Abstract
The nonrandom distribution of meiotic recombination shapes heredity and genetic diversification. Theoretically, hotspots — favored sites of recombination initiation — either evolve rapidly toward extinction or are conserved, especially if they are chromosomal features under selective constraint, such as promoters. We tested these theories by comparing genome-wide recombination initiation maps from widely divergent Saccharomyces species. We find that hotspots frequently overlap with promoters in the species tested and, consequently, hotspot positions are well conserved. Remarkably, the relative strength of individual hotspots is also highly conserved, as are larger-scale features of the distribution of recombination initiation. This stability, not predicted by prior models, suggests that the particular shape of the yeast recombination landscape is adaptive, and helps in understanding evolutionary dynamics of recombination in other species.
Introduction
DNA double-strand breaks (DSBs) generated by the Spo11 protein initiate meiotic recombination, which alters genetic linkage and promotes pairing and accurate chromosome segregation (1). DSBs are distributed nonrandomly across genomes, occurring often within narrow regions called hotspots (2). Theoretical work exploring evolutionary dynamics of recombination has led to a prevailing hypothesis, the “hotspot paradox”, that predicts rapid hotspot extinction (3–7). This view rests on biased gene conversion, in which the broken chromosome copies genetic information from its uncut homolog, possibly generating an extra copy of a genetic variant (Fig. 1A). Consequently, hotspot alleles with different DSB activity deviate from a Mendelian segregation ratio, with less recombinationally active alleles overrepresented among the offspring. This type of meiotic drive is observed in yeast (8) and humans (9) and predicts that mutations that reduce or eliminate hotspot activity will be rapidly fixed in populations, while hotspot-activating mutations are rapidly extinguished (3, 5, 10). The paradox is that hotspots exist at all despite this drive against them.
Fig. 1.
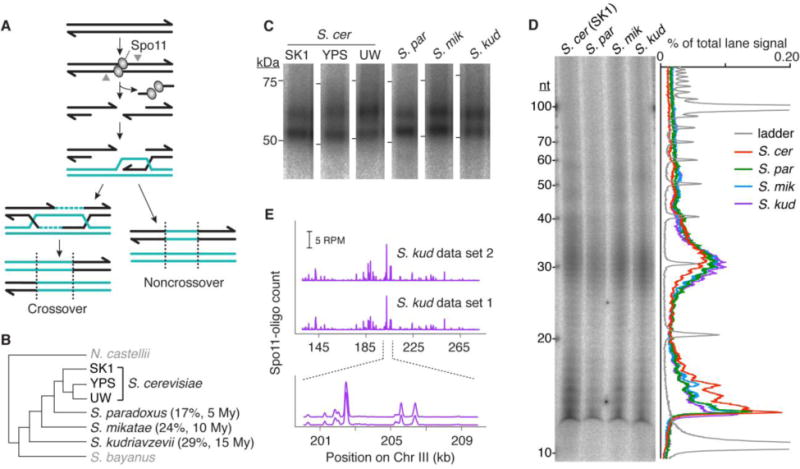
Generation of Spo11-oligo maps. (A) Meiotic recombination. Spo11 generates covalent protein-linked DSBs; endonucleolytic cleavage (grey arrowheads) liberates Spo11 bound to short oligos. DSB ends are resected and repaired to yield crossover or non-crossover products. The broken chromosome (black) copies information from the uncut allele (teal). (B) Schematic of Saccharomyces phylogeny (18, 20). Black, species/strains in this study; genic sequence divergence from S. cerevisiae (42) and estimated time since last common ancestor (20) are shown. YPS, YPS128; UW, UWOPS03-461.4. (C,D) Conserved sizes of Spo11 oligos. Immunoprecipitated, radiolabelled Spo11-oligo complexes were separated by SDS-PAGE (C), or were digested with proteinase K and resolved on a denaturing polyacrylamide gel (D). Autoradiographs (with lane traces in D) are shown. (E) Reproducibility of S. kudriavzevii Spo11-oligo maps. RPM, reads per million mapped; profiles were smoothed with 201-bp Hann window.
One answer to this paradox comes from PRDM9, a mammalian histone methyltransferase with an array of Zn-finger modules that rapidly evolve new DNA binding specificity (11). PRDM9 targets DSB formation near its binding sites, thus dictating hotspot positions. PRDM9 recognition motifs, which have no known intrinsic function, are lost quickly from genomes of humans and mice because of meiotic drive from biased gene conversion (12–14), but appearance of new PRDM9 alleles with different sequence specificity creates new hotspots and redraws the recombination landscape (11). This hotspot-targeting mechanism confirms the rapid extinction predicted by the hotspot paradox and explains how hotspots can nonetheless exist. However, most taxa (including yeast and some mammals) lack such a system, so it has remained unclear how generalizable this solution is.
An alternative view predicts that hotspot positions can be evolutionarily stable if Spo11 targets genomic features that are under selective constraint for functions unrelated to their roles as hotspots (8, 15). This hypothesis derives from correspondence of most hotspots in S. cerevisiae with promoter-containing intergenic regions (IGRs) (15). However, theoretical studies have considered this implausible as a mechanism to preserve hotspots (3, 10). Instead, many studies start from the assumption that hotspot lifespan must always be short and that the fine-scale recombination initiation landscape will always be highly dynamic over evolutionary scales (4, 6, 7, 16). This assumption is appropriate for primates and mice because they use PRDM9, but has not been evaluated for other taxa.
High resolution double-strand break maps in Saccharomycetes
To distinguish between these models, we asked whether the DSB landscape is conserved in yeast. Previously, population genetic data were used to deduce a recombination map in S. paradoxus and compare it to S. cerevisiae (17). Partial conservation was inferred, but the data had insufficient resolution to detect individual hotspots (15). We overcame these limitations by comparing high-resolution, whole-genome DSB maps between widely diverged Saccharomyces species and between S. cerevisiae strains (the laboratory strain SK1 and wild-derived strains YPS128 and UWOPS03-461.4) (18) (Fig. 1B, Table S1). DSB maps were generated by deep-sequencing of DNA oligonucleotides (oligos) covalently bound to Spo11 as a byproduct of DSB formation (15, 19) (Fig. 1A, Table S2).
The Saccharomyces sensu stricto clade last shared a common ancestor ~20 million years ago (20). We examined species ranging from S. paradoxus, with coding sequence divergence from S. cerevisiae comparable to that between humans and mice (~100 million years divergence), to S. kudriavzevii, roughly as distant as mammals from birds (~300 million years divergence) (21) (Fig. 1B). The S. cerevisiae strains chosen display 0.5–0.7% sequence divergence, comparable to the polymorphism density between humans and chimpanzees. Most differences are simple sequence polymorphisms (SNPs and small indels), with few large-scale structural differences aside from one discussed below (18, 22).
All yeasts examined underwent synchronous and efficient meiosis (fig. S1A), hence the strain SK1 is not anomalous in this regard. As in S. cerevisiae, two major size classes of Spo11-oligo complexes were observed (Figs. 1C, S1B), reflecting oligos of similar length distributions (Fig. 1D). Each oligo is a tag recording where Spo11 generated a DSB, and maps based on deep sequencing (23) agree spatially and quantitatively with direct detection of DSBs by Southern blot (15). Biological replicate maps were highly reproducible (Figs. 1E, S2) and most sequenced reads (>98%) were mapped uniquely (Table S2).
Targeting of breaks to promoters is conserved
We asked whether targeting of promoters is conserved among yeast. We mapped nucleosomes by sequencing micrococcal nuclease-resistant DNA (MNase-seq) from meiotic cultures (23). In S. cerevisiae, DSBs form preferentially in promoter-associated nucleosome-depleted regions (NDRs) (15, 24), and promoter chromatin structure during mitotic growth is conserved among other Saccharomyces species (25). Spo11 oligos were highly enriched in promoter NDRs in all species tested, whether examined at individual locations (Figs. 2A, S3A), or averaged across annotated genes (Figs. 2B, S3B). Many Spo11 oligos mapped to promoter-containing IGRs (i.e., IGRs flanked by divergent or tandemly oriented genes), whereas few mapped to convergent IGRs (i.e., lacking promoters) or within genes (Figs. 2D, S3D). We conclude that the Spo11 preference for promoters is a stable feature of the Saccharomyces DSB landscape.
Fig. 2.
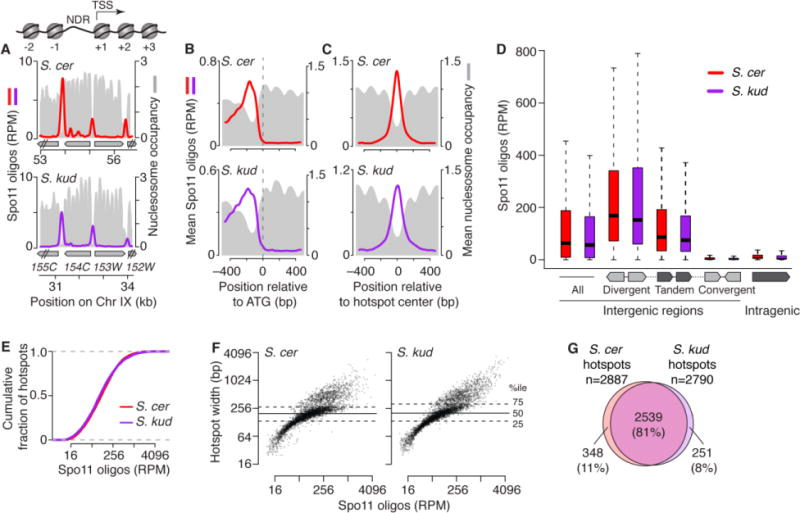
Conserved targeting of DSBs to promoters. (A) Overlap of DSB hotspots with promoter NDRs is evolutionarily conserved. The cartoon depicts typical yeast promoter chromatin structure, with an NDR upstream of the transcription start site (TSS). Sample region (around YIL154C) compares Spo11 oligos with the nucleosome map (MNase-seq read depth relative to genome average). (B) Average Spo11-oligo and nucleosome profiles around start codons (S. cer SK1, n=5766; S. kud, n=5578). (C) Average Spo11-oligo and nucleosome profiles at hotspots (S. cer SK1, n=4099; S. kud, n=3976). Spo11-oligo profiles were smoothed with 201-bp (A) or 75-bp (B,C) Hann window. (D) Spo11 oligos map preferentially to IGRs containing promoters. Box plots are as in fig. S3D. (E) Hotspot intensity varies over similar smooth continua in S. cerevisiae (SK1) and S. kudriavzevii. (F) Similar distributions of widths vs. Spo11-oligo counts in hotspots. (G) Conserved hotspot positions. Most promoter-containing IGRs hosting Spo11-oligo hotspots in S. cerevisiae (SK1) also had hotspots in S. kudriavzevii.
Similar numbers of Spo11-oligo hotspots (~4000) were identified in all species (Table S3). When ranked by Spo11-oligo count, hotspots formed a smooth continuum over a wide range, with nearly superimposable cumulative curves in all species (Figs. 2E, S3E). Hence, the distribution of DSBs among hotspots is the same. Hotspots had low average nucleosome occupancy (Figs. 2C, S3C) consistent with open chromatin structure providing a window of opportunity for Spo11 (26). The distribution of hotspot width was also nearly identical, with wider hotspots tending to have more Spo11 oligos (Figs. 2F, S3F). Conserved hotspot width agrees with conservation of NDR width observed previously (25). Importantly, most hotspots overlapped the same promoter-containing IGRs in all species examined (Figs. 2G, S3G). The low frequency of sex and outcrossing in yeasts could slow hotspot extinction compared to obligately outcrossed species (17), but the yeasts examined here have had ample sexual generations to allow biased gene conversion to erode hotspots. For example, there have been an estimated >200,000 outcrossed sexual generations since divergence of S. cerevisiae from S. kudriavzevii, comparable to the number of human sexual generations since divergence from chimpanzees (23). Thus, as predicted (8), DSB hotspot locations can be preserved when the targeted chromosome architecture is conserved.
Conservation of DSB frequency in hotspots
The hotspot paradox predicts that hotspot strength should vary widely even if their locations are conserved. Furthermore, the rate of hotspot extinction should scale with hotspot heat, because alleles that experience frequent DSBs provide more chances for loss (3–5, 10, 13). The selective constraint model is agnostic in this regard: if cis-acting sequence polymorphisms can quantitatively modulate DSB formation without ablating Spo11 targeting (which has been experimentally shown (e.g., 27)), then hotspot heats will change rapidly. On the other hand, if DSB frequency (not just position) is tied to selectively constrained features, or if DSB frequency is itself constrained, then hotspot heats will tend to be conserved.
To address this question, we summed Spo11 oligos within 3426 promoter-containing IGRs that could be stringently and unambiguously matched between species on the basis of conservation of flanking coding sequences (Fig. 3A, Table S4). This group contains 81% of divergent and tandem IGRs and accounts for 83% of promoter-proximal hotspots in S. cerevisiae, thus most of the relevant Spo11-targeted genomic space is included. An IGR-centric approach is preferable to relying on more arbitrary hotspot definitions (23). Within-IGR Spo11-oligo counts were highly similar between S. cerevisiae strains: we observed correlation coefficients (0.89–0.92) that were nearly as high as for comparisons between biological replicates (0.97–1.00) (Figs. 3B–D, S4A, Table S4). Thus, intra-species variation of DSB heat within these IGRs is low despite ~0.7–1% median sequence divergence.
Fig. 3.
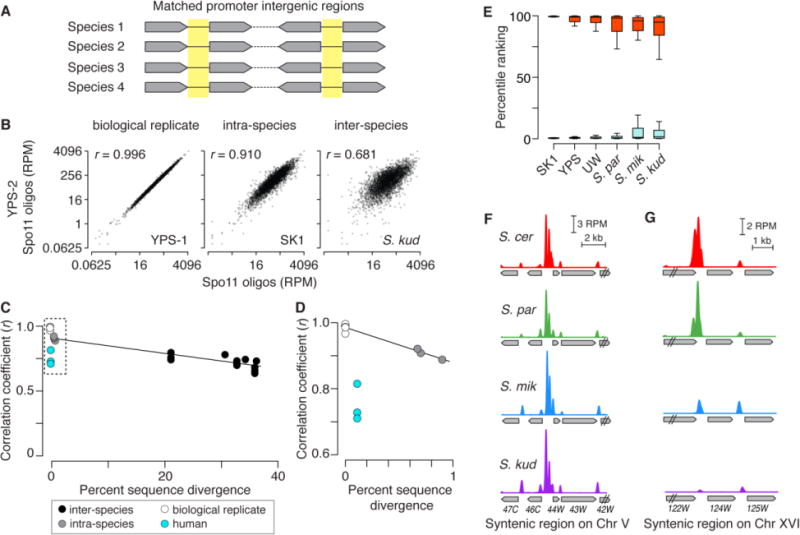
Conservation of hotspot strength. (A) Promoter-containing IGRs were matched between species using conservation of flanking genes. (B) Comparison of Spo11-oligo counts (log2 scale) within 3426 IGRs that were matched in all four species. Correlation coefficients for the log2-transformed data are shown (Pearson’s r). (C, D) Spo11-oligo counts in promoter IGRs remain highly correlated despite wide sequence divergence. Correlation coefficients (as in B) are plotted against the median sequence divergence within IGRs, which is substantially greater than the coding sequence divergence in Fig. 1B (22). D is a zoomed view of the boxed region in C. Black lines highlight the yeast comparisons; they are not regression lines. Human data (from ref. 28) are for three men with identical or similar PRDM9 alleles (37,345 hotspots (see fig. S4B); each had ~0.1% sequence difference from the reference genome (28)). (E) The hottest hotspots have stayed hot, and the coldest have stayed cold. Percentile rankings in other strains and species are shown for the matched promoter IGRs with the most (red) and least (cyan) Spo11 oligos in SK1 (top and bottom 1%). Box plots are as in fig. S3D. (F,G) Examples of a strong Spo11-oligo hotspot from SK1 whose heat is conserved (F, YEL046C) and one whose heat is not (G, YPR124W).
Strong correlations were also found between species, with little change in correlation strength over large evolutionary distances (Figs. 3B–D, S4A). Moreover, the hottest 1% of promoter IGRs in S. cerevisiae SK1 were enriched among the hottest IGRs in other species, with a median percentile ranking within the top 5% even in S. kudriavzevii (Fig. 3E, F). This was only modestly greater than the extent of conservation of the coldest IGRs (Fig. 3E). Theoretical modeling of biased gene conversion predicts that strong hotspots are less likely to be shared between species than weak ones (5). We found specific examples where strong hotspots in one species were substantially weaker in other species (Fig. 3G), so there is no absolute barrier to evolutionary changes. But the behavior of most IGRs leads us to conclude that the hottest hotspots present in the last common ancestor of Saccharomyces tended to retain high Spo11 target activity, and that it has been rare for ancestrally cold promoters to acquire strong hotspot activity.
This high degree of yeast hotspot conservation differs markedly from that in humans: DSB hotspot heat between men sharing the same or similar PRDM9 alleles (28) was less conserved than between S. cerevisiae strains despite much greater sequence identity (Fig. 3C, D, fig. S4B). This difference is consistent with PRDM9 motif erosion contributing to variation in hotspot strength between individuals (28).
Conservation of the DSB landscape over larger size scales
Hotspots are only one level of non-randomness in the DSB landscape in that they reside within larger domains of greater or lesser DSB potential (2, 15). In several taxa, conservation has been noted for the distribution of crossover recombination over broad genomic regions (16), but conservation of DSB distributions has not been evaluated. We therefore investigated if large-scale features of the DSB landscape are also conserved in yeast. Spo11-oligo maps demonstrated that DSB suppression observed near telomeres and centromeres (15, 26) is preserved (Figs. 4A, B, S5A, B). This result is not surprising, as recombination in these subchromosomal regions can interfere with genome integrity: subtelomeric regions are rife with repetitive DNA elements that can undergo nonallelic homologous recombination (29), and crossing over that occurs close to centromeres can cause segregation errors (30)).
Fig. 4.
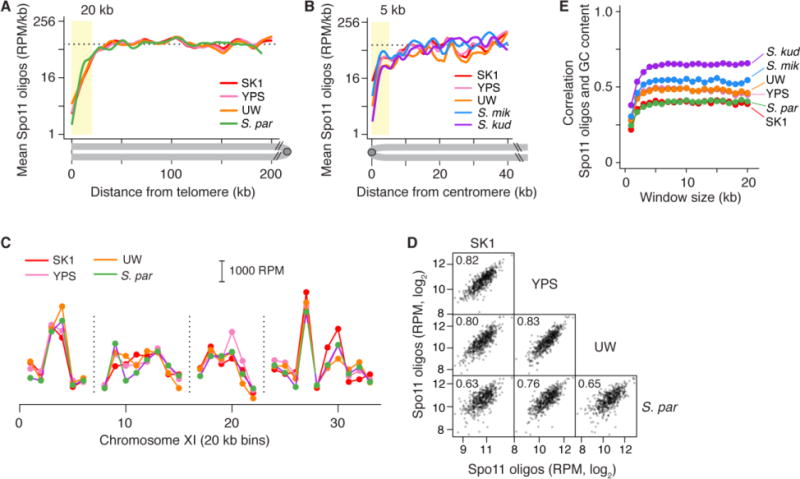
Conservation of large-scale features of the DSB landscape. (A,B) Telomere-proximal and pericentric DSB suppression. Lines are smoothed fit (Lowess) of Spo11-oligo densities in 500-bp bins averaged across 32 chromosome arms. Dashed line, genome average in SK1; yellow shading, DSB suppression zones. Genome assemblies are not complete enough to evaluate telomeres of S. mikatae or S. kudriavzevii. (C,D) Large-scale hot and cold interstitial domains are conserved. Interstitial segments (excluding subtelomeres and pericentromeres) were defined as syntenic between S. cerevisiae and S. paradoxus if orthologous genes were in the same order in both species. Spo11-oligo counts were summed in these syntenic segments divided into 20-kb bins (Table S5). A representative example is shown in panel C (vertical dashed lines denote synteny breaks, mostly from unresolved annotation discrepancies) and genome-wide scatter plots and correlation coefficients are in panel D. (E) Correlation (Pearson’s r) between mean Spo11-oligo counts and GC content binned in windows of varying size.
More remarkably, however, Spo11-oligo counts were also well correlated between species when we compared ~20-kb segments in syntenic regions across interstitial (i.e., non-telomeric and non-centromeric) portions of the chromosomes (Figs. 4C, D, S5C–F, Table S5). This scale is comparable to the average length of the chromatin loops of meiotic chromosomes, and DNA segments of this size typically encompass multiple hotspots (1, 15). These findings indicate that the larger-scale domain structure of the DSB landscape is also evolutionarily stable. Spo11-oligo counts were correlated with G+C content of DNA in each species tested, with weaker correlation over short distances (~1 kb) and stronger correlations over large distances (Fig. 4E). This scale-dependent pattern is consistent with the hypothesis that large-scale DSB domains, like hotspots, reflect selective constraint on the underlying chromosomal architecture (15). Furthermore, such large-scale domains presumably reflect factors — such as attachment of chromatin loops to chromosome axes — that work in cis but at a distance from DSB hotspots. Because such factors are too far to be frequently included in gene conversion tracts and are thus not subject to loss through biased gene conversion, they are not expected to evolve as rapidly as hotspots (3, 5, 16, 31).
Chromosome length affects DSB frequency
In S. cerevisiae, DSB density is anti-correlated with chromosome size, i.e., smaller chromosomes on average incur more DSBs per kb than larger ones (15). This relationship is conserved in other Saccharomycetes (Figs. 5A, S6). This whole-chromosome control of DSB density is in large part a patterning effect of a negative feedback circuit in which homologous chromosomes that have successfully engaged one another stop making additional DSBs (32). Perhaps smaller chromosomes tend to take more time to engage their homologs and thus enjoy a longer window of opportunity to make DSBs. It has been argued (15) that this form of DSB regulation can account for the earlier finding that smaller chromosomes undergo more crossing over per kb than larger chromosomes (33). However, whereas chromosome bisection and fusion experiments have demonstrated that difference in chromosome length is the cause of variation in crossover density (33), this has not been formally tested for DSBs.
Fig. 5.
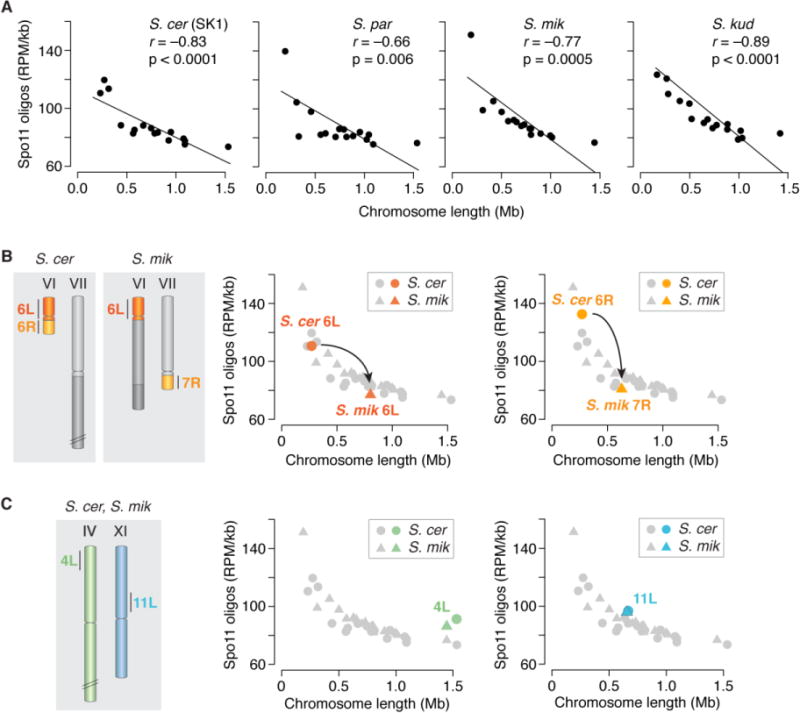
DSB density is influenced by chromosome length. (A) The anticorrelation between chromosome length and DSB density is conserved. Each point is one chromosome. (B) A natural experiment demonstrating chromosome length-dependent DSB control. The schematic illustrates syntenic segments on chromosomes of different size in S. cerevisiae and S. mikatae. The plots show that Spo11-oligo density is higher on these segments in S. cerevisiae (when on a short chromosome) than in S. mikatae (longer chromosomes). Gray symbols are whole-chromosome values from A for comparison. Note that the segments from ancestral chromosome VI display a Spo11-oligo density closely matched to the whole-chromosome value appropriate for the size of the chromosome on which they reside. (C) Control syntenic regions on similarly sized chromosomes have equivalent Spo11-oligo densities in both species.
S. mikatae provides a natural experiment, as reciprocal translocations have placed parts of ancestral chromosome VI onto longer chromosomes in that species (23) (Fig. 5B). DNA segments syntenic with the left and right arms of ancestral chromosome VI had a Spo11-oligo density predicted by their chromosome length: density was higher when the segments resided on the short chromosome VI in S. cerevisiae but lower when on longer chromosomes in S. mikatae (Fig. 5B). Syntenic segments on similar-length chromosomes exhibited matched Spo11-oligo densities (Fig. 5C). These findings indicate that whole-chromosome variation in DSB density is a direct consequence of chromosome size per se and is thus in large part extrinsic to the DNA sequence.
Conclusions
Our observations fit the hypothesis that hotspots tend to be stable if Spo11 targets functional genomic elements that are evolutionarily constrained (8). Conversely, evolutionary stability of DSB hotspots may indicate constrained function(s), even if that function is presently unknown. Interestingly, DSB hotspots are well conserved between the Schizosaccharomyces species S. pombe and S. kambucha (~0.5% sequence divergence) (34) despite mapping to sites without known function (35). In contrast, in Drosophila, which lacks a PRDM9-like system but also does not preferentially target recombination to promoters or known functional elements, the fine-scale distribution of recombination appears to evolve rapidly (36).
Strong conservation in Saccharomycetes of DSB frequencies within hotspots, across subchromosomal domains, and even across whole chromosomes supports the hypothesis that this conservation traces back to the DSB landscape being shaped by selectively constrained chromosomal features that work combinatorially, hierarchically, and over multiple size scales (15). For example, transcription, telomere and centromere function, and sister chromatid cohesion rely on and shape chromosome structures over scales ranging from tens to millions of base pairs. Because these structures in turn mold the DSB landscape, selective pressure to maintain them for gene expression, cell division, and other processes imposes a tendency to conserve the DSB landscape. However, the remarkable strength of conservation across millions of years of evolution leads us to speculate that the specific shape of the yeast DSB landscape may confer fitness benefits. The recombination distribution is a heritable trait subject to selection (16, 37), so we speculate that selective pressures may operate more directly on the DSB landscape genome-wide, perhaps related to accurate meiotic chromosome segregation and/or beneficial effects of disrupting or maintaining linkage groups at various size scales (37, 38).
Finally, we note that available evidence in plants, birds, and canids — all apparently lacking a PRDM9-like hotspot targeting mechanism — point to Spo11 acting preferentially at promoters, CpG islands, and/or other genomic elements that are under selective constraint to maintain functions separate from being Spo11 targets (39–41). In finches, high-resolution recombination maps inferred from population genetic data reveal evolutionary stability of recombination hotspots, analogous to Saccharomyces but wholly unlike PRDM9-reliant apes or mice (41). Thus, not only is it untrue that recombination initiation landscapes inevitably evolve rapidly, but conservation is likely to be a common pattern for many sexual species.
Supplementary Material
Acknowledgments
Sequencing data were deposited at the Gene Expression Omnibus (GEO) under accession number GSE71930. We are grateful to D. Greig, C. Hittinger, G. Liti, E. Louis, C. Müller, and J. Roop for providing strains and advice on culturing Saccharomyces species, A. Viale and the Integrated Genomics Operation (MSKCC) for sequencing, N. Socci (Bioinformatics Core Facility, MSKCC) for mapping Spo11-oligo and MNase-seq reads, S. Shuman for gifts of T4 RNA ligase, and S. Kim, J. Lange, N. Mohibullah, S. Tischfield, and X. Zhu for discussions. We thank M. Przeworski for discussions, communicating data prior to publication, and comments on the manuscript. This work was supported by NIH grant R01 GM058673. IL was supported in part by NIH fellowship F31 GM097861.
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Supplementary Materials:
Materials and Methods
References and Notes
- 1.de Massy B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu Rev Genet. 2013;47:563. doi: 10.1146/annurev-genet-110711-155423. [DOI] [PubMed] [Google Scholar]
- 2.Kauppi L, Jeffreys AJ, Keeney S. Where the crossovers are: recombination distributions in mammals. Nat Rev Genet. 2004;5:413. doi: 10.1038/nrg1346. [DOI] [PubMed] [Google Scholar]
- 3.Boulton A, Myers RS, Redfield RJ. The hotspot conversion paradox and the evolution of meiotic recombination. Proc Natl Acad Sci U S A. 1997;94:8058. doi: 10.1073/pnas.94.15.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calabrese P. A population genetics model with recombination hotspots that are heterogeneous across the population. Proc Natl Acad Sci U S A. 2007;104:4748. doi: 10.1073/pnas.0610195104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coop G, Myers SR. Live hot, die young: transmission distortion in recombination hotspots. PLoS Genet. 2007;3:e35. doi: 10.1371/journal.pgen.0030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friberg U, Rice WR. Cut thy neighbor: cyclic birth and death of recombination hotspots via genetic conflict. Genetics. 2008;179:2229. doi: 10.1534/genetics.107.085563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ubeda F, Wilkins JF. The Red Queen theory of recombination hotspots. J Evol Biol. 2011;24:541. doi: 10.1111/j.1420-9101.2010.02187.x. [DOI] [PubMed] [Google Scholar]
- 8.Nicolas A, Treco D, Schultes NP, Szostak JW. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature. 1989;338:35. doi: 10.1038/338035a0. [DOI] [PubMed] [Google Scholar]
- 9.Jeffreys AJ, Neumann R. Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nat Genet. 2002;31:267. doi: 10.1038/ng910. [DOI] [PubMed] [Google Scholar]
- 10.Pineda-Krch M, Redfield RJ. Persistence and loss of meiotic recombination hotspots. Genetics. 2005;169:2319. doi: 10.1534/genetics.104.034363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: localization and regulation. Nat Rev Genet. 2013;14:794. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- 12.Baker CL, et al. PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination. PLoS Genet. 2015;11:e1004916. doi: 10.1371/journal.pgen.1004916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole F, et al. Mouse tetrad analysis provides insights into recombination mechanisms and hotspot evolutionary dynamics. Nat Genet. 2014;46:1072. doi: 10.1038/ng.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Myers S, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan J, et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smukowski CS, Noor MA. Recombination rate variation in closely related species. Heredity. 2011;107:496. doi: 10.1038/hdy.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai IJ, Burt A, Koufopanou V. Conservation of recombination hotspots in yeast. Proc Natl Acad Sci U S A. 2010;107:7847. doi: 10.1073/pnas.0908774107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liti G, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Replansky T, Koufopanou V, Greig D, Bell G. Saccharomyces sensu stricto as a model system for evolution and ecology. Trends in ecology & evolution. 2008;23:494. doi: 10.1016/j.tree.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Dujon B. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 2006;22:375. doi: 10.1016/j.tig.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 22.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 23.Materials and methods are available as supporting material on Science Online.
- 24.Berchowitz LE, Hanlon SE, Lieb JD, Copenhaver GP. A positive but complex association between meiotic double-strand break hotspots and open chromatin in Saccharomyces cerevisiae. Genome Res. 2009;19:2245. doi: 10.1101/gr.096297.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsankov AM, Thompson DA, Socha A, Regev A, Rando OJ. The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol. 2010;8:e1000414. doi: 10.1371/journal.pbio.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lichten M. In: Recombination and Meiosis. Egel R, Lankenau DH, editors. Vol. 3. Springer-Verlag; Heidelberg: 2008. pp. 165–193. [Google Scholar]
- 27.White MA, Dominska M, Petes TD. Transcription factors are required for the meiotic recombination hotspot at the HIS4 locus in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1993;90:6621. doi: 10.1073/pnas.90.14.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pratto F, et al. DNA recombination. Recombination initiation maps of individual human genomes. Science. 2014;346:1256442. doi: 10.1126/science.1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louis EJ. The chromosome ends of Saccharomyces cerevisiae. Yeast. 1995;11:1553. doi: 10.1002/yea.320111604. [DOI] [PubMed] [Google Scholar]
- 30.Rockmill B, Voelkel-Meiman K, Roeder GS. Centromere-proximal crossovers are associated with precocious separation of sister chromatids during meiosis in Saccharomyces cerevisiae. Genetics. 2006;174:1745. doi: 10.1534/genetics.106.058933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters AD. A combination of cis and trans control can solve the hotspot conversion paradox. Genetics. 2008;178:1579. doi: 10.1534/genetics.107.084061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thacker D, Mohibullah N, Zhu X, Keeney S. Homologue engagement controls meiotic DNA break number and distribution. Nature. 2014;510:241. doi: 10.1038/nature13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaback DB, Guacci V, Barber D, Mahon JW. Chromosome size-dependent control of meiotic recombination. Science. 1992;256:228. doi: 10.1126/science.1566070. [DOI] [PubMed] [Google Scholar]
- 34.Zanders SE, et al. Genome rearrangements and pervasive meiotic drive cause hybrid infertility in fission yeast. eLife. 2014;3:e02630. doi: 10.7554/eLife.02630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fowler KR, Sasaki M, Milman N, Keeney S, Smith GR. Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res. 2014;24:1650. doi: 10.1101/gr.172122.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heil CS Smukowski, Ellison C, Dubin M, Noor MA. Recombining without hotspots: A comprehensive evolutionary portrait of recombination in two closely related species of Drosophila. bioRxiv. 2015 doi: 10.1101/016972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coop G, Przeworski M. An evolutionary view of human recombination. Nat Rev Genet. 2007;8:23. doi: 10.1038/nrg1947. [DOI] [PubMed] [Google Scholar]
- 38.Keller PJ, Knop M. Evolution of mutational robustness in the yeast genome: a link to essential genes and meiotic recombination hotspots. PLoS Genet. 2009;5:e1000533. doi: 10.1371/journal.pgen.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Auton A, et al. Genetic recombination is targeted towards gene promoter regions in dogs. PLoS Genet. 2013;9:e1003984. doi: 10.1371/journal.pgen.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi K, Henderson IR. Meiotic recombination hotspots – a comparative view. Plant J. 2015;83:52. doi: 10.1111/tpj.12870. [DOI] [PubMed] [Google Scholar]
- 41.Singhal S, et al. Stable recombination hotspots in birds. doi: 10.1126/science.aad0843. (co submission) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liti G, et al. High quality de novo sequencing and assembly of the Saccharomyces arboricolus genome. BMC genomics. 2013;14:69. doi: 10.1186/1471-2164-14-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kugou K, et al. Rec8 guides canonical Spo11 distribution along yeast meiotic chromosomes. Mol Biol Cell. 2009;20:3064. doi: 10.1091/mbc.E08-12-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scannell DR, et al. The Awesome Power of Yeast Evolutionary Genetics: New Genome Sequences and Strain Resources for the Saccharomyces sensu stricto Genus. G3. 2011;1:11. doi: 10.1534/g3.111.000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neale MJ, Keeney S. End-labeling and analysis of Spo11-oligonucleotide complexes in Saccharomyces cerevisiae. Methods Mol Biol. 2009;557:183. doi: 10.1007/978-1-59745-527-5_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murakami H, Keeney S. Temporospatial coordination of meiotic DNA replication and recombination via DDK recruitment to replisomes. Cell. 2014;158:861. doi: 10.1016/j.cell.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu X, Keeney S. High-Resolution Global Analysis of the Influences of Bas1 and Ino4 Transcription Factors on Meiotic DNA Break Distributions in Saccharomyces cerevisiae. Genetics. 2015 Aug 5; doi: 10.1534/genetics.115.178293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown CA, Murray AW, Verstrepen KJ. Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr Biol. 2010;20:895. doi: 10.1016/j.cub.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fischer G, James SA, Roberts IN, Oliver SG, Louis EJ. Chromosomal evolution in Saccharomyces. Nature. 2000;405:451. doi: 10.1038/35013058. [DOI] [PubMed] [Google Scholar]
- 50.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai IJ, Bensasson D, Burt A, Koufopanou V. Population genomics of the wild yeast Saccharomyces paradoxus: Quantifying the life cycle. Proc Natl Acad Sci U S A. 2008;105:4957. doi: 10.1073/pnas.0707314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liti G. The fascinating and secret wild life of the budding yeast S. cerevisiae. eLife. 2015;4 doi: 10.7554/eLife.05835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson LJ, et al. Population genetics of the wild yeast Saccharomyces paradoxus. Genetics. 2004;166:43. doi: 10.1534/genetics.166.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruderfer DM, Pratt SC, Seidel HS, Kruglyak L. Population genomic analysis of outcrossing and recombination in yeast. Nat Genet. 2006;38:1077. doi: 10.1038/ng1859. [DOI] [PubMed] [Google Scholar]
- 55.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khil PP, Smagulova F, Brick KM, Camerini-Otero RD, Petukhova GV. Sensitive mapping of recombination hotspots using sequencing-based detection of ssDNA. Genome Res. 2012;22:957. doi: 10.1101/gr.130583.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


