Abstract
Many oncogenic drivers related to the pathogenesis of OSCC have identified, but the discovery of new molecular markers for early detection of this cancer, remains one the main goals of clinical research. HOX genes regulate normal embryonic development, cell differentiation and other critical processes in eukaryotic cell life. Several studies have demonstrated that the deregulation of HOX genes play a significant role in cancer development and progression. In this study, we built a prognostic TMA with 119 OSCC samples, representative of deep and superficial part of the tumour, to investigate, the paralogous 13 HOX proteins expression, correlating them with clinicpathological parameters, outcomes and therapy information. Our results show an aberrant expression of HOX A13 and HOX D13 in OSCC pathogenesis and tumour progression. HOX A13 overexpression is related to an OSCC better prognosis (P=0.029) and better therapy response in patients treated with both radiotherapy and chemotherapy (P=0.015). HOX D13 overexpression is inversely related to an overall survival (P=0.004). These data highlight the potential prognostic role of HOX paralogous group 13 genes in OSCC.
Keywords: Paralogous 13 HOX genes, OSCC, tumour progression
Introduction
Oral squamous cell carcinoma (OSCC) is the sixth most common cancer worldwide with a 5-year survival rate of 60%. In particular, it represents 4% of all malignancies in men and 2% in women. The majority of OSCC patients present with an advanced stage of the disease, not only because the clinical manifestations are difficult to define but also for the absence of early diagnosis tools [1]. The understanding of the molecular mechanisms related to the pathogenesis and progression of this disease could allow to improve life expectancy, quality of life, disease-free survival of the patients and to establish new and more effective therapeutic strategies [2,3].
Homeobox genes regulate normal embryonic development, cell differentiation and other critical processes in eukaryotic cell life [4]. Several members of the Dlx family are essential for normal development of the jaw, skull, and inner ear [5], while Pax3 and Pax 7 homeodomain proteins are crucial for the differentiation of mesodermal precursors into muscle cells [6]. Several studies have demonstrated that in particular the genes belonging to Class I homeobox genes, defined HOX genes in humans, play a crucial role in neoplastic transformation in several human tissues [7,8]. Specifically, the genes belonging to HOX paralogous group 13 seem to carry out a relevant role in both tumour development and progression. We have recently demonstrated the aberrant expression of all paralogous group 13 HOX genes, HOX A13, HOX B13, HOX C13 and HOX D13, in thyroid cancer [9]. Moreover, we have identified a significant prognostic role of HOX D13 in pancreatic cancer [10], a HOX A13 gene deregulation in liver carcinogenesis [11] and an abnormal over-expression of HOX C13 in metastatic melanoma and in de-differentiated and well-differentiated liposarcoma [12,13]. Finally, we have also showed the aberrant expression of HOX B13 in bladder tumorigenesis and progression [14]. Data on the role played by HOX genes in oral cancers are still few, and most refer to epigenetic alterations, in particular hypermethylation and hypomethylation of HOX genes promoters [15].
In this study we have analyzed paralogous 13 HOX genes expression, by immunohistochemistry, in a series of 119 OSCC samples included in a prognostic Tissue Micro Array (TMA), highlighting, the main role of HOX A13 and HOX D13 in pathogenesis and tumor progression of OSCC.
Material and methods
OSCC patients
One hundred nineteen patients admitted to the National Cancer Institute “Giovanni Pascale” of Naples, between 1998 and 2011, were recruited in this study. All patients had provided written informed consent for the use of samples according to the institutional regulations and the study was approved by the ethics committee of the National Cancer Institute “G. Pascale”.
All OSCC cases have been reviewed according to WHO/ISUP 2007 classification criteria, using standard tissue sections. Medical records have been reviewed for clinical information, including histologic parameters assessed on standard H&E-stained slides.
TMA building
A Prognostic-Tumor Array was constructed using 119 tumor tissue samples. Two cores from different areas, one superficial and one representative of the deep invasion, were arrayed in a recipient block. All tumors and controls were reviewed by two experienced pathologists (RF and SL). Discrepancies for the same case were resolved in a joint analysis. Tissue cylinders with a diameter of 1 mm were punched from morphologically representative tissue areas of each ‘donor’ tissue block and brought into one recipient paraffin block (3 × 2.5. cm) using a semi-automated tissue array (Galileo TMA).
Immunohistochemistry analysis
Immunohistochemical staining was carried out on slides from formalin-fixed, paraffin embedded tissues, in order to evaluate the expression of HOX A13, HOX B13, HOX C13 and HOXD13. Paraffin slides was deparaffinized in xylene and rehydrated through graded alcohols. Antigen retrieval was performed with slides heated in 0.01 M citrate buffer (pH 6.0.) in a bath for 20 min at 97°C. After antigen retrieval, the slides were allowed to cool. The slides were rinsed with TBS and the endogenous peroxidase was inactivated with 3% hydrogen peroxide. After protein block (BSA 5% in PBS 1 ×), the slides were incubated with primary antibody to human HOX A13 (dilution 1:200, cod. Ab106503, Abcam, Cambridge, UK), HOX B13 (dilution 1:300, cod. ab28575, Abcam, Cambridge, UK), HOX C13 (dilution 1:1200, cod.ab55251, Abcam, Cambridge, UK), HOX D13 (dilution 1:100, cod. Ab19866, Abcam, Cambridge, UK) overnight. Sections were incubated with mouse anti-rabbit or goat anti-mouse secondary IgG biotinylated secondary antibody for 30 min. Immunoreactivity was visualized by means of avidin-biotin-peroxydase complex kit reagents (Novocastra, Newcastle, UK) as the chromogenic substrate. Finally, sections were weakly counterstained with haematoxylin and mounted.
Evaluation of immunostaining
Antigen expression was independently evaluated by two experienced pathologists (RF/SL) using light microscopy. For paralogous 13 HOX genes nuclear and cytoplasmic localization were considered. All values of immunostaining were expressed only in percentage terms of positive cells. The percentage of positive cancer cells was evaluated in each sample by counting the number of positive cells over the total cancer cells in 10 non-overlapping fields using × 400 magnification.
RNA extraction and analysis
Total RNA was isolated from selected FFPE samples collected from the National Cancer Institute “Fondazione G. Pascale” Institutional Bio-Bank. For RNA extraction, 4 sections at 10 μm thick were cut from each FFPE tissue block. Total RNA was extracted using High pure FFPE RNA Micro Kit (Roche Molecular Biochemicals, Mannheim, Germany) following the manufacturer’s instructions. A total of 1 μg RNA was subjected to cDNA synthesis for 1 hour at 37°C using the Ready To Go You-Primer First-Strand Beads kit (Amersham Biosciences Europe Gmbh, Freiburg, Germany) in a reaction mixture containing 0.5 μg random hexamers (GeneAmp RNA PCR Random Hexamers Set N808-0127 Applied Biosystems, Foster City, CA).
Quantitative real-time PCR
Quantitative RT-PCR was performed in a LightCycler system (Roche Molecular Biochemicals, Mannheim, Germany) using TaqMan® analysis. In this system, all reactions have been run in glass capillaries in a volume of 20 μl with 4 μl of The LightCyclerTaqMan Master Mix (Roche Molecular Biochemicals), 2 μl of cDNA and 1 μl of specific TaqMan Gene Expression Assays for human HOX A13, HOX B13, HOX C13, HOX D13 (RealTime Designer Assay, Roche Molecular Biochemicals) according to the manufacturer’s directions. All reactions were performed in triplicate. The thermal cycling conditions included a step of 20 sec at 95°C followed by a 40 cycles of 95°C for 1 sec and 60°C for 20 sec. The comparative C t method was employed to determine the human HOX genes variation, using TaqMan Endogenous Controls Human ACTB (β-actin) Endogenous Control (Real Time Designer Assay, Roche Molecular Biochemicals) as reference gene. Final amounts of target were determined as follows: target amount=2-Ct, where C t=(C t (HOX genes)-C t (ACTB))sample-(C t (HOX genes)-C t (ACTB))calibrator. Data were expressed as mean ± standard deviation (SD, n=3).
Statistical analysis
Only a percentage of immunoreactive cells was considered for the evaluation of paralogous 13 HOX IHC expression on TMA samples. Kruskal-Wallis test was applied to identify differences in median expression values of each marker between two groups of OSCC (superficial and deep side). Wilcoxon signed-rank test was used to study the correlation between the nuclear and cytoplasmic expression in the paralogous 13 HOX group genes because of nonparametric and paired values.
The association between HOX A13, HOX B13, HOX C13 and HOX D13 with the clinic-pathological data was conducted using the χ2 test considering the median of expression for each marker as cut-off. The Pearson χ2 test was used to determine whether a relationship exists between the variables included in the study, while the value Pearson’s R represents a measure of linear association between the variables. The level of significance was defined as P<0.05. Overall Survival (OS) curves were calculated using the Kaplan-Meier method. OS was defined as the time from diagnosis (first biopsy) to death by any cause or until the most recent follow-up.
All the statistical analyses were carried out using MedCalc 12.7.
Results
Clinic pathological features of OSCC patients
The main clinic pathological characteristics of the patients included in the TMA are reported in Table 1. 119 patients aged between 31 and 92 years (mean age 70 years). 78 patients had lymph node metastases at diagnosis and 1 patient had distant metastases. Furthermore, 29 patients were submitted to adjuvant chemotherapy, 60 to radiotherapy, and 27 to radio and chemotherapy. All selected patients were treated with chemotherapy after surgery and none of them had received the drug in the neoadjuvant therapy. Finally, the appearance of local recurrence was observed in 3 cases, 42 patients died over an average period of 24 months. The follow up of 37 patients was not available. Regarding the histopathological grading, 20 cases were well-differentiated squamous cell carcinoma (G1), 66 squamous cell carcinomas were moderately differentiated (G2) and 33 cases were poorly differentiated carcinomas (G3). The tongue, with 71 cases, was the most affected location, followed by oral floor with 12 cases, lip with 3 cases and other sites with 33 cases.
Table 1.
Main clinical features of the patients arranged in the prognostic OSCC TMA
| Overall Population (119) | |
|---|---|
| Age at the diagnosis | |
| <45 | 4\119 |
| >45 | 115\119 |
| Gender | |
| M | 83\119 |
| F | 36\119 |
| Grading | |
| G1 | 20\119 |
| G2 | 66\119 |
| G3 | 33\119 |
| Overall free disease survival | 40\85 |
| Death | 42\85 |
| No follow-up | 37\119 |
| Recurrence | 3\85 |
| Tumor staging | |
| T1-2 | 75\119 |
| T3-4 | 44\119 |
| Lymph Node metastases | 78\119 |
| Distant metastases | 1\119 |
| No metastasis | 40\119 |
| THERAPY | |
| Chemotherapy | 29\89 |
| Radiotherapy | 60\89 |
| Chemotherapy+Radiotherapy | 27\89 |
| No Therapy information | 20\119 |
| Primary anatomical Site | |
| Tongue | 71\119 |
| Other sites | 45\119 |
| Lips | 3\119 |
IHC paralogous 13 HOX expression in OSCC patients series
The immunohistochemical analysis mainly revealed a nuclear localization of paralogous 13 HOX, whereas a cytoplasmic localization was observed only in some areas. Generally, paralogous 13 HOX protein was detected in nucleus of normal cells. HOXA13 and HOXD13 were expressed in normal basal epithelium instead, HOXB13 stained normal peritumoral epithelium. HOXC13 is not expressed in normal tissue. We evaluated both the cytoplasmic and nuclear positivity and, for statistical associations, we considered them separately in the deep and superficial part of cancer (schematically shown in Figures 1 and 2). In the deep part of the lesion, the immunohistochemical analysis of HOXA13 showed an increased cytoplasmic and nuclear expression in 46% and 40% of cases respectively. Even in the superficial side of lesion HOXA13 was abundantly localized in cytoplasmic (45%) compare nuclear (33%) expression. HOXB13 showed a high nuclear detection in the deep portion of the cancer. In detail, in the deep side, HOXB13 expression was nuclear in 50% of cases and cytoplasmic in 30%; in the superficial side of tumor, HOXB13 expression was nuclear in 40% and cytoplasmic in 38% of cases. HOXC13 nuclear expression increased in the superficial portion of the lesion. In detail, in the superficial side of the tumor, it showed nuclear positivity in 46% of cases and cytoplasmic in 24%; in the deep side of the tumor, nuclear positivity was present in 43% of cases and cytoplasmic in 22%. HOXD13 showed a high nuclear expression. In detail, in the deep side of the cancer, HOXD13 expression was nuclear in 40% of cases and cytoplasmic in 22%; in the superficial side of tumor, its expression was nuclear in 37% of cases and cytoplasmic in 23%. The only cytoplasmic expression has been proven on selected samples by Real Time PCR (data not shown).
Figure 1.
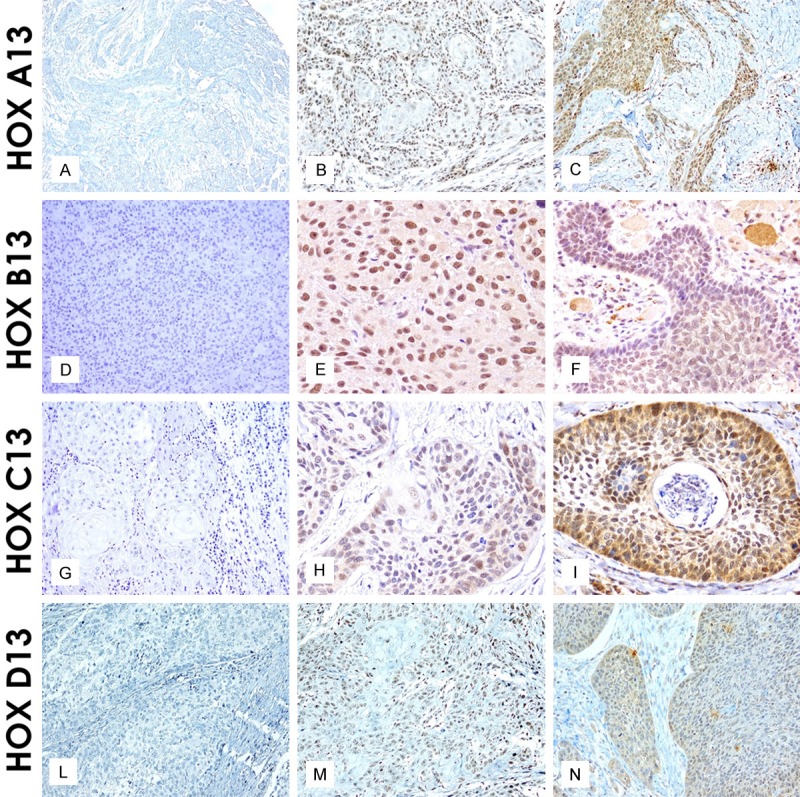
Paralogous group 13 HOX protein localization in OSCC samples: Negative, Weak and Strong HOX A13 nuclear and cytoplasmic expression (20 ×) respectively (A-C); Negative, Weak and strong HOX B13 nuclear and cytoplasmic expression (20 × and 40 ×) respectively (D-F); Negative, Weak and Strong HOX C13 nuclear and cytoplasmic expression (20 × and 40 ×) respectively (G-I); Negative, Weak and strong HOX D13 nuclear and cytoplasmic expression (40 ×) respectively (L-N).
Figure 2.
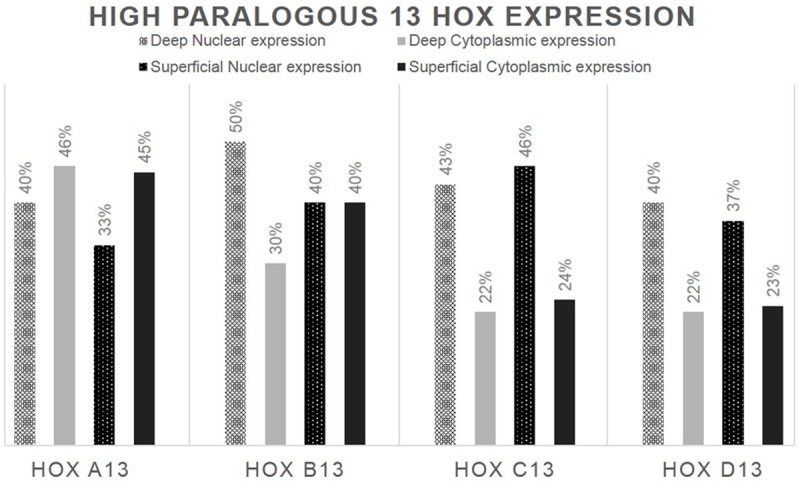
Percentage distribution of paralogous 13 HOX high expression in the deep and superficial margin of the lesion.
Relation between paralogous 13 HOX expression and clinic pathological features of OSCC patients series
We considered both cytoplasmic and nuclear positivity in the deep and superficial margin of the tumor for statistical associations. In general, tumor samples stained more consistently for paralogous 13 HOX proteins, compared to non-neoplastic areas.
All IHC HOX expression data were statistically analyzed and all elaborations are schematized in Supplementary Tables 1, 2, 3, 4.
At first, the Immunohistochemical HOX expression data were statistically analyzed using tumor deep and superficial parameters. In detail, aberrant HOX A13 cytoplasmic superficial expression appeared associated (P<0.0.44) with different anatomic sites of OSCC samples. Its expression increased in lips. There is a trend of statistical association between superficial cytoplasmic expression of HOXC13 and anatomic site. In fact, the tongue site showed an aberrant cytoplasmic expression in superficial tumor side. Finally, a strong statistical significance has been shown for cytoplasmic deep expression (P=0.0.01) of HOX D13, that significantly increases with tumor size (data not shown). Subsequently, we gathered in a single value the paralogous HOX expression of deep and superficial cores, as there were no significant differences between cytoplasmic and nuclear expressions related to the depth of the tumor. Table 2 reports the mean and median of cytoplasmic and nuclear paralogous HOX expression. We have also standardized the series. In fact, the new data encompassed only 94 patients with all paralogous protein expressions and all clinical information (missing expression data for 15 cases). The new HOX expression data were statistically analyzed and all elaborations are schematized in Table 3. This second analysis showed no significant differences with the previous one. Only HOX C13 expression was significantly correlated with male gender (P=0.0.06) and deep invasion (P=0.0.45).
Table 2.
Mean and median of cytoplasmic and nuclear paralogous HOX expression
| HOXA13Nucl | HOXA13Cyto | HOXB13Nucl | HOXB13Cyto | HOXC13Nucl | HOXC13Cyto | HOXD13Nucl | HOXD13Cyto | |
|---|---|---|---|---|---|---|---|---|
| N Valid | 94 | 94 | 94 | 94 | 94 | 94 | 94 | 94 |
| Missing | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mean | 37.8723 | 38.3777 | 27.0213 | 5.7979 | 20.1117 | 13.4574 | 27.1277 | 4.4415 |
| Median | 40.0000 | 45.0000 | 25.0000 | .0000 | 15.0000 | 10.0000 | 15.0000 | .0000 |
Table 3.
Correlation between paralogous group 13 (HOXA-HOXB-HOXC-HOXD) expression and main clinical features of 94 patients
| Main clinical-pathological characteristics | HOXA13 | Pearson Chi-Square | Pearson’s R | HOXB13 | Pearson Chi-Square | Pearson’s R | HOXC13 | Pearson Chi-Square | Pearson’s R | HOXD13 | Pearson Chi-Square | Pearson’s R | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||||||||||
| Weak or Negative expression | Positive expression | Weak or Negative expression | Positive expression | Weak or Negative expression | Positive expression | Weak or Negative expression | Positive expression | ||||||||||
| Age | ≤68 | 36.7% | 63.3% | 0.512 | 0.083 | 24.5% | 75.5% | 0.315 | 0.111 | 49.0% | 51.0% | 0.146 | 0.159 | 20.4% | 79.6% | 0.113 | -0.169 |
| >69 | 28.9% | 71.1% | 15.6% | 84.4% | 33.3% | 66.7% | 35.6% | 64.4% | |||||||||
| Gender | F | 35.7% | 64.3% | 0.811 | 0.038 | 21.4% | 78.6% | 1 | 0.020 | 64.3% | 35.7% | 0.006 | 0.301 | 17.9% | 82.1% | 0.212 | -0.143 |
| M | 31.8% | 68.2% | 19.7% | 80.3% | 31.8% | 68.2v | 31.8% | 68.2% | |||||||||
| Site | Lip | 0 | 100% | 0.624 | -0.061 | 0 | 100% | 0.319 | 0.072 | 0 | 100% | 0.345 | -0.029 | 0 | 100% | 0.427 | 0.038 |
| Tongue | 33.9% | 66.1% | 25% | 75% | 44.6% | 55.4% | 32.1% | 67.9% | |||||||||
| Other | 34.3% | 65.7% | 14.3% | 85.7% | 40% | 60% | 22.9% | 77.1% | |||||||||
| Deep invasion | ≤5 | 26.7% | 73.3% | 0.766 | -0.093 | 20% | 80% | 1 | -0.002 | 66.7% | 33.3% | 0.045 | 0.223 | 20% | 80% | 0.548 | -0.075 |
| >6 | 34.2% | 65.8% | 20.3% | 79.7% | 36.7% | 63.3% | 29.1% | 70.9% | |||||||||
| Grading | 1 | 16.7% | 83.3% | 0.236 | -0.093 | 27.8% | 72.2% | 0.541 | 0.114 | 38.9% | 61.1% | 1 | -0.114 | 27.8% | 72.2% | 1 | 0.004 |
| 2 | 38.9% | 61.1% | 20.4% | 79.6% | 42.6% | 57.4% | 27.8% | 72.2% | |||||||||
| 3 | 31.8% | 68.2% | 13.6% | 86.4% | 40.9% | 59.1% | 27.3% | 72.7% | |||||||||
| T | 1 | 31.3% | 68.8% | 0.406 | -0.003 | 25% | 75% | 0.544 | 0.144 | 50% | 50% | 0.284 | 0.031 | 43.8% | 56.3% | 0.261 | 0.204 |
| 2 | 37.5% | 62.5% | 25% | 75% | 42.5% | 57.5% | 30% | 70% | |||||||||
| 3 | 16.7% | 83.3% | 16.7% | 83.3% | 22.2% | 77.8% | 22.2% | 77.8% | |||||||||
| 4 | 40% | 60% | 10% | 90% | 50% | 50% | 15% | 85% | |||||||||
| Staging | 1 | 20% | 80% | 0.217 | -0.094 | 30% | 70% | 0.749 | 0.107 | 50% | 50% | 0.908 | 0.049 | 50% | 50% | 0.224 | 0.180 |
| 2 | 39.1% | 60.9% | 21.7% | 78.3% | 39.1% | 60.9% | 34.8% | 65.2% | |||||||||
| 3 | 18.2% | 81.8% | 22.7% | 77.3% | 45.5% | 54.5% | 18.2% | 81.8% | |||||||||
| 4 | 41% | 59% | 15.4% | 84.6% | 38.5% | 61.5% | 23.1% | 76.9% | |||||||||
Relation between paralogous 13 HOX expression and survival of OSCC patients series
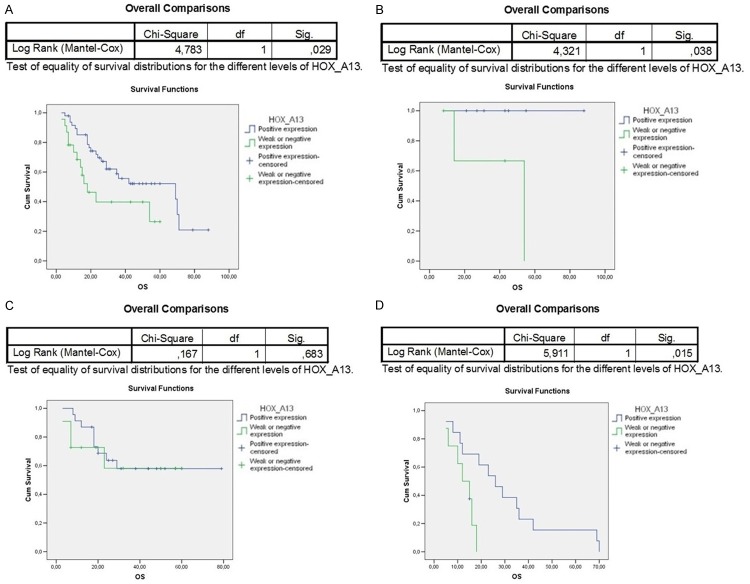
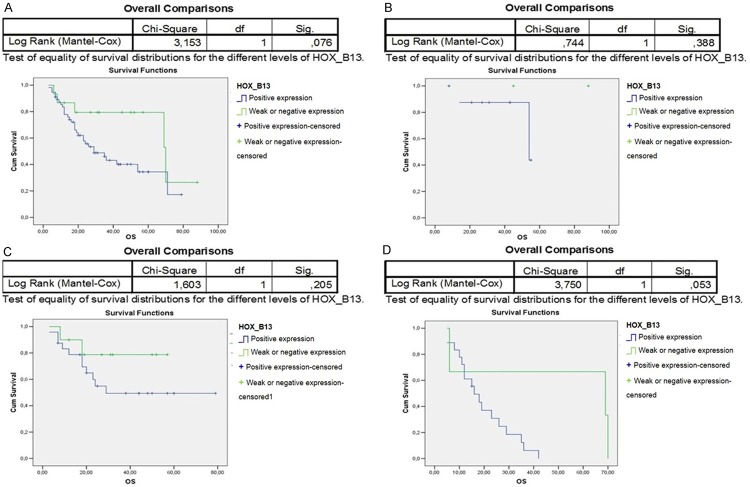
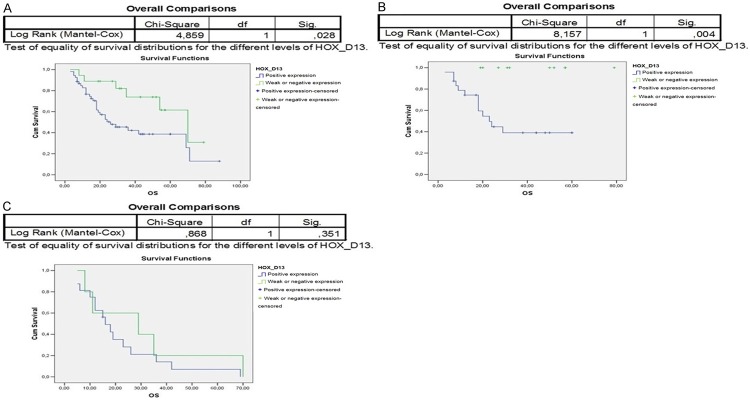
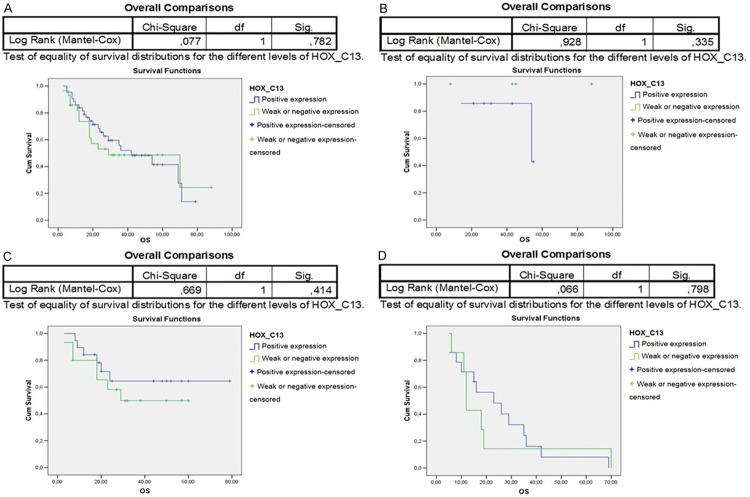
The paralogous 13 HOX expression influenced the prognosis of OSCC differently. Indeed, the overall survival of 94 OSCC patients associated to paralogous 13 HOX proteins positivity was statistically significant, in particular for HOXA13, HOXB13 and HOXD13 (Figures 3A, 4A, 6A). The first assessments showed that the overexpression of HOX A13 was significantly associated with a better prognosis at 60 months (P=0.029). (Figure 3A). In addition, patients with high HOXA13 expression, treated only with surgery, showed a better overall survival (P=0.038) (Figure 3B). Patients undergone chemo and radiotherapy with HOXA13 positivity were heavily associated with a better survival (P=0.015) (Figure 3D). Regarding HOXB13, its expression was associated with a low overall survival (trend of statistical association, P=0.076) (Figure 4A) and, in the same way, its expression in patients undergone chemo and radiotherapy was associated with a poor overall survival (Figure 4D). No significant changes of overall survival associated to HOXC13 expression were found, as shown in Figure 5A-D. Instead HOXD13 expression was significantly associated with a worse prognosis (P=0.029), as shown in Figure 6A and HOXD13 expression in patients undergone radio therapy was associated with a low overall survival (P=0.004).
Figure 3.
Kaplan-Meier curves analysis to evaluate overall survival according to HOXA13 expression in A. 94 OSCC patients; in B. Patients without treatment; C. Patients treated only with radiotherapy; D. Patients treated with chemo/radiotherapy.
Figure 4.
Kaplan-Meier curves analysis to evaluate overall survival according to HOXB13 expression in A. 94 OSCC patients; in B. Patients without treatment; C. Patients treated only with radiotherapy; D. Patients treated with chemo/radiotherapy.
Figure 6.
Kaplan-Meier curves analysis to evaluate overall survival according to HOXD13 expression in A. 94 OSCC patients; in B. Patients treated only with radiotherapy; C. Patients treated with chemo/radiotherapy.
Figure 5.
Kaplan-Meier curves analysis to evaluate overall survival according to HOXC13 expression in A. 94 OSCC patients; in B. Patients without treatment; C. Patients treated only with radiotherapy; D. Patients treated with chemo/radiotherapy.
Discussion
Squamous cell carcinoma of the oral cavity is one of the most common malignancies in the population, but its diagnosis is often late for the body location in which it occurs and for the irregularity with which patients yet consult specialists [1]. The molecular mechanisms associated with the pathogenesis and evolution of this disease are poorly understood, although some indications in the literature suggest to direct the attention on the role of HOX genes. Early studies, carried out on small numbers of cases, suggested the aberrant expression of several HOX genes in oral dysplasia and OSCC [16]. In particular, the overexpression of HOX B7 was significantly correlated to T and N stages and ki67 in OSCC patients [17]. Molecular and immunohistochemical studies also highlighted the progressive increase of expression of HOX A5 from non-tumor epithelium to carcinoma [18]. Moreover, Hunter et al., through transcriptomic analysis of cell cultures representative of OSCC development, showed a dysregulation of expression of several HOX genes, in particular of HOX D10 [19]. More recently, the transfection of HOX D10 gene in an OSCC cellular model caused a decrease in cell invasion but an increase of proliferation, adhesion and migration of tumor cells [20].
In this study we have focused the attention on HOX genes of paralogous group 13, which activity is correlated to cell proliferation and their deregulation is often associated with tumor evolution [9-14]. We analyzed paralogous 13 HOX proteins expression in a Prognostic-Tumor Array including 119 OSCC samples. HOX A13, HOX B13, HOX C13 and HOX D13 protein localization appeared prevalently nuclear but some areas showed also a cytoplasmic localization. This different sub cellular localization has been tested by analysis of mRNA transcripts for each of the 4 HOX genes. Cytoplasmic localization was previously described, and in some tumor types has represented an unfavorable prognostic factor during tumor progression [14]. In our OSCC series HOXB13 showed a high nuclear expression in the deep side of cancer, and only a trend of statistical association with a low overall survival of patients. HOXC13 nuclear expression increased in the superficial margin of the lesion. HOX B13 and HOXC13 do not display significant associations with other clinic pathological parameters in OSCC, despite HOX B13 has been associated with tumor evolution and progression of several hormone-dependent tumors, such as prostate, ovarian and breast cancers [21-23], while the prognostic role of HOX C13 was recently described in metastatic melanoma [12]. The most interesting results seem to be associated with HOX A13 and HOX D13 proteins expression in OSCC. Aberrant HOX A13 expression increased in lip, and in the superficial side of the lesion; HOXA13 was detected with a prevalent cytoplasmic (45%) compare to nuclear expression (33%). The most consistent data showed the strong HOX A13 expression significantly associated with a better prognosis at 60 months. This appears real in both patients treated only with surgery and those undergoing chemo and radiotherapy with an even stronger statistical significance. HOX A13 expression has been previously associated with progression of esophageal squamous cell carcinoma (ESCC), and it was involved in the pathogenesis of gastric cancer and hepatocarcinoma (HCC) [11,24]. More recently, the up-regulation of HOX A13 and the long non-coding RNA, HOTTIP, located in physical contiguity with HOXA13 and that directly controls the HOXA locus gene expression, were associated with HCC patients’ clinical progression and were able to predict clinic outcome and therapeutic response [25]. Moreover, HOXA13 knockdown reduces HOTTIP expression in liver cancer-derived cell lines [26]. To date all the evidence reported in literature emphasize the prognostic role of HOX A13, highlighting its overexpression associated with tumor progression. In OSCC, the role of HOX A13 seems to follow an opposite trend. The overexpression of the marker appears protective in patients with OSCC, making to hypothesize a potential role as tumor suppressor. Regarding HOX D13 expression, a strong cytoplasmic expression, in deep side of the tumor, was detected, that gradually increased with tumor size.
HOXD13 expression appeared significantly associated with a worse prognosis and in patients undergoing radiotherapy it was strongly associated with a low overall survival. In detail, the cytoplasmic HOX D13 expression appeared inversely related to overall survival of OSCC patients, highlighting its role as tumor progression marker. Also for this gene, its activity in tumor pathogenesis and evolution is often described in an opposite manner. In fact, the downregulation of HOX D13, during tumor progression, was previously demonstrated in pancreatic cancer. HOXD13 homeoprotein expression not only decreased from normal to pancreatic tumor tissues but its de-regulation was strongly associated with clinic outcome [10]. This effect on outcome was independent from the T or N stage of the patients at the time of diagnosis. On the contrary, as in OSCC, HOX D13 expression gradually increased from normal tissue until thyroid cancers [9]. In conclusion, our data strongly highlight that HOXA13 tumor expression could be a good prognosis marker, instead HOXD13 and HOXB13 expression could be the worst prognosis markers, suggesting the potential prognostic value of paralogous 13 HOX genes in squamous cell carcinoma of the oral cavity. Their deregulation, also in OSCC, supports the important role of HOX genes in tumor evolution and suggests the development of new potential therapies targeted against the activities of these genes.
Acknolwledgements
All authors of this manuscripts report NONE conflict of interest with respect to any financial or personal relationships with other people or organizations that could inappropriately influence our work. This study has no funding source and no other involvement.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Almholt K, Lund LR, Rygaard J, Nielsen BS, Danø K, Rømer J, Johnsen M. Reduced metastasis of transgenic mammary cancer in urokinase-deficient mice. Int J Cancer. 2005;113:525–32. doi: 10.1002/ijc.20631. [DOI] [PubMed] [Google Scholar]
- 2.Westra W. Handbook of Head and neck pathology. Vol Foundations in diagnostic pathology. LDR Thompson Elsevier; 2006. Malignant neoplasm of the oral cavity and oropharynx. [Google Scholar]
- 3.Bose P, Brockton NT, Dort JC. Head and neck cancer: from anatomy to biology. Int J Cancer. 2013;133:2013–2023. doi: 10.1002/ijc.28112. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein JM, Bernstein CR, West CM, Homer JJ. Molecular and cellular processes underlying the hallmarks of head and neck cancer. Eur Arch Otorhinolaryngol. 2013;270:2585–2593. doi: 10.1007/s00405-012-2323-x. [DOI] [PubMed] [Google Scholar]
- 5.Mallo M, Wellik DM, Deschamps J. Hox genes and regional patterning of the vertebrate body plan. Dev Biol. 2010;344:7–15. doi: 10.1016/j.ydbio.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kraus P, Lufkin T. Mammalian Dlx homeobox gene control of craniofacial and inner ear morphogenesis. J Cell Biochem. 1999;33:133–140. doi: 10.1002/(sici)1097-4644(1999)75:32+<133::aid-jcb16>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 7.Buckingham M, Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol. 2007;23:645–673. doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- 8.Cillo C, Faiella A, Cantile M, Boncinelli E. Homeobox genes and cancer. Exp Cell Res. 1999;248:1–9. doi: 10.1006/excr.1999.4451. [DOI] [PubMed] [Google Scholar]
- 9.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 10.Cantile M, Scognamiglio G, La Sala L, La Mantia E, Scaramuzza V, Valentino E, Tatangelo F, Losito S, Pezzullo L, Chiofalo MG, Fulciniti F, Franco R, Botti G. Aberrant expression of posterior HOX genes in well differentiated histotypes of thyroid cancers. Int J Mol Sci. 2013;14:21727–21740. doi: 10.3390/ijms141121727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantile M, Franco R, Tschan A, Baumhoer D, Zlobec I, Schiavo G, Forte I, Bihl M, Liguori G, Botti G, Tornillo L, Karamitopoulou-Diamantis E, Terracciano L, Cillo C. HOX D13 expression across 79 tumor tissue types. Int J Cancer. 2009;125:1532–1541. doi: 10.1002/ijc.24438. [DOI] [PubMed] [Google Scholar]
- 12.Cillo C, Schiavo G, Cantile M, Bihl MP, Sorrentino P, Carafa V, D’ Armiento M, Roncalli M, Sansano S, Vecchione R, Tornillo L, Mori L, De Libero G, Zucman-Rossi J, Terracciano L. The HOX gene network in hepatocellular carcinoma. Int J Cancer. 2011;129:2577–87. doi: 10.1002/ijc.25941. [DOI] [PubMed] [Google Scholar]
- 13.Cantile M, Scognamiglio G, Anniciello A, Farina M, Gentilcore G, Santonastaso C, Fulciniti F, Cillo C, Franco R, Ascierto PA, Botti G. Increased HOX C13 expression in metastatic melanoma progression. J Transl Med. 2012;10:91. doi: 10.1186/1479-5876-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cantile M, Galletta F, Franco R, Aquino G, Scognamiglio G, Marra L, Cerrone M, Malzone G, Manna A, Apice G, Fazioli F, Botti G, De Chiara A. Hyperexpression of HOXC13, located in the 12q13 chromosomal region, in well‑differentiated and dedifferentiated human liposarcomas. Oncol Rep. 2013;30:2579–2586. doi: 10.3892/or.2013.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marra L, Cantile M, Scognamiglio G, Perdonà S, La Mantia E, Cerrone M, Gigantino V, Cillo C, Caraglia M, Pignata S, Facchini G, Botti G, Chieffi S, Chieffi P, Franco R. Deregulation of HOX B13 expression in urinary bladder cancer progression. Curr Med Chem. 2013;20:833–839. [PubMed] [Google Scholar]
- 16.Marcinkiewicz KM, Gudas LJ. Altered epigenetic regulation of homeobox genes in human oral squamous cell carcinoma cells. Exp Cell Res. 2014;320:128–143. doi: 10.1016/j.yexcr.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassan NM, Hamada J, Murai T, Seino A, Takahashi Y, Tada M, Zhang X, Kashiwazaki H, Yamazaki Y, Inoue N, Moriuchi T. Aberrant expression of HOX genes in oral dysplasia and squamous cell carcinoma tissues. Oncol Res. 2006;16:217–24. doi: 10.3727/000000006783981080. [DOI] [PubMed] [Google Scholar]
- 18.De Souza Setubal Destro MF, Bitu CC, Zecchin KG, Graner E, Lopes MA, Kowalski LP, Coletta RD. Overexpression of HOXB7 homeobox gene in oral cancer induces cellular proliferation and is associated with poor prognosis. Int J Oncol. 2010;36:141–149. [PubMed] [Google Scholar]
- 19.Tucci R, Campos MS, Matizonkas-Antonio LF, Durazzo M, Pinto Junior Ddos S, Nunes FD. HOXB5 expression in oral squamous cell carcinoma. J Appl Oral Sci. 2011;19:12512–9. doi: 10.1590/S1678-77572011000200008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter KD, Thurlow JK, Fleming J, Drake PJ, Vass JK, Kalna G, Higham DJ, Herzyk P, Macdonald DG, Parkinson EK, Harrison PR. Divergent routes to oral cancer. Cancer Res. 2006;66:7405–7413. doi: 10.1158/0008-5472.CAN-06-0186. [DOI] [PubMed] [Google Scholar]
- 21.Hakami F, Darda L, Stafford P, Woll P, Lambert DW, Hunter KD. The roles of HOXD10 in the development and progression of head and neck squamous cell carcinoma (HNSCC) Br J Cancer. 2014;111:807–816. doi: 10.1038/bjc.2014.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miao J, Wang Z, Provencher H, Muir B, Dahiya S, Carney E, Leong CO, Sgroi DC, Orsulic S. HOXB13 promotes ovarian cancer progression. Proc Natl Acad Sci U S A. 2007;104:17093–17098. doi: 10.1073/pnas.0707938104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, Muir B, Mohapatra G, Salunga R, Tuggle JT, Tran Y, Tran D, Tassin A, Amon P, Wang W, Wang W, Enright E, Stecker K. A twogene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 24.McMullin RP, Mutton LN, Bieberich CJ. Hoxb13 regulatory elements mediate transgene expression during prostate organogenesis and carcinogenesis. Dev Dyn. 2009;238:664–672. doi: 10.1002/dvdy.21870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han Y, Tu WW, Wen YG, Li DP, Qiu GQ, Tang HM, Peng ZH, Zhou CZ. Identification and validation that up-expression of HOXA13 is a novel independent prognostic marker of a worse outcome in gastric cancer based on immunohistochemistry. Med Oncol. 2013;30:564. doi: 10.1007/s12032-013-0564-1. [DOI] [PubMed] [Google Scholar]
- 26.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z, Boldanova T, Andersen JB, Hämmerle M, Tornillo L, Heim MH, Diederichs S, Cillo C, Terracciano LM. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–23. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.