SUMMARY
Listeria monocytogenes is a food-borne pathogen that uses actin–dependent motility to spread between human cells. Cell-to-cell spread involves the formation by motile bacteria of plasma membrane-derived structures termed ‘protrusions’. In cultured enterocytes, the secreted Listeria protein InlC promotes protrusion formation by binding and inhibiting the human scaffolding protein Tuba. Here we demonstrate that protrusions are controlled by human COPII components that direct trafficking from the endoplasmic reticulum. Co-precipitation experiments indicated that the COPII proteins Sec31A and Sec13 interact directly with a Src Homology 3 domain in Tuba. This interaction was antagonized by InlC. Depletion of Sec31A or Sec13 restored normal protrusion formation to a Listeria mutant lacking inlC, without affecting spread of wild-type bacteria. Genetic impairment of the COPII component Sar1 or treatment of cells with brefeldin A affected protrusions similarly to Sec31A or Sec13 depletion. These findings indicated that InlC relieves a host-mediated restriction of Listeria spread otherwise imposed by COPII. Inhibition of Sec31A, Sec13, or Sar1 or brefeldin A treatment also perturbed the structure of cell-cell junctions. Collectively, these findings demonstrate an important role for COPII in controlling Listeria spread. We propose that COPII may act by delivering host proteins that generate tension at cell junctions.
INTRODUCTION
Many intracellular bacterial pathogens have evolved mechanisms to actively spread within human tissues (Gouin et al., 2005; Haglund and Welch, 2011; Ireton, 2013). One common mechanism involves the recruitment of filamentous (F)-actin in the host cytosol. F-actin forms a tail behind the microbe, propelling it through the human cell. The motile pathogen ultimately contacts and remodels the host plasma membrane into a protrusion. Finally, the pathogen-containing protrusion is engulfed by a neighboring cell. Bacteria that spread via F-actin–based motility include Shigella flexneri, Rickettsia spp., and Listeria monocytogenes. The mechanism of pathogen-directed F-actin assembly is well understood. In contrast, the process of protrusion formation is just beginning to be elucidated (Ireton, 2013; Ireton et al., 2014; Talman et al., 2014).
Listeria monocytogenes is a Gram-positive food-borne bacterium capable of causing gastroenteritis or invasive disease culminating in meningitis or abortion (Vazquez-Boland et al., 2001; Posfay-Barbe and Wald, 2009). Cell-to-cell spread of Listeria is initiated by the bacterial surface protein ActA, which induces the formation of F-actin ‘comet tails’ (Domann et al., 1992; Kocks et al., 1992). ActA is thought to be needed for spreading in all cell types. In cells that lack tight barriers, such as macrophages or fibroblasts, actin-dependent motility may be the sole factor governing bacterial protrusion formation and spread (Monack and Theriot, 2001). By contrast, in the polarized enterocyte cell line Caco-2 BBE1, the secreted Listeria protein InlC acts in conjunction with ActA to promote bacterial spread (Rajabian et al., 2009). InlC acts after F-actin tail formation to enhance the formation of bacterial protrusions (Rajabian et al., 2009; Rigano et al., 2014).
InlC promotes bacterial protrusions by physically interacting with and antagonizing a human scaffolding protein called Tuba (Rajabian et al., 2009). InlC binds to a Src Homology 3 (SH3) domain in the carboxyl terminus of Tuba, displacing the human actin regulatory protein N-WASP (Rajabian et al., 2009; Polle et al., 2014). Experiments involving RNA interference (RNAi)-mediated depletion of Tuba or N-WASP demonstrate that each of these proteins limits the spread of a Listeria mutant lacking inlC (ΔinlC), but not of wild-type bacteria (Rajabian et al., 2009). These findings indicate that InlC relieves the antagonism of protrusions normally imposed by Tuba/N-WASP complexes. Apart from Tuba and N-WASP, another host protein that regulates protrusions is the GTPase Cdc42. In uninfected cells, Cdc42 is activated by a Dbl Homology (DH) in Tuba (Salazar et al., 2003; Otani et al., 2006; Kodani et al., 2009; Rigano et al., 2014). During infection, wild-type Listeria inhibits Cdc42, an event critical for bacterial protrusion formation (Rigano et al., 2014). Impairment of host Cdc42 involves an incompletely characterized mechanism that is independent of InlC.
How bacterial antagonism of host Tuba, N-WASP, and Cdc42 promotes Listeria spread is not fully understood, but may involve perturbation of the host apical junction complex- a structure consisting of tight junctions and underlying adherens junctions (Miyoshi and Takai, 2005). Tuba, N-WASP, and Cdc42 are each needed to maintain the linearity of apical junctions in epithelial cells (Otani et al., 2006; Rajabian et al., 2009; Rigano et al., 2014). Such junctional linearity is thought to reflect cortical tension at the plasma membrane (Otani et al., 2006). Importantly, infection of host cells with wild-type Listeria induces slackening of apical junctions (Polle et al., 2014; Rajabian et al., 2009; Rigano et al., 2014). Bacteria deleted for the inlC gene fail to alter apical junctions, indicating that InlC is needed for this event. Collectively, these results led to the hypothesis that host Tuba, N-WASP, and Cdc42 have the potential to restrict bacterial protrusions by generating cortical tension that opposes the outward force exerted by motile bacteria on the host plasma membrane (Ireton, 2013; Rajabian et al., 2009; Rigano et al., 2014). By producing InlC, Listeria antagonizes Tuba and N-WASP, thereby relieving cortical tension and allowing efficient generation of bacterial protrusions.
Despite recent advances on the control of Listeria protrusion formation, the physiological processes that affect cell junctions and bacterial spread remain unknown. Tuba and its effectors N-WASP and Cdc42 promote many events in mammalian cells, including actin polymerization, cell motility, endocytic and exocytic trafficking of vesicles, the formation and maintenance of cell junctions, centrosome organization, and cell polarity (Otani et al., 2006; Kodani et al., 2009; Bryant et al., 2010; Harris and Tepass, 2010; Qin et al., 2010; Kovacs et al., 2011; Sato et al., 2012; Wu et al., 2014). Given the multiple physiological processes controlled by these effectors, dissecting which of these events impacts bacterial spread is challenging.
In this work, we report the identification of a novel Tuba interacting host protein that controls Listeria protrusions. Importantly, this Tuba ligand has an established function in the early secretory pathway, pointing to a host process that may regulate Listeria spread. We found that the carboxyl-terminal SH3 domain of Tuba, previously demonstrated to bind N-WASP (Salazar et al., 2003; Rajabian et al., 2009; Polle et al., 2014), also interacts directly with a complex containing the human proteins Sec31A and Sec13. Sec31A and Sec13 are components of the evolutionarily conserved COPII coat complex, which promotes trafficking from the endoplasmic reticulum to the cis-Golgi (Lord et al., 2013; Miller and Schekman, 2013). RNAi or dominant negative approaches demonstrate that Sec31A, Sec13, and other COPII components restrain protrusion formation of a ΔinlC mutant Listeria strain, but not of wild-type bacteria. Thus the negative role of COPII in Listeria spread resembles that previously reported for Tuba, N-WASP, and Cdc42, suggesting that COPII may act with these other host proteins to control bacterial protrusions. Biochemical experiments demonstrate that InlC displaces Sec31A from the Tuba carboxyl-terminal SH3 domain, indicating that this Listeria protein may disrupt Tuba/Sec31A complexes. Importantly, treatment of host cells with brefeldin A (BFA), a known inhibitor of ER-Golgi transport, restores normal protrusion production to a ΔinlC Listeria strain. Finally inhibition of Sec31A, Sec13, or other COPII components or treatment with BFA decreases the linearity of apical junctions, indicating a function for ER-Golgi transport in maintenance of junctional structure. Collectively, our findings support a model in which the bacterial protein InlC perturbs cell junctions and promotes Listeria spread by antagonizing COPII.
RESULTS
Tuba associates with human Sec31A and other COPII components
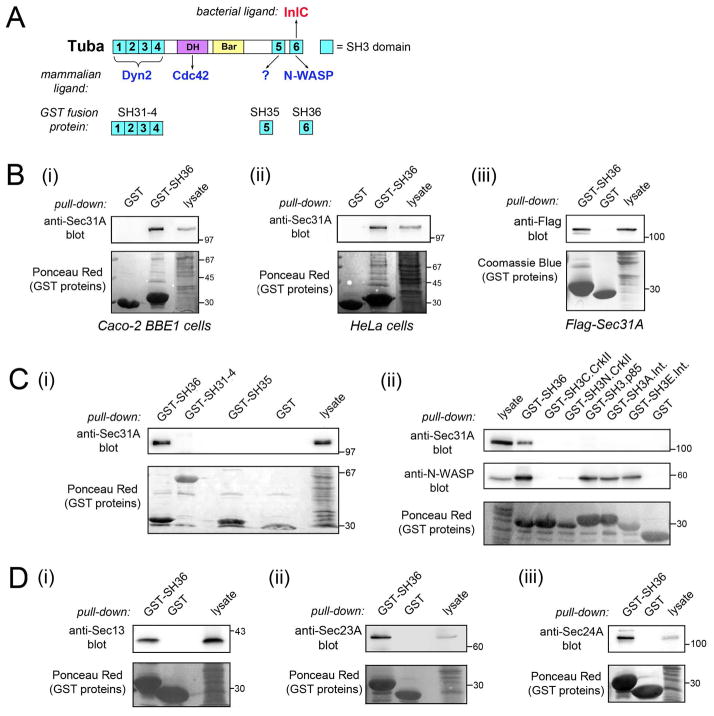
The Listeria protein InlC promotes cell-to-cell spread by binding and inhibiting the human scaffolding protein Tuba (Rajabian et al., 2009; Polle et al., 2014). Tuba has several functional domains, including a DH (Dbl Homology) domain, a Bar (Bin/Amphiphysin/Rvs) domain, and six SH3 domains (Fig. 1A) (Salazar et al., 2003). InlC interacts with the carboxyl-terminal SH3 domain, referred to as ‘SH36’ (Rajabian et al., 2009; Polle et al., 2014). This SH3 domain in Tuba is known to bind several human proteins, including the actin regulatory protein N-WASP (Salazar et al., 2003; Rajabian et al., 2009; Polle et al., 2014). By performing co-precipitation experiments with a glutathione S-transferase (GST) protein fused to SH36 (Figs. 1A, S1A), we identified human Sec31A as a novel ligand of the Tuba SH36 domain (Fig. 1B). Sec31A from the human cell lines Caco-2 BBE1 or HeLa associated with GST-SH36, but not with GST alone (Fig. 1Bi,ii). Flag-tagged human Sec31A expressed by transient transfection also interacted with GST-SH36 (Fig. 1Biii). By contrast, Sec31A did not co-precipitate with GST fused to other any other SH3 domain tested (Figs. 1C; S1A). These domains included the five remaining SH3 domains in Tuba or SH3 domains in the signaling proteins p85 (End et al., 1993), CrkII (Tanaka et al., 1995) or intersectin-1 (Hussain et al., 2001). Collectively, these results identify Sec31A as a previously unrecognized human ligand of the Tuba SH36 domain.
Figure 1. The Tuba SH36 domain associates with human Sec31A and other COPII components.
A. Diagram depicting functional domains in Tuba and known mammalian or bacterial ligands of these domains. DH (Dbl Homology) and Bar (Bin/Amphiphysin/Rvs) domains are indicated. SH3 domains are colored blue. ‘Dyn2’ is an abbreviation for Dynamin 2. SH3 domains present in GST fusion proteins used for co-precipitation experiments are shown below Tuba. B. Association of Sec31A with GST-SH36. Lysates from Caco-2 BBE1 cells (i) HeLa cells (ii), or HeLa cells transfected with a plasmid expressing Flag-tagged human Sec31A (iii) were incubated with GST-SH36 or GST alone, followed by precipitation of the GST protein. The presence of Sec31A was detected by Western blotting of precipitates with anti-Sec31A or anti-Flag antibodies. Lysates not subjected to precipitation were used as a positive control for detection of Sec31A. Stripped membranes were stained with Ponceau Red to confirm precipitation of GST proteins. Images in (i) and (ii) are of spliced gels in which irrelevant lanes had been excised. C. Specificity of interaction of Sec31A with various SH3 domains. Lysates of Caco-2 BBE1 cells were incubated with GST-SH36, GST alone, GST fused to other SH3 domains in Tuba (i) or GST fused to SH3 domains in mammalian proteins apart from Tuba (ii). Co-precipitation experiments were performed as described in B. Precipitates were Western blotted with anti-Sec31A antibodies. In ii, membranes were stripped and reacted with anti-N-WASP antibodies. N-WASP detection confirmed activity of some SH3 domains, since several of these domains are known to interact with N-WASP. SH3 domains tested in ii include a domain from the p85 regulatory subunit of type IA PI 3-kinase (SH3.p85), two domains from the adaptor protein CrkII (SH3N.CrkII and SH3C.CrkII), and two domains from the endocytic protein intersectin-l (SH3A.int and SH3E.int). D. Association of GST-SH36 with the COPII components Sec13. Sec23A, and Sec24A. Caco-2 BBE1 cell lysates were used for co-precipitation experiments and Western blotted with anti-Sec13 (i), Sec23A (ii), or anti-Sec24A (iii) antibodies. Experiments in parts B–D were performed at least three times, with similar results.
Sec31A is part of the evolutionarily conserved COPII complex, which mediates budding of vesicles from the ER (D’Arcangelo et al., 2013; Lord et al., 2013; Miller and Schekman, 2013). Apart from Sec31A, COPII contains the proteins Sar1, Sec23, Sec24, and Sec13 (Lord et al., 2013; Miller and Schekman, 2013). Sar1 is a GTPase which, when bound to GTP, initiates vesicle formation on ER membranes by recruiting the ‘inner coat’ proteins Sec23 and Sec24. Sec13 and Sec31A interact to form the vesicle ‘outer coat’. Importantly, we found that Sec13, Sec23A, and Sec24A from Caco-2 BBE1 cells each co-precipitated with GST fused to the Tuba SH36 domain (Fig. 1D). The ability of the Tuba SH36 domain to co-precipitate multiple COPII components may reflect an association between Tuba and a fully assembled COPII complex.
The Tuba SH3 domain binds directly to the Sec31A-Sec13 complex
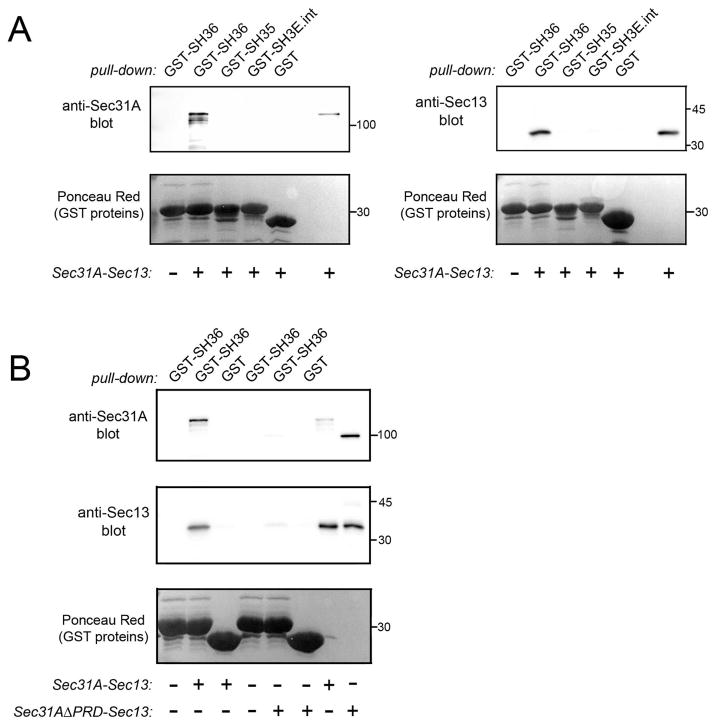
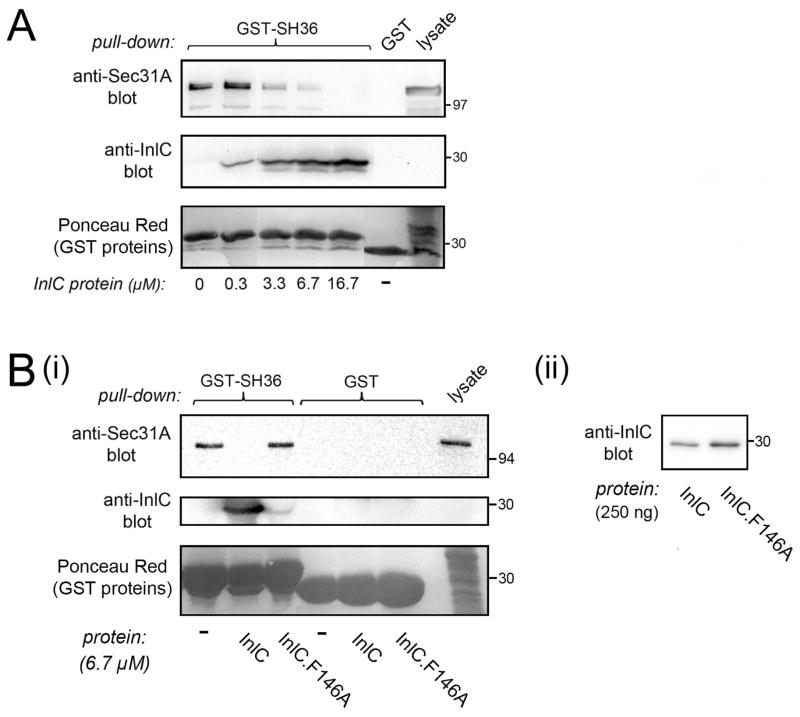
In order to determine if interaction between Tuba and COPII is direct, we employed a purified protein complex consisting solely of human Sec31A bound to Sec13 (Fig. S1Bi) (Stagg et al., 2006). The Sec31A-Sec13 complex was tested for binding to GST-SH36 in co-precipitation assays. As negative controls, GST alone or GST fused to other SH3 domains in Tuba (SH35) or intersectin-l (SH3E) were used. Importantly, GST-SH36 co-precipitated with the Sec31A-Sec13 complex, whereas the other GST proteins did not (Fig. 2A). These findings indicate that the Tuba SH36 domain binds directly to human Sec31A and/or Sec13.
Figure 2. The Tuba SH36 domain interacts directly with the Sec31A-Sec13 complex.
A. Binding of SH36 to wild-type Sec31A complexed with Sec13. Purified Sec31A-Sec13 complex was incubated with GST-SH36 and binding was assessed in co-precipitation assays as described in the Experimental Procedures. As negative controls, GST alone or GST fused to the SH35 domain of Tuba (GST-SH35) or the SH3E domain of intersectin-l (GST-SH3E.int) were used. Binding was detected by probing precipitates with antibodies against Sec31A (left panel) or anti-Sec13 (right panel). In the last lane, 100 ng of Sec31A-Sec13 protein was loaded as a control for antibody reactivity. B. Experiments with Sec31A deleted for its proline-rich domain (Sec31AΔPRD) complexed with Sec13. Co-precipitation experiments were performed with purified Sec31A-Sec13 or Sec31A-ΔPRD-Sec13 complexes and the indicated GST fusion proteins. The last two lanes contain 100 ng of purified Sec31A-Sec13 or Sec31AΔPRD-Sec13 complexes. Experiments in A and B were performed 3 times with similar results.
Most SH3 domains bind proline-rich peptides with the motif PxxP, where x is any amino acid (Mayer, 2001). Human Sec31A has a ~ 400 amino acid domain with five PxxP peptides (Tang et al., 2000). In order to determine if this proline-rich domain (PRD) in Sec31A contributes to binding Tuba, we tested the ability of SH36 to interact with a purified protein complex containing wild-type Sec13 and a mutant Sec31A protein deleted for the PRD (Sec31AΔPRD) (Fig. S1Bii). Compared to the complex of wild-type Sec31A and Sec13, the complex with Sec31AδPRD and Sec13 exhibited reduced binding to GST-SH36 (Fig. 2B). These data indicate that the PRD in Sec31A has an important role in interaction with the Tuba SH36 domain.
Sec31A and other COPII components control cell-to-cell spread of Listeria
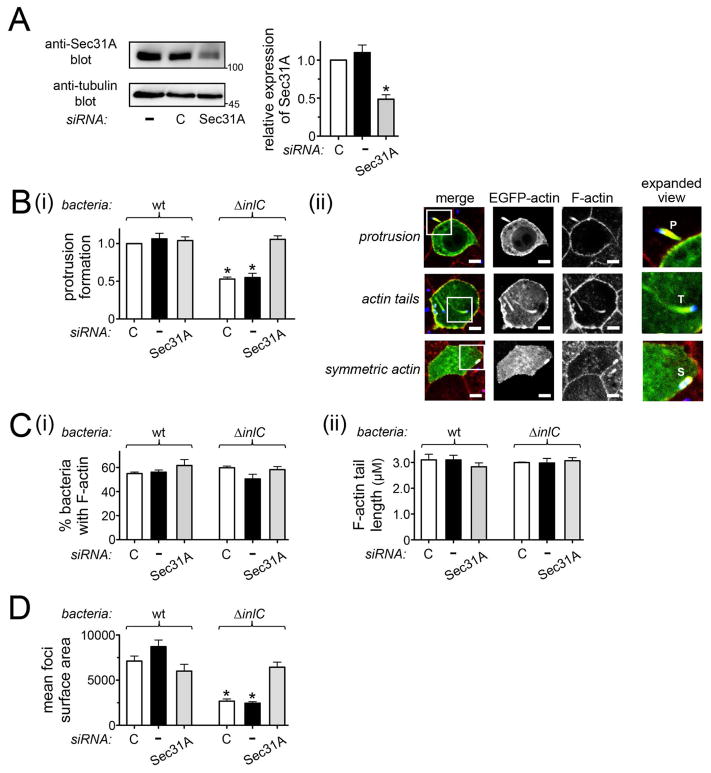
In order to investigate the role of Sec31A in Listeria dissemination, a short interfering RNA (siRNA) was used to inhibit Sec31A expression in Caco-2 BBE1 cells (Fig. 3A). The resulting effects on protrusion formation of wild-type bacteria or an isogenic mutant strain deleted for the inlC gene (ΔinlC) were assessed by a previously described confocal microscopy-based assay (Rajabian et al., 2009; Polle et al., 2014; Rigano et al., 2014) (Fig. 3B). Control conditions included mock transfection in the absence of siRNA or transfection with a control ‘non-targeting’ siRNA. As previously reported (Rajabian et al., 2009; Polle et al., 2014; Rigano et al., 2014), the efficiency of protrusion formation of the ΔinlC mutant was about 50% that of wild-type bacteria (Fig. 3B). These results indicate a role for InlC in the production of protrusions. Importantly, siRNA-mediated depletion of Sec31A restored normal protrusion formation to the ΔinlC mutant (Fig. 3B). This effect was observed with two different siRNAs targeting distinct sequences in Sec31A mRNA (Figs 3B and S2B). Sec31A depletion did not affect the proportion of ΔinlC bacteria in the main body of the cell that recruited F-actin or the length of F-actin comet tails (Fig. 3C). These latter findings indicate that Sec31A does not control actin-based motility of Listeria, and that the effect of this host protein on protrusion formation is therefore likely direct. In contrast to the situation observed with ΔinlC bacteria, Sec31A depletion did not significantly affect protrusions by wild-type Listeria (Fig. 3B). Taken together, the results in Figures 3A–C and S2B indicate that host Sec31A restrains protrusion formation by the ΔinlC mutant. By producing InlC, wild-type bacteria relieve this restraint.
Figure 3. Sec31A controls spread of Listeria.
A. RNAi-mediated depletion of Sec31A. Caco-2 BBE1 cells grown in transwells were either mock transfected in the absence of siRNA (−), or transfected with a control non-targeting siRNA (C) or an siRNA directed against Sec31A. Approximately 72 h after addition of siRNA, cells lysates were prepared and used for Western blotting analysis of Sec31A expression. A. Effect of Sec31A siRNA on target protein expression. A representative Western blot is in the left panel and quantification of Western blots from three experiments is presented in the right panel. B. Effect of Sec31A depletion on Listeria protrusion formation. After transfection with the indicated siRNA, Caco-2 BBE1 cells were infected with wild-type (wt) or ΔinlC strains of Listeria for 5.5 h prior to fixation and processing for confocal microscopy analysis. (i). Bar graph displaying mean relative protrusion formation values +/− SEM. (ii). Confocal microscopy images illustrating how protrusions were quantified. Protrusion efficiency is expressed as the proportion of total bacteria-associated F-actin structures in protrusions. These F-actin structures are defined as protrusions, actin tails, or symmetric actin. The top, middle, and bottom panels display human cells containing a Listeria protrusion, bacteria with F-actin tails, or bacteria decorated with symmetric F-actin, respectively. In the ‘merge’ image, EGFP-actin is green, total F-actin labeled with phalloidin is red, and bacteria are blue. Individual channels for EGFP-actin or F-actin (phalloidin labeling) are also shown. The regions in white boxes in the ‘merge’ images are expanded in the right panels. Protrusions (P) were identified as EGFP-positive actin tails projecting from transfected cells into neighboring EGFP-negative cells. Bacteria with actin tails (T) and symmetric F-actin (S) within the EGFP-positive cell body are shown. The scale bars indicate 5 micrometers. C. Bacterial-directed F-actin tail assembly in Sec31A-depleted cells. (i) The percentages of bacteria associated with F-actin (i) or the lengths of bacterial F-actin tails (ii) were quantified in the same samples used for protrusion analysis. D. Effect of Sec31A depletion on cell-to-cell spread of Listeria. Caco-2 BBE1 cells transfected with Sec31A or subjected to control conditions were infected with the indicated Listeria strains for 12 h, followed by processing for immunofluorescence and analysis by confocal microscopy. Spread was assessed by measuring surface areas of foci containing intracellular bacteria, as described in the Experimental Procedures. Mean relative surface areas +/− SEM of foci produced by wt or ΔinlC bacterial strains are presented. The data in A, B, and C are mean +/− SEM from three experiments, whereas results in D are from a single experiment, representative of three total experiments. *, P < 0.05 relative to control siRNA transfected cells infected with wt Listeria.
In addition to assessing bacterial protrusions, we also determined the effect of Sec31A on the final event of spreading. Cell-to-cell spread of wild-type and ΔinlC strains was evaluated by quantifying the surface area of foci resulting from infection of a monolayer of Caco-2 BBE1 cells, as previously described (Polle et al., 2014; Rigano et al., 2014). Consistent with the protrusion formation results, we found that siRNA-mediated depletion of Sec31A restored normal spreading to the ΔinlC mutant, without significantly affecting spread of wild-type Listeria (Fig. 3D).
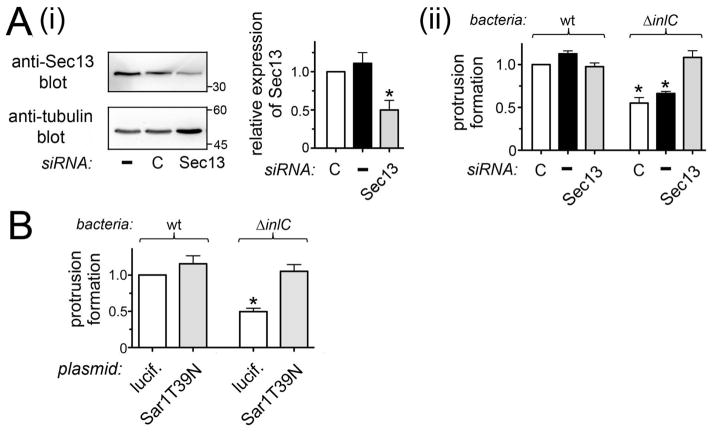
We next investigated the roles of other COPII components apart from Sec31A in Listeria protrusion formation. Importantly, siRNA-mediated depletion of Sec13 caused the same effect on Listeria protrusions as depletion of Sec31A. Specifically, inhibition of Sec13 expression restored normal protrusion efficiency to the ΔinlC bacterial mutant (Fig. 4A). Likewise, impairment of Sar1 using the GDP-restricted allele Sar1.T39N (Kuge et al., 1994) allowed the ΔinlC mutant to efficiently generate protrusions (Fig. 4B). Taken together, the results in Figures 3 and 4 indicate that the human COPII complex controls InlC-mediated spreading of Listeria.
Figure 4. The COPII components Sec13 and Sar1 control Listeria protrusion formation.
A. Effect of Sec13 depletion on Listeria protrusions. Caco-2 BBE1 cells were either mock transfected in the absence of siRNA (−), transfected with a control non-targeting siRNA (C), or transfected with an siRNA directed against Sec13. Approximately 72 h after addition of siRNA, cells lysates were prepared and used for analysis of Sec13 expression by Western blotting (i) or infected with wild-type (wt) or ΔinlC Listeria strains for 5.5 h followed by assessment of bacterial protrusions (ii). *, P < 0.05 relative to control siRNA transfected cells infected with wt Listeria. B. Impact of inhibition of Sar1 on Listeria protrusions. Caco-2 BBE1 cells were transfected with plasmids expressing an Ha-tagged dominant negative allele of human Sar1 (Sar1.T39N) or Ha-tagged luciferase as a control. About 72 h post-transfection, cells were infected with wt or ΔinlC Listeria strains and protrusion formation was quantified by confocal microscopy analysis. *, P < 0.05 relative to Ha-luciferase-expressing cells infected with wt bacteria. Data in A and B are each mean +/− SEM values from three experiments.
It is noteworthy that the protrusion phenotypes caused by depletion of Sec31A or Sec13 or inhibition of Sar1 (Figs. 3B, 4, S2B) are virtually identical to that produced by Tuba depletion (Rajabian et al., 2009). The similar roles of COPII and Tuba in bacterial protrusion formation combined with the interaction data in Figures 1 and 2 suggest that COPII and Tuba may act on the same pathway to control Listeria protrusions.
Inhibition of secretion with Brefeldin A affects Listeria spreading
A well-characterized function of COPII is to promote the formation of ER-derived vesicles that are subsequently delivered to the cis-Golgi (Lord et al., 2013; Miller and Schekman, 2013). If COPII controls Listeria spread by mediating ER-Golgi transport, then conditions that interfere with delivery of ER-derived vesicles to the Golgi, without directly affecting COPII, should impact bacterial protrusion formation. The fungal metabolite brefeldin A (BFA) causes fragmentation of the Golgi and inhibits ER-Golgi transport by blocking activation of the Golgi-associated GTPase Arf1 (Klausner et al., 1992; Zeghouf et al., 2005). Importantly, BFA does not target COPII components or affect the subcellular distribution of the COPII complex (Lippincott-Schwartz et al., 2000; Orci et al., 1993; Ward et al., 2001).
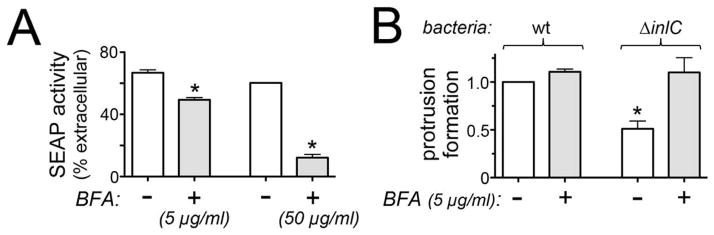
We investigated the effect of BFA on protrusion formation by Listeria. First, BFA was confirmed to inhibit secretion in Caco-2 BBE1 cells by measuring activity of secreted alkaline phosphatase (SEAP) (Berger et al., 1988; Kagan et al., 2004; Kim et al., 2007). When used at concentrations of 5 or 50 μg/ml, BFA caused an approximately 25% or 80% inhibition in SEAP secretion, respectively (Fig. 5A). Since lactate dehydrogenase assays indicated that 5 μg/ml BFA did not affect Caco-2 BBE1 cell viability (A. Gianfelice and K. Ireton, unpublished results), we used this lower concentration to examine the effect of BFA on Listeria protrusions. Caco-2 BBE1 cells were infected with Listeria for 1.5 h in the absence of BFA and then treated with BFA during the remaining 4.5 h of infection. The antibiotic gentamicin was included during the 4.5 h period, in order to ensure that any observed effects of BFA were due to post-entry events. Importantly, treatment of cells with 5 μg/ml BFA restored normal protrusion formation to the ΔinlC mutant Listeria without significantly affecting protrusions made by wild-type bacteria (Fig. 5B). These findings indicate that interference with protein trafficking is sufficient to affect Listeria protrusion formation.
Figure 5. Effects of brefeldin A (BFA) on protein secretion and Listeria protrusion formation.
Caco-2 BBE1 cells were transfected with a plasmid expressing Secreted Placental Alkaline Phosphatase (SEAP). Approximately 72 h later, cells were used for analysis of SEAP secretion or bacterial protrusion formation. A. SEAP activity. After treatment of Caco-2 BBE1 cells with the indicated concentrations of BFA (+) or with the vehicle DMSO for 1 h, levels of secreted (extracellular) SEAP were measured as described in the Experimental Procedures. *, P < 0.05 relative to cells treated with DMSO (−). B. Listeria protrusion formation. Caco-2 BBE1 cells were infected with wt or ΔinlC bacterial strains for 1 h followed by incubation for another 4.5 h in medium containing 5 μg/ml BFA (+) or DMSO (−). Samples of fixed and labeled cells were analyzed by confocal microscopy for quantification of bacterial protrusions. *, P < 0.05 relative to DMSO-treated cells (−) infected with wt Listeria. Data in A and B are mean +/− SEM from three experiments.
InlC displaces Sec31A from the Tuba SH36 domain
Results from RNAi experiments indicate that host COPII proteins or Tuba each inhibit protrusion formation of the ΔinlC Listeria mutant, but not of wild-type bacteria (Figs. 3B and 4) (Rajabian et al., 2009). These genetic data can be interpreted as meaning that InlC relieves the impairment in spread normally imposed by COPII and Tuba. The question remains as to how InlC antagonizes COPII. Since the Sec31A-Sec13 complex and InlC each bind directly to the Tuba SH36 domain (Polle et al., 2014; Rajabian et al., 2009) (Fig. 2), we determined if InlC was capable of disrupting Sec31A interaction with this domain. GST-SH36 was incubated with a constant amount of Caco-2 BBE1 cell lysate in the absence of InlC or in the presence of increasing concentrations of purified InlC protein. After precipitation of the GST fusion protein, the presence of Sec31A was detected by Western blotting. The results indicate that InlC inhibited association of Sec31A with the Tuba SH36 domain, when used at 3.3 μM or higher concentrations (Fig. 6A). As a specificity control, we employed a mutant InlC protein that contains a single amino acid substitution in phenylalanine 146 that compromises interaction with the Tuba SH36 domain (Polle et al., 2014). Whereas 6.7 μM InlC inhibited association of Sec31A with GST-SH36, the same molar concentration of InlC.F146A had no effect (Fig. 6B). Collectively, the results in Figure 6 indicate that wild-type InlC protein has the ability to displace human Sec31A from the Tuba SH36 domain.
Figure 6. InlC displaces Sec31A from the Tuba SH36 domain.
A. InlC-mediated inhibition in association of Sec31A with the SH36 domain. A constant amount of lysate from Caco-2 BBE1 cells was incubated with approximately 0.30 μM GST-SH36 and increasing concentrations (0–16.7 μM) of purified InlC. Sec31A in GST-SH36 precipitates was detected by Western blotting. The image is of a spliced gel in which irrelevant lanes had been excised. B. Control with the mutant protein InlC.F146A. (i). Caco-2 BBE1 cell lysates were incubated with approximately 0.30 μM GST-SH36 in the absence of competitor (−) or in the presence of 6.7 μM of InlC or InlC.F146A protein. GST alone (0.30 μM) was used as a negative control. Sec31A in precipitates was detected by Western blotting. Binding of InlC to GST-SH36 was confirmed by probing a stripped membrane with anti-InlC antibodies. (ii). Anti-InlC Western blot of purified wild-type and mutant InlC proteins. This experiment verified that the InlC.F146A protein does not have diminished reactivity to anti-InlC antibodies. Data in A and B are each representative of three experiments.
COPII components affect the morphology of tight junctions
The ability of InlC to promote cell-to-cell spread correlates with its effects on host apical junctions (Rajabian et al., 2009). Specifically, infection of Caco-2 BBE1 cells with wild-type Listeria or ectopic expression of InlC causes normally linear junctions to slacken, suggesting diminished cortical tension (Rajabian et al., 2009; Polle et al., 2014; Rigano et al., 2014). InlC-mediated perturbation of junctions appears to be due to inhibition of Tuba, since RNAi-mediated knockdown of this human protein affects junctions similarly to bacterial infection (Otani et al., 2006; Rajabian et al., 2009). The most direct interpretation of these data is that InlC promotes bacterial protrusion formation by relieving cortical tension that would otherwise be generated by Tuba (Rajabian et al., 2009).
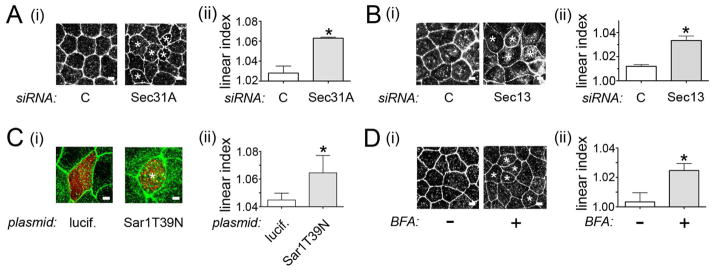
Since Sec31A binds to the Tuba SH36 domain in a manner that is inhibited by InlC (Fig. 6), we determined if COPII components control apical junction structure. RNAi-mediated depletion of Sec31A or Sec13 or dominant negative inhibition of Sar1 each increased the overall curvature of apical junctions in Caco-2 BBE1 cells (Figs. 7A–C, S2C). In addition, treatment of cells with 5 μg/ml BFA perturbed cell junctions in a similar manner (Fig. 7D). These data indicate a previously unrecognized function of the COPII complex and ER-Golgi trafficking in controlling apical junction structure. The results also suggest that the COPII complex may limit Listeria spread, at least in part, by contributing to junctional tension.
Figure 7. COPII components regulate apical junction morphology.
A. Effect of depletion of Sec31A on cell junctions. (i). About 72 h after transfection with control siRNA or an siRNA targeting Sec31A, Caco-2 BBE1 cells were fixed and labeled for the tight junction protein ZO-1. Images of ZO-1 labeling were acquired using a confocal microscope. (ii). Linear indexes were quantified as described in the Experimental Procedures. B. Impact of Sec13 depletion on cell junctions. Transfection, imaging of cell junctions (i), and measurement of linear indexes (ii) was performed as described in A. C. Perturbation of cell junctions by a dominant negative allele of Sar1. Caco-2 BBE1 cells transfected with plasmids expressing Ha-tagged Sar1.T39N or luciferase for 72 h were fixed and labeled for Ha (red) and ZO1 (green). Images of ZO-1 labeling (i) and linear index values (ii) are shown. D. Effect of BFA on cell junctions. Caco-2 BBE1 cells were treated with 5 μg/ml BFA for 2 h, followed by processing for confocal microscopy. Images of ZO-1 labeling (i) and quantification of linear indexes (ii) are shown. Asterisks in A–D indicate cells with curved junctions. Scale bars represent 3 μm. Linear index data in A–D are mean+/− SEM values from three experiments. *, P < 0.05 relative to control siRNA (C) treated cells.
DISCUSSION
In this work, we identify an important role for the host COPII complex in controlling cell-to-cell spread of Listeria in the polarized enterocyte cell line Caco-2 BBE1. Specifically, our findings indicate that COPII restrains protrusion formation by bacteria that lack the virulence protein InlC. Importantly, several COPII components including Sec31A, Sec13, Sec23A, and Sec24A, co-precipitate with a carboxyl-terminal SH3 domain in human Tuba, a scaffolding protein known to control Listeria spread (Rajabian et al., 2009). Two other lines of evidence suggest that InlC promotes Listeria protrusion formation, at least in part, through antagonism of COPII/Tuba complexes. First, biochemical data indicate that InlC can stimulate the disassociation of Sec31A from the Tuba SH36 domain. Second, genetic inhibition of several different COPII components affected Listeria spread similarly to a previously observed inhibition of Tuba (Rajabian et al., 2009). Specifically, depletion of Sec31A, Sec13, or dominant negative inhibition of Sar1 each restored normal protrusion formation to an inlC deletion mutant, without affecting protrusions made by wild-type bacteria. Collectively, these results support the idea that COPII and Tuba comprise a host pathway that negatively regulates Listeria spread in the absence of the InlC. By producing InlC, Listeria alleviates the inhibition in protrusions that would otherwise be imposed by COPII and Tuba.
Importantly, experiments with purified proteins indicate that the Sec31A-Sec13 complex, known to form the outer COPII coat (Miller and Schekman, 2013), directly interacts with the Tuba SH36 domain. Results with a Sec31A mutant protein deleted for its proline-rich domain (PRD) suggest that interaction with SH36 involves the PRD. The simplest interpretation of these results is that one or more PxxP peptides in the PRD mediate binding to the SH36 domain. In future work, it will be important to confirm this idea by testing individual PxxP peptides for interaction with SH36 and also by assessing the effects of mutation of these PxxP sequences on binding.
Although interaction of COPII components with the isolated Tuba SH36 domain was readily detected, it is worth mentioning that we were unable to confirm association of endogenous Tuba and Sec31A in co-immunoprecipitation experiments (P.H.B. Le and K. Ireton, unpublished results). There are several factors that could contribute to difficulties in detecting Tuba/Sec31A-Sec13 complexes. First, the SH36 domain in Tuba binds several human ligands apart from Sec31A-Sec13 (Salazar et al., 2003; Oda et al., 2014). Occupation of SH36 by these other ligands likely reduces detectability of Tuba/Sec31A-Sec13 complexes. Secondly, it is possible that full-length Tuba is subject to autoinhibition that controls binding of ligands to its SH36 domain. Tuba contains a Bar domain (Salazar et al., 2003). Several other Bar domain-containing proteins are autoinhibited through a mechanism involving interaction of their Bar and SH3 domains (Quan and Robinson, 2013). Finally, the weak nature of SH3 domain/ligand interactions would be expected to make detection by co-immunoprecipitation challenging. Ligands that engage SH3 domains through proline-rich peptides typically bind with affinities ranging from 1–100 μM (Mayer, 2001). Although we do not know the affinity of interaction of SH36 and Sec31A, it seems likely to be in the micromolar range. Experts have noted the difficulty in detecting interactions of micromolar affinities by co-immunoprecipitation (Castagnoli et al., 2004).
Interestingly, the role of COPII in Listeria protrusion formation resembles that previously described for the human actin regulatory protein N-WASP. Like the Sec31A-Sec13 complex, N-WASP binds directly to the Tuba SH36 domain and can be displaced from this domain by the InlC (Rajabian et al., 2009; Polle et al., 2014). In addition, RNAi-mediated depletion of N-WASP restores normal protrusion formation to ΔinlC mutant Listeria. The fact that the PRD in Sec31A contributes to binding SH36 suggests that Tuba might form separate (and perhaps sequential) complexes with N-WASP and COPII. This idea is based on the fact that SH3 domains cannot simultaneously accommodate more than one proline-rich peptide (Mayer, 2001). Future work is clearly required to understand the relationship between Tuba, N-WASP, and COPII.
How does COPII restrain bacterial spread in the absence of InlC? It seems plausible that COPII-mediated secretion in the host cell impairs the ability of ΔinlC mutant bacteria to make protrusions. This idea is supported by data indicating that treatment of cells with BFA, a widely used inhibitor of ER-Golgi transport (Klausner et al., 1992), restored normal protrusions to ΔinlC mutant bacteria. The lack of effect of COPII depletion or BFA treatment on protrusions of wild-type Listeria raises the possibility that InlC might attenuate ER-Golgi transport.
Although our genetic and pharmacological data imply that the early secretory pathway controls Listeria protrusions, direct evidence that InlC perturbs host secretion is currently lacking. Ectopic expression of an EGFP-InlC fusion protein that is functional in promoting Listeria protrusion formation failed to affect secretion of SEAP in Caco-2 BBE1 cells (Fig. S3A). Surprisingly, RNAi-mediated depletion of Sec31A also failed to reduce secretion of SEAP (Fig. S3B). A possible explanation for the apparent discrepancy between the bacterial spreading and host secretion data is that expression of EGFP-InlC or depletion of Sec31A might cause minor impairments in secretion that are difficult to detect in SEAP assays. Indeed, recent reports indicate that RNAi-mediated depletion of Sec31A, Sec13, or other COPII components cause little or no measurable inhibition in secretion of cargo thought to be trafficked through conventional sized COPII vesicles (Cutrona et al., 2013; Simpson et al., 2012; Townley et al., 2008). This is contrast to the strong impairment of trafficking of SEAP or other conventional cargo by mutant alleles of Sar1 (Kuge et al., 1994; Kagan et al., 2004) or BFA (de Silva et al., 1990; Irurzun et al., 1993; Kim et al., 2007; Clements et al., 2011). The reason for the lack of effect of depletion of COPII components is unclear, but may reflect functional redundancy among some component isoforms or incomplete knockdown of target proteins. Another potential explanation for our data is that COPII controls bacterial dissemination through a mechanism unrelated to ER-Golgi transport. To the best of our knowledge, the only proposed function for COPII apart from secretion is in the generation of autophagosomes (Ishihara et al., 2001; Zoppino et al., 2010; Ge et al., 2013; Graef et al., 2013). However, BFA does not impair autophagosome formation (Ge et al., 2013; Zoppino et al., 2010). Since BFA alters Listeria protrusion formation, it seems unlikely that COPII controls Listeria spread through autophagy. We favor the view that COPII mediates the secretion of one or more host plasma membrane proteins that control the generation of bacterial protrusions.
In addition to controlling Listeria spread, host COPII also regulated the structure of apical junctions in Caco-2 BBE1 cells. Inhibition of the COPII components, Sec31A, Sec13, or Sar1 resulted in cell junctions with increased curvature. These findings suggest a previously unappreciated function for COPII in maintaining cortical tension at apical junctions. How COPII affects cell junctions is not known, but may involve the exocytic delivery of one or more membrane protein involved in generating tension. Importantly, previous results demonstrate that depletion of Tuba or infection with wild-type, but not inlC mutant, Listeria increases junctional curvature (Rajabian et al., 2009; Polle et al., 2014; Rigano et al., 2014). In combination with these established findings, the results in the present study indicate that InlC likely perturbs apical junctions through inhibition of Tuba and COPII. This perturbation in cell junctions is thought to enhance bacterial spread by removing an inward tensile force that would otherwise counteract the outward force of motile bacteria in protrusions (Rajabian et al., 2009).
Apart from Listeria, several other microbial pathogens have aspects of infection that are impacted by the early secretory pathway (Pierini et al., 2009; von Bargen et al., 2012; Raymond et al., 2013). One notable case is enteropathogenic E. coli (EPEC), an extracellular bacterial pathogen that interferes with host ER-Golgi transport through injection of the microbial toxin NleA into human cells (Kim et al., 2007). NleA binds to Sec24 and impairs export of vesicles from the ER. NleA-Sec24 interaction is thought to contribute to diarrhea through disruption of tight junctions (Thanabalasuriar et al., 2013). Other microbes known to subvert trafficking from the ER or Golgi include poliovirus and the bacteria Legionella pneumophila, and Brucella spp. These pathogens each utilize Golgi- or ER-derived membrane to promote their intracellular replication. Poliovirus diverts host Golgi-derived membrane to form vesicles containing viral replication complexes (Belov et al., 2007). Legionella or Brucella replicate within vacuoles containing membrane derived from the ER (von Bargen et al., 2012) (Isberg et al., 2009; Hubber and Roy, 2010). Recruitment of ER membrane by these pathogens requires host COPII. A novel aspect of our work is the finding that COPII controls the intercellular dissemination of a microbial pathogen, rather than supporting its intracellular replication. Future studies should elucidate the molecular mechanism by which COPII controls Listeria protrusions. By focusing on the novel finding that Sec31A-Sec13 interacts with Tuba, such work also has the potential to lead to a better understanding of how mammalian COPII is regulated.
Experimental Procedures
Bacterial strains, mammalian cell lines, and media
The wild-type Listeria monocytogenes strain EGD and the isogenic ΔinlC strain with an in-frame deletion in the inlC gene were previously described (Engelbrecht et al., 1996; Rajabian et al., 2009). These strains were grown and prepared for infection of cultured human cells as described (Ireton et al., 1999). The human enterocyte cell line Caco-2 BBE1 (ATTC; CRL-2102) and the human epithelial cell line HeLa (ATTC CCL-2) were cultured as previously detailed (Dokainish et al., 2007; Rajabian et al., 2009; Rigano et al., 2014). Unless otherwise stated, Caco-2 BBE1 cells were grown on transwell permeable supports (Costar; 0.4 μm pore size) for 4–7 days, depending on the experiment. The passage number of these cells was always between 16 and 22.
Antibodies, siRNAs, plasmids, and other reagents
Rabbit polyclonal antibodies against recombinant InlC, human Sec31A, or human Sec13, were previously described (Tang et al., 2000; Rajabian et al., 2009). Commercially available primary antibodies used were rabbit anti- Listeria monocytogenes (Becton Dickenson; 223021), rabbit anti-Sec31A (Bethyl Laboratories; A302-336), rabbit anti-Sec13 (Bethyl Laboratories; A303-980A), rabbit anti-Sec23A (Cell Signaling; 8162), rabbit anti-Sec24A (Cell Signaling; 9678), rabbit anti-N-WASP (Cell Signaling; 4848), mouse anti-GM130 (BD Biosciences; 610822), mouse anti-Ha (Covance; clone 16B12), rabbit anti-Ha (Santa Cruz Biotechnology; sc-805), mouse anti-Flag (Sigma-Aldrich; F3165), mouse anti-GFP (Santa Cruz Biotechnology; sc-9996), mouse anti-occludin (Invitrogen; 33-1500), mouse anti-tubulin (Sigma; T5168), and mouse anti-ZO1 (Life Technologies; 33-9100). Secondary antibodies coupled to Alexa Fluor 488 or Alexa Fluor 647, and phalloidin conjugated to Alexa Fluor 555 were purchased from Life Technologies. siRNAs used to deplete human Sec31A were Sec31A (5′ ACACAGGAGAGGUGUUAUAUU -3′) and Sec31A-2 (5′-CCUGAAGUAUUCUGAUAAAUU- 3′). siRNA directed against Sec13 was Sec13 (5′-GUGAUGAUGCCUCAAGCAATT -3′). The negative, ‘non-targeting’ control siRNA molecule #1 (Catalogue no. D-001210-01) was from Dharmacon. This siRNA contains two or more mismatches with all sequences in the human genome, indicating that it should not target host mRNAs. The pEGFPC1 plasmid was from Clontech. The mammalian expression vectors encoding EGFP-tagged wild-type InlC, EGFP-tagged actin, or Flag-tagged human Sec31A (gift of M. Komada, Tokyo Institute of Technology) were previously described (Komada et al., 2006; Rajabian et al., 2009; Ong et al., 2010; Leung et al., 2013). The plasmid expressing Secreted Embryonic Alkaline Phosphatase (SEAP) under control of the CMV promoter (Addgene catalog number 24595) was from A. Cochrane (University of Toronto). pcDNA3.1 expressed Ha-tagged Sar1T39N (Celli et al., 2005) was a kind gift of J. Celli (Rocky Mountain Laboratories, NIAID). pcDNA3.1 expressing Ha-tagged luciferase was previously described (Sun et al., 2005). The pGex2T plasmid expressing glutathione S transferase (GST) alone was from GE Healthcare. Plasmids expressing GST fused to the first four (SH31-4), fifth (SH35), or sixth (SH36) Src Homology 3 (SH3) domains of human Tuba (Salazar et al., 2003; Rajabian et al., 2009), were gifts of P. De Camilli (Yale University). Plasmids expressing GST-InlC, 6 x histidine tagged InlC, and 6xhistidine tagged InlC.F146A were previously described (Rajabian et al., 2009; Polle et al. 2014). Plasmids allowing expression of GST fusion proteins containing the SH3 domain from bovine p85-alpha (End et al., 1993), the amino-terminal SH3 domain (SH3-N) of chicken CrkII (Tanaka et al., 1995), or the first or 5th SH3 domains (SH3A or SH3E) from human intersectin-l (Hussain et al., 2001) were gifts of M. Kasuga (Kobe University School of Medicine), B. Mayer (University of Connecticut), or P.S. McPherson (McGill University), respectively. A plasmid expressing GST fused to the carboxyl-terminal SH3 domain (SH3-C) of chicken CrkII was constructed through the polymerase chain reaction using Pfu DNA polymerase and pEBB-Ha.CrkII (Sun et al., 2005) as a template. Primers 5′-(CGC)GGATCCACCATGCTCCCTAACCTTCAG-3′ and 5′-(AATT)GCGGCCGCTCAGCTGAAGTCCTCATC-3′ (underlined sequences indicate BamHI and NotI sites, respectively). The resulting PCR products were subcloned into the BamHI/NotI sites of pGex2N (Tanaka et al., 1995), Brefeldin A (B7651) and the phosphatase substrate 4-nitrophenyl phosphate (P4744) were obtained from Sigma-Aldrich.
Plasmid DNA or siRNA transfection
Approximately 1×105 Caco-2 BBE1 cells were grown in transwells for ~ 24 hours prior to transfection. siRNA transfections were performed with 100 nM siRNA and 8–16 microliters of Lipofectamine LF2000 (LF2000) per transwell. Plasmid DNA transfection was performed as described (Rigano et al., 2014). In situations in which cells were transfected with both siRNA and plasmid DNA, siRNA transfections were performed first, and plasmid DNA transfections were carried out approximately 24 hours later. In cases involving co-transfection of two plasmids (Fig. 3B and 6C), 1 microgram of plasmid expressing Ha-Sar1T39N or Ha-luciferase and 2 micrograms of plasmid expressing EGFP-actin were used, respectively. About 72 hours after transfection, cells were solubilized for analysis of target protein expression or infected with Listeria strains.
Construction of cell lines stably expressing EGFP or EGFP-InlC
Caco-2 BBE1 cell lines stably expressing EGFP or EGFP-InlC were constructed by limiting dilution, essentially as described (Basar et al., 2005). Clones were selected using G418 at 1.0 mg/ml. Expression of EGFP-tagged constructs was confirmed by Western blotting with anti-GFP antibodies.
Protein purification
Recombinant 6 x histidine-tagged InlC, InlC.F146A, GST alone, or GST fused to InlC or the SH3 domains of Tuba, p85-alpha, CrkII, or intersectin-l were expressed and purified essentially as described (Ireton et al., 1999; Sun et al., 2005). In the case of InlC used in Fig. 6A, the GST tag was removed by thrombin-mediated proteolysis, according to the manufacturer’s protocol (GE Healthcare). Recombinant human wild-type Sec31A (Sec31L1) and his-Sec13 (GenBank accession codes NM_014933 and NM_183352, respectively) or mutant Sec31A deleted for its proline-rich domain (Sec31AΔPRD) and 6 x histidine-tagged Sec13 were expressed in baculovirus-infected insect cells and purified as previously described (Stagg et al., 2006).
GST fusion protein co-precipitations with cell lysates
Caco-2 BBE1 or HeLa cells (2 × 106) were grown in 75 cm2 flasks for approximately four days. Cells were then washed once in cold PBS and solubilized in lysis buffer (25 mM Hepes pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 10 mM sodium fluoride, 1 mM PMSF, 1 mM sodium orthovanadate, and 10 micrograms/ml each aprotinin and leupeptin). Insoluble material was removed by centrifugation for 10 min at 12,000 rpm at 4° C. In order to remove proteins that may interact with GST alone, a pre-clearance step was performed. Briefly, each milligram of Caco-2 BBE1 or HeLa cell supernatant was incubated with approximately 5 micrograms of GST coupled to glutathione sepharose 4B beads for 45 min with rotation at 4°C and then centrifuged at 6000 rpm for 5 min. Supernatants resulting from preclearance were then used for co-precipitation with glutathione sepharose 4B beads coupled to GST alone, or GST fused to SH3 domains from Tuba, p85-alpha, CrkII, or intersectin-l. Approximately 20 micrograms of GST protein was incubated with 2 milligrams of HeLa cell lysate or 4 milligrams of Caco-2 BBE1 cell lysate for 2 h with rotation at 4°C. GST protein precipitates were washed twice in lysis buffer and twice in wash buffer. The composition of wash buffer was identical to that of lysis buffer except 0.20% Triton X-100 was used. After washing, Laemmli sample buffer was added, precipitates were boiled, and samples were stored at −80°C before loading on SDS/polylacrylamide gels. Competition experiments in Fig. 6 were performed similarly to other co-precipitation experiments, except that InlC or InlC.F146A proteins were added to lysates 30 min prior to addition of 10 micrograms of GST-SH36 or GST proteins coupled to glutathione sepharose 4B.
Binding experiments with purified Sec31A-Sec13
The molar concentrations of proteins used in these experiments were approximately 0.50 μM of GST fusion protein and 0.0125 μM of Sec31A-Sec13 complex. In order to control for any non-specific interaction with GST, a pre-clearance step was performed. Briefly, 2.1 μg of Sec31A-Sec13 protein was incubated with 20 μg of GST prebound to glutathione sepharose 4B beads in 1.0 ml of cold binding buffer (1 x PBS pH 7.4, 1 mM CaCl2, 1 mM MgCl2, 0.2% Triton X-100, 0.2% BSA, 1 mM PMSF, 10 μg/ml each aprotinin and leupeptin) on a rotating wheel for 45 min at 4°C. Samples were then centrifuged for at 7000 rpm at 4°C to obtain GST precipitates. Supernatants were used for binding reactions with GST-SH36 or other GST proteins. GST-SH36, GST-SH35, or GST-SH3E (17.5 μg), or GST (15.0 μg) coupled to glutathione sepharose 4B beads were added to 1.0 ml of precleared supernatant containing Sec31A-Sec13 or Sec31AΔPRD-Sec13 proteins. Samples were incubated at 4°C on a rotating wheel for 1.5 h. Samples were then centrifuged at 7000 rpm and washed twice in cold binding buffer and three times in binding buffer lacking BSA. After addition of Laemmli sample buffer and boiling, samples were stored at −80°C.
Western blotting analysis
Western blotting of precipitates prepared with GST fusion proteins was performed by migration on 7.5% or 9% SDS/polyacrylamide gels and transfer to nitrocellulose or polyvinylidene difluoride (PVDF) membranes. Incubation of membranes with primary antibodies and secondary antibodies coupled to horseradish peroxidase, and detection with ECL or ECL Plus reagents were performed as described (Shen et al., 2000).
For assessment of siRNA-mediated depletion of Sec31A or Sec13, Caco-2 BBE1 cells were washed once in PBS, solubilized in RipA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, 10 mM NaF, 1 mM phenylmethylsulfonylfluoride [PMSF], 10 micrograms/ml each aprotinin and leupeptin), and supernatants remaining after centrifugation at 12,000 rpm were used for determination of protein concentrations using a bicinchoninic acid [BCA] kit (Pierce). Equal protein quantities from each sample were migrated on 7.5% SDS/polyacrylamide gels and transferred to PVDF membranes. After detection of signals corresponding to Sec31A or Sec13 with ECL Plus, membranes were stripped and re-probed with antibodies against tubulin to confirm equivalent sample loading. Quantification of relative Sec31A or Sec13 expression in samples was performed using Image J (version 1.45r) software. Integrated pixel densities in bands corresponding to Sec31A, Sec13, or tubulin were quantified and background was subtracted. The ratio of Sec31A:tubulin or Sec13:tubulin values were determined. Ratio values were then normalized to that of the no siRNA control in order to obtain relative Sec31A or Sec13 expression levels.
Secreted Embryonic Alkaline Phosphatase (SEAP) assays
Caco-2 BBE1 cells in transwells were transiently transfected with a plasmid expressing SEAP under control of the CMV promoter. In situations involving RNAi, siRNA transfections were performed about 24 h prior to transfection with the SEAP plasmid. SEAP activity was assessed 3–4 days after plasmid DNA transfection. SEAP activity was measured using the substrate 4-nitrophenylphosphate essentially as described (Berger et al., 1988). Extracellular SEAP activity produced over a 4 h period was assessed in culture supernatants from both the top and bottom chambers of transwells. Intracellular SEAP activity was determined using cell lysates prepared by incubation in DMEM with 0.2% Triton X-100 for 20 min, followed by centrifugation at 12,000 rpm for 3 min to remove insoluble debris. SEAP secretion was expressed as the percentage of total SEAP activity that was extracellular. In experiments involving brefeldin A (BFA), cells were treated with the indicated concentration of BFA or the vehicle DMSO for 1 h prior to assessment of SEAP activity and also during the 4 h period of SEAP accumulation in extracellular medium. A Varioskan Flash Multimode plate reader was used to measure absorbance at 405 nm. Absorbance measurements were performed in triplicate.
Quantification of bacterial protrusion formation
Protrusion efficiency was measured using a previously described assay involving the detection of protrusions projecting from human cells expressing EGFP or EGFP-tagged actin into surrounding EGFP-actin negative cells (Rajabian et al., 2009; Polle et al., 2014; Rigano et al., 2014). In the case of experiments in Figures 3B, 4, and 5B, Caco-2 BBE1 cells in transwells were transfected with a plasmid expressing EGFP-actin. Approximately 72 h after transfection, cells were infected with wild-type or ΔinlC strains of Listeria for 1.5 h in the absence of gentamicin and 4.0 h in the presence of gentamicin. In experiments involving BFA (Fig. 5B), cells were infected with Listeria strains in the absence of BFA for 1 h, followed by incubation in medium with 5 micrograms per ml BFA and gentamicin for another 4.5 h. Samples were fixed and mounted as described (Rajabian et al., 2009; Sun et al., 2005). Labelling was performed with anti-Listeria antibodies and phalloidin Alexa Fluor 555 to detect F-actin. Images of optical sections 1.0 μM in thickness were acquired using a Zeiss LSM 710 confocal microscope equipped with diode (405 nm), multiline argon (458/488/514 nm), helium-neon 1 (543 nm), and helium-neon 2 (633 nm) lasers. Protrusions were identified as F-actin comet tails emanating from EGFP-positive cells into neighboring EGFP-negative cells. Protrusions, F-actin comet tails in the cell body, and bacteria decorated with symmetric F-actin in the cell body of each EGFP-positive cell were quantified. Experiments in Figure 4B were performed similarly to those in Figures 3B, 4A, and 5B, except that Caco-2 BBE1 cells in transwells were co-transfected with plasmids expressing Ha-luciferase or Ha-Sar1T39N and EGFP-actin at a 1:2 ratio. Only cells positive for both Ha and EGFP-actin expression were used for analysis. For experiments involving cell lines stably expressing EGFP or EGFP-InlC (Fig. S3A), the stable cell lines were mixed with untransfected Caco-2 BBE1 cells at a ratio of 1:3 and seeded into transwells. Cells were grown for 4 days and used for bacterial infections as normal. Protrusions were identified as EGFP-positive structures emanating into surrounding EGFP-negative cells.
In all protrusion studies (Figs. 3B, 4, 5B, and S3A), approximately 50 protrusions were scored for the wild-type Listeria strain in control cells in each experiment. Control conditions were control (non-targeting) siRNA (Figs. 3B and 4A), Ha-luciferase expression (Fig. 4B), DMSO treatment in the absence of BFA (Fig. 5B), or EGFP expression (Fig. S3A). Corresponding numbers of total bacterial-associated actin structures were quantified for all other conditions. Protrusion efficiency was expressed as the percentage of total bacteria-associated F-actin structures in protrusions. Relative protrusion efficiencies were calculated by normalizing absolute percentage values to that of the wild-type strain.
Analysis of F-actin recruitment and tail length
Analyses of bacterial F-actin recruitment and tail length in control and Sec31A-depleted cells (Fig. 3C) were performed on the same samples used for evaluation of protrusion efficiencies. F-actin recruitment efficiencies were expressed as the percentage of total bacteria decorated with F-actin. Bacteria having symmetric F-actin or asymmetric comet tails were scored. In each experiment, 100–200 bacteria with F-actin were scored for cells treated with control siRNA and infected with wild-type Listeria. An equivalent number of bacteria were analyzed for the other conditions. Comet tail lengths were measured using Image J software as previously described (Rajabian et al., 2009; Rigano et al., 2014). The lengths of approximately 40 comet tails were measured for each condition in each experiment.
Measurement of cell-cell spread
Caco-2 BBE1 cell lines were transfected with control siRNA or an siRNA targeting Sec31A or mock transfected in the absence of siRNA. Approximately 72 h post-transfection, cells were infected with wild-type or ΔinlC strains of Listeria for 1.5 h in the absence of gentamicin followed by 10.5 h in the presence of gentamicin. After fixation in 3% PFA and permeabilization, samples were labeled with rabbit anti-Sec31A antibodies, anti-rabbit antibodies coupled to Alexa Fluor 488, and phalloidin conjugated to Alexa Fluor 555.
Spreading efficiencies of wild-type or ΔinlC bacterial strains were assessed by measuring the surface areas of foci containing bacteria decorated with F-actin (Fig. 3D), essentially as described (Rigano et al., 2014). Approximately 20 foci were measured for each condition in each experiment. Absolute surface areas were converted to relative surface areas by normalization to absolute values for EGFP-expressing cells infected with the wild-type Listeria strain.
Analysis of cell junction structure
Caco-2 BBE1 cells were transfected with control siRNA, siRNAs targeting Sec31A or Sec13, or with plasmids expressing Ha-tagged luciferase or Sar1T39N. Approximately 72 h after transfection, cells were fixed in 3% PFA and labeled with mouse antibodies against the tight junction proteins ZO-1 (Fig. 7) or occludin (Fig. S2B). Samples transfected with plasmids expressing Ha-tagged proteins were also incubated with rabbit antibodies against the Ha epitope. Secondary antibodies used were anti-mouse-Alexa Fluor 488 and anti-rabbit-Alexa Fluor 555. In experiments involving BFA treatment, 5 μg/ml of BFA or the vehicle DMSO were added to cells 2 hr before fixation and labeling. Images of optical sections 1.0 μM in thickness were obtained using a Zeiss LSM 710 confocal microscope. Cell junction morphology was evaluated by measuring the degree of junction linearity, as described (Otani et al., 2006; Rajabian et al., 2009; Polle et al., 2014; Rigano et al., 2014). This analysis involves the calculation of ‘linear index’ values, where an index of 1.0 indicates perfect linearity and values above 1.0 indicate curvature. In the case of samples subjected to plasma DNA transfection, only cells expressing Ha-tagged luciferase or Sar1.T39N were analyzed. Linear index values of approximately 100 cell junctions were calculated for each condition in each experiment.
Statistical analysis
Statistical analysis was performed using Prism (version 5.0a, GraphPad software). Analysis of variance (ANOVA) and the Tukey-Kramer posttest were employed when comparing more than two datasets. An unpaired Student’s t-test was used when comparing two data sets. A P value of 0.05 or lower was considered significant.
Supplementary Material
Figure S1. Purified proteins used in this work. A. GST fusion proteins containing various SH3 domains or GST alone. Approximately 20 micrograms of each protein was loaded. B. Purified Sec31A-Sec13 (i) or Sec31AΔPRD-Sec13 (ii) complexes. One microgram (Sec31A-Sec13) or 2.0 micrograms (Sec31AΔPRD-Sec13) of protein was loaded. Positions of Sec31A, Sec31AΔPRD, or Sec13 are indicated. C. InlC proteins used in competition experiments. (i). 2.0 micrograms of InlC used for experiments in Fig. 6A. (ii). 6 x histidine tagged InlC or InlC.F146A proteins used in Fig. 6B. The protein amounts loaded were 0.90 and 4.5 micrograms for InlC, and 0.80 and 4.0 micrograms for InlC.F146A. Proteins were migrated on 12% SDS/polyacrylamide gels and stained with Coomassie Brilliant Blue.
Figure S2. Effect of a second siRNA targeting Sec31A on Listeria protrusion formation and cell junction morphology. A. RNAi-mediated depletion of Sec31A. Caco-2 BBE1 cells in transwells were transfected with a control non-targeting siRNA (C) or an siRNA directed against Sec31A (Sec31A-2). Sec31A-2 siRNA was different from the siRNA used in Figure 3. Approximately 72 h post-transfection, cells lysates were prepared and used for analysis of Sec31A expression. A representative Western blot is shown in the left panel and quantification of Western blots from three experiments is in the right panel. B. Effect of the Sec31A-2 siRNA on Listeria protrusions. After siRNA transfection, Caco-2 BBE1 cells were infected with wild-type (wt) or ΔinlC strains of Listeria and processed for confocal microscopy analysis. The bar graph displays mean relative protrusion formation values +/− SEM. *, P < 0.05 relative to control siRNA treated cells infected with wild-type Listeria. C. Effect of Sec31A-2 siRNA on cell junctions. About 72 h after transfection with control (C) or Sec31A-2 siRNAs, Caco-2 BBE1 cells were fixed and labeled for the tight junction protein occludin. (i) Confocal microscopy images of occludin labeling. Scale bars indicate 3 micrometers. (ii). Quantification of linear indexes, as described in the Experimental Procedures. *, P < 0.05 relative to control siRNA treated cells (C) treated cells. Data in A and B are mean +/− SEM from three experiments.
Figure S3. Effect of InlC, Sec31A, or Tuba on protein secretion. A. Expression of EGFP-InlC does not alter secretion of SEAP. (i). Analysis of EGFP or EGFP-InlC expression in stable Caco-2 BBE1 cell lines by Western blotting. The arrow or arrowhead indicates EGFP-InlC or EGFP, respectively. (ii). Assessment of protrusion formation efficiency of wild-type (wt) or ΔinlC Listeria strains. (iii). Measurement of protein secretion through SEAP assays. B. RNAi-mediated depletion of Sec31A does not affect secretion of SEAP. (i). Confirmation of Sec31A depletion through Western blotting analysis (ii). Measurement of SEAP secretion in cells depleted for Sec31A, treated with control (C) siRNA, or mock transfected in the absence of siRNA (−). Results in A and B are mean +/− SEM from three experiments.
Acknowledgments
We thank Drs. J. Brumell, S.D. Gray-Owen, and P. Fineran for critically reviewing the manuscript. Yang Shen is gratefully acknowledged for constructing the GST-CrkII.SH3C plasmid, Yi Wang for help in cell line construction, Libby Mehrtens for purification of GST fusion proteins, and W.D Schubert for 6 x histidine tagged InlC constructs. This work was supported by grants from the NIH (R01GM086892) to S.S. and the Marsden fund of the Royal Society of New Zealand (UOO1003), Lottery Health Research New Zealand, the NIH (R01AI085072), and Canadian Institutes of Health Research (MT-15497) awarded to K.I.
References
- Basar T, Shen Y, Ireton K. Redundant roles for Met docking site tyrosines and the Gab1 pleckstrin homology domain in InlB-mediated entry of Listeria monocytogenes. Infect Immun. 2005;73:2061–2074. doi: 10.1128/IAI.73.4.2061-2074.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov GA, Altan-Bonnet N, Kovtunovych G, Jackson CL, Lippincott-Schwartz J, Ehrenfeld E. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J Virol. 2007;81:558–567. doi: 10.1128/JVI.01820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J, Hauber J, Hauber R, Geiger R, Cullen BR. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene. 1988;66:1–10. doi: 10.1016/0378-1119(88)90219-3. [DOI] [PubMed] [Google Scholar]
- Bergmann JE, Singer SJ. Immunoelectron microscopic studies of the intracellular transport of the membrane glycoprotein (G) of vesicular somatitis virus in infected Chinese hamster ovary cells. J Cell Biol. 1983;97:1777–1787. doi: 10.1083/jcb.97.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant DM, Datta A, Rodriguez-Fraticelli AE, Peranen J, Martin-Belmonte F, Mostov KE. A molecular network for de novo generation of the apical surface and lumen. Nat Cell Biol. 2010;12:1035–1045. doi: 10.1038/ncb2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagnoli L, Costantini A, Dall’Armi C, Gonfioni S, Montecchi-Palazzi L, Paoluzi S, Santonico E, Cesareni G. Selectivity and promiscuity in the interaction network mediated by protein recognition modules. FEBS Lett. 2004;567:74–79. doi: 10.1016/j.febslet.2004.03.116. [DOI] [PubMed] [Google Scholar]
- Celli J, Salcedo SP, Gorvel JP. Brucella coopts the small GTPase Sar1 for intracellular replication. Proc Natl Acad Sci USA. 2005;102:1673–1678. doi: 10.1073/pnas.0406873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements A, Smollet K, Lee SF, Hartland EL, Lowe M, Frankel G. EspG of enteropathogenic and enterohemorrhagic E. coli binds the Golgi matrix protein GM130 and disrupts Golgi structure and function. Cell Microbiol. 2011;13:1429–1439. doi: 10.1111/j.1462-5822.2011.01631.x. [DOI] [PubMed] [Google Scholar]
- Cutrona MB, Beznoussenko GV, Fusella A, Martella O, Moral P, Mironov AA. Silencing of mammalian Sar1 isoforms reveals COPII-independent protein sorting and transport. Traffic. 2013;14:691–708. doi: 10.1111/tra.12060. [DOI] [PubMed] [Google Scholar]
- D’Arcangelo JG, Stahmer KR, Miller EA. Vesicle-mediated export from the ER: COPII coat function and regulation. Biochim Biophys Acta. 2013;1833:2464–2472. doi: 10.1016/j.bbamcr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva AM, Balch WE, Helenius A. Quality control in the endoplasmic reticulum: folding and misfolding of vesicular stomatitis virus G protein in cells and in vitro. J Cell Biol. 1990;111:857–866. doi: 10.1083/jcb.111.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokainish H, Gavicherla B, Shen Y, Ireton K. The carboxyl-terminal SH3 domain of the mammalian adaptor protein CrkII promotes internalization of Listeria monocytogenes through activation of host phosphoinositide 3-kinase. Cell Microbiol. 2007;9:2497–24516. doi: 10.1111/j.1462-5822.2007.00976.x. [DOI] [PubMed] [Google Scholar]
- Domann E, Wehland J, Rohde M, Pistor S, Hartl M, Goebel W, et al. A novel bacterial gene in Listeria monocytogenes required for host cell microfilament interaction with homology to the proline-rich region of vinculin. EMBO J. 1992;11:1981–1990. doi: 10.1002/j.1460-2075.1992.tb05252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- End P, Gout I, Fry MJ, Panayotou G, Dhand R, Yonezawa K, et al. Biosensor approach to probe the structure and function of the p85α subunit of the phosphatidylinositol 3-kinase complex. J Biol Chem. 1993;268:10066–10075. [PubMed] [Google Scholar]
- Engelbrecht F, Chun SK, Ochs C, Hess J, Lottspeich F, Goebel W, Sokolovic Z. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol Microbiol. 1996;21:823–837. doi: 10.1046/j.1365-2958.1996.541414.x. [DOI] [PubMed] [Google Scholar]
- Ge L, Melville D, Zhang M, Schekman R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife. 2013;2:e00947. doi: 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouin E, Welch MD, Cossart P. Actin-based motility of intracellular pathogens. Curr Opin Microbiol. 2005;8:35–45. doi: 10.1016/j.mib.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Graef M, Friedman JR, Graham C, Babu M, Nunnari J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol Biol Cell. 2013;24:2918–2931. doi: 10.1091/mbc.E13-07-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund CM, Welch MD. Pathogens and polymers: Microbe-host interactions illuminate the cytoskeleton. J Cell Biol. 2011;195:7–17. doi: 10.1083/jcb.201103148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K, Tepass U. Cdc42 and vesicle trafficking in polarized cells. Traffic. 2010;11:1272–1279. doi: 10.1111/j.1600-0854.2010.01102.x. [DOI] [PubMed] [Google Scholar]
- Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- Hussain NK, Jenna S, Glogauer M, Quinn CC, Wasiak S, Guiponi M, et al. Endocytic protein intersectin-1 regulates actin assembly via Cdc42 and N-WASP. Nat Cell Biol. 2001;3:927–932. doi: 10.1038/ncb1001-927. [DOI] [PubMed] [Google Scholar]
- Ireton K. Molecular mechanisms of cell-cell spread of intracellular bacterial pathogens. Open Biol. 2013;3:130079. doi: 10.1098/rsob.130079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton K, Payrastre B, Cossart P. The Listeria monocytogenes protein InlB is an agonist of mammalian phosphoinositude-3-kinase. J Biol Chem. 1999;274:17025–17032. doi: 10.1074/jbc.274.24.17025. [DOI] [PubMed] [Google Scholar]
- Ireton K, Rigano LA, Polle L, Schubert WD. Molecular mechanism of protrusion formation during cell-to-cell spread of Listeria. Front Cell Infect Microbiol. 2014 Feb 21; doi: 10.3389/fcimb.2014.00021. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irurzun A, Perez L, Carrasco L. Brefeldin A blocks protein glycosylation and RNA replication of vesicular stomatitis virus. FEBS Lett. 1993;336:496–500. doi: 10.1016/0014-5793(93)80863-p. [DOI] [PubMed] [Google Scholar]
- Isberg RR, O’Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, et al. Autophagosome requires specific early Sec proteins for its formation. Mol Biol Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Pahuja KB, Wickliffe KE, Gorur A, Baumgärtel C, Schekman R, Rape M. Ubiquitin-dependent regulation of COPII coat size and function. Nature. 2012;482:495–500. doi: 10.1038/nature10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan JC, Stein MP, Pypaert M, Roy CR. Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J Exp Med. 2004;199:1201–1211. doi: 10.1084/jem.20031706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Thanabalasuriar A, Chaworth-Musters T, Fromme JC, Frey EA, Lario PI, et al. The bacterial virulence factor NleA inhibits cellular protein secretion by disrupting mammalian COPII function. Cell Host Microbe. 2007;2:160–171. doi: 10.1016/j.chom.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Kim SD, Pahuja KB, Ravazzola M, Yoon J, Boyadjiev SA, Hammamoto S, et al. The Sec23-Sec31 interface plays critical role for export of procollagen from the endoplasmic reticulum. J Biol Chem. 2012;287:10134–10144. doi: 10.1074/jbc.M111.283382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintscher C, Wuertenberger S, Eylenstein R, Uhlendorf T, Groemping Y. Autoinhibition of GEF activity in Intersectin 1 is mediated by the short SH3-DH domain linker. Protein Sci. 2010;19:2164–2174. doi: 10.1002/pro.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausner RD, Donaldson JG, Lippincott-Schwartz J. Brefeldin A: insights into the control of membrane traffic and organelle structure. J Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. Listeria monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell. 1992;68:521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- Kodani A, Kristensen I, Huang L, Sutterlin C. GM130-dependent control of Cdc42 activity at the Golgi regulates centrosome organization. Mol Biol Cell. 2009;20:1192–1200. doi: 10.1091/mbc.E08-08-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komada A, Tani K, Yamamoto A, Kitamura N, Komada M. The Ca2+-binding protein ALG-2 is recruited to endoplasmic reticulum exit sites by Sec31A and stabilizes the localization of Sec31A. Mol Biol Cell. 2006;17:4876–4887. doi: 10.1091/mbc.E06-05-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs EM, Verma S, Thomas SG, Yap AS. Tuba and N-WASP function cooperatively to position the central lumen during epithelial cyst morphogenesis. Cell Adh Migr. 2011;5:344–350. doi: 10.4161/cam.5.4.16717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge O, Dascher C, Orci L, Rowe T, Amherdt M, Plutner H, Ravazzola M, et al. Sar1 promotes vesicle budding from the endoplasmic reticulum but not Golgi compartments. J Cell Biol. 1994;125:51–65. doi: 10.1083/jcb.125.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung N, Gianfelice A, Gray-Owen SD, Ireton K. Impact of the Listeria monocytogenes protein InlC on infection in mice. Infec Immun. 2013;81:1334–1340. doi: 10.1128/IAI.01377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Roberts TH, Hirschberg K. Secretory protein trafficking and organelle dynamics in living cells. Annu Rev Cell Dev Biol. 2000;16:557–589. doi: 10.1146/annurev.cellbio.16.1.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Ferro-Novick S, Miller EA. The highly conserved COPII coat complex sorts cargo from the endoplasmic reticulum and targets it to the golgi. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a013367. pii: a013367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer BJ. SH3 domains: complexity in moderation. J Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- Miller EA, Schekman R. COPII- a flexible vesicle formation system. Curr Opin Cell Biol. 2013;25:420–427. doi: 10.1016/j.ceb.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi J, Takai Y. Molecular perspective on tight-junction assembly and epithelial polarity. Adv Drug Deliv Rev. 2005;57:815–855. doi: 10.1016/j.addr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Monack DM, Theriot JA. Actin-based motility is sufficient for bacterial membrane protrusion formation and host cell uptake. Cell Microbiol. 2001;3:633–647. doi: 10.1046/j.1462-5822.2001.00143.x. [DOI] [PubMed] [Google Scholar]
- Oda Y, Otani T, Ikenouchi J, Furuse M. Tricellulin regulates junctional tension of epithelial cells at tricellular contacts through Cdc42. J Cell Sci. 2014;127:4201–4212. doi: 10.1242/jcs.150607. [DOI] [PubMed] [Google Scholar]
- Ong YS, Tang BL, Loo LS, Hong W. p125a exists as part of the mammalian Sec13/Sec31 COPII subcomplex to facilitate ER-Golgi transport. J Cell Biol. 2010;190:331–345. doi: 10.1083/jcb.201003005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Perrelet A, Ravazzola M, Wieland FT, Schekman R, Rothman JE. ‘BFA bodies”: a subcompartment of the endoplasmic reticulum. Proc Natl Acad Sci USA. 1993;90:11089–11093. doi: 10.1073/pnas.90.23.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani T, Ichii T, Aono S, Takeichi M. Cdc42 GEF Tuba regulates the junctional configuration of simple epithelial cells. J Cell Biol. 2006;175:135–146. doi: 10.1083/jcb.200605012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierini R, Cottam E, Roberts R, Wileman T. Modulation of membrane traffic between endoplasmic reticulum, ERGIC and Golgi to generate compartments for the replication of bacteria and viruses. Semin Cell Dev Biol. 2009;20:828–833. doi: 10.1016/j.semcdb.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polle L, Rigano L, Julian R, Ireton K, Schubert WD. Structural details of human Tuba recruitment by InlC of Listeria monocytogenes elucidates bacterial cell-cell spreading. Structure. 2014;22:304–314. doi: 10.1016/j.str.2013.10.017. [DOI] [PubMed] [Google Scholar]
- Posfay-Barbe KM, Wald ER. Listeriosis. Semin Fetal Neonatal Med. 2009;14:228–233. doi: 10.1016/j.siny.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJM, Lippincott-Schwartz J. ER-to-golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Quan A, Robinson PJ. Syndapin- a membrane remodelling and endocytic F-BAR protein. FEBS J. 2013;280:5198–5212. doi: 10.1111/febs.12343. [DOI] [PubMed] [Google Scholar]
- Qin Y, Meisen WH, Macara IG. Tuba, a Cdc42 GEF, is required for polarized spindle orientation during epithelial cyst formation. J Cell Biol. 2010;189:661–669. doi: 10.1083/jcb.201002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajabian T, Gavicherla B, Heisig M, Muller-Altrock S, Goebel W, Gray-Owen SD, Ireton K. The bacterial virulence factor InlC perturbs apical cell junctions and promotes cell-to-cell spread of Listeria. Nat Cell Biol. 2009;11:1212–1218. doi: 10.1038/ncb1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond B, Young JC, Pallet M, Endres RB, Clements A, Frankel G. Subversion of trafficking, apoptosis, and innate immunity by type III secretion system effectors. Trends Microbiol. 2013;21:430–441. doi: 10.1016/j.tim.2013.06.008. [DOI] [PubMed] [Google Scholar]
- Rigano L, Dowd GC, Rigano L, Ireton K. Listeria monocytogenes antagonizes the human GTPase Cdc42 to promote bacterial spread. Cell Microbiol. 2014;16:1068–1079. doi: 10.1111/cmi.12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar MA, Kwiatkowski AV, Pellegrini L, Cestra G, Butler MH, Rossman KL, et al. Tuba, a novel protein containing Bin/Amphiphysin/Rvs and Dbl homology domains, links dynamin to regulation of the actin cytoskeleton. J Biol Chem. 2003;278:49031–49043. doi: 10.1074/jbc.M308104200. [DOI] [PubMed] [Google Scholar]
- Sato M, Kitaguchi T, Numano R, Ikematsu K, Kakeyama M, Murata M, et al. The small GTPase Cdc42 modulates the number of exocytosis-competent dense-core vesicles in PC12 cells. Biochem Biophys Res Commun. 2012;420:417–421. doi: 10.1016/j.bbrc.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Scales SJ, Pepperkok R, Kreis TE. Visualization of ER-to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell. 1997;90:1137–1148. doi: 10.1016/s0092-8674(00)80379-7. [DOI] [PubMed] [Google Scholar]
- Shen Y, Naujokas M, Park M, Ireton K. InlB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell. 2000;103:501–510. doi: 10.1016/s0092-8674(00)00141-0. [DOI] [PubMed] [Google Scholar]
- Simpson JC, Joggerst B, Laketa V, Verissimo F, Cetin C, Holger E, et al. Genome-wide RNAi screening identifies human proteins with a regulatory function in the early secretory pathway. Nat Cell Biol. 2012;14:764–774. doi: 10.1038/ncb2510. [DOI] [PubMed] [Google Scholar]
- Stagg SM, Gürkan C, Fowler DM, LaPointe P, Foss TR, Potter CS, Carragher B, Balch WE. Structure of the Sec13/31 COPII coat cage. Nature. 2006;439:234–238. doi: 10.1038/nature04339. [DOI] [PubMed] [Google Scholar]
- Sun H, Shen Y, Dokainish H, Holgado-Madruga M, Wong A, Ireton K. Host adaptor proteins Gab1 and CrkII promote InlB-dependent entry of Listeria monocytogenes. Cell Microbiol. 2005;7:443–457. doi: 10.1111/j.1462-5822.2004.00475.x. [DOI] [PubMed] [Google Scholar]
- Talman AM, Chong R, Chia J, Svitkina T, Agaisse H. Actin network disassembly powers dissemination of Listeria monocytogenes. J Cell Sci. 2014;127:240–249. doi: 10.1242/jcs.140038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Gupta R, Mayer BJ. Differential inhibition of signaling pathways by dominant-negative SH2/SH3 adapter proteins. Mol Biol Cell. 1995;15:6829–6837. doi: 10.1128/mcb.15.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang BL, Zhang T, Low DYH, Wong ET, Horstmann H, Hong W. Mammalian homologues of yeast Sec31p. An ubiquitously expressed form is localized to the endoplasmic reticulum (ER) exit sites and is essential for ER-Golgi transport. J Biol Chem. 2000;275:13597–13604. doi: 10.1074/jbc.275.18.13597. [DOI] [PubMed] [Google Scholar]
- Thanabalasuriar A, Kim J, Gruenheid S. The inhibition of COPII trafficking is important for intestinal epithelial tight junction disruption during enteropathogenic Escherichia coli and Citrobacter rodentium infection. Microbes Infect. 2013;15:738–744. doi: 10.1016/j.micinf.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Townley AK, Feng Y, Schmidt K, Carter DA, Porter R, Verkade P, Stephens DJ. Efficient coupling of Sec23-Sec24 to Sec13-Sec31 drives COPII-dependent collagen secretion and is essential for normal craniofacial development. J Cell Sci. 2008;121:3025–3034. doi: 10.1242/jcs.031070. [DOI] [PubMed] [Google Scholar]
- Vazquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, et al. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bargen K, Gorvel JP, Salcedo SP. Internal affairs: investigating the Brucella intracellular lifestyle. FEMS Microbiol Lett. 2012;36:533–562. doi: 10.1111/j.1574-6976.2012.00334.x. [DOI] [PubMed] [Google Scholar]
- Ward TH, Polishchuk RS, Caplan S, Hirschberg K, Lippincott-Schwartz J. Maintenance of Golgi structure and function depends on the integrity of ER export. J Cell Biol. 2001;155:557–570. doi: 10.1083/jcb.200107045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SK, Gomez GA, Michael M, Verma S, Cox HL, Lefevre JG, et al. Cortical F-actin stabilization generates apical-lateral patterns of junctional contractility that integrates cells in to epithelia. Nat Cell Biol. 2014;16:167–178. doi: 10.1038/ncb2900. [DOI] [PubMed] [Google Scholar]
- Zamanian JL, Kelly RB. Intersectin 1L guanine nucleotide exchange activity is regulated by adjacent Src homology 3 domains that are also involved in endocytosis. Mol Biol Cell. 2003;14:1624–1637. doi: 10.1091/mbc.E02-08-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeghouf M, Guibert B, Zeeh JC, Cherfils J. Arf, Sec7, and Brefeldin A: a model towards the therapeutic inhibition of guanine nucleotide-exchange factors. Biochem Soc Trans. 2005;33:1265–1268. doi: 10.1042/BST0331265. [DOI] [PubMed] [Google Scholar]
- Zoppino FC, Militello RD, Slavin I, Alvarez C, Colombo MI. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic. 2010;11:1246–1261. doi: 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Purified proteins used in this work. A. GST fusion proteins containing various SH3 domains or GST alone. Approximately 20 micrograms of each protein was loaded. B. Purified Sec31A-Sec13 (i) or Sec31AΔPRD-Sec13 (ii) complexes. One microgram (Sec31A-Sec13) or 2.0 micrograms (Sec31AΔPRD-Sec13) of protein was loaded. Positions of Sec31A, Sec31AΔPRD, or Sec13 are indicated. C. InlC proteins used in competition experiments. (i). 2.0 micrograms of InlC used for experiments in Fig. 6A. (ii). 6 x histidine tagged InlC or InlC.F146A proteins used in Fig. 6B. The protein amounts loaded were 0.90 and 4.5 micrograms for InlC, and 0.80 and 4.0 micrograms for InlC.F146A. Proteins were migrated on 12% SDS/polyacrylamide gels and stained with Coomassie Brilliant Blue.
Figure S2. Effect of a second siRNA targeting Sec31A on Listeria protrusion formation and cell junction morphology. A. RNAi-mediated depletion of Sec31A. Caco-2 BBE1 cells in transwells were transfected with a control non-targeting siRNA (C) or an siRNA directed against Sec31A (Sec31A-2). Sec31A-2 siRNA was different from the siRNA used in Figure 3. Approximately 72 h post-transfection, cells lysates were prepared and used for analysis of Sec31A expression. A representative Western blot is shown in the left panel and quantification of Western blots from three experiments is in the right panel. B. Effect of the Sec31A-2 siRNA on Listeria protrusions. After siRNA transfection, Caco-2 BBE1 cells were infected with wild-type (wt) or ΔinlC strains of Listeria and processed for confocal microscopy analysis. The bar graph displays mean relative protrusion formation values +/− SEM. *, P < 0.05 relative to control siRNA treated cells infected with wild-type Listeria. C. Effect of Sec31A-2 siRNA on cell junctions. About 72 h after transfection with control (C) or Sec31A-2 siRNAs, Caco-2 BBE1 cells were fixed and labeled for the tight junction protein occludin. (i) Confocal microscopy images of occludin labeling. Scale bars indicate 3 micrometers. (ii). Quantification of linear indexes, as described in the Experimental Procedures. *, P < 0.05 relative to control siRNA treated cells (C) treated cells. Data in A and B are mean +/− SEM from three experiments.
Figure S3. Effect of InlC, Sec31A, or Tuba on protein secretion. A. Expression of EGFP-InlC does not alter secretion of SEAP. (i). Analysis of EGFP or EGFP-InlC expression in stable Caco-2 BBE1 cell lines by Western blotting. The arrow or arrowhead indicates EGFP-InlC or EGFP, respectively. (ii). Assessment of protrusion formation efficiency of wild-type (wt) or ΔinlC Listeria strains. (iii). Measurement of protein secretion through SEAP assays. B. RNAi-mediated depletion of Sec31A does not affect secretion of SEAP. (i). Confirmation of Sec31A depletion through Western blotting analysis (ii). Measurement of SEAP secretion in cells depleted for Sec31A, treated with control (C) siRNA, or mock transfected in the absence of siRNA (−). Results in A and B are mean +/− SEM from three experiments.