Abstract
Cancer is a disease characterized by unrestrained cellular proliferation. In order to sustain growth, cancer cells undergo a complex metabolic rearrangement characterized by changes in metabolic pathways involved in energy production and biosynthetic processes. The relevance of the metabolic transformation of cancer cells has been recently included in the updated version of the review “Hallmarks of Cancer”, where the dysregulation of cellular metabolism was included as an emerging hallmark. While several lines of evidence suggest that metabolic rewiring is orchestrated by the concerted action of oncogenes and tumor suppressor genes, in some circumstances altered metabolism can play a primary role in oncogenesis. Recently, mutations of cytosolic and mitochondrial enzymes involved in key metabolic pathways have been associated with hereditary and sporadic forms of cancer. Together, these results suggest that aberrant metabolism, once seen just as an epiphenomenon of oncogenic reprogramming, plays a key role in oncogenesis with the power to control both genetic and epigenetic events in cells. In this review, we discuss the relationship between metabolism and cancer, as part of a larger effort to identify a broad-spectrum of therapeutic approaches. We focus on major alterations in nutrient metabolism and the emerging link between metabolism and epigenetics. Finally, we discuss potential strategies to manipulate metabolism in cancer and tradeoffs that should be considered. More research on the suite of metabolic alterations in cancer holds the potential to discover novel approaches to treat it.
Keywords: Cancer metabolism, mitochondria, Warburg, host metabolism, cancer therapy
1. Introduction
A non-profit organization called Getting to Know Cancer launched an initiative entitled “The Halifax Project” in 2011, which was charged with identifying synergistic molecular targets and/or small molecules for each of the areas that are widely considered to be hallmarks of cancer [1]. The rationale for this approach is based on the idea that cancers harbor significant genetic heterogeneity [2], which is often not addressed with monotherapeutic approaches. While efforts have been made to combine therapies to overcome resistance, rising drug costs, significant levels of toxicity, and a lack of overall success have stymied efforts to effectively treat cancer with multi-drug combinations [3].
Thus, the first aim of the Halifax Project was to produce a series of reviews, including this review on cancer metabolism, to broadly assess current knowledge on the biology of cancer. The overall goal of the Halifax Project is to identify biological targets and prospective lead compounds that could potentially be used to reach each prioritized area, and synergistically target multiple hallmarks of cancer. By building this rationale into the approach a priori, the problem of heterogeneity might be overcome. In theory, multiple low toxicity approaches could be experimentally combined, which then might lead to synergism within a given hallmark, such as cancer metabolism. Future studies will build upon these findings and test these hypotheses, as well as integrate these concepts into the approaches recommended in other hallmark areas in this special issue.
In this review, we first discuss the relationships between metabolism and cancer. We focus on major alterations in nutrient metabolism, as well as the emerging links between metabolism and epigenetics. Next, we discuss potential therapeutic strategies that could be used to manipulate metabolism in cancer cells or to manipulate host metabolism thereby influencing cancer metabolism. Finally, we describe tradeoffs that should be considered when leveraging these approaches. Together, this information will be the basis of significant future research to fully realize the potential of targeting metabolism in cancer.
2. Classic Metabolic Derangements
The first realization that metabolism is altered in cancer can trace its roots to the work of Otto Warburg. During the 1920s, Warburg found that unlike most normal tissues, cancer tissues fermented glucose to lactate at high rates regardless of the presence of oxygen [4, 5]. This was in contrast to the results that Pasteur had obtained previously studying fermentation in yeast, whereby O2 was found to inhibit fermentation [6, 7]. To study the metabolism of cancer in vivo, Warburg used Jensen sarcoma cells to form tumors within the abdomens of rats. By comparing arterial glucose and lactate concentrations to venous glucose and lactate concentrations, Warburg was able to infer the glucose uptake and lactate excretion by the tumor. Whereas normal tissues took up 2–18% of arterial glucose, tumors consumed 47–70%. Lactate was not significantly changed in blood after perfusion of normal tissues, but by Warburg’s calculations, tumors converted 66% of their consumed glucose into lactate. Thus, Warburg surmised that tumors take up much more glucose than normal tissues and convert a much larger percentage of it to lactate [4].
Warburg’s work on respiration and fermentation in cancer cells ultimately led him to propose that “the respiration of all cancer cells is damaged” [8]. In fact, he reasoned that known carcinogens, such as arsenic and hydrogen sulfide, likely worked by inhibiting respiration. He suggested that the primary oncogenic insult was an inability of cells to oxidize glucose carbons, and that X-rays were carcinogenic mainly due to their effect on mitochondria [8], which by this time had been shown to be the respiratory center of cells.
The exact molecular mechanisms leading to the Warburg effect and to altered metabolism in cancer remain a major unsolved question; for a review, see [9]. Subsequent studies have shown that while changes in mitochondrial respiration are sometimes seen in cancer cells, these alterations are not likely the driving lesion for most cancer cells. For example, Warburg’s follow-up work suggested that oxidative respiration was important in malignant tumors, and reported that placing rats in 5% O2 for 40 hours resulted in the death of most cancer cells, suggesting that oxygen was needed for viability of those cancer cells [4]. Similarly, the work of his contemporaries showed that oxygen consumption is intact in many cancers, thereby decoupling the Warburg effect from defective oxygen consumption [10, 11]. However, oxygen consumption cannot be a direct measurement of intact respiration, because mitochondrial coupling/uncoupling influences the efficiency of oxygen consumed to ATP produced. Nevertheless, many cancer cells display increased glucose uptake and elevated lactate production, irrespective of oxygen availability – also called “aerobic glycolysis” or the Warburg effect [12], and this observation remains a hallmark of altered metabolism in cancer cells.
3. Emerging Metabolic Derangements
While the mechanisms leading to the Warburg effect are under intense investigation, the general consensus of the field is that dysregulated metabolism and altered mitochondrial structure-function [13] is consistently found in several cancer cell types. These changes may occur before, as a result of, or in combination with, the genetic changes driving cancer, including oncogene expression or tumor suppressor loss; for recent comprehensive reviews on these concepts, see [14, 15]. For example, one well-studied link between oncogenesis and glucose metabolism is the phosphoinositide 3-kinase (PI3K) signaling pathway. Activating mutations in PI3K or overexpression of the AKT oncogenes, which lie downstream of PI3K, can induce high rates of aerobic glycolysis in non-transformed cells. This occurs in part by increasing expression and localization of the high-affinity glucose transporter, GLUT1, on the plasma membrane [16, 17]. In addition, activation of the PI3K pathway can accelerate flux through glycolysis by increasing the activity of hexokinase-2, phosphfructokinase-1 (PFK1), or phosphofructokinase-2 (PFK-2) [18–120]. The tumor suppressor p53, which has a well described role in DNA damage sensing, cell cycle control, and control of apoptosis, is also able to oppose the Warburg effect by stimulating respiration and reducing glycolytic flux [21–23]. Thus, loss of p53 in cancer cells is another event that can impact glucose metabolism in cancer cells.
Given the inextricable relationship between oncogenes, tumor suppressors, and the regulation of glycolysis, metabolic alterations including the Warburg effect could provide a selective advantage to rapidly proliferating cells [24]. Although fermentation produces almost 20 times less adenosine 5′-triphosphate (ATP) per glucose molecule than oxidative glucose metabolism, it has been suggested that ATP is never limiting in dividing cells [24, 25]. Instead, proliferating cells require macromolecular precursors and reducing power in the form of reduced nicotinamide adenine dinucleotide phosphate (NADPH) to synthesize new biomass. Therefore, one possible advantage of the Warburg effect is that high flux through glycolysis allows for more efficient use of glycolytic intermediates for NADPH production and biosynthetic pathways including lipid synthesis, nucleotide synthesis, and amino acid synthesis, which would be permissive for rapid cellular proliferation.
Although glucose metabolism is important for proliferating cells, it has recently been appreciated that other nutrient sources contribute as well. TCA cycle intermediates are required for biosynthetic processes, including the generation of citrate for lipid synthesis and aspartate for nucleotide synthesis. When these intermediates are removed from the TCA cycle, they must be replenished in a process known as anaplerosis. Glutamine has been shown to be a key contributor to anaplerotic flux in many cancer cells [26, 27]. Glutamine carbon entry into the TCA cycle supports both ATP generation and biosynthesis, and a wide variety of cancer cell types are sensitive to glutamine withdrawal [28, 29]. More recent work has suggested catabolism of extracellular protein can also contribute carbon to the TCA cycle [30], and lipids are scavenged to support proliferation of some cancer cells [31].
Collectively, it is becoming apparent that derangements to metabolism that enable the generation of biosynthetic precursors are a key feature in cancer initiation, development, and/or growth. Here, we discuss how alterations in key metabolic pathways contribute to biomass generation (Table 1). We discuss some prioritized targets within these pathways, their therapeutic potential, and strategies to manipulate metabolism for the prevention or treatment of cancer.
Table 1.
Prioritized pathways to target cancer metabolism
| Target | Known Pathways | Predicted Manipulation |
|---|---|---|
| Hexokinase 2 (HK2) | Glucose Metabolism | Reduce glucose uptake and metabolism |
| 6-Phosphofructo-2-Kinase/Fructose-2,6-Biphosphatase 3 (PFKFB3) | Glucose Metabolism | Reduce glycolysis |
| Pyruvate kinase isoform M2 (PKM2) | Glucose Metabolism | Activate pyruvate oxidation |
| Glutaminolysis | Amino Acid Metabolism | Inhibit glutamine anaplerosis |
| Reductive Carboxylation | Amino Acid Metabolism | Inhibit reductive carboxylation |
| O-GlcNAcylation | Epigenetics | Unknown |
| Methylation/One-carbon metabolism | Epigenetics | Unknown |
| Acetylation/Sirtuin deacylases (SIRTs) | Epigenetics | Unknown |
| Oncometabolites | Epigenetics | Inhibit oncometabolite formation and signaling |
| Lactate | Glucose Metabolism | Inhibit use of lactate as a fuel |
4. Glucose Metabolism
4.1 Hexokinase 2 (HK2)
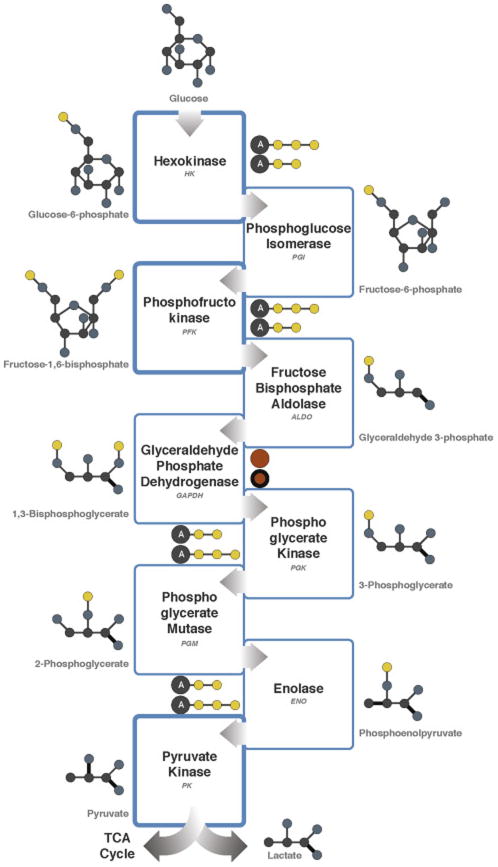
Regulation of glycolysis is exerted by the three important kinases that catalyze discrete phosphorylation reactions (Figure 1). Hexokinases are a family of enzymes that catalyze the first phosphorylation of glucose to glucose-6-phosphate. Early work on a highly malignant AS-30D hepatoma cell line showed that one isozyme of hexokinase was uniquely bound to the outer mitochondrial membrane [32, 33], which was later identified as Hexokinase 2 (HK2) [34].
Figure 1.
Schematic of glycolysis. Enzymes altered in cancer are shown in bold. Color scheme: carbon, gray; oxygen, blue; phosphate, yellow; adenosine, A; NAD+/NADH, orange; single bonds, thin; double bonds, bold.
In a series of experiments in this system, removal of malignant tumor mitochondria containing bound hexokinase from the cytoplasm by centrifugation markedly decreased the rate of glycolysis. Then, when tumor mitochondria containing bound hexokinase were added back to the tumor cytosol, the original glycolytic capacity of the cytoplasm was restored. Finally, when solubilized hexokinase alone was added to the liver cytosol, it markedly enhanced the glycolytic rate [34]. Therefore, HK2 could be a major contributor to high glycolysis and lactate production even in the presence of oxygen. One mechanism by which HK2 binds to mitochondria is via the outer membrane protein known as the voltage-dependent anion channel VDAC [35]. This binding interaction facilitates the immortalization of cancer cells [36, 37].
To identify compounds that could selectively inhibit the two main energy-producing pathways (glycolysis and mitochondrial oxidative phosphorylation), a limited screen was performed in cancer cells. The small molecule 3-bromopyruvate (3BP) was identified as an inhibitor of glycolysis and also as an inhibitor of mitochondrial energy production, which could be explained by inhibiting HK2 [38]. Follow-up work showed that 3BP has potent anticancer activity and has the capacity to eradicate cancers in different animal models [39]. Furthermore, 3BP was recently tested in a single human case study and showed promising results [40, 41]. Future work will be directed at the role of other HK isoforms in cancer, as well as the specificity of 3BP towards HK2. However, these early studies are one proof-of-principle that targeting energy metabolism of cancer cells can be an effective therapeutic approach.
4.2 6-Phosphofructo-2-Kinase/Fructose-2,6-Biphosphatase 3 (PFKFB3)
Another gluco-regulatory kinase is phosphofructokinase (PFK), which catalyzes the phosphorylation of fructose-6-phosphate to fructose-1,6-bisphosphate (Figure 1). PFK is regulated by several metabolites, including inhibition by high concentrations of ATP and activation by fructose 2,6-bisphosphate. Early studies indicated that phosphofructokinase activity correlated with the growth rate of Morris hepatomas transplanted in rats and also correlated with lactate production by slices of those tissues [42]. Those observations suggested that inhibition of phosphofructokinase activity represented a logical target for inhibition of malignant tumor growth. With the discovery that fructose 2,6-bisphosphate is an activator of phosphofructokinase 1 [43], the enzyme activity catalyzing the formation of fructose 2,6-bisphosphate became an alternative target for the inhibition of glycolysis and cancer growth. Steady state levels of fructose 2,6-bisphosphate are regulated by bifunctional enzymes that have both phosphofructo-2-kinase and fructose-2,6-bisphosphatase (PFKFBs). Of these enzymes, PFKFB3 has the highest phosphofructo-2-kinase activity [44]. Interestingly, PFKFB3 expression, but not other PFKFBs, is markedly elevated in multiple aggressive primary cancers, including colon, breast, ovarian and thyroid carcinomas [45].
The role of PFKFB3 in the regulation of glucose metabolism in cancer cells has been reviewed previously [46]. An early indication that PFKFB3 might be a regulatory enzyme was the identification of multiple copies of the AUUUA instability motif in its 3′ untranslated region [44]. Expression of the PFKFB3 gene is induced by hypoxia through hypoxia-inducible factor-1 [47]. Low pH, a common feature in malignant tumors, is another factor that results in upregulation of PFKFB3. This may be mediated through the metabolic stress sensor AMP-activated protein kinase (AMPK) resulting in an increase in serine phosphorylation [48]. In breast cancer cells, synthetic progestins activate PFKFB3 isoenzyme phosphorylation and a long-term sustained action due to increased PFKFB3 protein levels. An immediate early response occurs through the ERK/RSK pathway leading to phosphorylation on S461 followed by activation of transcription via cis-acting sequences on the PFKFB3 promoter [49]. In myeloproliferative neoplasms, PFKFB3 expression was upregulated via active JAK2 and STAT5 [50], suggesting that specific inhibitors of PFKFB3 might inhibit JAK2/STAT5-dependent malignancies.
Levels of PFKFB3 are regulated by ubiquitinylation during the cell cycle. PFKFB3 accumulates in mid to late G1 and breakdown in S phase occurs specifically via a distinct S273 residue within the conserved recognition site for SCF-beta-TrCP [51]. Others have noted that the activity of PFKFB3 is short lasting, coinciding with a peak in glycolysis in mid to late G1 in contrast to glutamine metabolism, which remains high throughout S phase [52]. Glycolysis is characteristically associated with cytosolic fractions of cells but PFKFB3 has a nuclear localization signal. Furthermore, nuclear localization of PFKFB3 was associated with increased proliferation by increased expression of cyclin-dependent kinases and decreased expression of the cell cycle inhibitor p27 [53]. This brings to mind the more recent observation with the pyruvate kinase isoenzyme PKM2 (described below) that has been shown to have potential transcription regulatory action in addition to its role in glycolysis [54]. Therefore, a dual function may be associated with some regulatory enzymes involved in glycolysis.
Molecular modeling studies were the basis for the identification of the first published report of a small molecule inhibitor of PFKFB3 [55]. This molecule, 3-(3-pyridinyl)-1-(4-pyridnyl)-2-propen-1-one known as 3PO became commercially available through Calbiochem, EMD Millipore in 2013. 3PO was shown to suppress glucose uptake and glycolytic flux and was selectively cytostatic to Ras-transformed human bronchial epithelial cells relative to normal cells. Growth of human erythroleukemia cells was inhibited by 3PO [50]. Treatment of tumor-bearing mice reduced the intracellular concentration of fructose 2,6-bisphosphate, glucose uptake and tumor growth. The observation that 3PO suppresses T-cell activation indicates a potential use for small molecule inhibitors of PFKFB3 in the treatment of autoimmune conditions but might be problematic in combination treatments with immunosuppressive cancer chemotherapeutic agents [56]. A more potent derivative of 3PO designated PFK15 has been identified and a phase I clinical trial is planned [57].
4.3 Pyruvate kinase isoform M2 (PKM2)
Pyruvate kinase (PK) catalyzes the final rate-limiting step in glycolysis (Figure 1), transferring a phosphate group from phosphoenolpyruvate (PEP) to ADP and thereby generating pyruvate and ATP [58]. Humans have four PK enzymes: PKR is restricted to erythrocytes; PKL is found predominantly in liver and kidney; PKM1 is expressed in differentiated somatic cells (e.g. muscle and brain) and PKM2 is found in fetal tissues and proliferating cells. In cancer cells, expression of PKM2 is up-regulated such that it becomes the most predominant PK isoform [59]. Accumulating evidence suggests that PKM2 expression is elevated in cancer cells, as its enzymatic activity can be regulated by various metabolic and signaling inputs [24, 60, 61]. PKM2 influences the fate of glucose carbons (Figure 1). In general, PKM2 activity is low in proliferating cells, which creates a bottleneck at the terminal step in glycolysis, which results in elevated concentrations of upstream glycolytic metabolites. As a consequence, such intermediates are available for the biosynthetic reactions that branch off of glycolysis, thereby increasing the generation of cellular building blocks needed for proliferation [24]
Current models describing the function of PKM2 in cellular proliferation focus mainly on mechanisms that regulate its pyruvate-generating activity as a cytoplasmic component of glycolysis [61, 62]. However, recent studies have provided evidence for activities of PKM2 that extend beyond this canonical role. Namely, several nuclear activities for PKM2, and mechanisms that enable the shuttling of PKM2 into the nucleus, have now been described [54, 63–70]. Nuclear activity promotes tumor growth through the direct transcriptional activation of genes involved in cancer metabolism, including PKM2 itself and the RNA splicing factors that repress PKM1 [54, 65, 68]. In this way, PKM2 acts in a feed forward loop to promote both its nuclear activities and its metabolic role in cancer metabolism. Ongoing studies seek to examine the relative contributions of these two functions in oncogensis and growth.
The central principle presented above asserts that inhibiting PK activity facilitates proliferation. This can be achieved through the expression of the less active and ‘regulatable’ PK enzyme, PKM2. Alternatively, PKM2 might be upregulated to eliminate PKM1 expression; PKM1 is a constitutively active enzyme, which reduces the generation of carbon for anabolic reactions and facilitates the generation of ATP in the mitochondria. Recent support for this line of thinking comes from the observation that PKM2 is not required for tumor maintenance, where short hairpin (sh)RNA-mediated depletion does not affect tumor growth [71]. Furthermore, genetic experiments, in which the PKM2-specific exon was deleted in a mouse model of breast cancer using Cre-lox technology, demonstrated that the absence of PKM2 increased oncogenesis and negatively affected survival. Strikingly, these tumors showed no tissue-specific PK expression. They did, however, still produce lactate from glucose, suggesting that alternative and non-canonical methods of pyruvate generation were functioning [72]. Collectively, these experiments demonstrate that PKM2 is not required for oncogenesis or growth, and support for the concept that cancer cells have a growth advantage by removing PKM1.
Despite potential mechanistic differences between these two models on the role of PK in cellular proliferation, they converge on a common point: constitutive PK activity is detrimental for proliferation. Consistent with this concept, the first study to examine the activity of PK directly in an isogenic context demonstrated that cancer cell lines engineered to express PKM1 produce less lactate, consume more oxygen and are less tumorigenic in nude mouse xenografts than those expressing PKM2 [73]. Importantly, these results demonstrated that less PK activity provided a selective growth advantage for cancer cells in vivo.
From a therapeutic standpoint, activation of PK may serve as a promising strategy to slow cancer growth. Indeed, numerous research teams in the public and private sectors have recently developed drug-like small molecule activators of PKM2 that make it behave more like PKM1 [74–79]. Studies using such compounds have shown that activation of PKM2 results in decreased accumulation of biosynthetic carbon building blocks and reduced cancer growth [74]. Interestingly, all compounds described thus far share a high degree of structural similarity and bind at the same interface in the PKM2 multimer. The small molecule activator binding pocket is distinct from the fructose bisphosphate (FBP) binding pocket, where PKM2 activators overcome negative regulation by phosphotyrosine peptides. Together, these two activities enable compounds to overcome mechanisms that negatively regulate PKM2.
Several of the compounds described above have been investigated in animal models [74, 75]. In one study, a compound called TEPP-46 activated PKM2 and impaired tumor seeding and growth. Subsequent metabolic analyses support the hypothesis that impaired anabolic metabolism is responsible for the growth inhibition. Future studies are now aimed at exploring the use of these agents in combination with cytotoxic chemotherapies that generate oxidative stress. Support for this concept comes from the observation that activating PKM2 impairs the metabolic control of redox – specifically the generation of reducing equivalents in the form of NADPH and GSH [80] – which sensitizes cancer cells to further oxidative stress. Finally, PKM2 activators may prove to be doubly effective, as multimer formation prevents the nuclear translocation of PKM2 and thus its activity as a protein kinase and activator of gene expression [67].
5. Amino Acid Metabolism
In addition to the well-established role for altered glucose metabolism in cancer, recent research highlights the involvement of amino acid metabolism in cancer, especially glutamine. In proliferating cells, the tricarboxylic acid (TCA) cycle functions as a source of precursors for macromolecular synthesis in addition to generating reducing equivalents for oxidative phosphorylation [81]. Citrate, for example, is both the canonical entry point of the TCA cycle and a precursor for the acetyl-CoA used in fatty acid/sterol synthesis and acetylation reactions. Normally, the withdrawal of citrate to supply these other pathways is matched by an influx of carbon into the cycle to yield oxaloacetate, refilling the pools of TCA cycle intermediates and maintaining function of the cycle during growth [26].
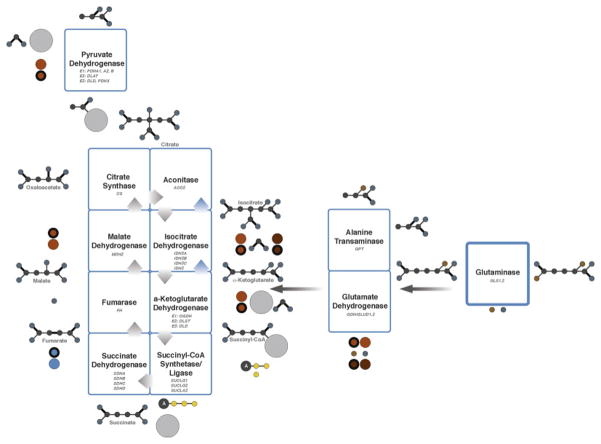
In culture, many cancer cells meet their anaplerotic demand through the oxidative metabolism of glutamine to oxaloacetate (Figure 2) [26]. Glutamine is an advantageous anaplerotic precursor because its conversion to α-ketoglutarate (α-KG) has the potential to disperse nitrogen to hexosamines, nucleotides, and non-essential amino acids, all of which are required for growth. Furthermore, oxidative metabolism of α-KG to oxaloacetate produces reducing equivalents that can be used to generate energy [82]. Alternative anaplerotic pathways, such as pyruvate carboxylation, also produce oxaloacetate but do not satisfy these other demands, positioning glutamine as a key player in mitochondrial metabolism.
Figure 2.
Schematic of the TCA cycle and glutaminolysis. Enzymes altered in cancer are shown in bold. Reductive direction of the TCA cycle shown with blue arrows. Color scheme: carbon, gray; oxygen, blue; phosphate, yellow; nitrogen, brown; adenosine, A; coenzyme A, light gray; NAD+/NADH, orange; NADP+/NADPH, brown; FAD/FADH2, blue; single bonds, thin; double bonds, bold.
5.1 Glutaminolysis
Glutamine is a non-essential amino acid with an amine functional group and is the most abundant amino acid in circulation [83]. Glutamine supplies nitrogen for nucleobase synthesis and carbon for the TCA cycle, lipid synthesis and nucleotide synthesis [84]. Glutamine is involved in both anabolic and catabolic processes. The catabolism of glutamine is called glutaminolysis, which can be converted into glutamate, aspartate, CO2, pyruvate, lactate, alanine and citrate. The first step of glutaminolysis is the conversion of glutamine to glutamate and ammonia via glutaminase (GLS). After glutamine is converted into glutamate, the glutamate is oxidized into α-KG. This most often occurs through the enzyme glutamate dehydrogenase (GLDH), concomitant with the generation of mitochondrial NADH or NADPH and ammonia [85]. This is the first energy-yielding step in glutaminolysis and is the link to the TCA cycle (Figure 2).
While glutaminolysis is a normal process for many cells, such as lymphocytes and adipocytes, many cancer cells have elevated glutamine flux. In cell culture, human glioma and HeLa cells were found to be completely dependent upon glutamine for their survival. The cells died in the absence of glutamine, despite being in glucose-rich media, which is now known as the complete dependence on glutamine for cell survival (a.k.a glutamine addiction) [28, 86]. Interestingly, not all cancer cells exhibit glutamine addiction. While the complete molecular mechanism of glutamine addiction is not known, several oncogenic mutations or alterations have been found to explain glutamine-dependence in cancers. For example, Myc is able to increase glutamine metabolism by upregulating GLS expression, which leads to glutamate entry into the TCA cycle as α-KG [87].
The ability to use exogenous glutamine is enhanced by the upregulation of glutamine transporters [88]. For example, one study found the increase in glutaminolysis was so profound that cancer cells accumulated more glutamine than was necessary to meet the energy and anabolic requirements of the cell; excess glutamine-derived carbon was secreted from the cells in the form of lactate or, to a lesser extent, alanine [28]. Further evidence suggests that overexpression of Myc is enough to induce glutamine addiction, due to the fact that this mutation causes the metabolism of the mitochondria to be altered in such a manner as to rely on glutamine despite available glucose [28]. Interestingly, these glutamine-addicted cells were found to be keenly sensitive to electron transport chain inhibitors. This observation could indicate that either the potential energy in glutamine is being used in the production of ATP [89] or that mitochondrial glutamate uptake by the glutamate/aspartate mitochondrial transporter requires a proton-motive force.
In human pancreatic ductal adenocarcinoma (PDAC), the oncogene KRAS contributed to glutamine dependency by using glutamine-derived aspartate to produce oxaloacetate via aspartate transaminase (GOT1) in the cytoplasm. Oxaloacetate was in turn converted into malate and then pyruvate to maintain a high NADPH/NADP+ ratio for redox homeostasis. Therefore, disrupting these reactions, and ultimately glutamine metabolism, led to suppression of PDAC growth in vitro and in vivo [90].
In a recent study, the use of Nuclear Magnetic Resonance (NMR) Spectroscopy and Mass Spectrometry (MS), as well as stable isotope-resolved metabolomics, allowed the fate of 13C and 15N from labeled glutamine in B lymphoma cells to be traced under aerobic and hypoxic glucose-deprived conditions [91]. In this study, glutamine is the fuel that drives the TCA cycle, which is completely independent of the glucose supply. This reprogramming of the TCA cycle is particularly advantageous to cancer cells under glucose-deprived and/or hypoxic conditions. Where glucose is preferentially converted to lactate under hypoxic conditions, glutamine metabolism serves to sustain ATP production and redox homeostasis in order to support cancer cell growth and survival. This implies that even hypoxic cancer cells can oxidize glutamine through the TCA cycle, and in the absence of glucose, glutamine metabolism alone can sustain the TCA cycle and thereby meet the anapleurotic and energetic demands of the proliferating cancer cells.
Supporting this idea, Myc-transformed cells were found to rely on a means of α-KG synthesis other than that involving GLDH. Aspartate and alanine transaminases reversibly convert glutamate into α-KG, along with oxaloacetate to aspartate or pyruvate to alanine, respectively (Figure 2). These reactions occur despite the presence of GLDH in the mitochondria of cancer cells, which catalyzes the direct conversion of glutamate to α-KG [92]. This is especially important considering it was found that Myc-transformed breast cancer cells undergo apoptosis when aspartate transaminase is inhibited. This implies that this cancer is heavily reliant upon transaminase reactions to produce α-KG [93, 94].
Due to the central nature of glutaminolysis in many cancers, it is becoming an increasingly prominent target in cancer therapy. One of the early strategies to suppress glutamine metabolism was to reduce the amount of available glutamine by using glutamine analogs. Compounds such as 6-diazo-5-oxo-l-norleucine (DON) and acivicin showed cytotoxic effects against several malignant tumors types, including leukemia and colorectal cancers; however these analogs are no longer clinically available due to patient toxicity [95].
More recent strategies have focused on targeting the enzymes of glutamine metabolism. For example, GLS is a potential target for the inhibition of glutaminolysis in cancer cells. The kidney isoform, GLS1, is found in many malignant tumors [96] while the liver isoform, GLS2, is less often expressed in cancers. The compound bis-2-(5-phenylacetimido-1,2,4,thiadiazol-2-yl)ethyl sulfide (BPTES) has been shown to inhibit growth of a variety of cancers in mouse models and in cell culture, including B lymphoma, which allosterically inhibits GLS1 by altering the conformation of the enzyme [97]. The effect is enhanced under hypoxic conditions, often inducing cancer cell death [91].
GLDH is another potential target of glutaminolysis, which when knocked-down by siRNA resulted in a marked decrease in the proliferation of glioblastoma cells that were glutamine dependent [98]. Green tea polyphenols, hexachlorophene, GW5074, and bithionol may inhibit GLDH function. These inhibitors work by restricting enzyme movement, either by forming rings around the enzyme or wedging between the enzyme’s subunits. Currently, green tea polyphenols have been shown to inhibit lung, colon, and prostate adenocarcinoma growth in xenograft models [99]. These compounds also had significant effects on glioblastoma, colon, lung and prostate adenocarcinoma cell proliferation [100].
Another strategy to inhibit glutaminolysis is to target alanine transaminase through L-cycloserine [101] or aspartate transaminase through amino oxyacetate [93] which nearly halted the growth of breast cancer in xenograft mice. Similarly, aspartate transaminase knockdown in pancreatic cancer is also dramatically growth inhibitory in vitro and in vivo and leads to a profound disruption of redox homeostasis [90]. In both of these cases, little to no toxicity was observed with transaminase inhibition in non-neoplastic cells. These studies suggest that aspartate aminotransferase is a promising cancer target. Finally, given that inhibition of aspartate transaminase also leads to a disruption of redox homeostasis, such inhibition may synergize with therapies that increase reactive oxygen species (ROS), such as chemotherapy and radiation [102].
5.2 Reductive Carboxylation
Not all cancer cells have the ability to perform glutaminolysis. Hypoxia limits the oxidative capacity of the TCA cycle, and in particular suppresses production of acetyl-CoA from glucose via pyruvate dehydrogenase (PDH) [103, 104]. Furthermore, some cancer cells contain severe mutations in TCA cycle enzymes (succinate dehydrogenase, fumarate hydratase) or in components of the electron transport chain that prevent efficient production of oxaloacetate from glutamine [105]. Yet, both hypoxic cells and cells with defective mitochondria require glutamine for growth [106–108]. To address this paradox, metabolic labeling experiments were performed using 13C, and revealed that these cells metabolize glutamine through an unusual pathway characterized by “reversal” of isocitrate dehydrogenase (IDH) enzyme activity (Figure 2, blue arrows) [106, 107, 109, 110]. IDH typically acts as an oxidative decarboxylase, converting isocitrate to α-KG and CO2 in the presence of an electron acceptor. Indeed, IDH3, the mitochondrial NAD+-dependent IDH isoform, functions exclusively in this manner. However, the two other mammalian IDH isoforms, IDH1 and IDH2, use NADP+/NADPH as cofactors, and can act either as oxidative decarboxylases or reductive carboxylases. In the latter reaction, α-KG is carboxylated to produce isocitrate, converting NADPH to NADP+. Isocitrate is readily converted to citrate, which can then be cleaved to produce acetyl-CoA. Under conditions of reductive carboxylation, glutamine becomes the major source of acetyl-CoA for fatty acid synthesis, greatly decreasing the need to produce acetyl-CoA from glucose. Furthermore, citrate cleavage yields oxaloacetate, which is converted to other 4-carbon intermediates, meaning that essentially the entire cellular pool of TCA cycle intermediates can be derived from reductive carboxylation, in reverse order to their conventional route of production (Figure 2).
Reductive carboxylation had previously been observed as a minor source of isocitrate and citrate in a number of non-transformed mammalian tissues [111–114]. Its importance in cancer cell biology is related to its ability to serve as the major source of citrate and acetyl-CoA when cellular circumstances conspire to inactivate pathways that normally produce these metabolites, including hypoxia and genetic reprogramming of mitochondrial metabolism. For example, the Von Hippel-Lindau (VHL) tumor suppressor normally functions in the oxygen-dependent degradation of the alpha subunits of the hypoxia inducible factor (HIF) transcriptional activators. Thus, in cells expressing VHL, oxygen facilitates the degradation of HIF1α and HIF2α so that HIF target genes are not expressed. In contrast, in malignant tumor cells lacking VHL, HIF target genes are expressed regardless of oxygen availability. These genes, which include glucose transporters and glycolytic enzymes, are part of the metabolic adaptation to hypoxia. Importantly, hypoxia stimulates the expression of PDH kinase-1 (PDK1), which phosphorylates and inactivates the PDH complex, impairing its ability to provide acetyl-CoA and citrate from glucose [103, 104]. Cancer cells lacking VHL produce a substantial fraction of citrate and fatty acids using glutamine-dependent reductive carboxylation in culture, and a small but detectable fraction of citrate in vivo [115]. Heterologous expression of wild-type Vhl, or silencing of PDK-1, partially reverts metabolism to a phenotype in which citrate and fatty acids are produced from glucose/PDH [115].
Regulation of reductive carboxylation is an area of active investigation. Importantly, the pathway is stimulated by mutations in the electron transport chain that impair recycling of mitochondrial NADH to NAD+, but importantly NADPH is the cofactor for reductive carboxylation. This suggested a model in which a low NAD+/NADH ratio in mitochondria provides a source of reducing equivalents which are transmitted to NADPH by nicotinamide nucleotide transhydrogenase (NNT) [107]. NNT is an inner mitochondrial membrane protein that uses the electrochemical proton gradient to transfer reducing equivalents from NADH to NADPH [116]. Silencing NNT expression in SkMel5 melanoma cells reduced the contribution of glutamine carbon to TCA cycle intermediates through both oxidative and reductive metabolism, and decreased the rate of growth of SkMel5-derived subcutaneous xenografts [117]. In VHL-deficient 786-O renal carcinoma cells, which produce a large fraction of citrate via reductive carboxylation, NNT silencing significantly suppressed reductive glutamine metabolism. These findings suggested that NADPH produced by NNT is required for reductive carboxylation in VHL-deficient cells.
Reductive carboxylation is also regulated by changes in the abundance of TCA cycle metabolites. Citrate levels tend to be low in cells with active reductive carboxylation, and maneuvers to suppress formation of citrate from glucose enhance the fraction of citrate derived from reductive carboxylation [106, 107, 109, 115]. By contrast, supplying cells with exogenous acetate or citrate increases the ratio of citrate/α-KG while reducing the overall contribution of reductive carboxylation to TCA cycle metabolism [115].
Compartmentalization of the reductive carboxylation reaction is not well understood. IDH1 is cytosolic while IDH2 is localized to the mitochondria, and data suggest that each isoform can participate in reductive carboxylation. IDH2 was found to be the crucial isoform for reductive carboxylation in hypoxic glioma cells [109], whereas other cell lines required IDH1 under hypoxia [106]. Genetic suppression of pyruvate dehydrogenase in lung cancer cells is sufficient to induce a net flux of reductive carboxylation, and under these circumstances the flux is completely dependent on IDH1 [118]. In cells with mutations in the electron transport chain, silencing either isoform reduced activity of the pathway, and silencing both together had a maximal effect [107]. The involvement of NNT as a source of NADPH argues for mitochondrial localization of the reductive carboxylation reaction, at least in those cells that require NNT expression to maintain citrate levels. However, cytosolic sources of NADPH, including the oxidative branch of the pentose phosphate pathway, may play a more prominent role in cells where IDH1 catalyzes reductive carboxylation. Presumably, the choice of isoform is also related to localization of an available source of the substrate α-KG, meaning that compartmentalization of glutamine metabolism could add another dimension to regulation of this pathway.
6. Lipid Metabolism
Cancers may use a wide variety of substrates and substrate sources to meet their catabolic and anabolic needs, including internally- and externally-derived fatty acids (FAs). Indeed, FAs are essential for cellular proliferation. In particular, FAs are used as cellular building blocks for lipid membrane synthesis, for energy storage and production, as well as for cellular signaling. Thus, limiting abundance of FAs could be a therapeutic strategy against cancer. Limiting cellular FAs could be achieved by: blocking synthesis (lipogenesis), increasing breakdown (oxidation), reducing availability from storage (lipolysis), or by increasing FA flux towards storage (re-esterification); these distinct strategies have been considered as chemotherapeutic strategies recently [119]. As with other metabolic shifts in cancer, characterizing the state of lipid metabolism in unique cancer types and cells lines is an important first step. Indeed, successful chemotherapeutic strategies will require understanding the specific abnormalities in lipid metabolism for a given cancer type. Targeting lipid metabolism in cancer is an emerging idea that warrants further investigation.
7. Epigenetics and Oncometabolites
In addition to alterations in metabolism, metabolic reprogramming mediated by specific oncogenes and tumor suppressors can impact dynamic regulation of chromatin via post-translational modifications. Indeed, increasing evidence indicates that altered metabolism can also lead to changes in nutrient-sensitive post-translational modifications. These chemical modifications, such as O-GlcNAcylation, methylation, and acetylation, can impact the activities of metabolic enzymes, signaling components, transcriptional regulators, and chromatin-associated proteins such as histones [120–124]. Furthermore, oncometabolites, such as 2-hydroxyglutarate (2-HG), can have profound consequences on the regulation of chromatin organization, gene expression, and genome integrity. Improved understanding of the links between metabolism and epigenetics is expected to have important therapeutic implications, and intense efforts are currently geared towards targeting both altered metabolism and epigenetics [125, 126].
7.1 O-GlcNAcylation
The hexosamine biosynthetic pathway (HBP), a branch of metabolism that diverges from glycolysis at fructose-6-phosphate, generates uridine diphosphate N-acetylglucosamine (UDP-GlcNAc), a key donor substrate for glycosylation reactions, including the O-GlcNAc modification, which is mediated by O-GlcNAc transferase (OGT) [122, 127, 128]. O-GlcNAcylation is generally elevated in cancer cells and has been linked to malignant tumor growth through direct modification of phosphofructokinase (PFK1) and indirect regulation of FoxM1 [129–131]. All four histones (H2A, H2B, H3, and H4) are modified by O-GlcNAc, which is dynamically regulated in response to nutrient availability [132–134]. O-GlcNAcylation of histone H3 on Ser10 is cell cycle regulated and suppresses mitosis-specific H3 phosphorylation [135], whereas H2B Ser112 O-GlcNAcylation facilitates monoubiquitylation of Lys120 of histone H2B, a mark associated with active transcription, indicating a role for O-GlcNAc in gene regulation through chromatin modification [134]. OGT can also associate with Ten-Eleven-translocation (TET) family 5-methylcytosine hydroxylases and participate in TET-mediated gene regulation [136–139]. Whether cancer cells exhibit altered histone O-GlcNAcylation patterns due to metabolic alterations is not yet clear. Notably, TET2 function is frequently disrupted in hematopoietic malignancies [140–142], and future studies will determine whether changes in histone O-GlcNAcylation participate in cancer development in these cases.
7.2 Methylation
Alterations in histone and DNA methylation are widely observed in cancer. Many cancer types display global DNA hypomethylation compared with normal tissue, while genes regulating cell-cycle and DNA damage response are frequently found to be hypermethylated, and thus silenced [143]. Histone and DNA methyltransferases utilize the methyl donor S-adenosyl methionine (SAM), which is synthesized from methionine and ATP. Hence, SAM availability for methylation reactions may be sensitive to levels of methionine taken up from the environment or synthesized through one-carbon metabolism pathways, discussed in more detail below. In yeast, SAM can serve as a sensor of amino acid availability, inhibiting autophagy and promoting growth [144]. While transformed cells require growth medium supplemented with methionine [145], knowledge of how cancer cells use methionine to regulate histone and DNA methylation is still limited. Notably, the enzyme nicotinamide N-methyltransferase (NNMT), which depletes SAM by catalyzing the transfer of SAM’s methyl group to nicotinamide, is frequently overexpressed in human cancers. A recent study demonstrated that NNMT levels impact methylation in cancer cells, with NNMT overexpression associated with reduced levels of SAM and histone methylation [146]. Interestingly, metabolic regulation of SAM production has also been implicated in modulating histone methylation levels and maintaining pluripotency in mouse embryonic stem cells [147]. In addition to overall cellular levels of SAM, localized pools of SAM may provide an additional layer of metabolic control. The enzyme methionine adenosyltransferase (MATIIα, which produces SAM, has been localized to gene promoters and implicated in gene regulation, indicating that enzymatic production of metabolites can be targeted to specific loci and coupled to transcriptional regulation [148]. DNA and histone methylation levels are also controlled by rates of demethylation. Metabolic control of demethylation is discussed below in the “Oncometabolites” sub-section. Given the substantial evidence that DNA and histone methylation abnormalities contribute to cancer initiation and growth, further investigation into the role of metabolism in determining methylation changes in cancer is needed.
7.3 Acetylation
Lysine acetylation levels are determined by the combined actions of lysine acetyltransferases (KATs) and deacetylases (HDACs), both of which may be influenced by metabolic state. Histone proteins are acetylated at multiple lysines, and these modifications are involved in chromatin-dependent processes, including gene transcription, DNA replication, and DNA damage repair. In mammalian cells, nuclear-cytosolic acetyl-CoA, the donor substrate for acetylation reactions, is generated either through the cleavage of citrate by ATP-citrate lyase (ACLY) or directly from acetate by acetyl-CoA synthetase (AceCS1) [122]. Histone acetylation can be regulated by the availability of glucose or acetyl CoA [149–152], and in mammalian cells glucose-dependent regulation of histone acetylation occurs in an ACLY-dependent manner [149]. Reciprocally, nuclear-cytoplasmic lysine deacetylation can be mediated by SIRT1, a member of the NAD+-dependent sirtuin family of lysine deacetylases [class III histone deacetylases (HDACs)] [153, 154]. NAD+ levels rise under nutrient-restricted conditions, in part mediated by AMP-activated protein kinase (AMPK) [155, 156]. In addition, the ketone body β-hydroxybutyrate (βOHB) was recently demonstrated to function as an endogenous inhibitor of class I HDACs under ketogenic conditions and influence the state of histone acetylation and gene transcription [157].
Metabolic control of histone acetylation is likely to impact cancer growth, although this has not yet been directly shown. In yeast, acetyl-CoA acts as a growth signal, promoting histone acetylation at and expression of growth-related genes [158]. Levels of ACLY are frequently elevated in cancer [159], although the impact of this on overall histone acetylation is not yet known. Global histone acetylation levels are highly heterogeneous among cancers, and several studies have shown correlations with cancer recurrence and patient survival [160–164]. Both KAT and HDAC inhibitors have shown promise in cancer therapy [125], although mechanisms of action remain poorly understood.
7.4 Sirtuin deacylases (SIRTs)
The sirtuins (SIRT1-7) are a class of conserved NAD+-dependent deacylases that control cellular metabolic processes and protect the cell against metabolic and genotoxic stresses. Not surprisingly, altered regulation of the sirtuins is associated with many diseases such as diabetes, neurodegenerative diseases, obesity, and cancer [165]. Rapidly proliferating cancer cells require shifts in metabolism that promote anabolic metabolism, as such the sirtuins play an important role in controlling cancer development and progression by maintaining normal cellular metabolism. Of the 7 mammalian sirtuins four of them, SIRT1, SIRT3, SIRT4, and SIRT6 have been associated with various types of cancer and are generally believed to be associated with cancer cell metabolism and control of the Warburg effect as well as DNA damage responses. SIRT1, SIRT3, and SIRT6 are strong deacetylases, while the enzymatic activity of SIRT4 is less clear but it has been shown to be a weak ADP-ribosyltransferase and possibly a weak deacetylase [166]. SIRT1 and SIRT6 serve to mainly deacetylate histones and transcription factors in the nucleus, while SIRT3 and SIRT4 are mitochondrial, where they act on metabolic proteins [167].
SIRT1 shares the most homology to Sir2 (S. cerevisiae), the first sirtuin identified, and has been shown to increase lifespan and protect against age-related diseases. The main targets of SIRT1 are the transcription factor peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1α), the FOXO family of transcription factors, and the tumor suppressor p53 [168]. Studies have found that SIRT1 expression is elevated in certain cancers and repressed in others, making the role of SIRT1 in cancer difficult to define [169]. It seems as though the main confusion underlying the role of SIRT1 in cancer is the complexity behind the SIRT1-mediated inhibitory deacetylation of p53. SIRT1 expression is also positively regulated by p53 in an apparent negative feedback loop where loss of p53 reduces SIRT1 expression, leading to an increase in p53 activity [170]. SIRT1 has been described as a tumor suppressor that promotes the DNA damage response, and SIRT1+/− p53+/− mice develop cancer in a variety of tissues, particularly sarcomas. Furthermore, SIRT1 overexpressing mice are more metabolically efficient, resistant to diabetes, and no cancer phenotype has been reported in these mice [171]. SIRT1 expression is reduced in various human cancers, particularly breast cancer and hepatocellular carcinoma [172]. Activation of SIRT1 has already proven beneficial in BRCA1-associated breast cancers as treatment with resveratrol activated SIRT1 and inhibited cell proliferation in vitro and tumor growth in vivo [173]. It seems as though activation of SIRT1 may have therapeutic potential in certain cancers in order to normalize cellular metabolism and improve the DNA damage response.
SIRT3 is the main mitochondrial deacetylase and as such it controls the activities of many metabolic enzymes in the mitochondria. SIRT3 deacetylates a long list of mitochondrial proteins acting at several nodes of mitochondrial metabolism, including fatty acid oxidation, glutamine metabolism, and mitochondrial ROS production. When SIRT3 is lost or ablated, metabolic derangements occur [174], which may be the source for the link between SIRT3 and cancer. The first described link between SIRT3 and cancer reported that SIRT3KO mice spontaneously develop mammary tumors after two years [175]. Furthermore, this study identified SIRT3 as a tumor suppressor by showing that SIRT3 KO MEFs could be transformed in vitro by the addition of a single oncogene, Myc or Ras [175]. A later study attributed the elevated level of cellular ROS seen with loss of SIRT3 to the stabilization of HIF-1α and activation of HIF-1α glycolytic target genes, leading to altered cellular metabolism towards glycolysis [176]. It is possible that SIRT3 deficiency leads to a cancer permissive state by coordinating a shift in metabolism to a Warburg phenotype.
Cancer cells also rely heavily on the metabolism of glutamine as a nitrogen source for protein and nucleotide synthesis necessary for proliferation [28]. SIRT4 was previously found to inhibit the activity of glutamate dehydrogenase (GDH) activity by ADP-ribosylation. More recently, one study showed that genotoxic stress causes an induction of SIRT4 expression leading to a repression of glutamine metabolism [177]. SIRT4 KO MEFs had higher glutamine uptake in response to UV damage and increased glutamine-dependent proliferation compared to wild type cells. Finally, SIRT4KO mice had increased incidence of various types of cancer, particularly lung cancer compared to wild type mice [177].
Finally, SIRT6 controls cancer metabolism and SIRT6 ablation activates aerobic glycolysis and leads to oncogenesis, in the absence of oncogene activation [178]. This study showed that when aerobic glycolysis was inhibited by inhibition of PDK1, an inhibitor of pyruvate dehydrogenase (PDH), the SIRT6 deficient cells failed to form tumors, showing a reliance on glycolytic metabolism of glucose. Further, SIRT6 expression is also low in pancreatic, colon, and liver cancers in humans [178, 179]. Overall, it seems as though SIRT6 suppresses cancer growth by regulating aerobic glycolysis and that a mutator phenotype is not required for SIRT6-dependent tumor formation.
Collectively, these studies show that the loss of an individual sirtuin can promote oncogenesis by creating a state that is favorable to cancer cells, such as increasing DNA damage, inhibiting DNA repair mechanisms, or altering cellular metabolism towards a Warburg-like phenotype. Modulating sirtuin expression may be a way to alter substrate use by cancer cells and slow or prevent their growth. Many of the sirtuins are known to be regulated by nutrient status, therefore it may be beneficial to modulate sirtuin expression by diet and/or exercise. SIRT1 and SIRT3 expression are induced with calorie restriction and fasting, and SIRT3 expression is reduced with high fat diet (HFD) feeding [180, 181]. Modulating sirtuin expression through diet, and possibly exercise, may be useful in combination with conventional cancer therapy. By combining chemotherapy with calorie restriction or exercise (described below), sirtuin expression may be elevated leading to a more normalized cellular metabolism and better control of the cancer, possibly increasing the efficacy of the drug therapy. Along these lines, efforts aimed at identifying calorie restriction mimetics, similar in action to resveratrol, have the overall goal of increasing the expression or activity of the sirtuins. However, resveratrol, other CR mimetics, and dietary manipulations are not selective for a particular sirtuin and may have undesired or off-target affects, and most importantly may not activate the sirtuins robustly. More investigation into the sirtuins in cancer is clearly needed, and better ways to selectively target individual sirtuins will be key moving forward.
7.5 One-carbon metabolism
The folate cycle in combination with the methionine cycle are collectively referred to as the one-carbon metabolism, since carbon units are circulated through multiple enzymatic reactions. The one-carbon cycle forms a critical component of the cellular metabolic network, which is linked to nearly all of the major biosynthetic pathways. The main sources that put carbon units into this cycle are serine and glycine biosynthetic pathways. Importantly, the fate of the carbons (i.e. outputs) of the one-carbon cycle consist of a variety of critical metabolic pathways; for example, some outputs include nucleotide metabolism [182], protein translation [183], lipid metabolism [184], methylation metabolism [121, 123, 185–187], protein sulfhydration, glutathione production. Therefore, the activity of the one-carbon cycle is important in regulating the biosynthesis of the building blocks of a cell as well as its epigenetic status. Furthermore, the redox state of the cell is regulated by this cycle at two levels: through the folate cycle by the function of tetrahydrofolate (THF), and also through the glutathione production via the transulfuration pathway that mediates the levels of ROS in cells. Both of these mechanisms serve to regulate the balance of NADPH/NADP+ levels in the cell [24]. Recently, a variety of cancers exhibiting the Warburg effect were shown to divert carbons from glycolytic metabolism into the one-carbon cycle, providing a link from the Warburg effect to the activity of one-carbon metabolism [188, 189].
Studies have shown that phosphoglycerate dehydrogenase (PHGDH) is hyperactivated in a subset of cancers, resulting in over-production of serine in these cancers [190, 191]. Furthermore, the gene encoding PHGDH was shown to have undergone significant copy number gain in some cancers and cancer cell lines, and these cell lines were dependent on the PHGDH amplification for rapid proliferation [192].
A second instance where one-carbon metabolism has shown correlations with cell transformation is the metabolism and generation of glycine. A metabolomics study on cancer cell lines reported a strong association between glycine uptake and catabolism with cancer cell proliferation [193]. Also another study found that ectopic expression of glycine dehydrogenase (GLDC, decarboxylating), phosphoserine aminotransferase (PSAT), and serine hydroxymethyltransferase (SHMT), all enzymes important in glycine uptake and catabolism, could induce cell growth in NIH 3T3 cells, thus conferring to these cells properties indicative of the hallmarks of cancer [194].
Additional levels at which the one-carbon cycle and its output pathways have been linked to cancer include nucleotide metabolism, epigenetic modifications, polyamine metabolism, and the regulation of the oxidative state of the cell. Multiple genes involved in nucleotide metabolism have been shown to be able to cause transformation by reducing the genomic integrity [195]. Also, through the transfer of methyl groups to proteins, DNA, and RNA, S-Adenosyl Methionine regulates protein activity as well as epigenetic marks on DNA and proteins in cells which are all broadly implicated in cancer.
Targeting one-carbon metabolism using folate antagonists has long been used as a major class of cancer therapeutic agents [196]. Historically, aminopterin was the first one of such drugs, and methotrexate and pemetrexed are still being used as common chemotherapeutic agents in a variety of cancers acting to inhibit di- and tetrahydrofolate reductase activities, thereby disrupting the one-carbon cycle. In addition to this class of compounds, another major group of chemotherapeutic agents linked to the one carbon cycle are inhibitors of nucleotide metabolism. These include 5-fluorouracil (5-FU) and gemcitabine [197].
Furthermore, several novel anti-cancer drugs that are currently being tested in clinical trials also target specific components of the one carbon cycle. These agents mostly target the epigenetic state of the cancer cells through inhibiting SAM or DNA methyltransferases or polyamine synthesis [198]. Difluromethylornithine (DFMO), methylglyoxal bis(guanylhydrazone) (MGBG), and an inhibitor of S-adenosylmethionine decarboxylase called SAM486A are examples of such agents.
One carbon metabolism consists of enzymes that are in principle druggable [199]. Therefore, several promising drug targets include PHGDH, PSAT, PSPH, GCAT, GLDC, and glycine N-methyltransferase (GNMT). A recent study has shown that reduction in serine and glycine intake in diet can significantly reduce cancer cell proliferation in mice, suggesting that one carbon metabolism can potentially be targeted through dietary adjustments [200]. Finally, due to the prevalence of anti-metabolite chemotherapeutic agents that are somehow involved with the one carbon cycle, expression levels of some of the components of this pathway could potentially be used as biomarkers for drug selection and outcome prediction in several types of cancers [201, 202].
7.6 Oncometabolites
IDH1 and IDH2 are frequently mutated in several cancer types, including glioma, acute myeloid leukemia, and chondrosarcoma [203–205], resulting in significant changes to the epigenetic landscape in cancers harboring these mutations [120, 141, 206, 207]. The products of wild type IDH1 and IDH2 enzymes, NADPH and α-KG, play broad functions in regulating cellular metabolism. The mutant IDH proteins lose their wild type function and instead convert α-KG into (R)-2-hydroxyglutarate [(R)-2HG], a structural analog of α-KG [208]. IDH mutant cancer cells can accumulate millimolar levels of this normally undetectable metabolite [208, 209]. Recently, mouse models with altered IDH2 activity have shown the capacity of IDH mutations to facilitate malignant tumor development and maintenance [210, 211].
(R)-2HG is thought to act by competitively inhibiting certain enzymes that require α-KG as a cofactor, including Jumonji-C domain histone demethylases (JHDMs) and TET proteins, which are implicated in DNA demethylation [212, 213]. IDH mutant tumors display a hypermethylation signature, associated with a block in cellular differentiation [120, 141, 212]. A similar hypermethylation signature is associated with IDH mutations and TET2 mutations, which occur in a mutually exclusive manner in AML, indicating that these mutations likely target the same pathway [141]. Moreover, either treatment with (R)-2HG or silencing of TET2 was sufficient to promote growth factor-independent growth of TF-1 leukemia cells [214]. However, the epigenetic alterations mediated by (R)-2HG may not fully explain its cancer promoting effects, since (S)-2HG, which is not produced by mutant IDH but serves as an even more potent inhibitor of TET2 than (R)-2HG, fails to transform TF-1 cells. One possible explanation is that (R)-2HG, but not (S)-2HG, can act as an agonist for EGLN prolyl hydroxylases, which promote hypoxia-inducible factor (HIF) degradation; hence regulation of HIF proteins may also be a part of the malignant tumor-promoting activity of (R)-2HG [214, 215]. Indeed, EGLN1 silencing impaired the ability of IDH1 R132H to transform TF-1 cells.
2HG has emerged as a useful diagnostic marker for patients with IDH mutant tumors. Patients with IDH mutant AML exhibit elevated serum 2HG levels, and those with the highest levels of 2HG had shorter overall survival [216]. Moreover, 2HG can be detected noninvasively by magnetic resonance spectroscopy in patients with IDH mutant glioma [217]. Substantial interest has also arisen in therapeutic targeting of mutant IDH enzymes, as a cancer-specific metabolic alteration. Inhibitors to mutant IDH1 and IDH2 were recently reported. For example, an IDH2/R140Q inhibitor promoted the differentiation of leukemia cells containing that mutation [218]. An IDH1/R132H inhibitor likewise promoted differentiation and reduced histone methylation in glioma cells. Growth of glioma xenografts was also impaired. Notably, the tumor growth inhibitory effects of the drug were observed at lower doses than were required to stimulate histone demethylation and differentiation, further suggesting that additional mechanisms may contribute to mutant IDH-mediated malignant tumor growth [219].
In addition to (R)-2HG, succinate and fumarate also have the potential to act as oncometabolites. In specific cancer types, loss-of-function fumarate hydratase (FH) and succinate dehydrogenase (SDH) mutations are observed, resulting in build-up of fumarate and succinate, respectively. Similar to 2HG, fumarate and succinate can competitively inhibit α-KG-dependent dioxygenases. As such, succinate has been known for several years to promote HIF1α stabilization via inhibition of prolyl hydroxylases [220]. More recently, it has been identified that succinate and fumarate also inhibit histone demethylases and TET proteins [221]. Both SDH mutant gastrointestinal stromal tumors and paraganglioma were shown to exhibit a hypermethylation phenotype [222, 223].
7.7 Lactate
As described above, lactate is made from pyruvate by the enzyme lactate dehydrogenase (LDH) during normal cellular metabolism (Figure 1). Cancer cells produce high levels of lactate, as described above. More recently, a new role for lactate has been described wherein some cancer cells use lactate to enable proliferation. For example, in a cancer microenvironment, excess lactate is secreted and contributes to an extracellular environment that promotes cancer progression [224]. Thus, lactate, which was previously considered a waste product of cancer cells, has now been identified as a key metabolite that plays a direct role in promoting cellular growth. The concept of the role of lactate as a signal or as a fuel in cancer is often called the “reverse Warburg effect” [225].
Lactate levels are governed by a number of factors, including differential expression of LDH isoforms, the lactate monocarboxylate transporter (MCT) levels, and oxidative capacity of tissues. LDH is the primary enzyme catalyzing lactate turnover, which interconverts pyruvate and lactate, with concomitant interconversion of NADH and NAD+, respectively. LDH is involved in the metabolism of both glucose and glutamine carbon, as well as in determining malignant tumor pH and the activity of the TCA cycle [226]. LDH is further classified into five different subtypes (LDH1-5), which structurally conform into homo- or hetero-tetramers of M protein subunits encoded by LDH isoform A (LDHA) and H protein subunits encoded by LDH isoform B (LDHB) genes. These subtypes vary based of tissue distribution. For example, in normal tissue, LDH1 has high expression in the brain, heart, and kidney; LDH5 is found in glycolytic tissues, such as liver or skeletal muscle [227].
Recently, LDHA was shown to be required for the maintenance and progression of many cancers [228–231], but the mechanisms by which LDHA facilitates cancer progression remain poorly understood. Indeed, targeting lactate metabolism is re-emerging as a therapeutic strategy [232]. Given the correlation between LDHA levels and poor outcomes, LDHA has attracted attention as a cancer target. Reducing LDHA levels (by shRNA) in cells leads to decreased proliferation and suppressed oncogenicity [228]. Furthermore, inhibition of LDHA pharmacologically (FX11) or by siRNA reduces ATP levels and results in cellular death [229]. A follow-up study examining the combination treatment of FX11 (LDHA inhibitor) and FK866 (an NAD+ synthesis inhibitor) resulted in lymphoma regression in a xenograft model [229]. Another study identified that LDHA plays a role in chemoresistance in breast cancer [233]. Specifically, paclitaxel-resistant breast cancer cells have high expression and activity of LDHA compared to paclitaxel-sensitive cells. Importantly, downregulation of LDHA restores chemosensitivity, indicating that lactate dehydrogenase, and more broadly lactate levels, play a key role in cancer drug resistance and may serve as a therapeutic target.
In addition to LDH, MCTs have come under increased scrutiny for their ability to import and export lactate. This family of transporters moves lactate and a proton down a concentration gradient, which determines the directionality of transport. Similar to LDH, different MCT isoforms are expressed in tissue types according to their oxidative capacity [234, 235]. For example, MCT1 is high in oxidative cell types like muscle sarcolemma [236], and increases with training. MCT4, on the other hand, is found in cells with high rates of glycolysis, such as fast-twitch muscle cells. Consistent with this idea, its expression is increased during hypoxia.
MCTs have also garnered interest as regulators of lactate metabolism. Indeed, inhibition of MCT1, either pharmacologically with α-cyano-4-hydroxycinnamate (CHC), or by RNA interference, shifted metabolism from lactate consumption to glucose consumption in normoxic cells. Interestingly, this also revealed a metabolic flexibility of cancer cells. Some studies have also tested MCT1 inhibition in vivo. In one mouse model of lung cancer and in a xenograft of human colorectal adenocarcinoma, CHC administration reduced hypoxia, induced tumor necrosis, and slowed overall growth. In another study, CHC was able to induce anti-tumoral and anti-angiogenic activity in gliomas, as well as to potentiate the effect of the alkylating agent temozolomide [237].
To conclude this section on emerging metabolic derangements contributing to cancer cell growth, we end with lactate – Warburg’s original observation. Collectively, these studies support a model where metabolites, like lactate, which facilitate malignant tumor development and/or survival, have the potential to be targeted therapeutically.
8. Therapeutic Strategies
Metabolism is a recent addition to the hallmarks of cancer, and the nature of these changes and their significance for the etiology of the disease are areas of intense investigation. As in other reviews of ‘hallmarks of cancer’ in this special issue, both molecular targets and small molecules were evaluated as lead candidates to influence different aspects of cancer metabolism. Each of the priority targets was outlined in the preceding sections and together are summarized in Table 1. These areas were chosen for their known or emerging mechanisms to contribute to cancer metabolism.
Next, a team of researchers consisting of specialists in each hallmark area performed an extensive literature search to determine if any studies had been performed addressing the effect of each molecular target on other hallmarks. The ‘Target Validation Team’ also looked for any possible reports of effects by the identified therapeutic approaches on other hallmarks. Given the interest in future research that will focus on combination approaches, this effort was primarily aimed at highlighting any evidence of contrary effects in other hallmark areas. However, any potential for complementarity or synergy were also considered as well.
Remarkably, many targets described above as having potential to manipulate different aspects of cancer metabolism have not been directly tested in the context of other hallmarks. In particular, compounds need to be tested for their influence on other hallmarks, independent of their effects on metabolism; this is the current major gap in knowledge. We primarily attribute this to the recent resurgence in metabolic studies, especially in the context of cancer. Indeed, less is known about if and how to effectively target cancer metabolism as a therapy than is known about targeting other hallmarks of cancer. Thus, our analysis at this time shows that prioritized targets and lead compounds that might affect these targets need further investigation. With more research, a broad-spectrum approach to target metabolism and to synergize with other hallmarks of cancer could be achieved.
In the interim, based on current knowledge, we consider from a drug development perspective the unique environment of mitochondria, which needs to be considered to target mitochondrial function, and ultimately manipulate metabolism. Because of the challenges associated with targeting mitochondria, we also take a step back from the cellular and biochemical mechanisms of metabolism and discuss how alterations in host metabolism could more broadly manipulate cancer metabolism in an efficacious way. Together, considering both cancer (direct) and host (indirect) metabolism could uncover novel therapeutic strategies or information that could be leveraged to target metabolism.
8.1 Strategies To Target Mitochondria In Cancer
Traditionally, mitochondria were not thought of as a ‘druggable target’ due to significant challenges unique to mitochondria, including selective delivery to this organelle, specific mitochondrial uptake within a targeted organ, and how to enhance the selective delivery to mitochondria within cancer cells; however, recent work is changing this view [238, 239]. In part, this is due to the growing realization that mitochondrial function contributes to multiple cellular functions, beyond energy metabolism, and these aspects could be targeted. The development of strategies to target different aspects of mitochondrial biology has led to the emergence of a new field called mitochondrial pharmacology [238, 239].
In mitochondrial pharmacology, development of drugs designed to affect mitochondria require the following considerations. Foremost, mitochondria originate from the coordination of two genomes, with 37 genes in the mitochondrial DNA and approximately 1,500 proteins being imported into the mitochondria following their transcription and translation from nuclear genes [240]. The expression of the two genomes is coordinated by a number of nuclear transcription factors [241]. Targeting nuclear- versus mitochondrial-derived proteins could be important. Furthermore, the mitochondria themselves continually join and separate through fission/fusion processes that are linked to the turnover and degradation of damaged, defective or surplus mitochondria [242]. Thus, one way for drugs to alter mitochondrial function is through interaction with the cellular machinery that controls overall mitochondrial biogenesis. For example, drugs such as 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) that interact with the AMP-activated protein kinase (AMPK) [243] may lead to upregulation of global mitochondrial biogenesis. Furthermore, agonists for PGC1α, a transcriptional co-activator that regulates gene expression of nuclear encoded mitochondrial proteins may do the same [244]. Such strategies may globally alter mitochondrial nutrient oxidation or function in therapeutically useful ways in cancer.
More direct strategies have been demonstrated by using drugs that act on particular targets within mitochondria. For example, dichloroacetate has been explored as an anticancer agent by blocking the activity of the inhibitory kinase that inactivates pyruvate dehydrogenase, thereby enhancing mitochondrial pyruvate oxidation [245]. Generally, in these examples, a drug acts on an aspect of metabolism that is mitochondrial and the selectivity comes from the interaction of the drug with its target, which happens to be in the mitochondrion. However, emerging methods are available to target compounds to mitochondria [238], and these may have a number of advantages. In particular, the concentration of a compound within mitochondria greatly enhances the potency of the therapy, meaning far less of a compound might be required. The localization of the compound within the mitochondria also minimizes metabolism and toxicity associated with reactions elsewhere in the body.
An important attribute in designing targeted therapies for mitochondria is the distinct mitochondrial compartments, with dramatically different properties. Most of energy metabolism occurs in the matrix and on the mitochondrial inner membrane. The intermembrane space has limited metabolic activity, although it is important in protein import; this compartment contains many of the proteins that are released into the cytoplasm during activation of apoptotic pathways. The mitochondrial outer membrane is also a key component in the regulation of apoptosis. Furthermore, many of the complexes involved in the propagation and stabilization of inflammatory pathways are assembled on the cytoplasmic-facing surface of the mitochondrial outer membrane [246].
Overall, the role of mitochondria in cancer is becoming better understood, suggesting that therapies targeted to the organelle may be useful [247, 248]. Drugs could be designed to affect mitochondrial function directly by binding to targets within mitochondria, or indirectly by altering mitochondrial biogenesis. Recent developments have also led to robust strategies to target bioactive compounds to mitochondria. These approaches may selectively modify mitochondrial metabolism in cancer cells to generate therapeutically beneficial outcomes, such as enhancing the selective killing or inhibiting growth of cancer cells. While many limitations and uncertainties with these approaches remain, modifying mitochondrial function could become a therapeutically useful extension of current cancer treatments.
8.2 Strategies To Target Metabolic Interactions Between Cancer And Host
Cancer metabolism might be manipulated specifically through host metabolism. This rationale is based on the concept that pharmacological mimetics of weight loss, calorie restriction, and/or targeting the associated hormonal systems and intracellular signaling pathways are emerging pathways for novel cancer therapeutics. Indeed, one of the possible mechanisms underlying known efficacy of fasting, CR, exercise, or weight loss on cancer could be multi-faceted, synergistic effects of these “treatments”. Thus, manipulating metabolism in these ways could be a successful therapeutic strategy. Additionally, further studying physiological approaches that influence a broad-spectrum of targets and/or hallmarks could inform more effective design of pharmacological approaches aimed at a broad-spectrum of targets.
8.3 Calorie Restriction, Fasting, and Cancer
Calorie restriction (CR), usually referring to a 20%–40% reduction in the daily intake of calories without malnutrition has been shown to extend lifespan and healthspan in multiple species, in part by delaying or preventing the occurrence of many age-related disorders including cancer [249–251]. CR has been also demonstrated to reduce the frequency of spontaneous and inducible tumors in mice and primates [252–257]. However, CR induces significant weight loss, which has limited its efficacy and has prevented its wide-spread clinical use. In some studies, CR delayed but did not stop tumor growth progression, and some tumors including those carrying a mutation in oncogenes such as PI3K and phosphatase and tensin homolog (PTEN) have been shown to be resistant to CR [258]. In addition, CR would be predicted to be incompatible with the treatment of most cancer patients including those malnourished or at risk for severe weight loss, or affected by immunosuppression or cachexia [259]. In fact, long-term CR can impair immune functions and delay wound healing repair [260, 261]. Thus, while CR might not be compatible with cancer therapy in humans, several lessons could be learned from the molecular pathways activated by CR and/or CR mimetics.
More recently, periodic fasting has emerged as a viable strategy to protect normal cells from toxicity associated with chemotherapy, while sensitizing cancer cells to treatment. Remarkably, fasting protects normal cells from oxidants, common chemotherapeutic agents, and radiotherapy in several in vitro and in vivo models [262–264]. Specifically, glucose and serum starvation in vitro and a 48–72 hour food deprivation (short-term starvation or STS) in mice activated several stress-response pathways that conferred resistance to oxidative stress induced by chemotherapy. The latter effect was termed “differential stress resistance” (DSR) [265], and is based on the concept that normal cells can adapt to stressors but not cancer cells [266]. The mechanisms responsible of starvation-induced DSR in mammals include reduced circulating insulin-like growth factor 1 (IGF-1) levels and possible down-regulation of growth pathways [265]. In addition, in normal mammalian cells, starvation has been demonstrated to induce AMPK which stabilized p53 and p21 thereby resulting in cell proliferation arrest [263].
In model organisms, including bacteria and yeast, the switch from nutrient rich media to water or buffer extends longevity and also increases resistance to a variety of toxins, including ROS [262, 267, 268]. In yeast cells, the molecular mechanisms responsible for fasting-mediated protection involve the reduced activity of components of the glucose-response Ras/cAMP/PKA pathway and of the amino acids response Tor/S6K pathway [267, 269, 270]. Similarly, fasting increases resistance to oxidative stress and prolongs life-span in model organisms, such as worms and flies, by down-regulating analogous nutrient-sensing pathways such as (PI3K/AKT/TOR) [271, 272].
Cancer cells are characterized by the presence of oncogenic mutations in the same growth signalling pathways including IGF1 receptor, PI3K, PTEN, Ras, and PKA that prevent the activation of protective systems in response to starvation (DSR) [1]. In addition to the absence of a stress resistant response to starvation, the set of mutations and chromosomal rearrangements prevent malignant cells from being able to adapt to a wide variety of conditions but particularly to those that involve broad and extreme changes such as starvation. Thus, “differential stress sensitization” (DSS) takes advantage of the fact that most mutations reduce the ability of cells to withstand changing environments. In one study, 48 to 60 hours of fasting retarded malignant tumor growth, similar to chemotherapy, and acted in synergy with chemotherapeutic drugs in killing a wide variety of cancer cells [265]. In mouse models for breast cancer, neuroblastoma, mesothelioma, and lung carcinoma, the combination of fasting and chemotherapy promoted 20–60% cancer free survival, whereas neither one of them alone resulted in any cancer free survival [263, 265]. Investigation of the molecular mechanisms underlying the STS-mediated biological effects indicate that breast cancer cells attempt to compensate for the nutrient deficiency caused by fasting by hyperactivating the AKT and mTOR/S6K pathways, increasing translation and, as a result, consuming even more energy and causing oxidation-dependent cell death [265]. A recent study in lung cancer cells, instead showed that starvation activated ATM/Chk2/p53 which sensitized cells to cisplatin treatment [263]. Many different mechanisms are likely responsible for the effects of fasting in the sensitization of different types of cancer cells. However, some of these mechanisms could to be shared by all cancer cell types.
The side effects caused by chemotherapeutic toxicity to normal cells and tissues are a major limitation of these agents. Consequently, these toxicities can limit chemotherapeutic dose intensity and compromise its overall efficacy. The need to maintain chemotherapeutic doses within this treatment range could be contributing to the survival of a sufficient number of cancer cells and lead to recurrence. Thus, reducing toxicity of chemotherapeutics by protection of normal cells and tissues, in combination with sensitizing of cancer cells, during fasting represents a promising strategy to enhance current treatments.
The safety of this intervention has been demonstrated in studies of a large cohort of patients affected by rheumatoid arthritis, who did not reveal any major adverse effect and presented clinical improvement after undergoing prolonged fasting [273]. Similarly, hypertensive patients, who fasted with only water for 10–11 days, showed normalization of systolic blood pressure values and no major side effects [274]. These studies provide a proof-of-principle for fasting in human studies.
The first clinical test of fasting in patients during chemotherapy was reported in a study where 10 patients affected by different malignancies voluntarily fasted 48–140 hours prior to and during chemotherapy. This study reported that fasting may reduce the side-effects induced by chemotherapeutic toxicity in humans [275]. Notably, these patients receiving different chemotherapeutic drugs in combination with fasting reported hunger and light-headedness as significant side-effects, and reported fewer toxicities associated with chemotherapy, including weakness, fatigue, and gastrointestinal distress [275]. Importantly, fasting did not appear to interfere with chemotherapeutic efficacy [275].
The combination of fasting with chemotherapy has gained clinical interest as a therapeutic approach for cancer patients [276]. Several clinical trials are now exploring fasting as a way to reduce the toxicity of chemotherapy and increase its efficacy in humans (e.g. ClinicalTrials.gov numbers, NCT00757094, NCT00936364, NCT01175837, NCT01304251). Taken together, fasting is a promising therapeutic strategy for cancer patients, with the potential to be useful in combination with several chemotherapeutic drugs and across many cancer types. Future studies will determine whether fasting may potentiate the efficacy of non-toxic cancer therapies, including antibody- or immune-based strategies.
8.4 Exercise and Cancer
Growing evidence shows that chronic exercise reduces the risk of certain forms of malignancies (i.e, prevention) compared with sedentary lifestyles [277, 278]. In addition, emerging evidence indicates that chronic exercise may also be inversely correlated with cancer-recurrence as well as cancer-specific mortality (i.e., prognosis) [279, 280]. While data are not currently available from randomized trials testing the role of exercise in either prevention or prognosis, the combination of the unique metabolic features of malignant tumor cells and exercise-induced perturbations in whole-body and intracellular metabolic substrate use suggests that exercise might directly influence cancer prognosis and survival [280, 281].
Investigation into the effects of metabolic perturbations in a chemically induced rat model breast cancer showed that both CR and voluntary wheel running reduced tumor incidence and burden [282]. This reduction occurred in conjunction with reductions in circulating plasma levels of metabolic factors such as IGF-1, insulin, leptin, and increased plasma levels of IGF-binding protein 3 (IGFBP-3) and adiponectin. Further studies using the same breast cancer model showed that CR combined with voluntary wheel running inhibited tumor growth, and was associated with downregulation of mTOR-related targets including pAkt [283]. Plasma insulin and leptin were strongly correlated with intratumoral pAkt levels. In contrast, another study found tumor growth was similar between sedentary control groups and voluntary wheel running in animals orthotopically implanted with human breast cancer or prostate cancer [284, 285]. More work needs to be done in this area to determine if and how exercise effects tumor growth in these models.
Several studies in humans have investigated the effects of structured exercise training interventions on changes in metabolic parameters in subjects at-risk or with a confirmed diagnosis of cancer. For example, the Physical Activity for Total Health (PATH) Study found that structured moderate-intensity exercise training had no effect on circulating levels of IGF-1, IGFBP-3, or the ratio [286]. However, in these at-risk patients, training was associated with improvements in insulin and leptin [287]. In patients diagnosed with cancer, several studies have found that exercise training is associated with favorable changes in circulating levels of these same metabolic growth factors. For example, moderate intensity aerobic training (defined as 60–75% of baseline VO2peak, 3 times per week, for 30–45 minutes per session, over 15 weeks) lead to alterations in IGF-1, IGFBP-3, and the IGF-1:IGFBP-3 ratio compared to a sedentary controls [288]. This is consistent with larger studies on aerobic exercise and resistance training in patients with breast cancer [289–291]. In another study, sedentary early-stage breast cancer patients were randomly assigned to a home-based aerobic and resistance exercise program for 16 weeks or a control group [292]. Another study using aerobic exercise found a trend toward decreased fasting insulin [293].
Indeed, the potential of exercise to alter host–tumor metabolic interactions may not only cause a shift towards a less aggressive phenotype but could also alter sensitivity to anticancer therapies. Based on current knowledge, it is difficult to predict the complex and multifaceted interaction between the host, exercise, tumor, and antineoplastic index of current therapeutics. However, exercise may affect tumor cell sensitivity in at least two ways. First, solid tumors have an abnormal vascular system that impairs effective oxygen and drug transport [294, 295], which is associated with therapeutic resistance [294, 296]. In contrast, exercise leads to favorable vascular adaptations in preclinical models of cardiac and hindlimb ischemia, as well as improvements in peripheral vascular function [297–301]. Similarly, tumors from exercised mice had significantly increased intratumoral perfusion and vascularization (i.e. physiologic angiogenesis) in orthotopic models of breast and prostate cancer [284, 285]. One study recently reported that the combination of exercise (voluntary wheel running) and chemotherapy (cyclophosphamide) was associated with significantly prolonged tumor growth delay compared with chemotherapy alone in a mouse model of murine breast cancer. Although the molecular mechanisms remain to be elucidated, hypoxia is intricately linked with tumor metabolism and exercise may effect vascularization and chemosensitivity. Second, as described above, short-term calorie restriction and/or fasting induces a stress response adaptation in normal cells that provides protection against oxidative injury from chemotherapy [262, 265]. Importantly, cancer cells have an impaired ability to respond to CR-induced stress, and may increase the therapeutic index. Exercise is also a potent form of both host and intracellular stress that may also confer similar effects to caloric restriction.
The data from these early studies, coupled with the powerful influence of exercise on metabolic homeostasis, suggest that more research in this area is needed. Specifically, investigations that adopt a translational approach are required to determine how exercise influences metabolic milieu in conjunction with biomarkers of tumor metabolic phenotypes using tissue and imaging modalities. Overall, careful elucidation of the molecular mechanisms by which exercise influences cancer metabolism could lead to rational pharmacologic agents to optimize clinical outcomes. Indeed, several lessons can be learned from interactions between cancer and fasting, cancer and CR, as well as cancer and exercise. Such research has the potential to provide new therapeutic opportunities.
8.5 Obesity and Cancer
At the other end of the metabolic spectrum, the obese phenotype is characterized by profound metabolic dysregulation, and can especially disrupt the endocrine system [302–305]. The 2007 World Cancer Research Fund/American Institute for Cancer Research (WCRF/AICR) report and subsequent epidemiological studies show that obesity is associated with a number of different cancers, including colorectal, kidney, liver/gall bladder, pancreatic, esophageal, stomach, prostate, postmenopausal breast, endometrial, uterine and ovarian cancers [306]. Factors associated with weight gain and obesity, excess calorie intake/or low energy expenditure, were concluded to increase obesity-related cancers, while those protecting against weight gain were also protected against developing cancers [306]. Recent statistical modeling of data from the American Cancer Society cohort, which provided early evidence of obesity-cancer links [307], estimated that 20% of cancer deaths are attributable to being overweight or obese [308]. Consequently, obesity and obesity-associated morbidities, such as diabetes, have been intensively investigated to determine the signaling pathways involved that lead to increased carcinogenesis [309–311] and if anti-obesogenic therapies are effective for cancer treatment and/or prevention.
Several manifestations of the obese phenotype overlap with cancer phenotypes and provide insights into approaches for metabolic modulation and possible strategies for cancer therapy. In particular, both cancer and obesity are associated with activation of cellular signaling pathways. Therefore, strategies to prevent synthesis or secretion of the signaling molecules dysregulated by obesity, or alternatively hindering their interaction with target receptors, may be a useful strategy for cancer preventative therapies. For example, therapeutic targeting of the intracellular pathways, such as JAK/STAT and PI3K/Akt/mTOR, which are often activated in both cancer and obesity, presents opportunities to intervene in carcinogenesis. Additionally, cancer and obesity activate anabolic processes, and small molecules that disrupt macromolecule biosynthesis, such as with metformin and orlistat, two drugs that lower glucose and inhibit cellular FA synthesis, respectively, might have the ability to restore homeostatic control and cellular defense systems with potential to impact cancer cells within the body [312, 313]. Indeed, metformin has received significant attention recently as a novel chemotherapeutic; for a recent review, see [314]. Finally, weight loss and reduced calorie consumption have been demonstrated to normalize altered adipokine profiles, reduce oxidative stress, and lower growth factors linking obesity and cancer, suggesting that synergistic manipulation of these pathways along with metabolism holds therapeutic promise.
In conclusion, most targets and/or compounds identified above as having potential to manipulate different aspects of cancer metabolism have not been directly tested in the context of other hallmarks. Thus, our analysis shows that more work needs to be done on cancer metabolism to identify targets or compounds that could have synergistic effects with other hallmarks against cancer. However, the emerging efficacy of physiological strategies to manipulate cancer outcomes, such as fasting/CR and exercise, and weight loss could be attributed, in part, to their ability to influence cancer metabolism as well as several other hallmarks of cancer. Future studies in this area will determine if and how dysregulated metabolism in cancer can be targeted directly or indirectly through host metabolism.
9. Tradeoffs
The recognition that cancer cells have altered metabolic pathways from their normal counterparts has opened a new opportunity to treat cancer [315]. Identification of altered metabolic enzymes and regulation of metabolism through oncogenic signals may render cancer cells highly dependent on specific pathways and metabolic programs. Mutations in some enzymes can force alternate metabolic pathways or oncogenes can drive specific metabolic programs while limiting alternatives. Targeting these pathways, therefore, may directly lead to cancer cell death or may provide opportunities to generate synthetic lethality. The challenge in targeting cancer metabolism, however, is that even if cancer cells are highly dependent on specific metabolic pathways, normal cells use these same pathways. To what extent cancer cells and normal cells differentially rely on specific metabolic pathways will be crucial to determine the therapeutic window of any treatment [126].
A key benefit of aerobic glycolysis is the generation of metabolic intermediates for biosynthesis and cell growth [24]. Of course, normal cells also undergo rapid cell growth in development and upon stimulation. It is now evident that oncogenic signals can mimic normal growth factor signaling mechanisms. That these signals are sufficient to directly promote aerobic glycolysis has implied that normal cells also utilize these same metabolic pathways. Indeed lymphocytes transition from a quiescent metabolic state that is characterized by oxidative metabolism to one that is highly glycolytic upon stimulation with antigen in appropriate inflammatory conditions. Key pathways to drive this transition include the PI3K/Akt/mTORC1 and cMyc pathways—each of which are well established to drive aerobic glycolysis in cancer, as described above. The purpose of this glycolytic phenotype in lymphocytes is, as in cancer cells, to drive cellular growth [316]. In addition, however, aerobic glycolysis may also have a signaling role to promote expression of inflammatory cytokines.
Interestingly, not all activated lymphocytes use aerobic glycolysis. Depending on the cytokine environment, some CD4 lymphocytes develop into inflammatory effectors while others develop into suppressive regulatory T cells (Treg). The signaling pathways that drive these different CD4 T cell fates are also key metabolic regulators, and mTORC1 is essential to develop effector T cells but represses Treg [317]. Accordingly, the metabolism of these subsets differs. Effector T cells, like cancer cells with activated mTORC1, are highly glycolytic [318, 319]. Treg, however, have low mTORC1 and instead have high levels of activity in the tumor-suppressive AMPK pathway, a key inhibitor of mTORC1 and driver of oxidative metabolic pathways. Rather than favor aerobic glycolysis, therefore, Treg utilize lipid oxidation as a primary metabolic mechanism [318]. Similarly, memory T cells that return to quiescence following an immune response revert from the glycolytic phenotype of effector T cells to a lipid oxidative metabolism [320]. Suppressing lymphocyte glycolysis and promoting oxidative metabolism, therefore, inhibits the primary effector response but can enhance the generation and survival of memory T cells [321].
The metabolic implications of these distinct metabolic transitions of cells moving from quiescence to proliferation are manifold and go well beyond fundamental aspects of immunology. Firstly, lymphocytes are not unique in these transitions. Rather, it appears to be a general feature of the transition from quiescence to proliferation that cells undergo a metabolic reprogramming. Endothelial cells also undergo metabolic reprogramming [322], as do smooth muscle cells [323] and a variety of other cell types. Second, lymphocytes in the tumor environment may have difficulty competing for nutrients such as glucose. This nutrient limitation, in turn, would suppress effector lymphocytes that may play roles in anti-tumor immunity while at the same time promoting the development and activity of Treg that can maintain malignant tumors. Thirdly, efforts to target metabolism in cancer metabolism-directed therapies may have inadvertent effects to suppress normal cell proliferation. In the case of the immune system, blocking aerobic glycolysis can suppress the immune system [319, 324]. While this strategy could potentially be helpful to relieve inflammatory diseases, targeting cancer metabolism could have a tradeoff of immune suppression. In this setting, immune suppression may increase susceptibility to infection, and inhibit the potential benefit of an anti-tumor immune response.
A key question now is not only to discover how cancer metabolism itself is programmed and functions to advance cancer, but also to understand the metabolism of normal proliferating cells. The similarities may be useful to exploit new developments in cancer metabolism to target cells in other settings. In particular, anti-inflammatory treatments may become evident from cancer metabolism drugs. Differences in the metabolic programs of normal and cancer cells will also be highly useful, as these distinctions will provide directions to target cancer metabolism, while sparing other highly proliferative cells. In some cancers, mutations will make it clear how to selectively target cancer metabolism. For most cases, however, it will be important to target cancer metabolism, while maintaining metabolic pathways regulating normal cellular function. This ongoing challenge will be critical for establishing the therapeutic window for cancer metabolism treatments as well as opening new opportunities to understand normal biology.
10. Conclusion and Future Perspectives
The energy requirements of cancer cells vary greatly from those of quiescent, terminally differentiated cells. Mammalian cells require external cues from growth factors to take up nutrients from their surroundings and in the absence of these signals they are programmed to die by apoptosis [325]. Cells must compete for limiting amounts of growth factors that direct nutrient uptake in order to survive. Under these conditions it is advantageous to maximize energy production from nutrients such as glucose, fatty acids and amino acids. This is accomplished by complete oxidation of nutrients to carbon dioxide (CO2) and efficient coupling of ATP production though oxidative phosphorylation by the electron transport chain.
In contrast, cancer cells are characterized by growth factor-independent nutrient uptake, apoptosis evasion, and uncontrolled growth and proliferation [1]. The metabolic profile of cancer cells reflect these changes with increased nutrient uptake supplying the raw materials to synthesize the lipids, nucleotides, and proteins necessary for cell growth and proliferation [326]. The metabolic reprogramming which occurs during oncogenesis is similar to that which occurs in other cells transitioning from a quiescent to proliferative state such as induced pluripotent stem cells, embryonic stem cells and immune cells [327, 328]. This process involves a dramatic reworking of intermediary metabolism from that of a catabolic to an anabolic state. A key feature of this transition is a shift away from oxidative metabolism towards glycolytic metabolism, the net effect of which is to provide additional biosynthetic precursors for macromolecule synthesis [24]. Increased glycolytic intermediates are used to generate nucleosides, amino acids and reducing equivalents in the form of NADPH. Additionally, while levels of oxidative metabolism may be reduced, functional mitochondria are essential to cancer cell survival. In cancer cells, mitochondria function not only in energy production but also in modulation of redox status, generation of ROS, maintenance of calcium homeostasis, inhibition of apoptosis and contribute biosynthetic precursors to fuel macromolecule synthesis [247].
An important question that remains about the metabolic reprogramming which occurs in cancer is whether it is a driving force or a secondary effect of oncogenic mutations. Until recently, it was believed that metabolic reprogramming occurred secondarily to mutations in oncogenes or tumor suppressors that activate proliferation and survival signals [1]. However, a number of recent findings highlight the importance of metabolic reprogramming in driving oncogenesis (see Oncometabolites section), which provide strong evidence that altered metabolism is a key event that is selected for early in the oncogenic process.
Metabolic reprogramming in cancer cells is characterized by altered expression, mutation, post-translational modification, and/or changes in enzyme isoform expression. Regardless of the stage of oncogenesis during which metabolic reprogramming occurs, these unique changes could be specifically targeted to treat cancer. For these strategies to be useful, the metabolic phenotypes of several cancer types will have to be determined; techniques to do so are being developed and becoming more common [329]. Once the metabolic phenotype of a cancer is known, therapies can indirectly target upstream regulatory pathways, or directly target aberrant biosynthetic, anabolic pathways. Importantly, many cancers become resistant to radiotherapies and chemotherapy because of their altered metabolism [330]. Thus, a targeted therapy that serves to revert cancer cell metabolism back to a more catabolic state may re-sensitize a tumor to other therapies.
Much additional work is required to make targeted metabolic cancer therapies a reality. Several recent analyses, ranging from biochemical to omic-levels, have greatly increased our understanding of cancer metabolism. However, these techniques are sometimes limited to use in cell culture systems that do not account for contributions from the tumor microenvironment, the immune system, or all hallmarks of cancer. Thus, future work will be directed at measured alterations in metabolism directly in the context of other hallmarks. Together, targeting metabolism specifically or synergistically holds the potential for effectively treating a variety of cancers.
Acknowledgments
We would like to thank Leroy Lowe for conceptualization of the Halifax project, the unnamed reviewers of this manuscript for their suggestions, and colleagues in the field who drive the science described in this review. We apologize to those whose work could not be included due to space constraints. Finally, we acknowledge all the co-authors of this review who were part of the Target Validation Team, including (in alphabetical order): Amedeo Amedei, PhD, University of Florence, Italy; Amr Amin, PhD, UAE University, United Arab Emirates; S. Salman Ashraf, PhD, UAE University, United Arab Emirates; Asfar S. Azmi, PhD, Wayne State University, Karmanos Cancer Institute, United States; Dipita Bhakta, M.Sc.,PhD., VIT University (Vellore Institute of Technology), India; Alan Bisland, University of Glasgow, Glasgow, UK; Chandra S. Boosani, PhD, Creighton University, United States; Sophie Chen, PhD, Ovarian and Prostate Cancer Research Trust Laboratory, United Kingdom; Hiromasa Fujii, MD.,PhD., Nara Medical University, Japan; Alexandros Georgakilas, PhD, National Technical University of Athens, Greece; Gunjan Guha, M.Sc.,PhD, Assistant Professor, SASTRA University, India; Dorota Halicka, MD,PhD, New York Medical College, United States; Bill Helferich, PhD, University of Illinois at Urbana Champaign, United States; Kanya Honoki, MD,PhD, Nara Medical University, Japan; W.N. Keith, University of Glasgow, Glasgow, UK; Sulma Mohammed, DVM,PhD, Purdue University Cancer for Cancer Research, United States; Elena Niccolai, University of Florence, Italy; Somaira Nowsheen, Mayo Graduate School, Mayo Clinic College of Medicine, Rochester, Minnesota, United States; Xujuan Yang, University of Illinois at Urbana Champaign, United States.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hanahan D, Weinberg Robert A. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ringash J, Au HJ, Siu LL, Shapiro JD, Jonker DJ, Zalcberg JR, et al. Quality of life in patients with K-RAS wild-type colorectal cancer: the CO.20 phase 3 randomized trial. Cancer. 2014;120:181–9. doi: 10.1002/cncr.28410. [DOI] [PubMed] [Google Scholar]
- 4.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. The Journal of general physiology. 1927;8:519–30. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warburg O, Posener K, Negelein E. Ueber den Stoffwechsel der Tumoren. Biochem Z. 1924:319–44. [Google Scholar]
- 6.Pasteur L. Experiences et vues nouvelles sur la nature des fermentations. Comp Rend Acad Sci. 1861;52:1260–4. [Google Scholar]
- 7.Keilin D, Keilin J. The History of Cell Respiration and Cytochrome. Cambridge University Press; 1966. [Google Scholar]
- 8.Warburg O. On the origin of cancer cells. Science. 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 9.Bensinger SJ, Christofk HR. New aspects of the Warburg effect in cancer cell biology. Semin Cell Dev Biol. 2012;23:352–61. doi: 10.1016/j.semcdb.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Weinhouse S. Oxidative Metabolism of Neoplastic Tissues. In: Jeese PG, Alexander H, editors. Advances in Cancer Research. Academic Press; 1955. pp. 269–325. [DOI] [PubMed] [Google Scholar]
- 11.Weinhouse S. On respiratory impairment in cancer cells. Science. 1956;124:267–9. doi: 10.1126/science.124.3215.267. [DOI] [PubMed] [Google Scholar]
- 12.Racker E. Bioenergetics and the problem of tumor growth. American scientist. 1972;60:56–63. [PubMed] [Google Scholar]
- 13.Pedersen PL. Tumor mitochondria and the bioenergetics of cancer cells. Progress in experimental tumor research. 1978;22:190–274. doi: 10.1159/000401202. [DOI] [PubMed] [Google Scholar]
- 14.Thompson CB. Metabolic enzymes as oncogenes or tumor suppressors. The New England journal of medicine. 2009;360:813–5. doi: 10.1056/NEJMe0810213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seyfried TN, Flores RE, Poff AM, D’Agostino DP. Cancer as a metabolic disease: implications for novel therapeutics. Carcinogenesis. 2014;35:515–27. doi: 10.1093/carcin/bgt480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–9. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 17.Wieman HL, Wofford JA, Rathmell JC. Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and trafficking. Molecular biology of the cell. 2007;18:1437–46. doi: 10.1091/mbc.E06-07-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottlieb E, Tomlinson IP. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat Rev Cancer. 2005;5:857–66. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- 19.Rathmell JC, Fox CJ, Plas DR, Hammerman PS, Cinalli RM, Thompson CB. Akt-directed glucose metabolism can prevent Bax conformation change and promote growth factor-independent survival. Molecular and cellular biology. 2003;23:7315–28. doi: 10.1128/MCB.23.20.7315-7328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deprez J, Vertommen D, Alessi DR, Hue L, Rider MH. Phosphorylation and activation of heart 6-phosphofructo-2-kinase by protein kinase B and other protein kinases of the insulin signaling cascades. J Biol Chem. 1997;272:17269–75. doi: 10.1074/jbc.272.28.17269. [DOI] [PubMed] [Google Scholar]
- 21.Efeyan A, Serrano M. p53: guardian of the genome and policeman of the oncogenes. Cell cycle. 2007;6:1006–10. doi: 10.4161/cc.6.9.4211. [DOI] [PubMed] [Google Scholar]
- 22.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–3. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 23.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–20. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 24.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 26.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–50. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Portais JC, Voisin P, Merle M, Canioni P. Glucose and glutamine metabolism in C6 glioma cells studied by carbon 13 NMR. Biochimie. 1996;78:155–64. doi: 10.1016/0300-9084(96)89500-9. [DOI] [PubMed] [Google Scholar]
- 28.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–7. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. The Journal of cell biology. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–7. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamphorst JJ, Cross JR, Fan J, de Stanchina E, Mathew R, White EP, et al. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci U S A. 2013;110:8882–7. doi: 10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bustamante E, Morris HP, Pedersen PL. Energy metabolism of tumor cells. Requirement for a form of hexokinase with a propensity for mitochondrial binding. J Biol Chem. 1981;256:8699–704. [PubMed] [Google Scholar]
- 33.Bustamante E, Pedersen PL. High aerobic glycolysis of rat hepatoma cells in culture: role of mitochondrial hexokinase. Proc Natl Acad Sci U S A. 1977;74:3735–9. doi: 10.1073/pnas.74.9.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakashima RA, Paggi MG, Scott LJ, Pedersen PL. Purification and characterization of a bindable form of mitochondrial bound hexokinase from the highly glycolytic AS-30D rat hepatoma cell line. Cancer Res. 1988;48:913–9. [PubMed] [Google Scholar]
- 35.Nakashima RA, Mangan PS, Colombini M, Pedersen PL. Hexokinase receptor complex in hepatoma mitochondria: evidence from N,N′-dicyclohexylcarbodiimide-labeling studies for the involvement of the pore-forming protein VDAC. Biochemistry. 1986;25:1015–21. doi: 10.1021/bi00353a010. [DOI] [PubMed] [Google Scholar]
- 36.Robey RB, Hay N. Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene. 2006;25:4683–96. doi: 10.1038/sj.onc.1209595. [DOI] [PubMed] [Google Scholar]
- 37.Pastorino JG, Hoek JB. Hexokinase II: the integration of energy metabolism and control of apoptosis. Curr Med Chem. 2003;10:1535–51. doi: 10.2174/0929867033457269. [DOI] [PubMed] [Google Scholar]
- 38.Ko YH, Pedersen PL, Geschwind JF. Glucose catabolism in the rabbit VX2 tumor model for liver cancer: characterization and targeting hexokinase. Cancer Lett. 2001;173:83–91. doi: 10.1016/s0304-3835(01)00667-x. [DOI] [PubMed] [Google Scholar]
- 39.Ko YH, Smith BL, Wang Y, Pomper MG, Rini DA, Torbenson MS, et al. Advanced cancers: eradication in all cases using 3-bromopyruvate therapy to deplete ATP. Biochem Biophys Res Commun. 2004;324:269–75. doi: 10.1016/j.bbrc.2004.09.047. [DOI] [PubMed] [Google Scholar]
- 40.Pedersen PL. 3-Bromopyruvate (3BP) a fast acting, promising, powerful, specific, and effective “small molecule” anti-cancer agent taken from labside to bedside: introduction to a special issue. Journal of bioenergetics and biomembranes. 2012;44:1–6. doi: 10.1007/s10863-012-9425-4. [DOI] [PubMed] [Google Scholar]
- 41.Ko YH, Verhoeven HA, Lee MJ, Corbin DJ, Vogl TJ, Pedersen PL. A translational study “case report” on the small molecule “energy blocker” 3-bromopyruvate (3BP) as a potent anticancer agent: from bench side to bedside. Journal of bioenergetics and biomembranes. 2012;44:163–70. doi: 10.1007/s10863-012-9417-4. [DOI] [PubMed] [Google Scholar]
- 42.Weber G, Lea MA. The molecular correlation concept of neoplasia. Adv Enzyme Regul. 1966;4:115–45. doi: 10.1016/0065-2571(66)90011-2. [DOI] [PubMed] [Google Scholar]
- 43.Van Schaftingen E, Jett MF, Hue L, Hers HG. Control of liver 6-phosphofructokinase by fructose 2,6-bisphosphate and other effectors. Proc Natl Acad Sci U S A. 1981;78:3483–6. doi: 10.1073/pnas.78.6.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chesney J, Mitchell R, Benigni F, Bacher M, Spiegel L, Al-Abed Y, et al. An inducible gene product for 6-phosphofructo-2-kinase with an AU-rich instability element: role in tumor cell glycolysis and the Warburg effect. Proc Natl Acad Sci U S A. 1999;96:3047–52. doi: 10.1073/pnas.96.6.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atsumi T, Chesney J, Metz C, Leng L, Donnelly S, Makita Z, et al. High expression of inducible 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (iPFK-2; PFKFB3) in human cancers. Cancer Res. 2002;62:5881–7. [PubMed] [Google Scholar]
- 46.Yalcin A, Telang S, Clem B, Chesney J. Regulation of glucose metabolism by 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatases in cancer. Experimental and molecular pathology. 2009;86:174–9. doi: 10.1016/j.yexmp.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 47.Minchenko A, Leshchinsky I, Opentanova I, Sang N, Srinivas V, Armstead V, et al. Hypoxia-inducible factor-1-mediated expression of the 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (PFKFB3) gene. Its possible role in the Warburg effect. J Biol Chem. 2002;277:6183–7. doi: 10.1074/jbc.M110978200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mendoza EE, Pocceschi MG, Kong X, Leeper DB, Caro J, Limesand KH, et al. Control of Glycolytic Flux by AMP-Activated Protein Kinase in Tumor Cells Adapted to Low pH. Transl Oncol. 2012;5:208–16. doi: 10.1593/tlo.11319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Novellasdemunt L, Obach M, Millan-Arino L, Manzano A, Ventura F, Rosa JL, et al. Progestins activate 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3) in breast cancer cells. Biochem J. 2012;442:345–56. doi: 10.1042/BJ20111418. [DOI] [PubMed] [Google Scholar]
- 50.Reddy MM, Fernandes MS, Deshpande A, Weisberg E, Inguilizian HV, Abdel-Wahab O, et al. The JAK2V617F oncogene requires expression of inducible phosphofructokinase/fructose-bisphosphatase 3 for cell growth and increased metabolic activity. Leukemia. 2012;26:481–9. doi: 10.1038/leu.2011.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Colombo SL, Palacios-Callender M, Frakich N, Carcamo S, Kovacs I, Tudzarova S, et al. Molecular basis for the differential use of glucose and glutamine in cell proliferation as revealed by synchronized HeLa cells. Proc Natl Acad Sci U S A. 2011;108:21069–74. doi: 10.1073/pnas.1117500108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moncada S, Higgs EA, Colombo SL. Fulfilling the metabolic requirements for cell proliferation. Biochem J. 2012;446:1–7. doi: 10.1042/BJ20120427. [DOI] [PubMed] [Google Scholar]
- 53.Yalcin A, Clem BF, Simmons A, Lane A, Nelson K, Clem AL, et al. Nuclear targeting of 6-phosphofructo-2-kinase (PFKFB3) increases proliferation via cyclin-dependent kinases. J Biol Chem. 2009;284:24223–32. doi: 10.1074/jbc.M109.016816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14:1295–304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clem B, Telang S, Clem A, Yalcin A, Meier J, Simmons A, et al. Small-molecule inhibition of 6-phosphofructo-2-kinase activity suppresses glycolytic flux and tumor growth. Mol Cancer Ther. 2008;7:110–20. doi: 10.1158/1535-7163.MCT-07-0482. [DOI] [PubMed] [Google Scholar]
- 56.Telang S, Clem BF, Klarer AC, Clem AL, Trent JO, Bucala R, et al. Small molecule inhibition of 6-phosphofructo-2-kinase suppresses t cell activation. Journal of translational medicine. 2012;10:95. doi: 10.1186/1479-5876-10-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clem BF, O’Neal J, Tapolsky G, Clem AL, Imbert-Fernandez Y, Kerr DA, 2nd, et al. Targeting 6-phosphofructo-2-kinase (PFKFB3) as a therapeutic strategy against cancer. Mol Cancer Ther. 2013;12:1461–70. doi: 10.1158/1535-7163.MCT-13-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15:300–8. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 59.Bluemlein K, Gruning NM, Feichtinger RG, Lehrach H, Kofler B, Ralser M. No evidence for a shift in pyruvate kinase PKM1 to PKM2 expression during tumorigenesis. Oncotarget. 2011;2:393–400. doi: 10.18632/oncotarget.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. The international journal of biochemistry & cell biology. 2011;43:969–80. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 61.Chaneton B, Gottlieb E. Rocking cell metabolism: revised functions of the key glycolytic regulator PKM2 in cancer. Trends in biochemical sciences. 2012;37:309–16. doi: 10.1016/j.tibs.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 62.Lyssiotis CA, Anastasiou D, JWL, Vander Heiden MG, Christofk HR, Cantley LC. Cellular Control Mechanisms that Regulate Pyruvate Kinase M2 Activity and Promote Cancer Growth. Biomedical Research. 2012;23:213–7. [Google Scholar]
- 63.Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoshino A, Hirst JA, Fujii H. Regulation of cell proliferation by interleukin-3-induced nuclear translocation of pyruvate kinase. J Biol Chem. 2007;282:17706–11. doi: 10.1074/jbc.M700094200. [DOI] [PubMed] [Google Scholar]
- 65.Luo W, Hu H, Chang R, Zhong J, Knabel M, O’Meally R, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–44. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stetak A, Veress R, Ovadi J, Csermely P, Keri G, Ullrich A. Nuclear translocation of the tumor marker pyruvate kinase M2 induces programmed cell death. Cancer Res. 2007;67:1602–8. doi: 10.1158/0008-5472.CAN-06-2870. [DOI] [PubMed] [Google Scholar]
- 67.Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685–96. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–22. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ignacak J, Stachurska MB. The dual activity of pyruvate kinase type M2 from chromatin extracts of neoplastic cells. Comp Biochem Physiol B Biochem Mol Biol. 2003;134:425–33. doi: 10.1016/s1096-4959(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 70.Diaz-Jullien C, Moreira D, Sarandeses CS, Covelo G, Barbeito P, Freire M. The M2-type isoenzyme of pyruvate kinase phosphorylates prothymosin alpha in proliferating lymphocytes. Biochim Biophys Acta. 2011;1814:355–65. doi: 10.1016/j.bbapap.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 71.Cortes-Cros M, Hemmerlin C, Ferretti S, Zhang J, Gounarides JS, Yin H, et al. M2 isoform of pyruvate kinase is dispensable for tumor maintenance and growth. Proc Natl Acad Sci U S A. 2013;110:489–94. doi: 10.1073/pnas.1212780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vander Heiden MG. Targeting cell metabolism in cancer patients. Sci Transl Med. 2010;2:31ed1. doi: 10.1126/scitranslmed.3001210. [DOI] [PubMed] [Google Scholar]
- 73.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–3. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 74.Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8:839–47. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kung C, Hixon J, Choe S, Marks K, Gross S, Murphy E, et al. Small molecule activation of PKM2 in cancer cells induces serine auxotrophy. Chemistry & biology. 2012;19:1187–98. doi: 10.1016/j.chembiol.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yacovan A, Ozeri R, Kehat T, Mirilashvili S, Sherman D, Aizikovich A, et al. 1-(sulfonyl)-5-(arylsulfonyl)indoline as activators of the tumor cell specific M2 isoform of pyruvate kinase. Bioorg Med Chem Lett. 2012;22:6460–8. doi: 10.1016/j.bmcl.2012.08.054. [DOI] [PubMed] [Google Scholar]
- 77.Jiang JK, Boxer MB, Vander Heiden MG, Shen M, Skoumbourdis AP, Southall N, et al. Evaluation of thieno[3,2-b]pyrrole[3,2-d]pyridazinones as activators of the tumor cell specific M2 isoform of pyruvate kinase. Bioorg Med Chem Lett. 2010;20:3387–93. doi: 10.1016/j.bmcl.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boxer MB, Jiang JK, Vander Heiden MG, Shen M, Skoumbourdis AP, Southall N, et al. Evaluation of substituted N,N′-diarylsulfonamides as activators of the tumor cell specific M2 isoform of pyruvate kinase. Journal of medicinal chemistry. 2010;53:1048–55. doi: 10.1021/jm901577g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Walsh MJ, Brimacombe KR, Veith H, Bougie JM, Daniel T, Leister W, et al. 2-Oxo-N-aryl-1,2,3,4-tetrahydroquinoline-6-sulfonamides as activators of the tumor cell specific M2 isoform of pyruvate kinase. Bioorg Med Chem Lett. 2011;21:6322–7. doi: 10.1016/j.bmcl.2011.08.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–83. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002;277:30409–12. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 82.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–24. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scriver CRRL. Amino Acid Metabolism and Its Disorders. Philadelpha: WB Saunders; 1973. [PubMed] [Google Scholar]
- 84.Fan TWM, Tan JL, McKinney MM, Lane AN. Stable isotope resolved metabolomics analysis of ribonucleotide and RNA metabolism in human lung cancer cells. Metabolomics. 2012;8:517–27. doi: 10.1007/s11306-011-0337-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Frieden C. Glutamate Dehydrogenase .5. Relation of 5nzyme Structure to Catalytic Function. J Biol Chem. 1963;238:3286. [PubMed] [Google Scholar]
- 86.Reitzer LJ, Wice BM, Kennell D. Evidence That Glutamine, Not Sugar, Is the Major Energy-Source for Cultured Hela-Cells. J Biol Chem. 1979;254:2669–76. [PubMed] [Google Scholar]
- 87.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–U100. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nicklin P, Bergman P, Zhang BL, Triantafellow E, Wang H, Nyfeler B, et al. Bidirectional Transport of Amino Acids Regulates mTOR and Autophagy. Cell. 2009;136:521–34. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morrish F, Neretti N, Sedivy JM, Hockenbery DM. The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell cycle. 2008;7:1054–66. doi: 10.4161/cc.7.8.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–5. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–21. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moreadith RW, Lehninger AL. The Pathways of Glutamate and Glutamine Oxidation by Tumor-Cell Mitochondria - Role of Mitochondrial Nad(P)+-Dependent Malic Enzyme. J Biol Chem. 1984;259:6215–21. [PubMed] [Google Scholar]
- 93.Thornburg JM, Nelson KK, Clem BF, Lane AN, Arumugam S, Simmons A, et al. Targeting aspartate aminotransferase in breast cancer. Breast Cancer Res. 2008;10:R84. doi: 10.1186/bcr2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gaglio D, Metallo CM, Gameiro PA, Hiller K, Danna LS, Balestrieri C, et al. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Molecular systems biology. 2011;7:523. doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ahluwalia GS, Grem JL, Hao Z, Cooney DA. Metabolism and Action of Amino-Acid Analog Anticancer Agents. Pharmacology & Therapeutics. 1990;46:243–71. doi: 10.1016/0163-7258(90)90094-i. [DOI] [PubMed] [Google Scholar]
- 96.Elgadi KM, Meguid RA, Qian M, Souba WW, Abcouwer SF. Cloning and analysis of unique human glutaminase isoforms generated by tissue-specific alternative splicing. Physiol Genomics. 1999;1:51–62. doi: 10.1152/physiolgenomics.1999.1.2.51. [DOI] [PubMed] [Google Scholar]
- 97.Robinson MM, McBryant SJ, Tsukamoto T, Rojas C, Ferraris DV, Hamilton SK, et al. Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES) Biochem J. 2007;406:407–14. doi: 10.1042/BJ20070039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang CD, Sudderth J, Dang TY, Bachoo RG, McDonald JG, DeBerardinis RJ. Glioblastoma Cells Require Glutamate Dehydrogenase to Survive Impairments of Glucose Metabolism or Akt Signaling. Cancer Res. 2009;69:7986–93. doi: 10.1158/0008-5472.CAN-09-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang CS, Wang X, Lu G, Picinich SC. Cancer prevention by tea: animal studies, molecular mechanisms and human relevance. Nat Rev Cancer. 2009;9:429–39. doi: 10.1038/nrc2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li M, Smith CJ, Walker MT, Smith TJ. Novel inhibitors complexed with glutamate dehydrogenase: allosteric regulation by control of protein dynamics. J Biol Chem. 2009;284:22988–3000. doi: 10.1074/jbc.M109.020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Beuster G, Zarse K, Kaleta C, Thierbach R, Kiehntopf M, Steinberg P, et al. Inhibition of alanine aminotransferase in silico and in vivo promotes mitochondrial metabolism to impair malignant growth. J Biol Chem. 286:22323–30. doi: 10.1074/jbc.M110.205229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lyssiotis CA, Son J, Cantley LC, Kimmelman AC. Pancreatic cancers rely on a novel glutamine metabolism pathway to maintain redox balance. Cell cycle. 2013;12:1987–8. doi: 10.4161/cc.25307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–97. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 104.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–85. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 105.Mullen AR, DeBerardinis RJ. Genetically-defined metabolic reprogramming in cancer. Trends Endocrinol Metab. 2012;23:552–9. doi: 10.1016/j.tem.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–4. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–8. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–93. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108:19611–6. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Scott DA, Richardson AD, Filipp FV, Knutzen CA, Chiang GG, Ronai ZA, et al. Comparative metabolic flux profiling of melanoma cell lines: beyond the Warburg effect. J Biol Chem. 2011;286:42626–34. doi: 10.1074/jbc.M111.282046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Des Rosiers C, Fernandez CA, David F, Brunengraber H. Reversibility of the mitochondrial isocitrate dehydrogenase reaction in the perfused rat liver. Evidence from isotopomer analysis of citric acid cycle intermediates. J Biol Chem. 1994;269:27179–82. [PubMed] [Google Scholar]
- 112.Yoo H, Antoniewicz MR, Stephanopoulos G, Kelleher JK. Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J Biol Chem. 2008;283:20621–7. doi: 10.1074/jbc.M706494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Comte B, Vincent G, Bouchard B, Benderdour M, Des Rosiers C. Reverse flux through cardiac NADP(+)-isocitrate dehydrogenase under normoxia and ischemia. Am J Physiol Heart Circ Physiol. 2002;283:H1505–14. doi: 10.1152/ajpheart.00287.2002. [DOI] [PubMed] [Google Scholar]
- 114.Lemons JM, Feng XJ, Bennett BD, Legesse-Miller A, Johnson EL, Raitman I, et al. Quiescent fibroblasts exhibit high metabolic activity. PLoS Biol. 2010;8:e1000514. doi: 10.1371/journal.pbio.1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gameiro PA, Yang J, Metelo AM, Perez-Carro R, Baker R, Wang Z, et al. In Vivo HIF-Mediated Reductive Carboxylation Is Regulated by Citrate Levels and Sensitizes VHL-Deficient Cells to Glutamine Deprivation. Cell Metab. 2013;17:372–85. doi: 10.1016/j.cmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pedersen A, Karlsson GB, Rydstrom J. Proton-translocating transhydrogenase: an update of unsolved and controversial issues. Journal of bioenergetics and biomembranes. 2008;40:463–73. doi: 10.1007/s10863-008-9170-x. [DOI] [PubMed] [Google Scholar]
- 117.Gameiro PA, Laviolette LA, Kelleher JK, Iliopoulos O, Stephanopoulos G. Cofactor balance by nicotinamide nucleotide transhydrogenase (NNT) coordinates reductive carboxylation and glucose catabolism in the TCA cycle. J Biol Chem. 2013 doi: 10.1074/jbc.M112.396796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rajagopalan KN, Egnatchik RA, Calvaruso MA, Wasti AT, Padanad MS, Boroughs LK, et al. Metabolic plasticity maintains proliferation in pyruvate dehydrogenase deficient cells. Cancer & metabolism. 2015;3:7. doi: 10.1186/s40170-015-0134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Currie E, Schulze A, Zechner R, Walther TC, Farese RV., Jr Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–61. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16:9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Katada S, Imhof A, Sassone-Corsi P. Connecting threads: epigenetics and metabolism. Cell. 2012;148:24–8. doi: 10.1016/j.cell.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 122.Wellen KE, Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol. 2012;13:270–6. doi: 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- 123.Yun J, Johnson JL, Hanigan CL, Locasale JW. Interactions between epigenetics and metabolism in cancers. Front Oncol. 2012;2:163. doi: 10.3389/fonc.2012.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kaelin WG, McKnight SL. Influence of Metabolism on Epigenetics and Disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 126.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10:671–84. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 127.Love DC, Hanover JA. The hexosamine signaling pathway: deciphering the “O-GlcNAc code”. Sci STKE. 2005;2005:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- 128.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annual review of biochemistry. 2011;80:825–58. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lynch TP, Ferrer CM, Jackson SR, Shahriari KS, Vosseller K, Reginato MJ. Critical role of O-Linked beta-N-acetylglucosamine transferase in prostate cancer invasion, angiogenesis, and metastasis. J Biol Chem. 2012;287:11070–81. doi: 10.1074/jbc.M111.302547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Caldwell SA, Jackson SR, Shahriari KS, Lynch TP, Sethi G, Walker S, et al. Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene. 2010;29:2831–42. doi: 10.1038/onc.2010.41. [DOI] [PubMed] [Google Scholar]
- 131.Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA, 3rd, et al. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 2012;337:975–80. doi: 10.1126/science.1222278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang S, Roche K, Nasheuer HP, Lowndes NF. Modification of histones by sugar beta-N-acetylglucosamine (GlcNAc) occurs on multiple residues, including histone H3 serine 10, and is cell cycle-regulated. J Biol Chem. 2011;286:37483–95. doi: 10.1074/jbc.M111.284885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sakabe K, Wang Z, Hart GW. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci U S A. 2010;107:19915–20. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fujiki R, Hashiba W, Sekine H, Yokoyama A, Chikanishi T, Ito S, et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011;480:557–60. doi: 10.1038/nature10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fong JJ, Nguyen BL, Bridger R, Medrano EE, Wells L, Pan S, et al. beta-N-Acetylglucosamine (O-GlcNAc) is a novel regulator of mitosis-specific phosphorylations on histone H3. J Biol Chem. 2012;287:12195–203. doi: 10.1074/jbc.M111.315804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2012 doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vella P, Scelfo A, Jammula S, Chiacchiera F, Williams K, Cuomo A, et al. Tet Proteins Connect the O-Linked N-acetylglucosamine Transferase Ogt to Chromatin in Embryonic Stem Cells. Mol Cell. 2013;49:645–56. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 138.Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. Embo J. 2013;32:645–55. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang Q, Liu X, Gao W, Li P, Hou J, Li J, et al. Differential regulation of the ten-eleven translocation (TET) family of dioxygenases by O-linked beta-N-acetylglucosamine transferase (OGT) J Biol Chem. 2014;289:5986–96. doi: 10.1074/jbc.M113.524140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 141.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer cell. 2010;18:553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sutter BM, Wu X, Laxman S, Tu BP. Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell. 2013;154:403–15. doi: 10.1016/j.cell.2013.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cavuoto P, Fenech MF. A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension. Cancer Treat Rev. 2012;38:726–36. doi: 10.1016/j.ctrv.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 146.Ulanovskaya OA, Zuhl AM, Cravatt BF. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat Chem Biol. 2013;9:300–6. doi: 10.1038/nchembio.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–6. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Katoh Y, Ikura T, Hoshikawa Y, Tashiro S, Ito T, Ohta M, et al. Methionine adenosyltransferase II serves as a transcriptional corepressor of Maf oncoprotein. Mol Cell. 2011;41:554–66. doi: 10.1016/j.molcel.2011.02.018. [DOI] [PubMed] [Google Scholar]
- 149.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–80. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Friis RM, Wu BP, Reinke SN, Hockman DJ, Sykes BD, Schultz MC. A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA. Nucleic Acids Res. 2009;37:3969–80. doi: 10.1093/nar/gkp270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Molecular cell. 2011;42:426–37. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell. 2006;23:207–17. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 153.Sassone-Corsi P. When metabolism and epigenetics converge. Science. 2013;339:148–50. doi: 10.1126/science.1233423. [DOI] [PubMed] [Google Scholar]
- 154.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–38. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Developmental cell. 2008;14:661–73. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–60. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Shimazu T, Hirschey MD, Newman J, He W, Shirakawa K, Le Moan N, et al. Suppression of Oxidative Stress by beta-Hydroxybutyrate, an Endogenous Histone Deacetylase Inhibitor. Science. 2013;339:211–4. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell. 2011;42:426–37. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chypre M, Zaidi N, Smans K. ATP-citrate lyase: a mini-review. Biochem Biophys Res Commun. 2012;422:1–4. doi: 10.1016/j.bbrc.2012.04.144. [DOI] [PubMed] [Google Scholar]
- 160.Seligson DB, Horvath S, McBrian MA, Mah V, Yu H, Tze S, et al. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol. 2009;174:1619–28. doi: 10.2353/ajpath.2009.080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Barlesi F, Giaccone G, Gallegos-Ruiz MI, Loundou A, Span SW, Lefesvre P, et al. Global histone modifications predict prognosis of resected non small-cell lung cancer. J Clin Oncol. 2007;25:4358–64. doi: 10.1200/JCO.2007.11.2599. [DOI] [PubMed] [Google Scholar]
- 162.Elsheikh SE, Green AR, Rakha EA, Powe DG, Ahmed RA, Collins HM, et al. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 2009;69:3802–9. doi: 10.1158/0008-5472.CAN-08-3907. [DOI] [PubMed] [Google Scholar]
- 163.Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–6. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 164.Bianco-Miotto T, Chiam K, Buchanan G, Jindal S, Day TK, Thomas M, et al. Global levels of specific histone modifications and an epigenetic gene signature predict prostate cancer progression and development. Cancer Epidemiol Biomarkers Prev. 2010;19:2611–22. doi: 10.1158/1055-9965.EPI-10-0555. [DOI] [PubMed] [Google Scholar]
- 165.Carafa V, Nebbioso A, Altucci L. Sirtuins and disease: the road ahead. Frontiers in pharmacology. 2012;3:4. doi: 10.3389/fphar.2012.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Anderson KA, Green MF, Huynh FK, Wagner GR, Hirschey MD. SnapShot: Mammalian Sirtuins. Cell. 2014;159:956–e1. doi: 10.1016/j.cell.2014.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Molecular and cellular biology. 2007;27:8807–14. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–8. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 169.Stunkel W, Campbell RM. Sirtuin 1 (SIRT1): the misunderstood HDAC. Journal of biomolecular screening. 2011;16:1153–69. doi: 10.1177/1087057111422103. [DOI] [PubMed] [Google Scholar]
- 170.Deng CX. SIRT1, is it a tumor promoter or tumor suppressor? International journal of biological sciences. 2009;5:147–52. doi: 10.7150/ijbs.5.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Banks AS, Kon N, Knight C, Matsumoto M, Gutierrez-Juarez R, Rossetti L, et al. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–41. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–23. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang RH, Zheng Y, Kim HS, Xu X, Cao L, Luhasen T, et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008;32:11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–5. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19:416–28. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL, Pierce K, et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell. 2013;23:450–63. doi: 10.1016/j.ccr.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151:1185–99. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Marquardt JU, Fischer K, Baus K, Kashyap A, Ma S, Krupp M, et al. SIRT6 dependent genetic and epigenetic alterations are associated with poor clinical outcome in HCC patients. Hepatology. 2013 doi: 10.1002/hep.26413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–2. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 181.Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, et al. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell. 2011;44:177–90. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Current opinion in genetics & development. 2009;19:32–7. doi: 10.1016/j.gde.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Finkelstein JD. Methionine metabolism in mammals. J Nutr Biochem. 1990;1:228–37. doi: 10.1016/0955-2863(90)90070-2. [DOI] [PubMed] [Google Scholar]
- 184.Zatz M, Dudley PA, Kloog Y, Markey SP. Nonpolar lipid methylation. Biosynthesis of fatty acid methyl esters by rat lung membranes using S-adenosylmethionine. J Biol Chem. 1981;256:10028–32. [PubMed] [Google Scholar]
- 185.Anderson OS, Sant KE, Dolinoy DC. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J Nutr Biochem. 2012;23:853–9. doi: 10.1016/j.jnutbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Teperino R, Schoonjans K, Auwerx J. Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell Metab. 2010;12:321–7. doi: 10.1016/j.cmet.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 188.Snell K. Enzymes of serine metabolism in normal, developing and neoplastic rat tissues. Advances in enzyme regulation. Adv Enzyme Regul. 1984;22:325–400. doi: 10.1016/0065-2571(84)90021-9. [DOI] [PubMed] [Google Scholar]
- 189.Snell K, Natsumeda Y, Weber G. The modulation of serine metabolism in hepatoma 3924A during different phases of cellular proliferation in culture. Biochem J. 1987;245:609–12. doi: 10.1042/bj2450609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–74. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–50. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Pollari S, Kakonen SM, Edgren H, Wolf M, Kohonen P, Sara H, et al. Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res Treat. 2011;125:421–30. doi: 10.1007/s10549-010-0848-5. [DOI] [PubMed] [Google Scholar]
- 193.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–4. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–72. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 195.Hu CM, Yeh MT, Tsao N, Chen CW, Gao QZ, Chang CY, et al. Tumor cells require thymidylate kinase to prevent dUTP incorporation during DNA repair. Cancer Cell. 2012;22:36–50. doi: 10.1016/j.ccr.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 196.Chabner BA, Roberts TG., Jr Timeline: Chemotherapy and the war on cancer. Nat Rev Cancer. 2005;5:65–72. doi: 10.1038/nrc1529. [DOI] [PubMed] [Google Scholar]
- 197.Diasio RB, Harris BE. Clinical pharmacology of 5-fluorouracil. Clin Pharmacokinet. 1989;16:215–37. doi: 10.2165/00003088-198916040-00002. [DOI] [PubMed] [Google Scholar]
- 198.Casero RA, Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov. 2007;6:373–90. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- 199.Cheong H, Lu C, Lindsten T, Thompson CB. Therapeutic targets in cancer cell metabolism and autophagy. Nat Biotechnol. 2012;30:671–8. doi: 10.1038/nbt.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–6. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–4. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 202.Vazquez A, Tedeschi PM, Bertino JR. Overexpression of the mitochondrial folate and glycine-serine pathway: a new determinant of methotrexate selectivity in tumors. Cancer Res. 2013;73:478–82. doi: 10.1158/0008-5472.CAN-12-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Pansuriya TC, van Eijk R, d’Adamo P, van Ruler MA, Kuijjer ML, Oosting J, et al. Somatic mosaic IDH1 and IDH2 mutations are associated with enchondroma and spindle cell hemangioma in Ollier disease and Maffucci syndrome. Nat Genet. 2011;43:1256–61. doi: 10.1038/ng.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Parsons DW, Jones Sn, Zhang X, Lin JC-H, Leary RJ, Angenendt P, et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. The Journal of experimental medicine. 2010;207:339–44. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Losman JA, Kaelin WG., Jr What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes & development. 2013;27:836–52. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer cell. 2010;17:225–34. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kats LM, Reschke M, Taulli R, Pozdnyakova O, Burgess K, Bhargava P, et al. Proto-Oncogenic Role of Mutant IDH2 in Leukemia Initiation and Maintenance. Cell stem cell. 2014 doi: 10.1016/j.stem.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lu C, Venneti S, Akalin A, Fang F, Ward PS, Dematteo RG, et al. Induction of sarcomas by mutant IDH2. Genes & development. 2013;27:1986–98. doi: 10.1101/gad.226753.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO reports. 2011;12:463–9. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339:1621–5. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–8. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.DiNardo CD, Propert KJ, Loren AW, Paietta E, Sun Z, Levine RL, et al. Serum 2-hydroxyglutarate levels predict isocitrate dehydrogenase mutations and clinical outcome in acute myeloid leukemia. Blood. 2013;121:4917–24. doi: 10.1182/blood-2013-03-493197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18:624–9. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wang F, Travins J, Delabarre B, Penard-Lacronique V, Schalm S, Hansen E, et al. Targeted Inhibition of Mutant IDH2 in Leukemia Cells Induces Cellular Differentiation. Science (New York, NY) 2013;340:622–6. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 219.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, et al. An Inhibitor of Mutant IDH1 Delays Growth and Promotes Differentiation of Glioma Cells. Science (New York, NY) 2013 doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 221.Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes & development. 2012;26:1326–38. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Killian JK, Kim SY, Miettinen M, Smith C, Merino M, Tsokos M, et al. Succinate dehydrogenase mutation underlies global epigenomic divergence in gastrointestinal stromal tumor. Cancer Discov. 2013;3:648–57. doi: 10.1158/2159-8290.CD-13-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Letouze E, Martinelli C, Loriot C, Burnichon N, Abermil N, Ottolenghi C, et al. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–52. doi: 10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 224.Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, Jairam V, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513:559–63. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Pavlides S, Whitaker-Menezes D, Castello-Cros R, Flomenberg N, Witkiewicz AK, Frank PG, et al. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell cycle. 2009;8:3984–4001. doi: 10.4161/cc.8.23.10238. [DOI] [PubMed] [Google Scholar]
- 226.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–82. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 227.Granchi C, Bertini S, Macchia M, Minutolo F. Inhibitors of lactate dehydrogenase isoforms and their therapeutic potentials. Curr Med Chem. 2010;17:672–97. doi: 10.2174/092986710790416263. [DOI] [PubMed] [Google Scholar]
- 228.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–34. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 229.Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107:2037–42. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Seth P, Grant A, Tang J, Vinogradov E, Wang X, Lenkinski R, et al. On-target inhibition of tumor fermentative glycolysis as visualized by hyperpolarized pyruvate. Neoplasia. 2011;13:60–71. doi: 10.1593/neo.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997;94:6658–63. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Zhao Y, Liu H, Riker AI, Fodstad O, Ledoux SP, Wilson GL, et al. Emerging metabolic targets in cancer therapy. Frontiers in bioscience. 2011;16:1844–60. doi: 10.2741/3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Zhou M, Zhao Y, Ding Y, Liu H, Liu Z, Fodstad O, et al. Warburg effect in chemosensitivity: targeting lactate dehydrogenase-A re-sensitizes taxol-resistant cancer cells to taxol. Molecular cancer. 2010;9:33. doi: 10.1186/1476-4598-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Bonen A. Lactate transporters (MCT proteins) in heart and skeletal muscles. Medicine and science in sports and exercise. 2000;32:778–89. doi: 10.1097/00005768-200004000-00010. [DOI] [PubMed] [Google Scholar]
- 235.Hussien R, Brooks GA. Mitochondrial and plasma membrane lactate transporter and lactate dehydrogenase isoform expression in breast cancer cell lines. Physiol Genomics. 2011;43:255–64. doi: 10.1152/physiolgenomics.00177.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sonveaux P, Vegran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest. 2008;118:3930–42. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Miranda-Goncalves V, Honavar M, Pinheiro C, Martinho O, Pires MM, Pinheiro C, et al. Monocarboxylate transporters (MCTs) in gliomas: expression and exploitation as therapeutic targets. Neuro-oncology. 2013;15:172–88. doi: 10.1093/neuonc/nos298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Smith RA, Hartley RC, Cocheme HM, Murphy MP. Mitochondrial pharmacology. Trends Pharmacol Sci. 2012;33:341–52. doi: 10.1016/j.tips.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 239.Smith RA, Hartley RC, Murphy MP. Mitochondria-targeted small molecule therapeutics and probes. Antioxidants & redox signaling. 2011;15:3021–38. doi: 10.1089/ars.2011.3969. [DOI] [PubMed] [Google Scholar]
- 240.Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Calvo SE, Mootha VK. The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Green DR, Galluzzi L, Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333:1109–12. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Viscomi C, Bottani E, Civiletto G, Cerutti R, Moggio M, Fagiolari G, et al. In vivo correction of COX deficiency by activation of the AMPK/PGC-1alpha axis. Cell Metab. 2011;14:80–90. doi: 10.1016/j.cmet.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ventura-Clapier R, Garnier A, Veksler V. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1alpha. Cardiovascular research. 2008;79:208–17. doi: 10.1093/cvr/cvn098. [DOI] [PubMed] [Google Scholar]
- 245.Michelakis ED, Webster L, Mackey JR. Dichloroacetate (DCA) as a potential metabolic-targeting therapy for cancer. British journal of cancer. 2008;99:989–94. doi: 10.1038/sj.bjc.6604554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Lopez-Armada MJ, Riveiro-Naveira RR, Vaamonde-Garcia C, Valcarcel-Ares MN. Mitochondrial dysfunction and the inflammatory response. Mitochondrion. 2013;13:106–18. doi: 10.1016/j.mito.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 247.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–98. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Fulda S, Galluzzi L, Kroemer G. Targeting mitochondria for cancer therapy. Nat Rev Drug Discov. 2010;9:447–64. doi: 10.1038/nrd3137. [DOI] [PubMed] [Google Scholar]
- 249.Longo VD, Fontana L. Calorie restriction and cancer prevention: metabolic and molecular mechanisms. Trends Pharmacol Sci. 2010;31:89–98. doi: 10.1016/j.tips.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Lee C, Safdie FM, Raffaghello L, Wei M, Madia F, Parrella E, et al. Reduced levels of IGF-I mediate differential protection of normal and cancer cells in response to fasting and improve chemotherapeutic index. Cancer Res. 2010;70:1564–72. doi: 10.1158/0008-5472.CAN-09-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Pallavi R, Giorgio M, Pelicci PG. Insights into the beneficial effect of caloric/dietary restriction for a healthy and prolonged life. Front Physiol. 2012;3:318. doi: 10.3389/fphys.2012.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Tannenbaum A, Silverstone H. The influence of the degree of caloric restriction on the formation of skin tumors and hepatomas in mice. Cancer Res. 1949;9:724–7. [PubMed] [Google Scholar]
- 253.Silverstone H, Tannenbaum A. Proportion of dietary protein and the formation of spontaneous hepatomas in the mouse. Cancer Res. 1951;11:442–6. [PubMed] [Google Scholar]
- 254.Tannenbaum A, Silverstone H. Effect of limited food intake on survival of mice bearing spontaneous mammary carcinoma and on the incidence of lung metastases. Cancer Res. 1953;13:532–6. [PubMed] [Google Scholar]
- 255.Ruggeri BA, Klurfeld DM, Kritchevsky D, Furlanetto RW. Caloric restriction and 7,12-dimethylbenz(a)anthracene-induced mammary tumor growth in rats: alterations in circulating insulin, insulin-like growth factors I and II, and epidermal growth factor. Cancer Res. 1989;49:4130–4. [PubMed] [Google Scholar]
- 256.Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–4. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Mattison JA, Roth GS, Beasley TM, Tilmont EM, Handy AM, Herbert RL, et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012;489:318–21. doi: 10.1038/nature11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Kalaany NY, Sabatini DM. Tumours with PI3K activation are resistant to dietary restriction. Nature. 2009;458:725–31. doi: 10.1038/nature07782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Laviano A, Seelaender M, Sanchez-Lara K, Gioulbasanis I, Molfino A, Rossi Fanelli F. Beyond anorexia-cachexia. Nutrition and modulation of cancer patients’ metabolism: supplementary, complementary or alternative anti-neoplastic therapy? Eur J Pharmacol. 2011;668(Suppl 1):S87–90. doi: 10.1016/j.ejphar.2011.06.060. [DOI] [PubMed] [Google Scholar]
- 260.Kubo C, Day NK, Good RA. Influence of early or late dietary restriction on life span and immunological parameters in MRL/Mp-lpr/lpr mice. Proc Natl Acad Sci U S A. 1984;81:5831–5. doi: 10.1073/pnas.81.18.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Hunt ND, Li GD, Zhu M, Miller M, Levette A, Chachich ME, et al. Effect of calorie restriction and refeeding on skin wound healing in the rat. Age (Dordr) 2012;34:1453–8. doi: 10.1007/s11357-011-9321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Raffaghello L, Lee C, Safdie FM, Wei M, Madia F, Bianchi G, et al. Starvation-dependent differential stress resistance protects normal but not cancer cells against high-dose chemotherapy. Proc Natl Acad Sci U S A. 2008;105:8215–20. doi: 10.1073/pnas.0708100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Shi Y, Felley-Bosco E, Marti TM, Orlowski K, Pruschy M, Stahel RA. Starvation-induced activation of ATM/Chk2/p53 signaling sensitizes cancer cells to cisplatin. BMC cancer. 2012;12:571. doi: 10.1186/1471-2407-12-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Safdie F, Brandhorst S, Wei M, Wang W, Lee C, Hwang S, et al. Fasting enhances the response of glioma to chemo- and radiotherapy. PLoS One. 2012;7:e44603. doi: 10.1371/journal.pone.0044603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Lee C, Raffaghello L, Brandhorst S, Safdie FM, Bianchi G, Martin-Montalvo A, et al. Fasting cycles retard growth of tumors and sensitize a range of cancer cell types to chemotherapy. Sci Transl Med. 2012;4:124ra27. doi: 10.1126/scitranslmed.3003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Seyfried TN, Mukherjee P. Targeting energy metabolism in brain cancer: review and hypothesis. Nutrition & metabolism. 2005;2:30. doi: 10.1186/1743-7075-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Longo VD, Ellerby LM, Bredesen DE, Valentine JS, Gralla EB. Human Bcl-2 reverses survival defects in yeast lacking superoxide dismutase and delays death of wild-type yeast. The Journal of cell biology. 1997;137:1581–8. doi: 10.1083/jcb.137.7.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Gonidakis S, Finkel SE, Longo VD. Lifespan extension and paraquat resistance in a ubiquinone-deficient Escherichia coli mutant depend on transcription factors ArcA and TdcA. Aging. 2011;3:291–303. doi: 10.18632/aging.100301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Wei M, Fabrizio P, Hu J, Ge H, Cheng C, Li L, et al. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 2008;4:e13. doi: 10.1371/journal.pgen.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Wei M, Fabrizio P, Madia F, Hu J, Ge H, Li LM, et al. Tor1/Sch9-regulated carbon source substitution is as effective as calorie restriction in life span extension. PLoS Genet. 2009;5:e1000467. doi: 10.1371/journal.pgen.1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Weinkove D, Halstead JR, Gems D, Divecha N. Long-term starvation and ageing induce AGE-1/PI 3-kinase-dependent translocation of DAF-16/FOXO to the cytoplasm. BMC Biol. 2006;4:1. doi: 10.1186/1741-7007-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Tettweiler G, Miron M, Jenkins M, Sonenberg N, Lasko PF. Starvation and oxidative stress resistance in Drosophila are mediated through the eIF4E-binding protein, d4E-BP. Genes & development. 2005;19:1840–3. doi: 10.1101/gad.1311805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Michalsen A, Riegert M, Ludtke R, Backer M, Langhorst J, Schwickert M, et al. Mediterranean diet or extended fasting’s influence on changing the intestinal microflora, immunoglobulin A secretion and clinical outcome in patients with rheumatoid arthritis and fibromyalgia: an observational study. BMC Complement Altern Med. 2005;5:22. doi: 10.1186/1472-6882-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Goldhamer A, Lisle D, Parpia B, Anderson SV, Campbell TC. Medically supervised water-only fasting in the treatment of hypertension. J Manipulative Physiol Ther. 2001;24:335–9. doi: 10.1067/mmt.2001.115263. [DOI] [PubMed] [Google Scholar]
- 275.Safdie FM, Dorff T, Quinn D, Fontana L, Wei M, Lee C, et al. Fasting and cancer treatment in humans: A case series report. Aging. 2009;1:988–1007. doi: 10.18632/aging.100114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Laviano A, Rossi Fanelli F. Toxicity in chemotherapy--when less is more. The New England journal of medicine. 2012;366:2319–20. doi: 10.1056/NEJMcibr1202395. [DOI] [PubMed] [Google Scholar]
- 277.Kushi LH, Doyle C, McCullough M, Rock CL, Demark-Wahnefried W, Bandera EV, et al. American Cancer Society Guidelines on nutrition and physical activity for cancer prevention: reducing the risk of cancer with healthy food choices and physical activity. CA Cancer J Clin. 2012;62:30–67. doi: 10.3322/caac.20140. [DOI] [PubMed] [Google Scholar]
- 278.Byers T, Nestle M, McTiernan A, Doyle C, Currie-Williams A, Gansler T, et al. American Cancer Society guidelines on nutrition and physical activity for cancer prevention: Reducing the risk of cancer with healthy food choices and physical activity. CA Cancer J Clin. 2002;52:92–119. doi: 10.3322/canjclin.52.2.92. [DOI] [PubMed] [Google Scholar]
- 279.Ballard-Barbash R, Friedenreich CM, Courneya KS, Siddiqi SM, McTiernan A, Alfano CM. Physical activity, biomarkers, and disease outcomes in cancer survivors: a systematic review. Journal of the National Cancer Institute. 2012;104:815–40. doi: 10.1093/jnci/djs207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Betof AS, Dewhirst MW, Jones LW. Effects and potential mechanisms of exercise training on cancer progression: a translational perspective. Brain Behav Immun. 2013;30(Suppl):S75–87. doi: 10.1016/j.bbi.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.McTiernan A. Mechanisms linking physical activity with cancer. Nat Rev Cancer. 2008;8:205–11. doi: 10.1038/nrc2325. [DOI] [PubMed] [Google Scholar]
- 282.Zhu Z, Jiang W, Zacher JH, Neil ES, McGinley JN, Thompson HJ. Effects of energy restriction and wheel running on mammary carcinogenesis and host systemic factors in a rat model. Cancer Prev Res (Phila) 2012;5:414–22. doi: 10.1158/1940-6207.CAPR-11-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Jiang W, Zhu Z, Thompson HJ. Effects of limiting energy availability via diet and physical activity on mammalian target of rapamycin-related signaling in rat mammary carcinomas. Carcinogenesis. 2013;34:378–87. doi: 10.1093/carcin/bgs350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Jones LW, Antonelli J, Masko EM, Broadwater G, Lascola CD, Fels D, et al. Exercise modulation of the host-tumor interaction in an orthotopic model of murine prostate cancer. Journal of applied physiology. 2012;113:263–72. doi: 10.1152/japplphysiol.01575.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Jones LW, Viglianti BL, Tashjian JA, Kothadia SM, Keir ST, Freedland SJ, et al. Effect of aerobic exercise on tumor physiology in an animal model of human breast cancer. Journal of applied physiology. 2010;108:343–8. doi: 10.1152/japplphysiol.00424.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.McTiernan A, Sorensen B, Yasui Y, Tworoger SS, Ulrich CM, Irwin ML, et al. No effect of exercise on insulin-like growth factor 1 and insulin-like growth factor binding protein 3 in postmenopausal women: a 12-month randomized clinical trial. Cancer Epidemiol Biomarkers Prev. 2005;14:1020–1. doi: 10.1158/1055-9965.EPI-04-0834. [DOI] [PubMed] [Google Scholar]
- 287.Frank LL, Sorensen BE, Yasui Y, Tworoger SS, Schwartz RS, Ulrich CM, et al. Effects of exercise on metabolic risk variables in overweight postmenopausal women: a randomized clinical trial. Obesity research. 2005;13:615–25. doi: 10.1038/oby.2005.66. [DOI] [PubMed] [Google Scholar]
- 288.Fairey AS, Courneya KS, Field CJ, Bell GJ, Jones LW, Mackey JR. Effects of exercise training on fasting insulin, insulin resistance, insulin-like growth factors, and insulin-like growth factor binding proteins in postmenopausal breast cancer survivors: a randomized controlled trial. Cancer Epidemiol Biomarkers Prev. 2003;12:721–7. [PubMed] [Google Scholar]
- 289.Irwin ML, McTiernan A, Bernstein L, Gilliland FD, Baumgartner R, Baumgartner K, et al. Relationship of obesity and physical activity with C-peptide, leptin, and insulin-like growth factors in breast cancer survivors. Cancer Epidemiol Biomarkers Prev. 2005;14:2881–8. doi: 10.1158/1055-9965.EPI-05-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Schmitz KH, Ahmed RL, Hannan PJ, Yee D. Safety and efficacy of weight training in recent breast cancer survivors to alter body composition, insulin, and insulin-like growth factor axis proteins. Cancer Epidemiol Biomarkers Prev. 2005;14:1672–80. doi: 10.1158/1055-9965.EPI-04-0736. [DOI] [PubMed] [Google Scholar]
- 291.Janelsins MC, Davis PG, Wideman L, Katula JA, Sprod LK, Peppone LJ, et al. Effects of Tai Chi Chuan on insulin and cytokine levels in a randomized controlled pilot study on breast cancer survivors. Clin Breast Cancer. 2011;11:161–70. doi: 10.1016/j.clbc.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Ligibel JA, Campbell N, Partridge A, Chen WY, Salinardi T, Chen H, et al. Impact of a mixed strength and endurance exercise intervention on insulin levels in breast cancer survivors. J Clin Oncol. 2008;26:907–12. doi: 10.1200/JCO.2007.12.7357. [DOI] [PubMed] [Google Scholar]
- 293.Irwin ML, Varma K, Alvarez-Reeves M, Cadmus L, Wiley A, Chung GG, et al. Randomized controlled trial of aerobic exercise on insulin and insulin-like growth factors in breast cancer survivors: the Yale Exercise and Survivorship study. Cancer Epidemiol Biomarkers Prev. 2009;18:306–13. doi: 10.1158/1055-9965.EPI-08-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Taghian AG, Abi-Raad R, Assaad SI, Casty A, Ancukiewicz M, Yeh E, et al. Paclitaxel decreases the interstitial fluid pressure and improves oxygenation in breast cancers in patients treated with neoadjuvant chemotherapy: clinical implications. J Clin Oncol. 2005;23:1951–61. doi: 10.1200/JCO.2005.08.119. [DOI] [PubMed] [Google Scholar]
- 295.Hockel M, Schlenger K, Knoop C, Vaupel P. Oxygenation of carcinomas of the uterine cervix: evaluation by computerized O2 tension measurements. Cancer Res. 1991;51:6098–102. [PubMed] [Google Scholar]
- 296.Moon EJ, Brizel DM, Chi JT, Dewhirst MW. The potential role of intrinsic hypoxia markers as prognostic variables in cancer. Antioxidants & redox signaling. 2007;9:1237–94. doi: 10.1089/ars.2007.1623. [DOI] [PubMed] [Google Scholar]
- 297.Edwards DG, Schofield RS, Lennon SL, Pierce GL, Nichols WW, Braith RW. Effect of exercise training on endothelial function in men with coronary artery disease. Am J Cardiol. 2004;93:617–20. doi: 10.1016/j.amjcard.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 298.Hambrecht R, Wolf A, Gielen S, Linke A, Hofer J, Erbs S, et al. Effect of exercise on coronary endothelial function in patients with coronary artery disease. The New England journal of medicine. 2000;342:454–60. doi: 10.1056/NEJM200002173420702. [DOI] [PubMed] [Google Scholar]
- 299.Allen JD, Geaghan JP, Greenway F, Welsch MA. Time course of improved flow-mediated dilation after short-term exercise training. Medicine and science in sports and exercise. 2003;35:847–53. doi: 10.1249/01.MSS.0000064931.62916.8A. [DOI] [PubMed] [Google Scholar]
- 300.Allen JD, Welsch M, Aucoin N, Wood R, Lee M, LeBlanc KE. Forearm vasoreactivity in type 1diabetic subjects. Can J Appl Physiol. 2001;26:34–43. doi: 10.1139/h01-003. [DOI] [PubMed] [Google Scholar]
- 301.Hambrecht R, Fiehn E, Weigl C, Gielen S, Hamann C, Kaiser R, et al. Regular physical exercise corrects endothelial dysfunction and improves exercise capacity in patients with chronic heart failure. Circulation. 1998;98:2709–15. doi: 10.1161/01.cir.98.24.2709. [DOI] [PubMed] [Google Scholar]
- 302.Ostlund RE, Jr, Yang JW, Klein S, Gingerich R. Relation between plasma leptin concentration and body fat, gender, diet, age, and metabolic covariates. The Journal of clinical endocrinology and metabolism. 1996;81:3909–13. doi: 10.1210/jcem.81.11.8923837. [DOI] [PubMed] [Google Scholar]
- 303.Hu E, Liang P, Spiegelman BM. AdipoQ is a novel adipose-specific gene dysregulated in obesity. J Biol Chem. 1996;271:10697–703. doi: 10.1074/jbc.271.18.10697. [DOI] [PubMed] [Google Scholar]
- 304.Wellen KE, Hotamisligil GS. Obesity-induced inflammatory changes in adipose tissue. J Clin Invest. 2003;112:1785–8. doi: 10.1172/JCI20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Yildiz BO, Suchard MA, Wong ML, McCann SM, Licinio J. Alterations in the dynamics of circulating ghrelin, adiponectin, and leptin in human obesity. Proc Natl Acad Sci U S A. 2004;101:10434–9. doi: 10.1073/pnas.0403465101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Research WCRFAIfC. Food Nutrition, Physical Activity, and the Prevention of Cancer: a Global Perspective. Washington DC: AICR; 2007. [Google Scholar]
- 307.Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. The New England journal of medicine. 2003;348:1625–38. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 308.Colditz GA, Wolin KY, Gehlert S. Applying what we know to accelerate cancer prevention. Sci Transl Med. 2012;4:127rv4. doi: 10.1126/scitranslmed.3003218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat Rev Cancer. 2004;4:579–91. doi: 10.1038/nrc1408. [DOI] [PubMed] [Google Scholar]
- 310.Hursting SD, Dunlap SM. Obesity, metabolic dysregulation, and cancer: a growing concern and an inflammatory (and microenvironmental) issue. Annals of the New York Academy of Sciences. 2012;1271:82–7. doi: 10.1111/j.1749-6632.2012.06737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Vucenik I, Stains JP. Obesity and cancer risk: evidence, mechanisms, and recommendations. Annals of the New York Academy of Sciences. 2012;1271:37–43. doi: 10.1111/j.1749-6632.2012.06750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Pollak MN. Investigating metformin for cancer prevention and treatment: the end of the beginning. Cancer Discov. 2012;2:778–90. doi: 10.1158/2159-8290.CD-12-0263. [DOI] [PubMed] [Google Scholar]
- 313.Sankaranarayanapillai M, Zhang N, Baggerly KA, Gelovani JG. Metabolic shifts induced by fatty acid synthase inhibitor orlistat in non-small cell lung carcinoma cells provide novel pharmacodynamic biomarkers for positron emission tomography and magnetic resonance spectroscopy. Molecular imaging and biology : MIB : the official publication of the Academy of Molecular Imaging. 2013;15:136–47. doi: 10.1007/s11307-012-0587-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Luengo A, Sullivan LB, Heiden MG. Understanding the complex-I-ty of metformin action: limiting mitochondrial respiration to improve cancer therapy. BMC Biol. 2014;12:82. doi: 10.1186/s12915-014-0082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Madhok BM, Yeluri S, Perry SL, Hughes TA, Jayne DG. Targeting glucose metabolism: an emerging concept for anticancer therapy. American journal of clinical oncology. 2011;34:628–35. doi: 10.1097/COC.0b013e3181e84dec. [DOI] [PubMed] [Google Scholar]
- 316.Jacobs SR, Herman CE, Maciver NJ, Wofford JA, Wieman HL, Hammen JJ, et al. Glucose Uptake Is Limiting in T Cell Activation and Requires CD28-Mediated Akt-Dependent and Independent Pathways. J Immunol. 2008;180:4476–86. doi: 10.4049/jimmunol.180.7.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–44. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, Maciver NJ, Mason EF, et al. Cutting Edge: Distinct Glycolytic and Lipid Oxidative Metabolic Programs Are Essential for Effector and Regulatory CD4+ T Cell Subsets. J Immunol. 2011;186:3299–303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. The Journal of experimental medicine. 2011;208:1367–76. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–7. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.van der Windt GJ, Everts B, Chang CH, Curtis JD, Freitas TC, Amiel E, et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Harjes U, Bensaad K, Harris AL. Endothelial cell metabolism and implications for cancer therapy. British journal of cancer. 2012;107:1207–12. doi: 10.1038/bjc.2012.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Chiong M, Morales P, Torres G, Gutierrez T, Garcia L, Ibacache M, et al. Influence of glucose metabolism on vascular smooth muscle cell proliferation. VASA Zeitschrift fur Gefasskrankheiten. 2013;42:8–16. doi: 10.1024/0301-1526/a000243. [DOI] [PubMed] [Google Scholar]
- 324.Ostroukhova M, Goplen N, Karim MZ, Michalec L, Guo L, Liang Q, et al. The role of low-level lactate production in airway inflammation in asthma. American journal of physiology Lung cellular and molecular physiology. 2012;302:L300–7. doi: 10.1152/ajplung.00221.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Raff MC. Social controls on cell survival and cell death. Nature. 1992;356:397–400. doi: 10.1038/356397a0. [DOI] [PubMed] [Google Scholar]
- 326.Ward Patrick S, Thompson Craig B. Metabolic Reprogramming: A Cancer Hallmark Even Warburg Did Not Anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Panopoulos AD, Yanes O, Ruiz S, Kida YS, Diep D, Tautenhahn R, et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168–77. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Rathmell JC, Vander Heiden MG, Harris MH, Frauwirth KA, Thompson CB. In the absence of extrinsic signals, nutrient utilization by lymphocytes is insufficient to maintain either cell size or viability. Mol Cell. 2000;6:683–92. doi: 10.1016/s1097-2765(00)00066-6. [DOI] [PubMed] [Google Scholar]
- 329.Nicholson JK, Holmes E, Kinross JM, Darzi AW, Takats Z, Lindon JC. Metabolic phenotyping in clinical and surgical environments. Nature. 2012;491:384–92. doi: 10.1038/nature11708. [DOI] [PubMed] [Google Scholar]
- 330.Tennant DA, Duran RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267–77. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]