Abstract
AIM: To investigate the effect of transfected survivin antisense oligonucleotide (ASODN) on proliferation and apoptosis of gastric cancer cells.
METHODS: The authors designed ASODNs targeting different regions of survivin mRNA, including surviving ASODN1, ASODN2 and ASODN3. ASODNs were transfected into gastric cancer cell line SGC 7901, cell growth was detected by MTT assay. Cells exposed to the potent oligonucleotide were also examined for apoptosis induction by FCM and fluorescence microscopy. Semiquantitive RT-PCR and Western blot examinations were carried for expression of survivin mRNA and protein.
RESULTS: ASODN3 caused a statistically significant reduction of cell viability to 60.6% (± 2.9%) (P < 0.01), while ASODN1 and ASODN2 had no such changes (P > 0.05). The cell growth was also significantly inhibited by ASODN3, compared with reversal and scrambled sequence. A significant loss of survivin mRNA was presented in ASODN3 treated cells and this was not seen in treatment with sense ODN or scramble ODN. Protein level was significantly decreased 48 h after survivin ASODN trasfected by approximately 2-fold decrease compared with untreated controls. However, ASODN3 did not induce significant apoptosis response until 48 h after transfection (P > 0.05).
CONCLUSION: ASODN3, which targets translation initiation part, can be identified as a most potent antisense compound. Srvivin ASODN3 may provide a novel approach to therapy of gastric cancer.
INTRODUCTION
Disordered regulatory apoptosis plays an important role in tumorigenesis[1]. Several proteins that inhibit apoptosis have been identified, such as some members of bcl-2, HSP and IAP (inhibition of apoptosis). IAP gene family controls a downstream step in cell death by suppressing the activity of Caspases, the executers of cell death programs[2]. Survivin is a newly identified gene in IAP family, and it is characterized by a unique structure with a single, baculovirus IAP repeat and no zinc-binding domain known as Ring finger[3]. Its another characteristic is that it expresses in most human cancers but is essentially absent in normal tissues, which makes it an exciting potential therapeutic target in cancer treatment[4].
Gastric cancer is one of the leading causes of cancer deaths in China, and it still has poor prognosis in spite of some progresses in treatment. It has been demonstrated from immunohistochemicl analysis that about 34.5% of gastric cancers overexpress survivin, and are significantly associated with reduced apoptosis compared with survivin negative tumors[5]. Recently, manipulating apoptosis gene by antisense oligodeoxynucleotide (ASOND) might provide new therapeutic strategies in treating human diseases[6]. ASODN is a single-strand DNA which is complementary to specific regions of mRNA and capable of inhibiting the antiapoptotic gene, and it is noticeable that ASOND holds great promise as a pharmaceutical agent. Therefore, we investigated whether growth inhibition and induced apoptosis of gastric cancer cell line SGC7901 could be achieved by targeting survivin with ASODN.
MATERIALS AND METHODS
Cell culture
SGC7901 was maintained in RPMI 1640 and supplemented with 100 mL/L calf serum at 37 °C in humidified atmosphere containing 50 mL/L CO2.
Immunohistochemistry
Cells were plated in 60 mm tissue culture dishes containing coverglasses overnight. The coverglasses were washed in phosphate-buffered saline (PBS), fixed for 30 min in 70 mL/L ethanol, washed twice in PBS. Quenching of the endogenous perixidase activity was obtained by treatment with 0.3 mL/L H2O2 in methanol. The sections were blocked with 10 mL/L goat serum in PBS and incubated with 3 μg anti -survivin polyclonal antibody (gift from Dr. Katsuya Shiraki, First Department of Internal Medicine, Mie University School of Medicine, Tsu, Japan) at 4 overnight, then incubated with HRP-conjuncted antibody for 30 min at room temperature. After three washes of 5 min in PBS, they were developed in a substrate solution of horseradish peroxidase. Negative control omitting survivin antibody was also performed to confirm the absence of non-specific reactions. Expression of survivin was classified into three levels: (-) no or weak staining, (+) less than 50% positive nuclei staining, and (++) greater than 50% positive nuclei staining of the total nuclei.
Oligodeoxynucleotide
ASODN1 and ASODN2 were designed by a computer program and could theoretically access survivin mRNA. ASODN3 was complementary to the initiation codon and 5 downstream codons, and could empirically access survivin mRNA as shown in Table 1. In addition, the scramble sequence of survivin ASODN3 was as follows: 5’-GGA CCA CGC TAT CAG CCG-3’, and reverse anitsense (sense): 5’-ATG GGT GCC CCG ACG TTG-3’. Phosphothioalate oligodexynucleotides were synthesized using an applied biosystems 3 900 DNA synthesizer (Shenggong, Shanghai, China). After the synthesis, ODNs were purified by use of high-pressure liquid chromatography system, dissolved with PBS, and frozen in aliquots at -20 °C until use.
Table 1.
Sequence of ASODNS and their target site
| Name | Sequence | Position |
| ASODN1 | 5’-GTT CTT GGA TGT AGA GAT GC-3’ | 102-121 |
| ASODN2 | 5’-GCT TCT TGA CAG AAA GGA A-3’ | 305-323 |
| ASODN3 | 5’-CAA CGT CGG GGC ACC CAT-3’ | 50-67 |
Transfection
According to the manufacture’s instructions, ODNs were delivered into cells in the form of complexes with Lipofection regents (GIBCO, MD).
Measurement of cell growth
In vitro growth inhibitory effects of ODNs on SGC7901 were assessed by MTT assay performed according to the previously described protocol[7] with a slight modification. Two × 104 cells were seeded in each well of 96-well micrometer plates and allowed to attach overnight. The cells were then treated with 400 nmol/L ODN-Lipofection complex for 24 h and incubated for another 48 h. Subsequently, 20 μL of MTT (5 g/L, Sigma) in PBS was added to each well, followed by incubation for 4 h at 37 °C. Formazan crystals were dissolved in DMSO. Absorbance was determined with an enzyme-linked immunosorbent assay reader (model 318, Shanghai, China) at 540 nm. Each assay was performed nine times. The controls received medium alone. The results were expressed as mean ± SE of controls with no ODN.
Reverse transcriptase-PCR (RT-PCR) analysis of survivin mRNA
Total RNA was extracted from SGC7901 cells by modification of guanidium-thiocyanate acid phenol method[8] and quantified based on the measured absorbance at 260 nm. cDNA was synthesized using 2 μg of RNA, 106 U/ L reverse transcripatase (GIBCO, Inc), 0.5 g/L Oligod T, in a total volume of 20 μL. Reaction was performed at 42 °C for 60 min, and terminated by heating at 99 °C for 5 min. The sequences of oligodexynucleotides primes for RT-PCR were as follows: survivin-S 5’-ATG GGT GCC CCG ACG TTG CC-3’, survivin-A 5’-GCA GCT CCG GCC AGA GGC CT-3’, β-actin-S 5’-GGC GGC ACC ACC TGT ACC CT-3’ and β-actin-A 5’-AGG GGC CGG ACT CGT CAT ACT-3’, respectively. For amplification of cDNA, 2 μL cDNA product was subjected to PCR-based technique using 2.5 U of Taq DNA Polymerase, 1 μL each of forward and reverse primers, and 200 μmol/L each of dNTPs. PCR consisted of 35 cycles at 94 °C for 1 min, at 62 °C for 1 min and at 72 °C for 1 min followed by a final extension at 72 °C for 5 min.
Western blot analysis
Cells were washed in ice-cold PBS and lysed in a buffer using standard methods. After centrifugation at 10 000 g for 10 min, the supernatants were stored at -70 °C. Lysate equalized for protein content was separated in 150 g/L SDS-PAGE and transferred onto a PVDF membrane. The membranes were blocked for 1 h at room temperature in 10 g/L BSA, and incubated overnight at 4 °C with survivin antibody, followed by incubation with sheep anti-rabbit second antibody conjugated to horseradish peroxidase (Dako, Denmark) for 120 min. Finally the membranes were developed with DAB and incubated until color developed sufficiently.
Measurement of apoptosis by flow cytometry and staining with Hchest 33342
In preparation of flow cytometry (FCM), SGC 7901 cells were centrifuged 72 h after transfection. The cells were washed with PBS, and fixed in 70 mL/L cold ethanol. Samples were treated with RNase (10 g/L), resuspended and stained with 10 g/L propidium iodine. After 30 min at room temperature in the dark, the cells were analyzed using a FCM scan flow cytometer. Apoptotic cells appeared in the cell cycle distribution as cells with DNA contents less than G1 cells, and the percentage of apoptotic cells was calculated. In other experiments, transduced cells were cultivated for 24 h at 37 °C and assayed for nuclei staining. Both attached cells and detached cells were collected and resuspended. One drop of cell suspension was added to one drop of Hoechst 33 342 solution (10 mg/L in PBS), mixed gently on a slide, and immediately examined with Olympus fluorescence microscopy. Blue fluorescent condensed nuclei were interpreted as the number of apoptotic cells. Viable cells were interpreted as cells which exhibited green, diffusely stained intact nuclei. Cell ghosts with weak staining were not included in the cell count.
Statistical analysis
The data were expressed as mean ± SE. The results were analyzed by ANOVA test. P < 0.05 was considered statistically significant.
RESULTS
Expression of survivin in SGC7901
We examined the expression of survivin gene in SGC-7901 using immunohistochemistry. Expression of survivin was observed primarily in cytoplasm, and mild staining in nuclei (data not shown).
Cell proliferation assay
As shown in Figure 1, ASODN3 caused a statistically significant reduction of cell viability to 60.6 ± 2.9% (P < 0.01), while ASODN1 and ASODN2 had not such changes (P > 0.05). We also investigated whether growth inhibition was changed by control sequences including reversal antisense (i.e. sense) and scrambled sequence with the same composition. As shown in Figure 2, neither had any changes in activity (P > 0.05).
Figure 1.
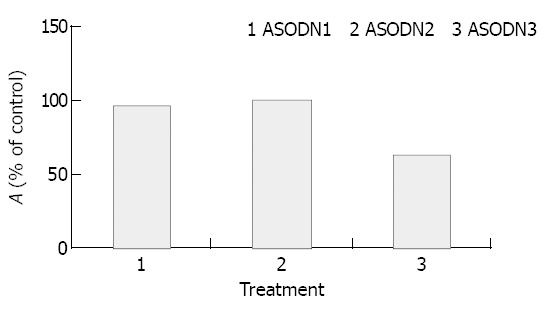
Cytotoxic effect of ASODN targeting different regions of survivin mRNA. Cells were cultured in plastic 96-well plates and quantitated using MTT assay as described in “Materials and Methods.”
Figure 2.
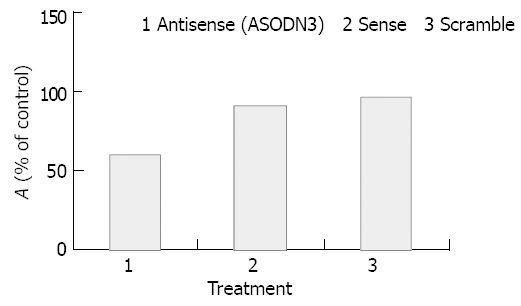
Cytotoxic effect of ASODN3 and control sequences.
Induction of apoptosis by antisense oligodexynucleotide
Cells exposed to the oligonucleotides were examined for apoptosis induction by FCM and fluorescence microscopy. As shown in Table 2, positive control 5-FU significantly induced apoptotic response, about 6.12 ± 3.25% for 24 h (P < 0.01) and 11.35 ± 1.50% for 48 h (P < 0.01). However, ASODN3 did not induce any significant apoptotic response until 48 h after transfection (P > 0.05). Fluorescence microscopy also did not show any increase of condensed nuclei (data not shown).
Table 2.
Apoptosis rates of each group 24 and 48 h after transfection (n = 4)
| 24 h (%) | 48 h (%) | |
| Untreated | 1.98±0.40 | 3.74±0.19 |
| survivin ASODN | 2.27±0.48 | 4.57±0.61 |
| 5-FU | 6.12±3.25 | 11.35±1.50 |
Changes of survivin mRNA expression after transfection
As shown in Figure 3, a significant loss of survivin mRNA was detected in antisense treated cells and this was not seen in either sense ODN treated or scramble ODN treated cells.
Figure 3.
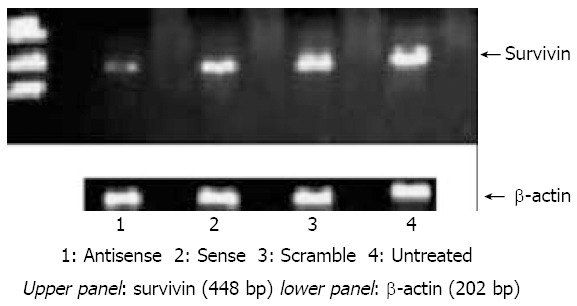
Survivin and β-actin (loading control) mRNA ex-pression following incubation with antisense, sense,scramble and no ODN control. It shows ethidium bromide staining of conventional agarose gel electrophoresis after amplification.
Changes of survivin protein expression after transfetion
We aimed to investigate whether survivin ASODN could downregulate survivin protein in SGC7901, and further determined whether it was in sequence-specific way. Figure 4 shows a typical change of survivin protein expression. Protein level was significantly decreased 48 h after survivin ASODN trasfection (by approximately 2-fold decrease compared with untreated control), whereas either sense ODN treated or scramble ODN treated cells did not modulate survivin expression levels. To sum up, these results provided strong evidence of sequence specific mechanism of survivin ASODN.
Figure 4.
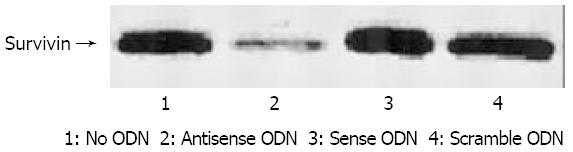
Survivin protein expression following incubation with surviivn, antisense, sense, scramble and no ODN control. Strong decrease of survivin protein could be detected in the ASODN group.
DISCUSSION
ASODN represents a useful experimental approach for manipulating gene expression, and recently some antisense compounds have shown anticancer efficacy in numerous preclinical studies[9-12]. Survivin was detectable in most types of cancer, and its presence was associated with a poor prognosis in many malignant tumors[13-16]. In this study, we chose survivin as the molecular target and designed ASODN to observe its influence on growth and apoptosis of gastric cancer cell line SGC7901. An immunohistochemical study showed that SGC7901 could positively express survivin, prior to ASODN transfection.
Then, it should be considered to design an optimal site for antisense blocking, which is the key factor of antisense research. Previous researches showed not all areas of a mRNA molecular were amenable to antisense hybridization. Therefore, among the possible ASODNs against a given target nucleic acid, only a small number of sequences seemed to give rise to satisfactorily strong inhibition of target gene expression. The reason is unclear and probably involves mRNA secondary structure. Therefore, computer program was used to design ASODN, but sometimes its usefulness was elusive[17]. On the other hand, empirically, ASODN targeting AUG starting site was most frequently used in antisense experiments[18], such as bcl-2 ASODN application in various cancer cell lines[19-23]. So far we still could not predict accurately the secondary structure of mRNA, and it is rather difficult to predict which is the best ASODN sequence unless through some screening experiments. In our experiment, ASODN1 and ASODN2 were designed by computer program and selected as candidate sequences. ASODN3 targeting the translation initiation codon region, was selected as another candidate sequence to screen the most potent sequence.
Survivin could regulate G2/M phase of the cell cycle by associating it with mitotic spindle microtubules[24,25], and its expression was positively correlated with proliferation of hepatic[26] and pancreatic[27] carcinoma cells. Therefore, we tried to determine which sequence was potent by observing cell growth as previous studies[7,28,29]. As a result, ASODN3 was able to suppress cell viability of approximate 40% decrease of the number of viable cells (P < 0.01), while ASODN1 and ASODN2 did not affect cell viability significantly (P > 0.05). On the other hand, the most widely recognized mechanism of ASODN involved RNase H mediated destruction of the target mRNA. Therefor, we also performed RT-PCR and Western blot to confirm the occurrence of antisense effect of ASODN3. As shown in Figures 3 and 4, ASODN3 caused a reduction in the mRNA and protein level 48 h after transduction. However, two different controls for ASODN3 did not modulate survivin mRNA and protein expression levels. Therefore, further evidences for sequence-specific antisense effect of ASODN3 were provided. The exact mechanism of ASODN3 is still unknown, and we suspect that the translation initiation codon region of survivin mRNA is probably a single strand.
In the present study, we identified that ASODN3 had a potent sequence to inhibit cell growth, then we detected apoptosis by FCM and nuclei staining when survivin ASOND3 was transfected. However, it demonstrated that transfecting survivin ASODN solely was not sufficient to trigger apoptosis, indicating that decrease of cell viability was not due to apoptosis. Our results were different from the report by Olie RA[30]. It seemed plausible that the effects of ASODN varied depending on the expression profile of treated cells. It was also probably due to the complexity of apoptotic pathway in which other antiapoptotic genes might play more important roles in gastric cancer cell line SGC-7901.
In conclusion, ASODN3 can inhibit the growth of gastric cancer cell line but can not induce apoptosis by itself. From a clinical standpoint, anti-cancer therapy still relies heavily on cytotoxic agents, in spite of some advances in identifying molecular target by ASODN treatment[31]. So, it is meaningful to investigate further whether ASODN targeting survivin can enhance the sensitivity of chemotherapeutic drugs by decreasing apoptosis thresholds.
ACKNOWLEDGMENTS
We are grateful to Dr. Katsuya Shiraki (First Department of Internal Medcine, Mie University School of Medicine, Tsu, Japan) for providing anti- survivin antibody.
Footnotes
Supported by Shanghai Key Research Project Grant in Medical Science Development (99ZD003).
Edited by Wang XL Proofread by Xu FM
References
- 1.Reed JC. Dysregulation of apoptosis in cancer. J Clin Oncol. 1999;17:2941–2953. doi: 10.1200/JCO.1999.17.9.2941. [DOI] [PubMed] [Google Scholar]
- 2.Deveraux QL, Reed JC. IAP family proteins--suppressors of apoptosis. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 3.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 4.Altieri DC, Marchisio PC. Survivin apoptosis: an interloper between cell death and cell proliferation in cancer. Lab Invest. 1999;79:1327–1333. [PubMed] [Google Scholar]
- 5.Lu CD, Altieri DC, Tanigawa N. Expression of a novel antiapoptosis gene, survivin, correlated with tumor cell apoptosis and p53 accumulation in gastric carcinomas. Cancer Res. 1998;58:1808–1812. [PubMed] [Google Scholar]
- 6.Nicholson DW. From bench to clinic with apoptosis-based therapeutic agents. Nature. 2000;407:810–816. doi: 10.1038/35037747. [DOI] [PubMed] [Google Scholar]
- 7.Ziegler A, Luedke GH, Fabbro D, Altmann KH, Stahel RA, Zangemeister-Wittke U. Induction of apoptosis in small-cell lung cancer cells by an antisense oligodeoxynucleotide targeting the Bcl-2 coding sequence. J Natl Cancer Inst. 1997;89:1027–1036. doi: 10.1093/jnci/89.14.1027. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isola-tion by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Yuen AR, Sikic BI. Clinical studies of antisense therapy in cancer. Front Biosci. 2000;5:D588–D593. doi: 10.2741/yuen. [DOI] [PubMed] [Google Scholar]
- 10.Jansen B, Wacheck V, Heere-Ress E, Schlagbauer-Wadl H, Hoeller C, Lucas T, Hoermann M, Hollenstein U, Wolff K, Pehamberger H. Chemosensitisation of malignant melanoma by BCL2 antisense therapy. Lancet. 2000;356:1728–1733. doi: 10.1016/S0140-6736(00)03207-4. [DOI] [PubMed] [Google Scholar]
- 11.Chi KN, Gleave ME, Klasa R, Murray N, Bryce C, Lopes de Menezes DE, D'Aloisio S, Tolcher AW. A phase I dose-finding study of combined treatment with an antisense Bcl-2 oligonucleotide (Genasense) and mitoxantrone in patients with metastatic hormone-refractory prostate cancer. Clin Cancer Res. 2001;7:3920–3927. [PubMed] [Google Scholar]
- 12.Rudin CM, Otterson GA, Mauer AM, Villalona-Calero MA, Tomek R, Prange B, George CM, Szeto L, Vokes EE. A pilot trial of G3139, a bcl-2 antisense oligonucleotide, and paclitaxel in patients with chemorefractory small-cell lung cancer. Ann Oncol. 2002;13:539–545. doi: 10.1093/annonc/mdf124. [DOI] [PubMed] [Google Scholar]
- 13.Ikehara M, Oshita F, Kameda Y, Ito H, Ohgane N, Suzuki R, Saito H, Yamada K, Noda K, Mitsuda A. Expression of survivin correlated with vessel invasion is a marker of poor prognosis in small adenocarcinoma of the lung. Oncol Rep. 2002;9:835–838. [PubMed] [Google Scholar]
- 14.Dong Y, Sui L, Watanabe Y, Sugimoto K, Tokuda M. Survivin expression in laryngeal squamous cell carcinomas and its prognostic implications. Anticancer Res. 2002;22:2377–2383. [PubMed] [Google Scholar]
- 15.Takai N, Miyazaki T, Nishida M, Nasu K, Miyakawa I. Expression of survivin is associated with malignant potential in epithelial ovarian carcinoma. Int J Mol Med. 2002;10:211–216. [PubMed] [Google Scholar]
- 16.Takai N, Miyazaki T, Nishida M, Nasu K, Miyakawa I. Survivin expression correlates with clinical stage, histological grade, invasive behavior and survival rate in endometrial carcinoma. Cancer Lett. 2002;184:105–116. doi: 10.1016/s0304-3835(02)00190-8. [DOI] [PubMed] [Google Scholar]
- 17.Reed JC. Promise and problems of Bcl-2 antisense therapy. J Natl Cancer Inst. 1997;89:988–990. doi: 10.1093/jnci/89.14.988. [DOI] [PubMed] [Google Scholar]
- 18.Cotter FE. Antisense therapy for lymphomas. Hematol Oncol. 1997;15:3–11. doi: 10.1002/(sici)1099-1069(199702)15:1<3::aid-hon583>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 19.Lopes de Menezes DE, Mayer LD. Pharmacokinetics of Bcl-2 antisense oligonucleotide (G3139) combined with doxorubicin in SCID mice bearing human breast cancer solid tumor xenografts. Cancer Chemother Pharmacol. 2002;49:57–68. doi: 10.1007/s00280-001-0385-3. [DOI] [PubMed] [Google Scholar]
- 20.Duggan BJ, Maxwell P, Kelly JD, Canning P, Anderson NH, Keane PF, Johnston SR, Williamson KE. The effect of antisense Bcl-2 oligonucleotides on Bcl-2 protein expression and apoptosis in human bladder transitional cell carcinoma. J Urol. 2001;166:1098–1105. [PubMed] [Google Scholar]
- 21.Wacheck V, Heere-Ress E, Halaschek-Wiener J, Lucas T, Meyer H, Eichler HG, Jansen B. Bcl-2 antisense oligonucleotides chemosensitize human gastric cancer in a SCID mouse xenotransplantation model. J Mol Med (Berl) 2001;79:587–593. doi: 10.1007/s001090100251. [DOI] [PubMed] [Google Scholar]
- 22.Hu Q, Bally MB, Madden TD. Subcellular trafficking of antisense oligonucleotides and down-regulation of bcl-2 gene expression in human melanoma cells using a fusogenic liposome delivery system. Nucleic Acids Res. 2002;30:3632–3641. doi: 10.1093/nar/gkf448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van de Donk NW, Kamphuis MM, van Dijk M, Borst HP, Bloem AC, Lokhorst HM. Chemosensitization of myeloma plasma cells by an antisense-mediated downregulation of Bcl-2 protein. Leukemia. 2003;17:211–219. doi: 10.1038/sj.leu.2402768. [DOI] [PubMed] [Google Scholar]
- 24.Ambrosini G, Adida C, Sirugo G, Altieri DC. Induction of apoptosis and inhibition of cell proliferation by survivin gene targeting. J Biol Chem. 1998;273:11177–11182. doi: 10.1074/jbc.273.18.11177. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto T, Tanigawa N. The role of survivin as a new target of diagnosis and treatment in human cancer. Med Electron Microsc. 2001;34:207–212. doi: 10.1007/s007950100017. [DOI] [PubMed] [Google Scholar]
- 26.Ito T, Shiraki K, Sugimoto K, Yamanaka T, Fujikawa K, Ito M, Takase K, Moriyama M, Kawano H, Hayashida M, et al. Survivin promotes cell proliferation in human hepatocellular carcinoma. Hepatology. 2000;31:1080–1085. doi: 10.1053/he.2000.6496. [DOI] [PubMed] [Google Scholar]
- 27.Sarela AI, Verbeke CS, Ramsdale J, Davies CL, Markham AF, Guillou PJ. Expression of survivin, a novel inhibitor of apoptosis and cell cycle regulatory protein, in pancreatic adenocarcinoma. Br J Cancer. 2002;86:886–892. doi: 10.1038/sj.bjc.6600133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shankar SL, Mani S, O'Guin KN, Kandimalla ER, Agrawal S, Shafit-Zagardo B. Survivin inhibition induces human neural tumor cell death through caspase-independent and -dependent pathways. J Neurochem. 2001;79:426–436. doi: 10.1046/j.1471-4159.2001.00596.x. [DOI] [PubMed] [Google Scholar]
- 29.Chen J, Wu W, Tahir SK, Kroeger PE, Rosenberg SH, Cowsert LM, Bennett F, Krajewski S, Krajewska M, Welsh K, et al. Down-regulation of survivin by antisense oligonucleotides increases apoptosis, inhibits cytokinesis and anchorage-independent growth. Neoplasia. 2000;2:235–241. doi: 10.1038/sj.neo.7900091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olie RA, Simões-Wüst AP, Baumann B, Leech SH, Fabbro D, Stahel RA, Zangemeister-Wittke U. A novel antisense oligonucleotide targeting survivin expression induces apoptosis and sensitizes lung cancer cells to chemotherapy. Cancer Res. 2000;60:2805–2809. [PubMed] [Google Scholar]
- 31.Narayanan R. Harnessing the power of antisense technology for combination chemotherapy. J Natl Cancer Inst. 1997;89:107–108. doi: 10.1093/jnci/89.2.107. [DOI] [PubMed] [Google Scholar]


