Abstract
Lung adenocarcinoma (LAD) and lung squamous cell cancer (LSCC) are two most common histological types of lung cancer, while they differ in many aspects. Recent evidence shows that long non-coding RNAs (lncRNAs) play an important role in the process of cancer initiation and progression. Thus, characterization of LAD and LSCC associated lncRNAs may help understand the difference between LAD and LSCC. Here, we analyzed three sets of RNA-seq data, including LAD RNA-seq data from TCGA project. We identified a novel lncRNA, long intergenic non-protein coding RNA 1207 (LINC01207) which was significantly up-regulated in LAD tissues compared with paired non-tumor tissues (5.78 fold increase, P<0.05), while there was no significant differences between LSCC tissues and adjacent non-tumor tissues. The expression level of LINC01207 was associated with TNM stage of LAD patients, and higher LINC01207 level indicated advanced TNM stage (P<0.05) and shorter survival (HR=2.53, P<0.05). By small interfering RNA (siRNA) mediated knockdown of LINC01207, we determined the biological function of LINC01207 in A549 cell line. After knockdown of LINC01207, cell proliferation ability was inhibited. Further analysis showed that after silence of LINC01207, the percentage of apoptotic cells significantly increased. By RNA immunoprecipitation and Chromatin immunoprecipitation assay, we demonstrated that LINC01207 could bind with EZH2 and mediated trimethylation of histone 3 lysine 27 at the promoter region of Bad, an important pro-apoptotic gene. Finally, we developed xenograft tumor models in nude mice and xenograft tumors derived from A549 cells transfected with siRNA-LINC01207 had significantly lower tumor weight and smaller tumor volume. In summary, the novel lncRNA, LINC01207 is specifically up-regulated in LAD but not in LSCC; and LINC01207 could promote LAD cell growth both in vivo and in vitro.
Keywords: Lung cancer, lung adenocarcinoma, LINC01207, apoptosis, bad
Introduction
Lung cancer is one of the most common causes of cancer-related mortality [1] and the five-year survival is still poor, about 16.6% [2,3]. In spite of recent advances in clinical and experimental oncology, the prognosis of lung cancer is still dissatisfying [4]. For lung cancer, about 85% newly diagnosed lung cancer cases are non-small-cell lung cancer (NSCLC). Non-small cell lung cancer consists of several histology types, and the most common two types are lung adenocarcinoma (LAD) and lung squamous cell cancer (LSCC) [5]. LAD and LSCC differs greatly in many aspects, therefore, it is essential to discover the molecular basis of LSCC and LAD.
Recently, mounting evidence shows that long noncoding RNA (lncRNA), which has been considered as transcriptome noise, exerts vital biological function [6-8]. In the pathological process of cancer initiation and progression, many lncRNAs are widely involved and play important roles. For lung cancer, many functional lncRNAs have been well characterized, such as MALAT1 [9], LET [10], TARID [11]. Due to the advances of high-throughout technology, such as microarray and RNA-sequencing, numerous lncRNAs have been detected and profiled in lung cancer [12-14]. For example, White and colleagues [15] have characterized hundreds of differentially expressed lncRNAs in lung cancer. In the study by White et al [15], they also identified various lncRNAs which show differential expression profiles between different pathological types of lung cancer. Considering the heterogeneity between LAD and LSCC, it is highly possible that these histological type specific lncRNAs may have important function. The high-throughout technology, especially RNA-sequencing, can yield huge amount of data, and numerous data are available for data mining. The landmark TCGA (the cancer genome atlas) project has released RNA-seq data of many cancers, including LAD, and data mining of TCGA RNA-seq data is an effective method.
Long intergenic non-protein coding RNA 1207 (LINC01207) is locates in the genomic 4q32 locus, includes 3 exons and 2 introns, and transcribes an lncRNA of 3212nt. In this study, we characterized that LINC01207 was a novel LAD specific lncRNA, which was significantly up-regulated in LAD and the expression level of LINC01207 was correlated with TNM stage. Both in vivo and in vitro experiments showed that LINC01207 could promote LAD cell proliferation.
Methods
Patients and tissue samples
This study was approved by the Ethics Committee of Shandong University. Paired NSCLC tissues and adjacent non-tumor tissues were obtained from patients who received surgical resection of NSCLC between 2005 and 2012. All surgical specimens were snap-frozen and stored in liquid nitrogen immediately after resection until total RNA extraction. All tumor and paired non-tumor tissues were confirmed by experienced pathologists, as well as the pathological stage, grade, and nodal status. Clinical and pathological characteristics were also collected for each patient. Informed written consents were obtained from all patients included in this study.
Cell lines and culture conditions
A549 cell line purchased from the Institute of Biochemistry and cell biology of Chinese academy of science (Shanghai, China). A549 cell line was cultured in RPMI 1640 medium (GIBCO) supplemented with 10% fetal bovine serum (10% FBS, GIBCO), 100 U/ml penicillin, and 100 mg/ml streptomycin in humidified air at 37°C with 5% CO2.
Total RNA extraction and qRT-PCR analysis
Total RNA was isolated with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacture’s protocol. 1000 ng total RNA was reverse transcribed in a final volume of 20 μl using random primers under standard conditions using the PrimerScript RT Master Mix (Takara, RR036A). The reverse transcription reaction was carried out under the following conditions: 37°C for 30 min; 85°C for 5 sec; and then hold on 4°C.
The quantitative real-time polymerase chain reaction (qRT-PCR) was performed using the SYBR Select Master Mix (Applied Biosystems, cat: 4472908) on ABI 7900 system (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. The relative levels of CCAT2 were determined by qPCR using gene specific primers. GAPDH was measured as an internal control. After the reverse transcription, 0.5 μl of the complementary DNA was used for subsequent qRT-PCR reaction. The PCR primers used were as follow: 5’-CCACATCGCTCAGACACCAT-3’ (sense) and 5’-ACCAGGCGCCCAATACG-3’ (antisense) for GAPDH and 5’-CAGACACAGGCCATTCAGTC-3’ (sense) and 5’-CTTCTTCACCAGAAGCATTCC-3’ (antisense) for LINC01207 and 5’-CCCAGAGTTTGAGCCGAGTG-3’ (sense) and 5’-CCCATCCCTTCGTCGTCCT-3’ (antisense) for Bad and 5’-GTTTTCCGCAGCTACGTTTTT-3’ (sense) and 5’-GCAGAGGTAAGGTGACCATCTC-3’ (antisense) for Bak and 5’-CCCGAGAGGTCTTTTTCCGAG-3’ (sense) and 5’-CCAGCCCATGATGGTTCTGAT-3’ (antisense) for Bax and 5’-GGTGGGGTCATGTGTGTGG-3’ (sense) and 5’-CGGTTCAGGTACTCAGTCATCC-3’ (antisense) for Bcl2. The qRT-PCR reaction was conducted under the following conditions: 95°C for 30 sec, 40 cycles of 95°C for 5 sec and 60°C for 60 sec. For cell expression, each sample was run in quintuplicate and each sample was run in triplicate for tissues expression. The Ct-value for each sample was calculated with the ΔΔCt-method, and the results were expressed as 2-ΔΔCT to analyze the fold change (tumor vs. normal): ΔΔCT=(CTtarget gene-CTactin)normal-(CTtarget gene-CTactin)tumor.
siRNA and transfection of NSCLC cells
A549 cells cultured on six-well plate were transfected with small interfering RNA (siRNA) or negative control using Lipofectamine 2000 (Invitrogen, Shanghai, China) according to the manufacturer’s instructions. Cells were harvested after 24 hours for qRT-PCR and other experiment. The siRNA sequences of LINC01207 were as follows: 5’-CCAGCTAAGACATTAGTAA-3’ for siRNA1, and 5’-GCAGGAAGGAATCCACAAT-3’ for siRNA2. Sequences of EZH2 siRNA were 5’-GAGGUUCAGACGAGCUGAUUU-3’ and non-specific siRNA (si-NC) was purchased from Invitrogen.
Cell proliferation assay
A549 cells were seeded into 96-well plates (2×104/well) and incubated in RPMI 1640 at 37°C and 5% CO2 atmosphere for 96 hours. The Cell Counting Kit-8 (CCK8) assay was used to determine relative cell growth according to the manufacturer’s instructions. The absorbance was measured at 450 nm with an ELx-800 Universal Microplate Reader. Each experiment was repeated at least three times independently.
Colony formation assay
For colony formation assay, a total of 500 cells were placed in a fresh six-well plate and maintained in media containing 10% FBS, replacing the medium every 4 days. After 14 days, cells were fixed with methanol and stained with 0.1% crystal violet. Visible colonies were manually counted. Triplicate wells were measured for each treatment group.
Flow-cytometric analysis
For cell cycle analysis, transfected cells were harvested after transfection by trypsinization. After the double staining with fluorescein isothiocyanate (FITC)-Annexin V and propidium iodide was done by the FITC Annexin V Apoptosis Detection Kit (BD Biosciences) according to the manufacturer’s recommendations. The cells were analyzed with a flow cytometry (FACScan; BD Biosciences) equipped with a Cell Quest software (BD Biosciences). Cells were discriminated into viable cells, dead cells, early apoptotic cells, and apoptotic cells. The percentage of early apoptotic cells were compared to control groups from each experiment. Cells for cell-cycle analysis were stained with propidium oxide by the Cycle TEST PLUS DNA Reagent Kit (BD Biosciences) following the protocol and analyzed by FACScan. The percentage of the cells in G0-G1, S, and G2-M phase were counted and compared.
RNA immunoprecipitation (RIP)
RIP experiments were performed using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer’s instructions. Antibody for RIP assays of EZH2 was from Abcam.
Chromatin immunoprecipitation assay (ChIP)
The ChIP assays were performed using EZ-CHIP KIT according to the manufacturer’s instruction (Millipore, Billerica, MA, USA). The EZH2 antibodies were obtained from Abcam. H3 trimethyl Lys 27 antibody was from Millipore. The ChIP primer sequences forword 1#-GGAGCTGTCACTATCCCCAC, reverse 1#-CAATCACCTCTCGGAACGTC, forword 2#-TCTGCCTCCTGTCCTCGTAA, and reverse 2#-CGCCAATTCTCGTACGGTTTC were used for the promoter region of Bad.
Western blot assay
Cells were lysed using mammalian protein extraction reagent RIPA (Beyotime china) supplemented with protease inhibitors cocktail (Roche. Switzerland) and PMSF (Roche, Switzerland). Protein concentration was measured with the Bio-Rad protein assay kit. 50 μg protein extractions were separated by 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), then transferred to 0.22 μm nitrocellulose membranes (Sigma-Aldrich. USA) and incubated with specific antibodies. ECL chromogenic substrate was used to visualize the bands. Anti-β-actin and anti-Caspase3 were from Abcam (Hong Kong, China). Anti-EZH2 and Bad was from Abcam (Hong Kong, China).
Xenograft experiment
A549 cells were transfected with siRNA-LINC01207 or negative control sequence using Lipofectamine 2000 (Invitrogen). After 48 hours of transfection, the cells were collected and injected into either side of the posterior flank of the same male BALB/c nude mouse. The tumor volumes and weights were measured every 4 days in the mice; the tumor volumes were measured as length×width2×0.5. 20 days after injection, the mice were sacrificed, the tumor weights were measured, and the tumors were collected for further analysis.
Immunohistochemistry
Xenograft tumor tissues derived from A549 cells transfected with negative control or si-LINC01207 siRNA were immunostained for Bad. Anti-Ki67 was from Santa Cruz Biotechnology.
Data source and statistical analysis
TCGA data was downloaded from the lncRNAtor website (http://lncrnator.ewha.ac.kr/index.htm). Student’s t-test, Spearman test, Pearson correlation analysis, Kaplan-Meier, and Cox-regression were performed to analyze the data using SPSS 18.0 software. A P Values less than 0.05 were considered statistically significant.
Results
LINC01207 is upregulated in LAD
To investigate lncRNA expression profile and identify functional lncRNAs, we first downloaded lncRNA expression data from the TCGA project. By lncRNAtor website, a comprehensive resource for functional investigation of lncRNA, lncRNA expression data of LAD patients were downloaded. Two sets of data were retrieved: the first set of data consists of 245 LAD tissues and 46 normal lung tissues, and the second set of data were from 59 LAD patients, of which 31 received cisplatin and 28 did not. Then, the third lncRNA expression data in LAD by White NM was also retrieved. Thus, 3 data sets were collected and two of them provided information about differential expression between LAD tissues and normal lung tissues, and one provided the information about response to chemotherapy. A venny plot was generated (Figure 1A), and we found 6 lncRNAs (LINC01207, LINC01426, LINC00152, CHKB-AS1, TBX5-AS1, and FAM95B1) were differentially expressed in all the 3 data sets.
Figure 1.
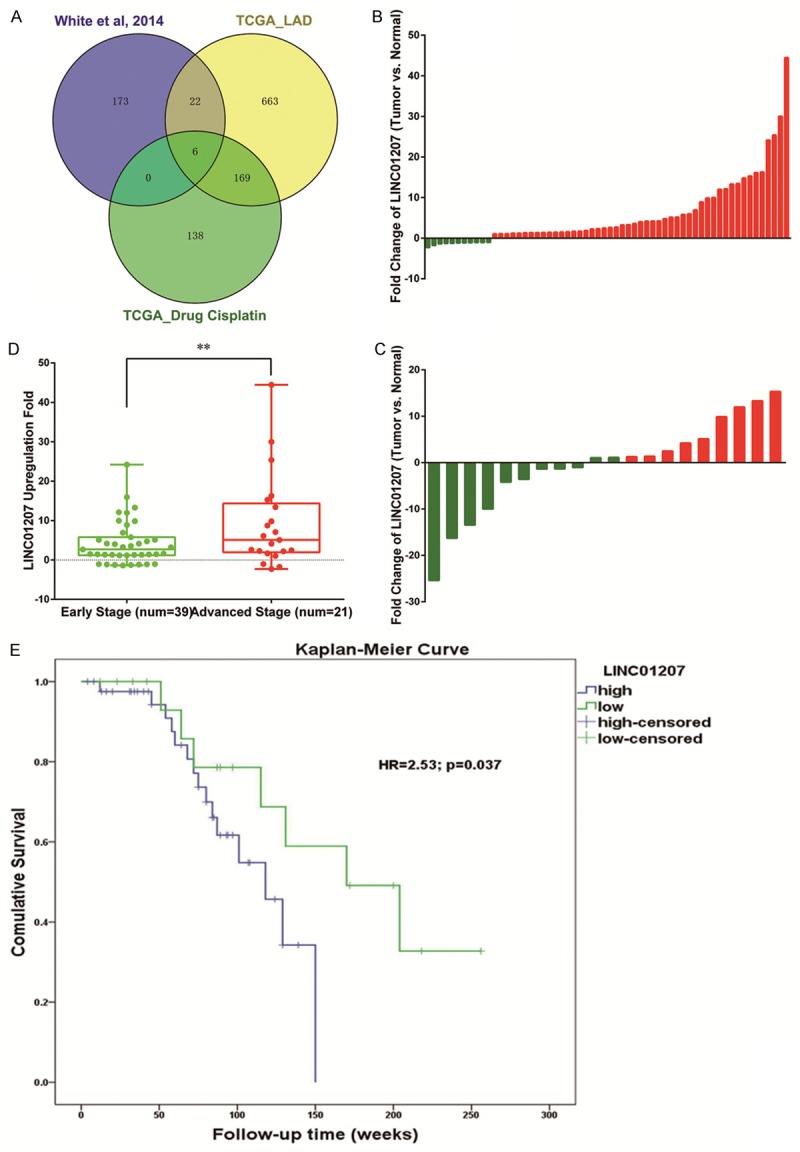
Venny plot show that 6 lncRNAs were differentially expressed in all the 3 data sets (A). Expression of LINC01207 in lung adenocarcinoma patients (B) and lung squamous cell cancer patients (C). Red: up-regulation; green: down-regulation. In LAD patients with advanced TNM stage, LINC01207 expression level was higher (D). Compared with LAD patients with low expression of LINC01207, patients with high expression had poor survival (E). **P<0.01.
The novel long intergenic noncoding RNA 1207, LINC01207 was chosen for further investigation. We first profiled the expression of LINC01207 in 80 pairs of NSCLC tissues and paired adjacent non-tumor tissues, including 60 LAD and 20 LSCC patients. In consistence with White NM [15], compared with paired non-tumor tissues, LINC01207 was significantly up-regualted in LAD (P<0.05) but no significant difference was observed in LSCC (Figure 1B and 1C). As shown, LINC01207 is up-reuglated in 49 of 60 LAD patients with an average increasing fold of 5.78. We further analyzed the correlation between LINC01207 expression level and clinical features. As shown in Table 1, we found that the expression level of LINC01207 was associated with TNM stage, and higher expression level indicated advanced TNM stage (P<0.05, Figure 1D). Kaplan-Meier curve and Cox regression were further applied to assess whether LINC01207 was associated survival of LAD patients. As shown, compared with patients with low expression level of LINC01207, those with high expression level of LINC01207 had significantly shorter survival time (Figure 1E, HR=2.53, 95% CI: 1.36-5.49; P=0.037).
Table 1.
Correlation between LINC01207 expression level and clinical characteristics
| Characteristics | Num of Patients (60) | Percentage | Fold Change | P Values |
|---|---|---|---|---|
| Age (years) | ||||
| <50 | 7 | 11.67% | 8.68 | 0.403 |
| 50-70 | 45 | 75.00% | 4.48 | |
| >70 | 8 | 13.33% | 10.55 | |
| Smoking | ||||
| Never | 38 | 63.33% | 7.45 | 0.294 |
| Ever | 22 | 16.67% | 2.89 | |
| Gender | ||||
| Male | 29 | 48.33% | 5.69 | 0.650 |
| Female | 31 | 51.67% | 5.86 | |
| Lymph Node Metastasis | ||||
| Negative | 40 | 66.67% | 6.71 | 0.081 |
| Positive | 20 | 33.33% | 3.92 | |
| TNM Stage | ||||
| I-II | 44 | 73.33% | 2.66 | 0.002* |
| III-IV | 16 | 26.67% | 14.36 |
Significant correlation.
Silence of LINC01207 inhibited proliferation in vivo
To probe the biological function of LINC01207, we designed small interfering RNAs (siRNAs) that can specifically target LINC01207. As shown, LINC01207 was effectively knockdown by siRNAs (Figure 2A). Then, we assessed whether the knockdown of LINC01207 could affect the biological function of A549, a LAD cell line. CCK8 assay showed that siRNA treatment significantly inhibited proliferation of A549 cell line (Figure 2B). Colony formation assay also showed that colony formation ability was decreased after silence of LINC01207 (Figure 2C).
Figure 2.
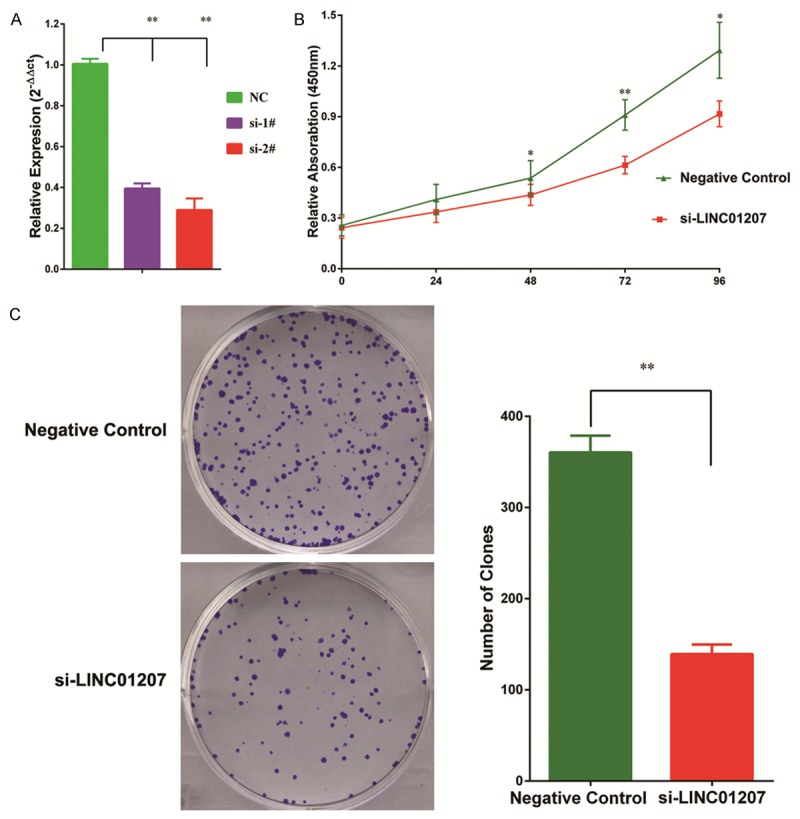
LINC01207 was efficiently knocked down by siRNA in A549 cell (A). CCK8 assay showed that knockdown of LINC01207 significantly inhibited cell proliferation (B). Knockdown of LINC01207 also inhibited colony formation ability of A549 cell (C). *P<0.05; **P<0.01.
Silence of LINC01207 promoted apoptosis
Flow cytometry was used to test whether cell cycle or apoptosis was affected by silence of LINC01207. We found that after knockdown of LINC01207, there was no significant change in cell cycle (Figure 3A). However, we observed that after silence of LINC01207, the percentage of apoptotic cells was significantly increased (Figure 3B). Consistent with FACIS analysis, western blot confirmed that the activated Caspase 3, an apoptosis marker, was up-regualted after LINC01207 silence. These lines of evidence showed that LINC01207 could promote LAD cell proliferation by inhibiting apoptosis.
Figure 3.
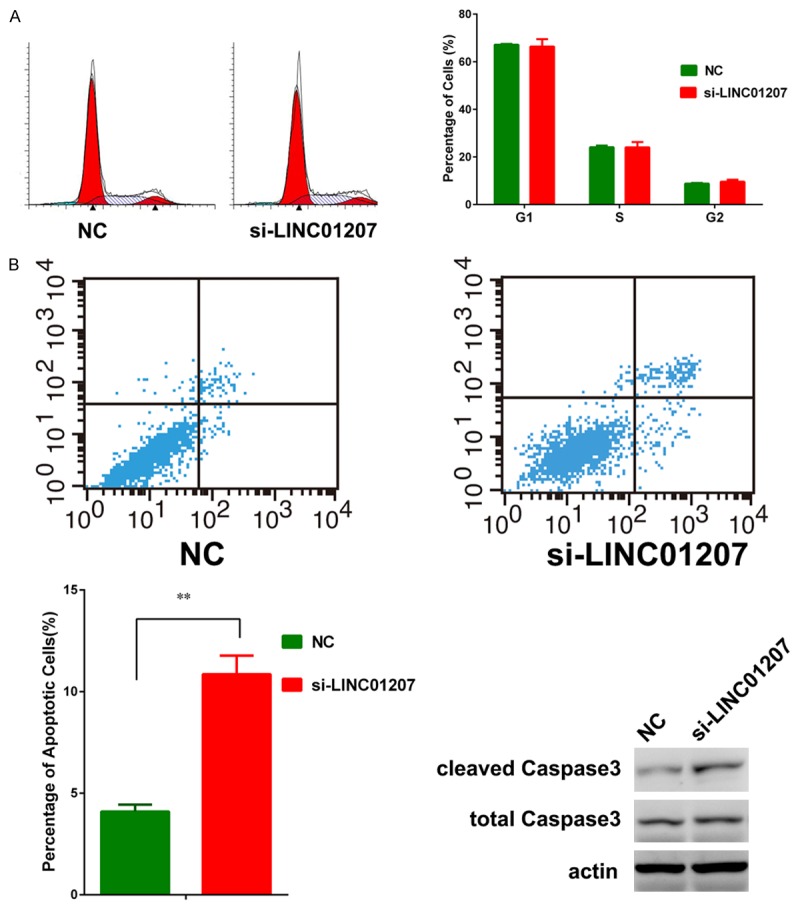
Cell cycle was not significantly affected by silence of LINC01207 (A). The percentage of apoptotic cells were significantly increased after knockdown of LINC01207 and the cleaved Caspase 3 was also up-regualted after knockdown of LINC01207 (B).
LINC01207 induced EZH2-mediated silence of Bad
To determine the downstream target of LINC01207, by which LINC01207 modulate apoptosis, we examined the expression level of Bcl2 family genes after silence of LINC01207. As shown, the pro-apoptotic gene, Bad was mostly up-regulated after knockdown of LINC01207, and the up-regulation was also confirmed by western blot assay (Figure 4A, 4B). Currently, most lncRNAs regulated gene expression on transcription and post-transcription levels and location of lncRNA could provide information about the molecular function of lncRNA. Thus, we determined the distribution of LINC01207 between cytoplasm and nucleus, and most LINC01207 transcripts located in nucleus (Figure 4C), indicating that LINC01207 may regulate Bad on transcription level. Almost all lncRNA exert their function by binding to RNA binding proteins. By RNA immunoprecipitation (RIP) assay, we demonstrated that LINC01207 could bind with EZH2 (Figure 4D), a core component of plolycomb repress complex 2 (PRC2). PRC2 is an important regulator for histone modification, and mediate trimethylation at histone 3 lysine 27 (H3K27). Previous evidence has showed that EZH2 might be associated with the H3K27-me3 at the promoter of Bad [16]. Using siRNA specific to EZH2, we demonstrated that Bad was up-regulated after silence of EZH2 (Figure 4E). Thus, it is possible that LINC017 can repress Bad by binding to EZH2 and mediate the H3K27-me3 at promoter region of Bad. To probe this hypothesis, we performed chromatin immunoprecipitation (ChIP) assay and the results revealed that after silence of LINC01207 the enrichment of EZH2 was decreased at the promoter region of Bad and the H3K27-me3 level was also decreased (Figure 4F). We further confirmed the negative correlation between LINC01207 and Bad in a set of 20 lung cancer patients. These lines of evidence showed that LINC01207 inhibited apoptosis by binding to EZH2 and mediating H3K27-me3 at the promoter region of Bad.
Figure 4.
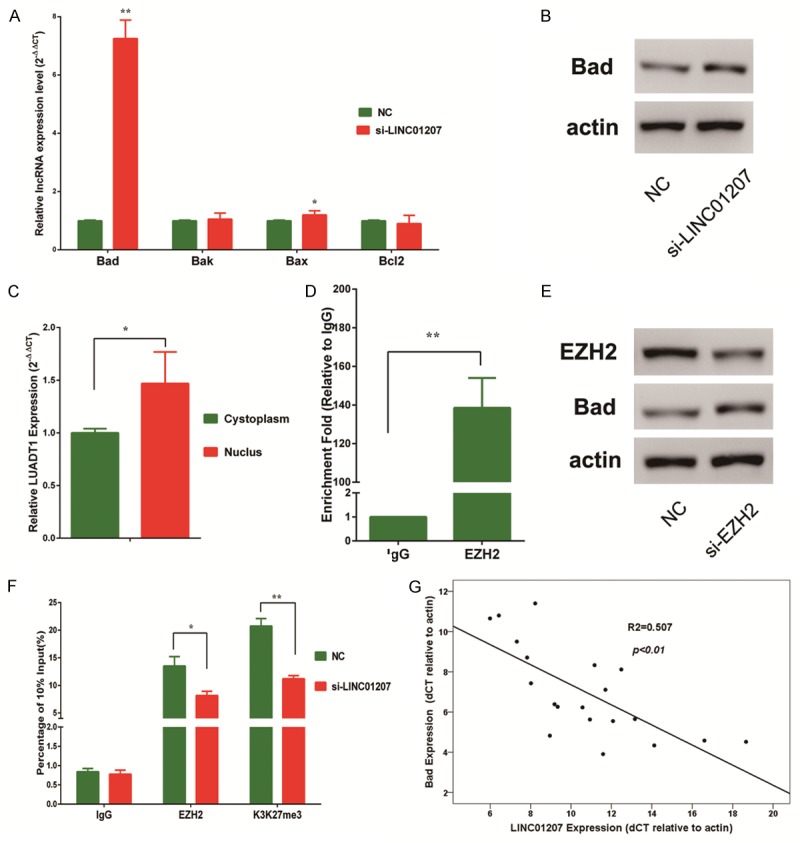
After silence of LINC01207, Bad was significantly up-regulated (A, B). Cell fraction analysis revealed that most LINC01207 transcripts located in nucleus (C). RIP assay showed that compared with IgG, LINC01207 could significantly bind with EZH2 (D). Western blot showed that after silence of EZH2, Bad was upregulated (E). ChIP assay revealed that after silence of LINC01207, the enrichment of EZH2 and H3K27-me3 level were decreased at the promoter region of Bad (F). In a set of 20 LAD patients, the expression levels of LINC01207 and Bad were negatively correlated (G). *P<0.05; **P<0.01.
LINC01207 inhibited LAD in vivo
To test whether LINC01207 could inhibit LAD cell proliferation in vivo, we developed xenograft tumor models in nude mice. After transfection with negative control siRNA or siRNA-LINC01207, A549 cells were injected to nude mice. As shown, siRNA-LINC01207 significantly inhibited tumor growth in vivo (Figure 5A, 5B). Tumors derived from siRNA-LINC01207 transfected A549 cells had significantly smaller tumor volume and lower tumor weight (Figure 5C, 5D). Ki-67 staining also confirmed that tumor proliferation was inhibited by siRNA treatment. IHC assay by Bad antibody also demonstrated that compared with tumors derived from negative control group, in the siRNA treated group Bad staining was stronger, which was consistent with in vitro experiments. Thus, this confirmed that LINC01207 could promote tumor growth in vivo.
Figure 5.
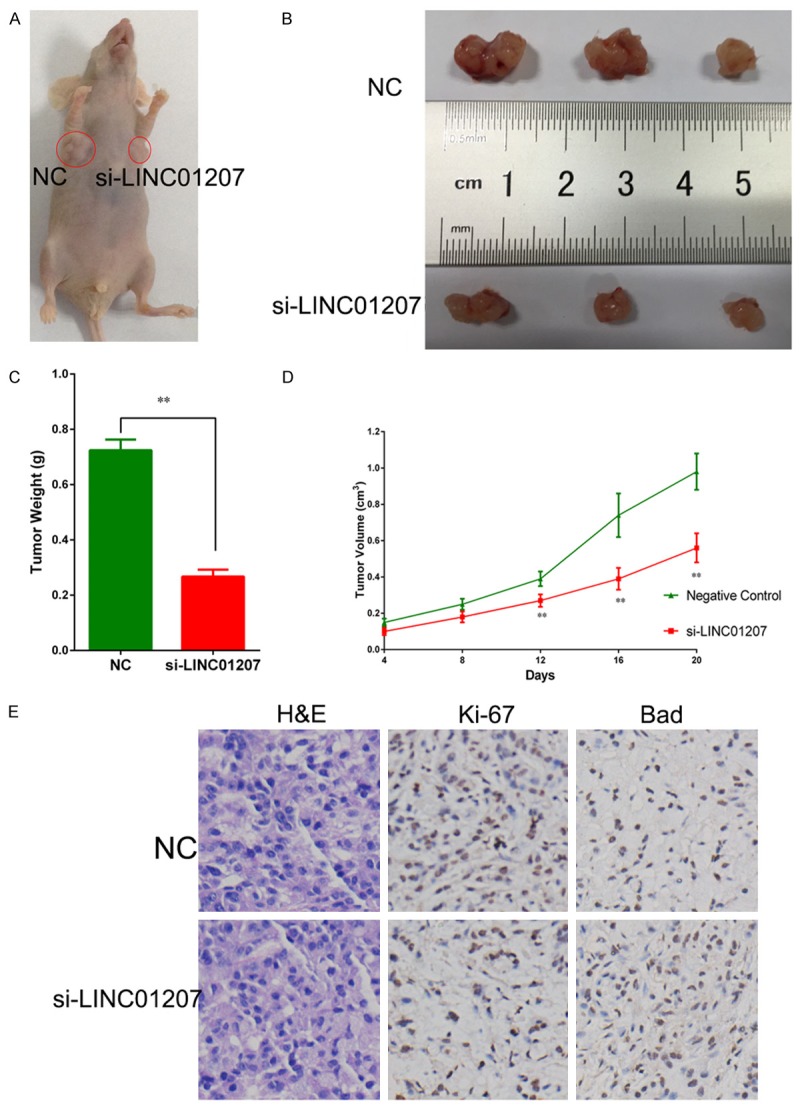
Knockdown of LINC01207 inhibited xenograft tumor growth in nude mice (A, B). Xenograft tumors derived from siRNA-LINC01207 transfected A549 cells had significantly smaller tumor volume (C) and lower tumor weight (D). IHC showed that Ki-67 staining was weaker in si-LINC01207 group and bad staining was higher in si-LINC01207 group. **P<0.01.
Discussion
It has been proved that mammalian genomes encode millions of lncRNAs, which was considered “transcription noise” or “junk”. Now, increasing evidence has demonstrated that these lncRNAs are junk no more and on the opposite, these lncRNAs have important biological functions [17-19]. Dysregulation of lncRNAs has been found in various diseases [8,20,21]. For cancer research, many cancer-associated lncRNAs have been identified and characterized and these lncRNAs help further understand the molecular mechanism of cancer [22]. In addition, most of these cancer-associated lncRNAs could be effective prognostic biomarkers and even therapeutic targets [22,23]. Therefore, identification and characterization of cancer-associated lncRNA is essential for better understanding and treatment of cancer.
For lung cancer, many works have been done to identify NSCLC associated lncRNAs, especially LAD and LSCC associated lncRNAs. By microarray and RNA-sequencing, high throughout technology has provided huge number of data [12,24,25]. By analysis of RNA-seq data from NSCLC tissues, White NM [15] have reported various LAD and LSCC related lncRNAs. Together with TCGA project, we are able to determine the expression profile of lncRNAs in lung cancer. Among these lncRNAs, we focused on LAD specific lncRNAs, which were specifically highly expressed in LAD tissues but not in LSCC. Thus, we found a novel lncRNA, LINC01207. LINC01207 is a 3212nt intergenic lncRNA, locates in the genomic 4q32 locus, and consists of 3 exons and 2 introns. We first validate the expression of LINC01207 in LAD and LSCC tissues. In consistence with White NM [15], LINC01207 was significantly up-regualted in LAD tissues but not in LSCC tissues, compared with adjacent non-tumor tissues. In addition, the expression of LINC01207 in LAD patients was quite homogeneity, since LINC01207 was up-regulated in 49 of 60 LAD patients. Further statistical analysis revealed that the expression level of LINC01207 was correlated with TNM stage of LAD, i.e., higher level of LINC01207 indicated advanced TNM stage. Survival analysis showed that LINC01207 could be a prognostic biomarker for LAD, and high LINC01207 expression level indicated poor survival (HR=2.53, P=0.057). Therefore, LINC01207 could be a LAD specific lncRNA.
We further analyzed the biological function of LINC01207 in vitro by siRNA-mediated silence. In A549 cells, a LAD cell line, we observed that after knockdown of LINC01207, proliferation ability was significantly decreased while cell migration and invasion ability was not affected. By flow cytometric analysis, we found that the percentage of apoptotic cells was increased after knockdown of LINC01207. Western blot also confirmed cleaved Caspase 3, biomarker of apoptosis, increased after knockdown of LINC01207. To further determine the role of LINC01207 in vivo, we developed xenograft tumor models in nude mice. Tumors developed from A549 cell transfected with siRNA-LINC01207 had smaller tumor volume and lower weight. Therefore, LINC01207 could inhibit proliferation of LAD both in vivo and in vitro.
Many lncRNA could bind with PRC2 and mediate the H3K27-me3 at the promoter of specific genes. According to Khalil AM and colleagues, 20% lincRNA could bind with PRC2 and there are many reports of these lncRNAs [26-28]. For apoptosis and Bad, few studies have characterized the regulatory role of lncRNA. In this study, we found LINC01207 inhibited the expression of Bad by recruiting EZH2 and mediated the subsequent H3K27-me3 at promoter region of Bad. In 2015, Deng HP [29] reported that the lncRNA HOTTIP may regulated Bad but the molecular mechanism was not clearly clarified. In the current study, we demonstrated that the lncRNA LINC01207 could regulate Bad via EZH2.
By searching published literature, we have found no reports about the biological function of LINC01207 in cancer or other diseases. In current study, we demonstrated that the novel lncRNA LINC01207 is up-regulated in LAD but not in LSCC tissues, and the expression level of LINC01207 is correlated with TNM stage and survival. By siRNA-mediated knockdown of LINC01207, we demonstrate that LINC01207 could promote LAD proliferation both in vivo and in vitro.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Ettinger DS, Akerley W, Bepler G, Blum MG, Chang A, Cheney RT, Chirieac LR, D’Amico TA, Demmy TL, Ganti AK, Govindan R, Grannis FW Jr, Jahan T, Jahanzeb M, Johnson DH, Kessinger A, Komaki R, Kong FM, Kris MG, Krug LM, Le QT, Lennes IT, Martins R, O’Malley J, Osarogiagbon RU, Otterson GA, Patel JD, Pisters KM, Reckamp K, Riely GJ, Rohren E, Simon GR, Swanson SJ, Wood DE, Yang SC NCCN Non-Small Cell Lung Cancer Panel Members. Non-small cell lung cancer. J Natl Compr Canc Netw. 2010;8:740–801. doi: 10.6004/jnccn.2010.0056. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 4.Thomson CS, Forman D. Cancer survival in England and the influence of early diagnosis: what can we learn from recent EUROCARE results? Br J Cancer. 2009;101(Suppl 2):S102–109. doi: 10.1038/sj.bjc.6605399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu L, Todd NW, Xing L, Xie Y, Zhang H, Liu Z, Fang H, Zhang J, Katz RL, Jiang F. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer. 2010;127:2870–2878. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Roosbroeck K, Pollet J, Calin GA. miRNAs and long noncoding RNAs as biomarkers in human diseases. Expert Rev Mol Diagn. 2013;13:183–204. doi: 10.1586/erm.12.134. [DOI] [PubMed] [Google Scholar]
- 7.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Wilusz JE, Freier SM, Spector DL. 3’ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Arab K, Park YJ, Lindroth AM, Schafer A, Oakes C, Weichenhan D, Lukanova A, Lundin E, Risch A, Meister M, Dienemann H, Dyckhoff G, Herold-Mende C, Grummt I, Niehrs C, Plass C. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell. 2014;55:604–614. doi: 10.1016/j.molcel.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 12.Wang P, Lu S, Mao H, Bai Y, Ma T, Cheng Z, Zhang H, Jin Q, Zhao J, Mao H. Identification of biomarkers for the detection of early stage lung adenocarcinoma by microarray profiling of long noncoding RNAs. Lung Cancer. 2015;88:147–153. doi: 10.1016/j.lungcan.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Morton ML, Bai X, Merry CR, Linden PA, Khalil AM, Leidner RS, Thompson CL. Identification of mRNAs and lincRNAs associated with lung cancer progression using next-generation RNA sequencing from laser micro-dissected archival FFPE tissue specimens. Lung Cancer. 2014;85:31–39. doi: 10.1016/j.lungcan.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen G, Yin K, Shi L, Fang Y, Qi Y, Li P, Luo J, He B, Liu M, Shi T. Comparative analysis of human protein-coding and noncoding RNAs between brain and 10 mixed cell lines by RNASeq. PLoS One. 2011;6:e28318. doi: 10.1371/journal.pone.0028318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White NM, Cabanski CR, Silva-Fisher JM, Dang HX, Govindan R, Maher CA. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol. 2014;15:429. doi: 10.1186/s13059-014-0429-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi JY, Lee S, Hwang S, Jo SA, Kim M, Kim YJ, Pang MG, Jo I. Histone H3 lysine 27 and 9 hypermethylation within the Bad promoter region mediates 5-Aza-2’-deoxycytidine-induced Leydig cell apoptosis: implications of 5-Aza-2’-deoxycytidine toxicity to male reproduction. Apoptosis. 2013;18:99–109. doi: 10.1007/s10495-012-0768-4. [DOI] [PubMed] [Google Scholar]
- 17.Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, Manos PD, Datta S, Lander ES, Schlaeger TM, Daley GQ, Rinn JL. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 20.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y, Park PK, Qin L, Wei Y, Hawke DH, Hung MC, Lin C, Yang L. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159:1110–1125. doi: 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P, Yang Z, Liu W, Zhang H, Chen N, Wang H, Wang H, Qin H. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/beta-catenin signalling pathway via suppression of activator protein 2α. Gut. 2015 doi: 10.1136/gutjnl-2014-308392. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Zhu N, Chen X. A novel long noncoding RNA LINC01133 is upregulated in lung squamous cell cancer and predicts survival. Tumour Biol. 2015;36:7465–71. doi: 10.1007/s13277-015-3460-9. [DOI] [PubMed] [Google Scholar]
- 25.Yu H, Xu Q, Liu F, Ye X, Wang J, Meng X. Identification and validation of long noncoding RNA biomarkers in human non-small-cell lung carcinomas. J Thorac Oncol. 2015;10:645–654. doi: 10.1097/JTO.0000000000000470. [DOI] [PubMed] [Google Scholar]
- 26.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 28.He W, Cai Q, Sun F, Zhong G, Wang P, Liu H, Luo J, Yu H, Huang J, Lin T. linc-UBC1 physically associates with polycomb repressive complex 2 (PRC2) and acts as a negative prognostic factor for lymph node metastasis and survival in bladder cancer. Biochim Biophys Acta. 2013;1832:1528–1537. doi: 10.1016/j.bbadis.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Deng HP, Chen L, Fan T, Zhang B, Xu Y, Geng Q. Long non-coding RNA HOTTIP promotes tumor growth and inhibits cell apoptosis in lung cancer. Cell Mol Biol. 2015;61:34–40. [PubMed] [Google Scholar]


