone chaperone. It associates with the regulatory MCM5 subunit of the replicative helicase.
INTRODUCTION
Nucleosomal histones are subject to an extraordinary array of chemical modifications, many of which have profound effects on chromatin structure and gene expression (Campos and Reinberg, 2009). While such posttranslational modifications (PTMs) can alter the epigenetic landscape, histones are systematically displaced from chromatin as DNA is transcribed, replicated or repaired (Annunziato, 2012; Campos et al., 2014). A number of histone deposition pathways are relatively well understood, but we have yet to fully understand the means by which nuclear histones are evicted and chaperoned as biological processes take place on chromatin.
While human histone H4 is invariant, the genome encodes a number of histone H3 variants. Studies identified three principal somatic forms of which the ‘replication-coupled’ H3.1 and H3.2 are highly transcribed in S-phase (Wu and Bonner, 1981; Wu et al., 1982) and serve to complement the dilution of nucleosomal histones that segregate onto nascent DNA. ‘Replication-independent’ H3.3 histones are expressed throughout interphase (Wu and Bonner, 1981; Wu et al., 1982) and thus viewed as ‘replacement histones’. Recent seminal studies described molecular pathways overseeing the deposition of these variants (Drane et al., 2010; Goldberg et al., 2010; Tagami et al., 2004; Wong et al.). In the cytoplasm, heat-shock proteins fold and pair newly translated H3-H4 histone polypeptides (Alvarez et al.; Campos et al.). The resultant histone dimer is then bound by the somatic Nuclear Autoantigenic Sperm Protein (sNASP) histone chaperone (Campos et al., 2010) to protect histones from targeted proteolysis (Cook et al., 2011), and to promote the di-acetylation of histone H4 at lysines 5 and 12 by the Histone Acetyltransferase 1 (HAT1) holoenzyme (Campos et al., 2010; Kleff et al., 1995; Parthun et al., 1996; Verreault et al., 1998). These di-acetyl marks, concomitant with the monomethylation of H3 lysine 9 (H3K9me1), constitute a key signature of newly synthesized histones (Loyola et al., 2006). The mature histone pair is finally shuttled into the nucleus while bound to the Anti-silencing Factor 1 (ASF1) histone chaperone and the importin-4 karyopherin (Alvarez et al., 2011; Campos et al., 2010).
In the nucleus, replication-coupled H3-H4 dimers are transferred from ASF1 to the Chromatin Assembly Factor 1 (CAF-1) histone chaperone for (H3-H4)2 tetramer formation (Liu et al., 2012; Winkler et al., 2012) and deposition (Mello et al., 2002; Tagami et al., 2004; Tyler et al., 2001). CAF-1 binds PCNA and is found directly behind the replicative helicase on replicating DNA (Shibahara and Stillman, 1999). Tetrasomes are subsequently flanked by two H2A-H2B histone dimers to complete the mature nucleosome (Annunziato, 2012). Replication-independent histones are channeled through a number of distinct histone chaperones including HIRA (Tagami et al., 2004) and DAXX-ATRX (Drane et al., 2010; Goldberg et al., 2010; Wong et al., 2010). Despite these important developments, it remains unclear how nucleosomal histones carrying potential epigenetic information are chaperoned upon eviction from chromatin, especially as they encounter the replicative helicase.
In eukaryotes, the processive replicative helicase is composed minimally of the CDC45/MCM2-7/GINS (CMG) complex (Gambus et al., 2006; Ilves et al., 2010; Kang et al., 2012; Pacek et al., 2006). GINS/CDC45 activates the MCM2-7 helicase activity by bridging a gap between the MCM2 and MCM5 subunits (Costa et al., 2011). The MCM2 subunit of the CMG complex has been shown to cooperatively bind histones and the FACT histone chaperone (Foltman et al., 2013; Tan et al., 2006). This likely constitutes an early step in nucleosome disassembly at replication forks (Campos et al., 2014). The H3-H4 histone chaperone ASF1 has also been proposed to associate with the replicative helicase (Groth et al., 2007). While ASF1 dissociates (H3-H4)2 tetramers into H3-H4 dimers (English et al., 2006; Natsume et al., 2007), the bulk of H3.1-H4 histones segregate at the fork as tetrameric units (Xu et al., 2010). This suggests that histones are either transiently dissociated into dimers at the fork, or are alternatively segregated independently of ASF1.
To better understand replication-coupled histone eviction and the stable protein complexes that ensue, we performed a comprehensive biochemical purification resulting in a detailed description of the soluble replication-coupled human H3.1 interactome. H3.1 protein complexes were analyzed for intrinsic enzymatic activities, with a particular focus on complexes formed in S-phase. We further surveyed the purified fractions for their ability to disassemble nucleosomal histones and identified TONSL-MMS22L as a histone chaperone with a preference for histone H3 monomethylated at lysine 9. TONSL-MMS22L was previously shown to promote homologous recombination (HR) during S-phase (Duro et al., 2010; O’Connell et al., 2010; O’Donnell et al., 2010; Piwko et al., 2010). Disruption of either TONSL or MMS22L results in the accumulation of DNA damage, activation of the ATM/ATR checkpoint response, and accumulation of cells in G2 (Duro et al., 2010; O’Connell et al., 2010; O’Donnell et al., 2010; Piwko et al., 2010). In this study we further demonstrate that purified fractions containing TONSL-MMS22L enhance histone eviction. Importantly, TONSL interacts with the regulatory MCM5 subunit of the replicative helicase, normally employed by GINS/CDC45 to bridge the MCM2–MCM5 junction and activate the MCM2-7 helicase (Costa et al., 2011).
RESULTS
Quantitative MS analysis of the Soluble Human H3.1 Proteome
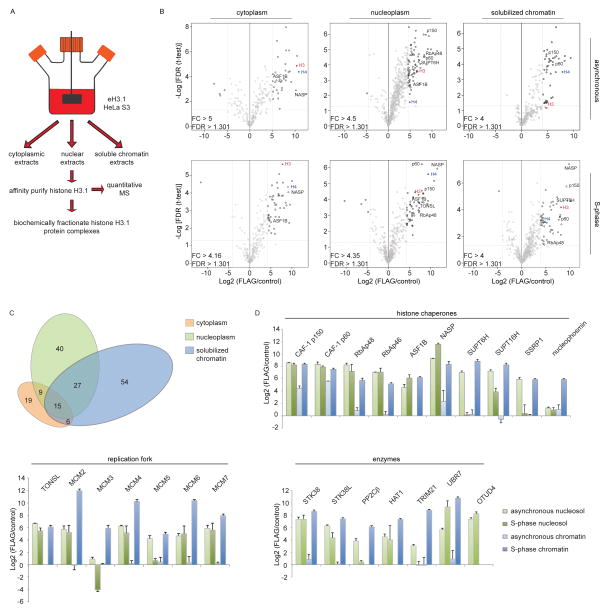
A comprehensive biochemical analysis of the soluble H3.1 proteome was performed from HeLa S3 spinner cultures (Figure 1A). The cells expressed low levels of epitope-tagged H3.1 (eH3.1) thus averting the co-purification of closely related histone variants (Tagami et al., 2004). In order to gain confidence in the validity of putative interacting partners, a label-free quantitative mass spectrometry (MS) approach (Smits et al., 2013) was first exploited to compare affinity-purified eH3.1 to mock purifications from different subcellular compartments in S-phase as well as from an asynchronous state. Protein hits were considered to be of interest if reproduced across at least three independent immunoprecipitations and statistically enriched over matching mock purifications (Figure 1B, Table S1). Two thirds of the 170 statistically enriched proteins were unique to one of the three examined subcellular compartments (Figure 1C), highlighting variances in the subcellular handling of histones. As expected, ontology-based classification of the associated proteins revealed a prevalence of proteins involved in complex assembly and organization in the cytoplasm, whereas ontology terms relating to transcription, and chromatin assembly, organization and modification abounded in the nuclear compartments (Figure S1A).
Figure 1.
Quantitative mass spectrometry (MS) analyses of H3.1-interacting proteins. (A) Purification Scheme. (B) Quantitative MS analysis of affinity-purified eH3.1 isolated from asynchronous or synchronized, replicating cells. Each volcano plot represents three independent eH3.1 (FLAG) pull-downs plotted against matching mock purifications. The x axis denotes the eH3.1 over mock ratio of MS intensity whereas a false discovery rate (FDR) adapted t-test is plotted on the y axis. (C) Distribution of H3.1-interacting proteins across subcellular compartments. (D) Relative enrichment of histone chaperones, components of the replication fork, and proteins with enzymatic activity within the eH3.1 immunoprecipitates. Data is presented as mean Label-free Quantification (LFQ) ratio +/− SD.
MCM2, sNASP, and UBR7 were the most enriched proteins co-purifying with nuclear H3.1, exhibiting over a thousand-fold enrichment in label-free quantification intensities relative to mock IPs, closely followed by an array of histone chaperones and transcription factors (Table S1). UBR7 belongs to the N-recognin family of E3 ligases but harbors a PHD domain in lieu of a RING domain (Tasaki et al., 2005). Unlike the other highly enriched proteins, UBR7 did not stably co-purify with H3.1 upon further fractionation (see below), suggesting that the interaction is abundant but unstable. Nonetheless, the observation is interesting given that the related UBR2 protein targets H2A-H2B for ubiquitination (An et al., 2012) and that the UBR7-related Mlo2 protein is involved in mitotic chromosome segregation in S. pombe (Javerzat et al., 1996).
In line with our report on cytosolic histone protein complexes (Campos et al., 2010), sNASP was the most enriched histone chaperone co-purifying with H3.1, an observation that was particularly evident in S-phase (Figure 1B, D). This was closely followed by SUPTH6, CAF-1, FACT, and TONSL, all of which co-purified with over a hundred-fold enrichment over mock IPs. In all, a total of ten known histone chaperone polypeptides were identified with H3.1.
A direct comparison between the H3.1 proteome from asynchronously growing cells and a synchronized population in S-phase yielded surprisingly little differences (Figure S1 B–C). Specifically, MCM subunits of the replicative helicase were enriched with H3.1 in S-phase, along with the NASP and ASF1B chaperones. In comparison, the SUPT6H histone chaperone that is implicated in transcription (Bortvin and Winston, 1996) was inversely enriched in asynchronous cells (Figure S1C). While transient interactions are widely expected to change through the cell cycle and in response to different stimuli, the overall stable interactions formed with histone H3.1 underwent little change. By implication, the observation underscores the close coupling of soluble histone complexes and the replication machinery. The noisy distribution of proteins in and out of S-phase may, however, also have hindered our ability to discern statistical changes over the H3.1 interactions.
Biochemical Characterization of Soluble H3.1 Protein Interactions
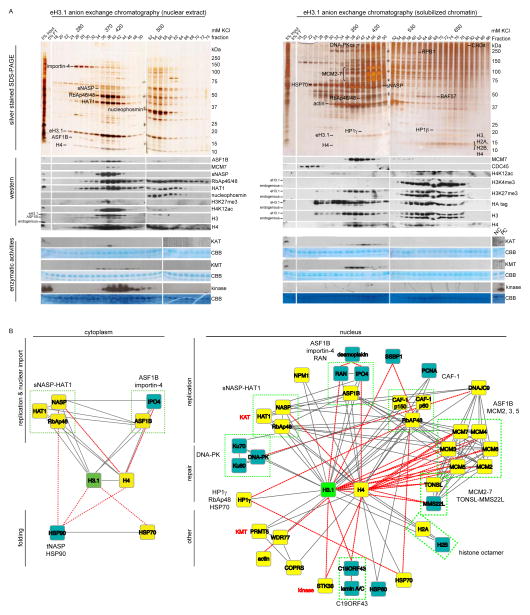
To further substantiate the identified H3.1 interactions and to dissect these into separate complexes, we performed successive biochemical fractionations on the affinity purified eH3.1 extracts. The eH3.1 affinity purified material was first resolved based on charge over an anion exchange chromatography column, and the protein fractions analyzed over silver stained SDS-PAGE and western blots (Figure 2A, top panels). These fractions were also assayed for H3.1-associated enzymatic activities (acetyltransferase, methyltransferase, kinase, ATPase) with free or nucleosomal histones as substrates (Figure 2A, bottom panels). Polypeptides co-eluting with H3.1 peak fractions, or with these enzymatic activities, were subjected to further biochemical fractionations based on the solubility and size of the protein complexes to reveal stable H3.1 interactions and identify the enzymes acting on histones (Figure S2, Figure S3). Proteins were identified by MS after excision from silver stained gels, and confirmed by western blotting when possible. No less than 39 soluble nuclear proteins remained stably associated with eH3.1 following a number of chromatography steps, the majority of which were detected in the quantitative MS analyses above. These polypeptides distributed over 20 putative histone protein complexes (Figure 2B, Table S2).
Figure 2.
Biochemical isolation of soluble eH3.1 protein complexes. (A) Anion exchange chromatography of eH3.1 affinity purified from either nuclear extracts (left panel) or solubilized chromatin (right panel). Silver stained SDS-PAGE and western analyses revealed co-eluting proteins (top panels), whereas the extracts were further essayed for intrinsic enzymatic activities towards histones using radiolabeled acetyl-CoA, SAM and ATP (bottom panels). The elution points above the gels denote fractions where eH3.1 protein levels peaked. These fractions, as well as fractions containing enzymatic activities towards histones were further fractionated (see Figure S2, Figure S3). (B) H3.1 interactome based on the biochemical purification of soluble eH3.1 protein complexes. Proteins validated in the quantitative MS analysis are highlighted in yellow. Solid grey and dashed red lines respectively represent interactions reported in STRING, and interactions found through the biochemical purification of soluble eH3.1. NC: negative control, PC: positive control, KAT: lysine acetyltransferase, KMT: lysine methyltransferase, CBB: Coomassie Brilliant Blue.
Nearly half of these protein complexes harbored proteins implicated in DNA replication despite being processed from asynchronous cells (Figure 2B). Few polypeptides pertained to transcription, suggesting that the most stable interactions formed with histone H3.1 primarily concerned cell cycle regulation. We observed H3.1 in association with an assortment of prominent MCM protein complexes, likely involved at different stages of the cell cycle. However, in spite of being detected in the input extracts, CDC45 and the GINS sub-complex, both of which are required to activate the MCM2-7 helicase (Gambus et al., 2006; Ilves et al., 2010; Kang et al., 2012; Pacek et al., 2006), did not co-purify with histone H3.1 nor were they observed in the quantitative MS analyses of the H3.1 S-phase proteome (Figure S2C–D, Figure S3 B–C). This finding suggests that histone interactions with active CMG helicase complexes, as those reported in yeast (Foltman et al., 2013), are biochemically unstable if occurring in mammalian cells.
Excluding enzymatic activities, eH3.1 co-eluted either with protein complexes that contained a known histone chaperone, with TONSL-MMS22L, or with the uncharacterized C19ORF43 proteins (Table S2, Figure S2, Figure S3). The one fraction with seemingly unchaperoned histones represents minute amounts of nucleosomes (Figure S3G), as histones would not have strongly bound our anion exchange resin in the absence of DNA. Indeed, free histones are seldom left unchaperoned in vivo (Campos et al., 2010) as their charged nature would result in adverse electrostatic interactions. Akin to cytosolic purifications (Alvarez et al., 2011; Campos et al., 2010), nuclear eH3.1 co-eluted with a broad range of interacting proteins with no free histones left to passively diffuse inside of cells (Figure 2, Figure S2, Figure S3).
Methyltransferase, kinase, and acetyltransferase activities towards core nucleosomal histones also co-fractionated with the soluble, nuclear, eH3.1 (Figure 2A). Despite the statistical enrichment of NuRD and BAF components in the quantitative MS analyses (Table S1), ATPase activity towards nucleosomal substrates was not detected (Figure S3H) nor did the complexes co-purify with soluble eH3.1 (Figure S2, Figure S3). The methyltransferase activity directed towards histone H4 (Figure 2A) originated from PRMT5 (Figure S3B–C), a contaminant common to FLAG affinity purifications (Mellacheruvu et al., 2013). In contrast, the acetyltransferase activity (Figure 2A) originated from nuclear HAT1 and associated with the sNASP histone chaperone (discussed below). Kinase activity towards histones (Figure 2A) in turn emanated from the STK38 (NDR1) protein kinase (Figure S2G). Although STK38 is a contaminant in FLAG purifications (Mellacheruvu et al., 2013), the recombinant protein does phosphorylate soluble but not nucleosomal histones in vitro (Figure S2H). The kinase however preferentially targeted H2A-H2B dimers for phosphorylation (Figure S2I).
Fractions Containing ASF1B, TONSL-MMS22L, or Nucleophosmin Promote Histone Eviction
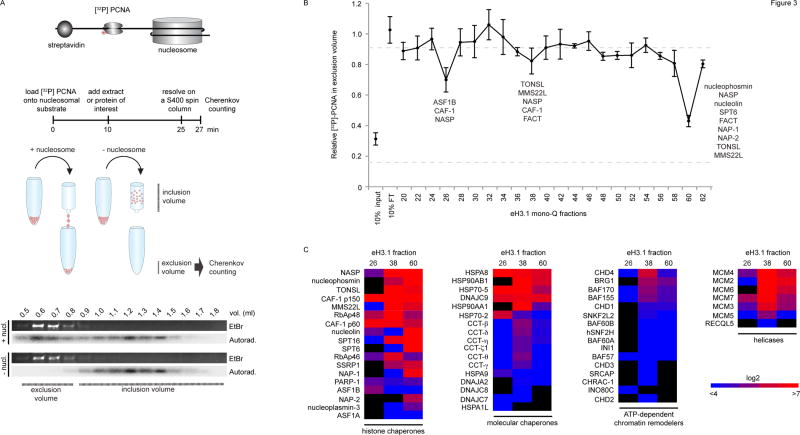
To further understand the role of the eH3.1 protein-protein interactions, a quantitative nucleosome disassembly test was designed to screen for fractions capable of histone eviction in vitro, in the absence of ATP (Figure 3A, Figure S4). In this experiment, a [32P]-labeled PCNA ring was loaded onto a linear DNA fragment flanked by a single nucleosome at one extremity and a streptavidin-biotin block on the opposite end. The radiolabeled ring was free to slide on DNA but was locked onto the template by the nucleosome and the streptavidin (Figure 3A, Figure S4F). The PCNA probe co-eluted from a size exclusion spin column with the bulky DNA if the nucleosome was intact. The small radioactive ring however remained in the column when histones were removed. Thus, histone retention correlated with Cherenkov counts from the exclusion volume.
Figure 3.
Nucleosome disassembly. (A) A radiolabeled PCNA probe is loaded onto a linear DNA template, flanked by a single nucleosome at one extremity and a biotin-streptavidin block at the other. DNA flows into the exclusion volume when applied onto a gel filtration spin column, along with the radiolabeled PCNA trapped by the nucleosome. The radioactive probe is free to slide on DNA but falls off and remains in the inclusion volume if histones are removed. The assay quantifies the amount of radioactive PCNA in the exclusion volume by Cherenkov counting. (B) Testing nucleosome disassembly activity within affinity-purified eH3.1 from nuclear fractions (Figure 2). Data represents mean [32P]-PCNA retention +/− SD. (C) Mass spectrometry analysis (peptide counts) of the three fractions exhibiting histone eviction.
Surprisingly, a titration of eH3.1-purified proteins revealed that the nuclear extracts evicted nucleosomal histones more aptly than chromatin-associated proteins (Figure S4H). The activity reflected an active loss of nucleosomes as no nuclease or phosphatase activity was detected in these extracts, under the same conditions (Figure S4I–J). Three of the eH3.1 mono Q fractions shown in Figure 2, consistently yielded a reproducible nucleosome loss (Figure 3B). These fractions were re-analysed by MS (Figure 3C) and corresponded to eH3.1 bound by either: ASF1B, RAN, importin-4 (Figure S2A); MCM2-7, TONSL-MMS22L (Figure S2C); or nucleophosmin (Figure S2F). Since the assay was performed in the absence of ATP, the resultant disassembly likely arises from the action of histone chaperones rather than molecular chaperones present in the samples. Amongst the former, FACT and NASP were also detected within the MCM2-7-TONSL-MMS22L-containing fraction.
Focus on Histone Chaperone Complexes
Nuclear sNASP
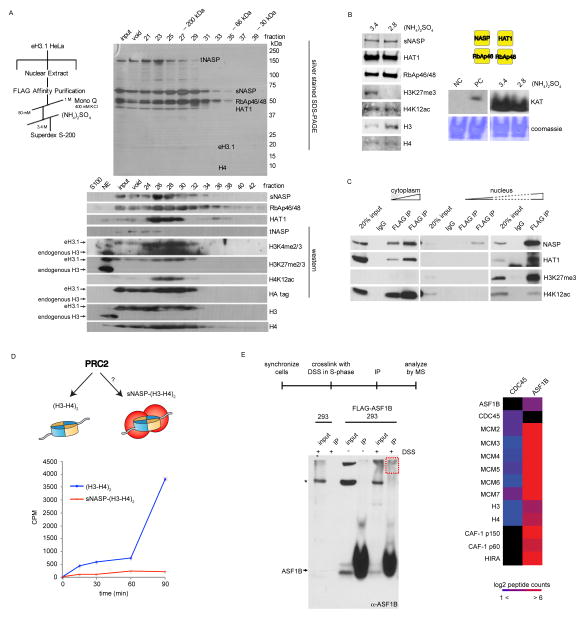
We previously found sNASP to be a major H3-H4 histone chaperone associating with the histone acetyltransferase (HAT) holoenzyme (HAT1 and RbAp46) in the cytosol (Campos et al., 2010). The complex was again isolated from the nuclear compartment (Figure 4A). Two distinct forms of the sNASP-HAT1 complex co-eluted by ion exchange chromatography but were resolved based on their solubility in the presence of ammonium sulfate (Figure S2E, Figure 4A). The first was identical to the abundant cytoplasmic complex in that it comprised di-acetylated newly-synthesized histones that were otherwise largely unmodified, and likely leached from the cytosol (Figure S2E). The second, however, contained histones heavily decorated with PTMs typical of both eu- and hetero-chromatin (Figure 4A). Both protein complexes harbored catalytically active HAT1 and were the source of the nuclear acetyltransferase activity (Figure 4B), demonstrating that H4 di-acetylation is not restricted to newly synthesized histones. Remarkably, nearly all of the soluble eH3.1 complexes isolated from the nucleus harbored H4K12ac regardless of whether they physically co-purified with HAT1 or whether they encompassed other histone PTMs (Table S2, Figure S2). The ubiquitous HAT1 activity from soluble nuclear extracts suggests that evicted histones are re-acetylated when bound and recycled by sNASP.
Figure 4.
Molecular functions of nuclear sNASP and ASF1B. (A) Nuclear sNASP co-elutes with HAT1 and evicted histones. (B) Both sNASP-HAT1 complexes (with or without evicted histones) are enzymatically active. Left panel: Western analysis of peak eH3.1 fractions containing sNASP with evicted or new histones (precipitated at 3.4 and 2.8 M ammonium sulfate, respectively). Right panel: Acetyltransferase activity towards histones demonstrated by autoradiography of acetyl-[3H] incorporation. (C) Nuclear, but not cytosolic sNASP co-precipitates histones with marks characteristic of eu- and hetero-chromatin. (D) In vitro PRC2 methyltransferase assay. sNASP-bound histones are poor substrates for the PRC2 complex compared to free soluble (H3-H4)2 tetramers. (E) In vivo crosslinking of replicating 293 cells (left panel), and mass spectrometry analysis of crosslinked, immunoprecipitated, ASF1B.
Association of sNASP with evicted histones was confirmed by immunoprecipitating epitope tagged sNASP from 293 cell extracts and immunoblotting against tri-methyl marks on histone H3 (Figure 4C). sNASP protects soluble histones from degradation in vivo (Cook et al., 2011) and likely stores a soluble pool of histones for deposition. Inhibition of DNA replication or transcription by targeting the appropriate polymerase with aphidicolin or α–amanitin, respectively, reduced H3 trimethyl marks but not acetylation of histone H4 on sNASP-bound histones (Figure S5), suggesting that histones can be captured by sNASP during both transcription and DNA replication. To further ensure that the di-/tri-methylation on sNASP-bound histones were acquired in the context of chromatin and not modified on the chaperone proper, we tested for the ability of the PRC2 methyltransferase to methylate sNASP-bound histone H3, given that its preferred substrates are nucleosomal histones (Cao et al., 2002). sNASP-bound histones were poor substrates for PRC2, even when compared to unbound counterparts, further confirming histone eviction as the source of sNASP-bound histone PTMs (Figure 4D). Altogether these findings suggest a model in which evicted histones are captured and protected from proteolytic degradation by sNASP (Cook et al., 2011) and re-acetylated by HAT1 prior to re-incorporation onto chromatin. We cannot however rule out the possibility that sNASP-HAT also directly acts on histones during deposition and eviction events as they occur on chromatin.
ASF1 and the replicative helicase
Two ASF1B-containing H3.1 complexes were purified in this study. We and others have reported that cytosolic histones associate with importin-4 karyopherin along with ASF1 (Alvarez et al., 2011; Campos et al., 2010) and now observe the ASF1-importin-4 complex in nuclear extracts with the addition of sub-stoichiometric levels of the RAN GTPase (Figure S2A). Furthermore, we isolated the ASF1B isoform along with the MCM2, -3, -5 proteins in association with eH3.1 (Figure S2D) but not with the CDC45 and GINS subunits of the CMG helicase. It is known that ASF1 co-localizes and co-purifies with MCM proteins (Groth et al., 2007; Schulz and Tyler, 2006) and this interaction increases with replicative stress (Jasencakova et al., 2010). Therefore, the detection of ASF1 with the eH3.1 fractions containing MCM proteins but not CDC45/GINS, suggests that the H3.1-associated pool of ASF1B does not associate with the active CMG complex. To ensure that this was not due to the experimental processing of the samples, 293 cells expressing epitope tagged ASF1B were synchronized, released in S-phase, and treated with the cell-permeable amine-reactive crosslinking agent disuccinimidyl suberate (DSS). The DNA was then digested with benzonase and ASF1B was immunoprecipitated. Slow-migrating (crosslinked) species were excised from SDS-PAGE gels and the interacting proteins identified by MS (Figure 4E). Although MCM proteins were well represented in ASF1B immunoprecipitates, ASF1B did not interact with CDC45 and vice-versa, indicating that it does not interact with the replicative helicase. ASF1B did however interact with the HIRA and CAF-1 chaperones (Figure 4E), as expected through its role in histone deposition (Tagami et al., 2004). Thus, while the ASF1B chaperone would be well positioned to recycle histones through CAF-1 (Mello et al., 2002; Tyler et al., 2001) and the complex does form in S-phase (Groth et al., 2007), our data implicates ASF1B with an inactive phase of the replicative helicase.
TONSL-MMS22L: A Histone Chaperone at Stalled Replication Forks
In addition to purifying soluble eH3.1 with known histone chaperones, we were quite intrigued in the isolation of histones with MCM2-7 and TONSL-MMS22L (Figure S2C). The TONSL-MMS22L complex promotes homologous recombination at stalled or stressed replication forks (Duro et al., 2010; O’Connell et al., 2010; O’Donnell et al., 2010; Piwko et al., 2010). Knockdown of either TONSL or MMS22L results in spontaneous DNA double-strand breaks in S-phase, checkpoint activation and accumulation of cells in G2 (Duro et al., 2010; O’Connell et al., 2010; O’Donnell et al., 2010; Piwko et al., 2010). Our TONSL-MMS22L-MCM2-7 complex harbored both endogenous and exogenous H3.1 as well as PTMs normally found on chromatin, suggesting that the proteins bind evicted (H3-H4)2 tetramers (Table S2). In comparison to the replication-dependent CAF-1 histone deposition machinery, which was enriched in MS but barely visible by silver staining (Figure S2F), TONSL-MMS22L was relatively abundant and stable. Despite its association with MCM2-7, the H3.1-associated complex was readily detected in soluble nuclear extracts but was not found in chromatin extracts nor with GINS/CDC45 (Figure S2C–D). To rigorously test whether MMS22L-TONSL interacts with CMG, a crosslinking experiment was again performed: replicating cells were exposed to the DSS cell permeable crosslinker, the DNA digested, and TONSL immunoprecipitated for MS analysis. Again, we observed a TONSL-MMS22L interaction with MCM2-7 but failed to detect peptides derived from CDC45/GINS (Table S3), indicating that like ASF1, TONSL-MMS22L does not associate with an active replicative helicase or the association is of very low abundance.
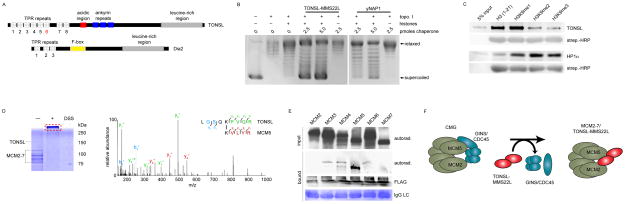
Since the soluble eH3.1 fractions containing TONSL-MMS22L poorly co-eluted with known histone chaperones, we sought to determine whether TONSL-MMS22L has histone chaperone activity. While MMS22L lacks recognizable histone binding domains, TONSL contains many protein modules including an acidic region—a feature that is common to histone chaperones (Figure 5A). The presence of an acidic region, the co-elution of TONSL with histones, and its enrichment in fractions that disassemble nucleosomes, led us to explore the possibility that TONSL-MMS22L functions as a histone chaperone. In order to be considered a histone chaperone, the proteins must be capable of binding histones and promote nucleosomal assembly in vitro in the absence of ATPase activity. Isopycnic centrifugation over a sucrose gradient revealed that recombinant TONSL-MMS22L was capable of binding all four core nucleosomal histones (Figure S6A). Histone-binding was mediated through two domains: the short acidic patch and the neighboring ankyrin repeats (Figure S6B). Furthermore, TONSL-MMS22L was able to assemble chromatin in a supercoiling assay used to gauge histone deposition in vitro (Figure 5B). The full-length TONSL protein and a protein fragment encompassing the acidic region and ankyrin repeats of TONSL bound histones equally well (Figure S6C, left panel). However, only the full-length TONSL protein exhibited histone chaperone activity in vitro (Figure S6C, right panel). Since neither TONSL nor MMS22L possess a recognizable ATPase domain, our results indicate that TONSL-MMS22L is a bona fide histone chaperone.
Figure 5.
TONSL is a histone chaperone that binds H3K9me1. (A) Primary structures of human TONSL and the similar yeast protein, Dia2. (B) Supercoiling assay demonstrating the histone chaperone ability of TONSL. (C) Pull-down assay utilizing immobilized histone peptides testing binding preference by recombinant TONSL and HP1. (D) MS/MS HCD spectrum of the (M + H)+4 ion of cross-linked peptides between TONSL (TRP repeat) and the C-terminus of MCM5. N-terminal fragment ions (b) are indicated in blue and C-terminal fragment ions (y) are indicated in green and red. The mass accuracy for precursor ion is better than 1 ppm and mass accuracy of all the fragment ions is better than 10 ppm. (E) Immobilized TONSL binds to in vitro translated MCM5. (F) Model for TONSL-MMS22L at the replication fork: Upon recruitment to stalled replication forks TONSL-MMS22L may maintain the CMG helicase inactive by binding to MCM5.
The ankyrin repeats of the GLP and G9a methyltransferase proteins were recently found to form an aromatic cage that binds mono and di-methylated H3K9 (Collins et al., 2008). We therefore assessed whether TONSL displayed any binding preference towards methylated histones. We found that recombinant TONSL-MMS22L binds to unmodified recombinant histones but preferentially bound to hyper-methylated H3 when incubated with histones purified from HeLa cells (Figure S6D). A similar result was obtained with recombinant TONSL in the absence of MMS22L (Figure S6E). Furthermore, we found that TONSL immunoprecipitates both methyl-H3 and -H4 in vivo and binding of hyper-methylated H3 was augmented upon treating U2OS cells with DNA-damaging doses of camptothecin (Figure S6F), a drug previously shown to mobilize the TONSL-MMS22L proteins onto chromatin (Duro et al., 2010; O’Connell et al., 2010; O’Donnell et al., 2010; Piwko et al., 2010). Recombinant TONSL was therefore incubated with an array of H3 histone peptides, spanning all known methyl-lysine modifications. TONSL exhibited preferential binding for H3K9me1 (Figure 5C, Figure S6G), an observation further confirmed by immunoprecipitation of TONSL from U2OS cells (Figure S6H).
While monomethylation of histone H3 on lysine 9 has not been implicated in the DNA damage response, recent studies suggest that the H3K9me2/3 demethyltransferase KMD4B is mobilized within seconds to sustain DNA repair, whereas the SUV39H1 methyltransferase that is responsible for H3K9me1 catalysis is downregulated at the transcriptional level (Young et al., 2013; Zheng et al., 2014). Laser microirradiation of cells confirmed a rapid recruitment of TONSL on damaged chromatin with peak accumulation occurring 10–20 minutes after irradiation, followed by a constant decline in chromatin-bound TONSL levels thereafter (Figure S6I). This suggests a dynamic mode of action, as TONSL promptly and transiently acts on chromatin to maintain the integrity of replicating DNA. However, we did not observe H3K9me1 accumulating on irradiated chromatin (Figure S6J). Instead, H3K9me1 levels markedly increased on the soluble pool of TONSL as evidenced by TONSL immunoprecipitation after induction of DNA damage (Figure S7A). This indicates that methyl H3 alone is not a critical component of TONSL recruitment to DNA damage.
TONSL was highly enriched in two of the three eH3.1-purified nuclear extracts exhibiting nucleosome disassembly (Figure 3B–C). We therefore repeated the assay shown in Figure 3, but directly titrating recombinant TONSL over the nucleosomal substrate. The addition of TONSL in itself had little effect on the stability of nucleosomes (Figure S7B, left panel). Recombinant FACT also lacked any effect by itself. A similar titration of FACT over the nucleosome substrate resulted in negligible disassembly (Figure S7B, right panel), meaning that any electrostatic interactions between nucleosomal histones and these histone chaperones is not sufficient to dissolve nucleosomes. Since H3.1-purified nuclear extracts contained nucleosome disassembly activities (Figure S4H), we re-titrated TONSL over 1.5 μg of the H3.1-purified nuclear extracts—the amount needed to dissociate roughly half of the nucleosomes (Figure S4H). The effect was modest, but TONSL enhanced disassembly until a 1:1 stoichiometric point was reached between TONSL and the nucleosomal octamers (100 fmoles), after which a plateau was reached (Figure S7B, left panel). This suggests that TONSL in itself lacks chromatin disassembly activities but may indirectly support the process.
To explore the role of TONSL at stalled forks, we further sought to identify the TONSL-MCM protein-protein interactions. The recombinant TONSL-MCM2-7 complex was pre-incubated, crosslinked in vitro using an amine-reactive crosslinker (Figure 5D), digested with trypsin, and the peptides analyzed by MS to map interacting surfaces (Yang et al., 2012). Numerous intra-protein crosslinked peptides were observed, along with crosslinks between MCM5-MCM3, MCM7-MCM4, MCM4-MCM6 and MCM6-MCM2 (Figure S7C), which is consistent with electron microscopy tomography analyses of the MCM2-7 complex (Costa et al., 2011). Of note, the sixth TPR repeat of TONSL crosslinked to the extreme carboxyl terminus of MCM5 near its AAA+ ATPase domain (Figure 5D). The finding is important since the same surface is utilized by CDC45/GINS (Costa et al., 2011) and suggests that TONSL can potentially maintain MCM2-7 in an inactive state. To confirm the TONSL-MCM5 interaction, immobilized TONSL was incubated with in vitro translated MCM2-7 subunits. The MCM5 subunit clearly remained bound to TONSL (Figure 5E). In a similar experiment, we again reproduced the TONSL-MCM5 interaction using bacteria-expressed MCM subunits (Figure S7D). Weak binding to MCM3 was also observed (Figures 5E, Figure S7D). Altogether these results illuminate the need for specialized histone chaperones taking on distinct functions through the course of DNA replication for histone processing, deposition, eviction and repair.
DISCUSSION
The pathway leading to the deposition of newly-synthesized histones on replicated DNA intimately involves histone chaperones (Annunziato, 2012; Campos et al., 2014). Thus, the hypothesis emerged that histones confronting a replication fork are similarly chaperoned, as nucleosomes are being dismantled and histones are being recycled within replication bubbles. Indeed, the cooperative histone binding by FACT and MCM2-7 (Foltman et al., 2013; Tan et al., 2006) as well the interaction between ASF1 and MCM proteins (Groth et al., 2007) substantiate this hypothesis. However, the question remains as to how histones are chaperoned at the forefront of the replication fork and throughout the remainder of the cell cycle. In this study we surveyed stable interactions between the replication-coupled histone H3.1 and histone chaperone protein complexes. We identified 11 histone chaperones that interact with histone H3.1, including the histone chaperone TONSL (Figure 1D). The results showcase the diverse roles of chaperones at the forefront of replication.
We previously identified sNASP as a key H3–H4 histone chaperone in the processing of cytoplasmic histones (Campos et al., 2010). Here, we demonstrated that regardless of its subcellular localization, sNASP stably associates with the catalytically-active HAT1 holoenzyme (Campos et al., 2010) (Figure 4A). Albeit ubiquitous in the cytosol, a pool of the type B acetyltransferase is also found in the nucleus in S-phase (Parthun et al., 1996; Verreault et al., 1998). Nuclear localization of sNASP has also been reported (Richardson et al., 2000), but we now observed a pool of nuclear sNASP in association with HAT1 and evicted histones (Figure 4A–C). Our quantitative MS analyses confirmed the ubiquitous and abundant nature of this histone chaperone (Figure 1B, D). The catalytic activity of the purified sNASP-histone complex (Figure 4B), along with the acetylation of bound histone H4 (Figure 4A–C), argues for a re-acetylation of evicted histone H4 prior to re-deposition. Like sNASP (Cook et al., 2011), loss of HAT1 has a limited impact on cell cycle dynamics (Nagarajan et al., 2013). However, absence of HAT1 results in hypersensitivity to DNA damage and increased global genomic instability (Nagarajan et al., 2013), stressing the significance of its histone acetylation activity. Thus, the pool of evicted sNASP-bound acetylated histones may help sustain genomic stability.
Evidence strongly suggests that H2A-H2B histone chaperone FACT initiates nucleosome disassembly at replication forks. FACT was identified as a factor that facilitates transcription through chromatin in vitro (LeRoy et al., 1998; Orphanides et al., 1998). It does so by interacting with the DNA-binding surfaces of H2A-H2B to displace histone dimers (Hsieh et al., 2013). Mammalian FACT directly interacts with MCM4 (Tan et al., 2006), and the yeast CMG cooperatively binds histones via Mcm2, in the presence of FACT (Foltman et al., 2013). FACT is also enriched in mammalian replication foci (Hertel et al., 1999), and loss of FACT results in S-phase delays across species (Abe et al., 2011; Okuhara et al., 1999; Schlesinger and Formosa, 2000), likely due to impaired replication fork progression (Abe et al., 2011). While mechanistic evidence of H2A-H2B segregation accumulates, the mechanistic segregation of nucleosomal histones (H3-H4)2 remains elusive.
We purified two H3-H4 histone chaperones in stable association with MCM proteins (Figure 2B). However, H3.1–CMG interactions were biochemically unstable in our conditions. Only upon chemical crosslinking were histones detected by MS in association with CDC45 (Figure 4E). Eukaryotic MCM proteins are abundant in cycling cells, and only a minute population is incorporated into the CMG complex (Moyer et al., 2006)—a severe limitation to our biochemical approach. Indeed, the lack of histone chaperone–CMG interaction could suggests that such transaction is transient and scant. Other factors may alternatively contribute to the recycling of nucleosomal H3.1-H4 histones within replication bubbles averting detectable histone-CMG interactions. These could be mediated by other components of the replication machinery, perhaps aided by the force of the helicase, ATP-dependent chromatin remodelers, or other transiently binding proteins. Therefore histone chaperone–MCM2-7 interactions hold specialized functions that need not be connected to the normal elongation phase of the CMG helicase, a notion well exemplified by ASF1 and TONSL.
Histone H3.1 purified with two ASF1B protein complexes, the first co-eluted with a karyopherin, importin-4, and RAN GTPase (Figure S2A). RAN modulates cargo binding in the nucleus (Pemberton and Paschal, 2005) and hence, presumably releases the ASF1B-H3-H4 complex coming from the cytoplasm. The latter protein complex, in association with MCM proteins (Figure S2D), is intriguing. ASF1 has been singled as a candidate to recycle histones encountering replication forks (Groth et al., 2007). Like FACT, evidence does intimately associate ASF1 with the replication process. ASF1 is detected over DNA replication foci in both mammals and fruit flies, and a loss of ASF1 results in delayed S-phase progression (Groth et al., 2007; Schulz and Tyler, 2006). The role of ASF1 in the transport (Alvarez et al., 2011; Campos et al., 2010) and handover of new histones to CAF-1 (Mello et al., 2002; Tyler et al., 2001) is well described and required for normal fork progression (Mejlvang et al., 2014). However, ASF1B interactions ahead of the replication fork are less clear, and whether ASF1 travels with active CMG helicases has remained controversial.
H3.1-associated ASF1B lacked components of the CMG other than MCM proteins (Figure S2D). We further used cell permeable DSS crosslinker and MS to univocally answer the question of whether ASF1 binds to an active CMG or inactive MCM2-7 helicase in the absence of stress. As such, MCM proteins crosslinked and were detected by MS from both immunoprecipitated ASF1B and CDC45 (Figure 4E). However, ASF1 and CDC45 did not crosslink to the same protein complexes regardless of whether ASF1B or CDC45 was immunoprecipitated. The results demonstrate that under normal growth conditions, ASF1–MCM interactions are limited to inactive helicase complexes. Since MCM proteins levels greatly exceed the number of replication origins, a biological function for this interaction remains to be fully determined.
Unlike ASF1, TONSL-MMS22L is not required for normal replication fork progression (O’Donnell et al., 2010). It instead promotes homologous recombination at stressed replication forks (Duro et al., 2010; O’Donnell et al., 2010). We found TONSL to have histone chaperone activity (Figure 5B, Figure S6A–C), and speculate a further supportive role in nucleosome disassembly (Figure 3B–C, Figure S7B, left panel). An attractive model entails TONSL recruitment to stalled or colliding replication bubbles, perhaps aided by transient accumulation of H3K9me1 alongside other factors associating with ssDNA. Once recruited, TONSL would subsequently contribute to the resolution of the stressed forks by binding to surfaces on MCM5/3, normally used by GINS/CDC45, resulting in the inactivation of CMG helicase activity (Figure 5F). A related role has been demonstrated for the yeast Dia2 protein, which shares a similar structure with TONSL but harbors an F-box in lieu of the ankyrin repeats (Figure 5A). The yeast protein interacts with MCM proteins via its TPR repeats (Mimura et al., 2009; Morohashi et al., 2009), and acts as a substrate-targeting subunit of the Skp1-Cdc53-F-box SCFDia2 E3 ligase complex (Koepp et al., 2006). The complex targets MCM7, which in turn causes the Cdc48/p97 segregase protein to dismantle the CMG complex at the end of replication (Maric et al., 2014). While TONSL-MMS22L was not part of an E3 ubiquitin-conjugating complex, we did detect a co-elution of TONSL with the CDC48/VCP ATPase (Figure S2F), which may help dismantle stalled replication forks as well. Furthermore, like TONSL, Dia2-defective cells are susceptible to DNA damage in S-phase (Blake et al., 2006; Koepp et al., 2006). Regardless of whether TONSL is involved in the inactivation of the CMG complex, its interaction with the MCM5 subunit of MCM2-7 is highly intriguing. Further experiments will now be required to determine whether there is competition between TONSL and CDC45/GINS over the same binding surfaces of the MCM hexameric ring (Figure 5F), and what it entails during homologous recombination.
Biochemical purification schemes are limited by the abundance of the material. While nucleosomal histones play profuse structural and regulatory epigenetic roles, the amount of soluble proteins complexes containing non-nucleosomal histones is highly limiting. The technicality severely impedes our ability to follow chromatin transactions in an unbiased manner. The large-scale analysis of the replication-coupled H3.1 protein revealed a number of unexpected roles for new and old histone chaperones. The purifications and experiments described herein not only provide a reference to the large number of soluble H3.1 protein complexes that exist in vivo, but also insights into the diverse functions undertaken by histone chaperones.
EXPERIMENTAL PROCEDURES
Biochemical Fractionation
The fractionation of HeLa S3 cells proceeded at 4°C, as described (Campos et al., 2010). Chromatin pellets were solubilized as described in the Extended Experimental Procedures.
Label-free Quantitative Mass Spectrometry
Briefly, raw MS data was analyzed with MaxQuant 1.3.0.5, with the options ‘label-free quantification’ (LFQ), ‘match-between-runs’ and ‘iBAQ’ enabled. The identified proteins were filtered for contaminants and reverse hits. Additionally, proteins were filtered to be at least detected in all replicates of the Flag IP or control. Significant interactors were identified based on the difference of LFQ intensity between Flag and control IPs using a permutation-based false discovery rate (FDR) adapted t-test (Perseus). The resulting FDR (-log10) was plotted against the Flag/control ratio (log2) in volcano plots and significance thresholds (minimal FDR and fold change (FC)) were empirically set for every experiment. Full experimental details are found in the Extended Experimental Procedures.
Nucleosome Disassembly
BsmBI cut pS601x1 plasmid was filled-in with biotinylated dNTPs (Thermo Scientific), and the gel purified ScaI-BsmBI fragment nicked with Nb.BbvCI. Nucleosomes were assembled by salt dialysis (Extended Experimental Procedures) after which 4 molar equivalents of streptavidin (NEB) was added. Three molar ratios of radiolabeled PCNA were loaded per mole DNA as previously described (Zhang et al., 1999), after which ATP was quenched with glucose and hexokinase. The radiolabeled substrate was then incubated at 37°C for 15 min with 1 μl of the eH3.1 fractions after which it was applied on top of a 2 ml sephacryl S-400 (GE Life Sciences) gel filtration column (packed by gravity flow on disposable 2 ml columns (Pierce) pre-equilibrated with Gel Filtration buffer (50 mM tris, pH 7.6, 150 mM NaCl, 50 mM EDTA, 0.1% NP-40 (v/v), 0.5 mM DTT, 1 mM NaF, 0.5 mM Na3VO4)). Samples were immediately centrifuged for 30 sec at 800 × g. 800 μl of Gel Filtration buffer was then added and samples re-centrifuged to collect the exclusion volume. An additional 1.2 ml of buffer was added to collect the inclusion volume. Radioactive signals were measured by Cherenkov counting.
Supplementary Material
Acknowledgments
We are thankful to Drs. Karim-Jean Armache, Vladimir Bermudez, Rachel Szilard, Wiebke Galal and Lynne Vales for insightful discussions and help with the research and manuscript. We also thank Dr. Jim Kadonaga for generously providing us with the ND423 construct. Additional support was provided to D.R. by the National Institute of Health (NIH) (GM-64844 and R37-37120) and the Howard Hughes Medical Institute. Work in the laboratory of M.V. is supported by the Netherlands Organization for Scientific Research (NWO-VIDI, 864.09.003 and Cancer Genomics Netherlands).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe T, Sugimura K, Hosono Y, Takami Y, Akita M, Yoshimura A, Tada S, Nakayama T, Murofushi H, Okumura K, et al. The histone chaperone facilitates chromatin transcription (FACT) protein maintains normal replication fork rates. J Biol Chem. 2011;286:30504–30512. doi: 10.1074/jbc.M111.264721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez F, Munoz F, Schilcher P, Imhof A, Almouzni G, Loyola A. Sequential establishment of marks on soluble histones H3 and H4. J Biol Chem. 2011;286:17714–17721. doi: 10.1074/jbc.M111.223453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JY, Kim E, Zakrzewska A, Yoo YD, Jang JM, Han DH, Lee MJ, Seo JW, Lee YJ, Kim TY, et al. UBR2 of the N-end rule pathway is required for chromosome stability via histone ubiquitylation in spermatocytes and somatic cells. PLoS One. 2012;7:e37414. doi: 10.1371/journal.pone.0037414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato AT. Assembling chromatin: The long and winding road. Biochim Biophys Acta. 2012;1819:196–210. doi: 10.1016/j.bbagrm.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Blake D, Luke B, Kanellis P, Jorgensen P, Goh T, Penfold S, Breitkreutz BJ, Durocher D, Peter M, Tyers M. The F-box protein Dia2 overcomes replication impedance to promote genome stability in Saccharomyces cerevisiae. Genetics. 2006;174:1709–1727. doi: 10.1534/genetics.106.057836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- Campos EI, Fillingham J, Li G, Zheng H, Voigt P, Kuo WH, Seepany H, Gao Z, Day LA, Greenblatt JF, et al. The program for processing newly synthesized histones H3.1 and H4. Nat Struct Mol Biol. 2010;17:1343–1351. doi: 10.1038/nsmb.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Campos EI, Stafford JM, Reinberg D. Epigenetic inheritance: histone bookmarks across generations. Trends in cell biology. 2014 doi: 10.1016/j.tcb.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Collins RE, Northrop JP, Horton JR, Lee DY, Zhang X, Stallcup MR, Cheng X. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat Struct Mol Biol. 2008;15:245–250. doi: 10.1038/nsmb.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook AJ, Gurard-Levin ZA, Vassias I, Almouzni G. A specific function for the histone chaperone NASP to fine-tune a reservoir of soluble H3-H4 in the histone supply chain. Mol Cell. 2011;44:918–927. doi: 10.1016/j.molcel.2011.11.021. [DOI] [PubMed] [Google Scholar]
- Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18:471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duro E, Lundin C, Ask K, Sanchez-Pulido L, MacArtney TJ, Toth R, Ponting CP, Groth A, Helleday T, Rouse J. Identification of the MMS22L-TONSL complex that promotes homologous recombination. Mol Cell. 2010;40:632–644. doi: 10.1016/j.molcel.2010.10.023. [DOI] [PubMed] [Google Scholar]
- English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltman M, Evrin C, De Piccoli G, Jones RC, Edmondson RD, Katou Y, Nakato R, Shirahige K, Labib K. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell reports. 2013;3:892–904. doi: 10.1016/j.celrep.2013.02.028. [DOI] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, Corpet A, Cook AJ, Roche D, Bartek J, Lukas J, Almouzni G. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- Hertel L, De Andrea M, Bellomo G, Santoro P, Landolfo S, Gariglio M. The HMG protein T160 colocalizes with DNA replication foci and is down-regulated during cell differentiation. Exp Cell Res. 1999;250:313–328. doi: 10.1006/excr.1999.4495. [DOI] [PubMed] [Google Scholar]
- Hsieh FK, Kulaeva OI, Patel SS, Dyer PN, Luger K, Reinberg D, Studitsky VM. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc Natl Acad Sci U S A. 2013;110:7654–7659. doi: 10.1073/pnas.1222198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasencakova Z, Scharf AN, Ask K, Corpet A, Imhof A, Almouzni G, Groth A. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol Cell. 2010;37:736–743. doi: 10.1016/j.molcel.2010.01.033. [DOI] [PubMed] [Google Scholar]
- Javerzat JP, Cranston G, Allshire RC. Fission yeast genes which disrupt mitotic chromosome segregation when overexpressed. Nucleic Acids Res. 1996;24:4676–4683. doi: 10.1093/nar/24.23.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YH, Galal WC, Farina A, Tappin I, Hurwitz J. Properties of the human Cdc45/Mcm2-7/GINS helicase complex and its action with DNA polymerase epsilon in rolling circle DNA synthesis. Proc Natl Acad Sci U S A. 2012;109:6042–6047. doi: 10.1073/pnas.1203734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleff S, Andrulis ED, Anderson CW, Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem. 1995;270:24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- Koepp DM, Kile AC, Swaminathan S, Rodriguez-Rivera V. The F-box protein Dia2 regulates DNA replication. Mol Biol Cell. 2006;17:1540–1548. doi: 10.1091/mbc.E05-09-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoy G, Orphanides G, Lane WS, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- Liu WH, Roemer SC, Port AM, Churchill ME. CAF-1-induced oligomerization of histones H3/H4 and mutually exclusive interactions with Asf1 guide H3/H4 transitions among histone chaperones and DNA. Nucleic Acids Res. 2012;40:11229–11239. doi: 10.1093/nar/gks906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Maric M, Maculins T, De Piccoli G, Labib K. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science. 2014;346:1253596. doi: 10.1126/science.1253596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejlvang J, Feng Y, Alabert C, Neelsen KJ, Jasencakova Z, Zhao X, Lees M, Sandelin A, Pasero P, Lopes M, et al. New histone supply regulates replication fork speed and PCNA unloading. J Cell Biol. 2014;204:29–43. doi: 10.1083/jcb.201305017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello JA, Sillje HH, Roche DM, Kirschner DB, Nigg EA, Almouzni G. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 2002;3:329–334. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura S, Komata M, Kishi T, Shirahige K, Kamura T. SCF(Dia2) regulates DNA replication forks during S-phase in budding yeast. EMBO J. 2009;28:3693–3705. doi: 10.1038/emboj.2009.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi H, Maculins T, Labib K. The amino-terminal TPR domain of Dia2 tethers SCF(Dia2) to the replisome progression complex. Curr Biol. 2009;19:1943–1949. doi: 10.1016/j.cub.2009.09.062. [DOI] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan P, Ge Z, Sirbu B, Doughty C, Agudelo Garcia PA, Schlederer M, Annunziato AT, Cortez D, Kenner L, Parthun MR. Histone acetyl transferase 1 is essential for mammalian development, genome stability, and the processing of newly synthesized histones H3 and H4. PLoS Genet. 2013;9:e1003518. doi: 10.1371/journal.pgen.1003518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- O’Connell BC, Adamson B, Lydeard JR, Sowa ME, Ciccia A, Bredemeyer AL, Schlabach M, Gygi SP, Elledge SJ, Harper JW. A genome-wide camptothecin sensitivity screen identifies a mammalian MMS22L-NFKBIL2 complex required for genomic stability. Mol Cell. 2010;40:645–657. doi: 10.1016/j.molcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell L, Panier S, Wildenhain J, Tkach JM, Al-Hakim A, Landry MC, Escribano-Diaz C, Szilard RK, Young JT, Munro M, et al. The MMS22L-TONSL complex mediates recovery from replication stress and homologous recombination. Mol Cell. 2010;40:619–631. doi: 10.1016/j.molcel.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuhara K, Ohta K, Seo H, Shioda M, Yamada T, Tanaka Y, Dohmae N, Seyama Y, Shibata T, Murofushi H. A DNA unwinding factor involved in DNA replication in cell-free extracts of Xenopus eggs. Curr Biol. 1999;9:341–350. doi: 10.1016/s0960-9822(99)80160-2. [DOI] [PubMed] [Google Scholar]
- Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- Piwko W, Olma MH, Held M, Bianco JN, Pedrioli PG, Hofmann K, Pasero P, Gerlich DW, Peter M. RNAi-based screening identifies the Mms22L-Nfkbil2 complex as a novel regulator of DNA replication in human cells. EMBO J. 2010;29:4210–4222. doi: 10.1038/emboj.2010.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RT, Batova IN, Widgren EE, Zheng LX, Whitfield M, Marzluff WF, O’Rand MG. Characterization of the histone H1-binding protein, NASP, as a cell cycle-regulated somatic protein. J Biol Chem. 2000;275:30378–30386. doi: 10.1074/jbc.M003781200. [DOI] [PubMed] [Google Scholar]
- Schlesinger MB, Formosa T. POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics. 2000;155:1593–1606. doi: 10.1093/genetics/155.4.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz LL, Tyler JK. The histone chaperone ASF1 localizes to active DNA replication forks to mediate efficient DNA replication. FASEB J. 2006;20:488–490. doi: 10.1096/fj.05-5020fje. [DOI] [PubMed] [Google Scholar]
- Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- Smits AH, Jansen PW, Poser I, Hyman AA, Vermeulen M. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res. 2013;41:e28. doi: 10.1093/nar/gks941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Tan BC, Chien CT, Hirose S, Lee SC. Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J. 2006;25:3975–3985. doi: 10.1038/sj.emboj.7601271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki T, Mulder LC, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, Muesing M, Kwon YT. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JK, Collins KA, Prasad-Sinha J, Amiott E, Bulger M, Harte PJ, Kobayashi R, Kadonaga JT. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol Cell Biol. 2001;21:6574–6584. doi: 10.1128/MCB.21.19.6574-6584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- Winkler DD, Zhou H, Dar MA, Zhang Z, Luger K. Yeast CAF-1 assembles histone (H3-H4)2 tetramers prior to DNA deposition. Nucleic Acids Res. 2012;40:10139–10149. doi: 10.1093/nar/gks812. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, George AJ, Morgan KA, Mann JR, Choo KH. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351–360. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RS, Bonner WM. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell. 1981;27:321–330. doi: 10.1016/0092-8674(81)90415-3. [DOI] [PubMed] [Google Scholar]
- Wu RS, Tsai S, Bonner WM. Patterns of histone variant synthesis can distinguish G0 from G1 cells. Cell. 1982;31:367–374. doi: 10.1016/0092-8674(82)90130-1. [DOI] [PubMed] [Google Scholar]
- Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328:94–98. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, Li S, Chi H, Li YX, Chen HF, et al. Identification of cross-linked peptides from complex samples. Nat Methods. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- Young LC, McDonald DW, Hendzel MJ. Kdm4b histone demethylase is a DNA damage response protein and confers a survival advantage following gamma-irradiation. J Biol Chem. 2013;288:21376–21388. doi: 10.1074/jbc.M113.491514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Gibbs E, Kelman Z, O’Donnell M, Hurwitz J. Studies on the interactions between human replication factor C and human proliferating cell nuclear antigen. Proc Natl Acad Sci U S A. 1999;96:1869–1874. doi: 10.1073/pnas.96.5.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Chen L, Pledger WJ, Fang J, Chen J. p53 promotes repair of heterochromatin DNA by regulating JMJD2b and SUV39H1 expression. Oncogene. 2014;33:734–744. doi: 10.1038/onc.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.