Abstract
The apoptotic pathway is important in the control of vital processes of hepatocellular carcinoma (HCC). In the current study, we aimed to determine whether apoptotic gene-related polymorphisms modified HCC prognosis. We genotyped 16 single nucleotide polymorphisms (SNPs) in 10 core genes (TP53, TP53INP1, TP53BP1, CDKN2A, CDKN1A, CDKN1B, MDM2, BAX, CCDN1 and BCL2) in the apoptotic pathway by using DNA from blood samples of 362 HCC patients receiving surgical resection of HCC tumor. The associations between genotypes/haplotypes of the 10 genes and overall survival (OS) of HCC patients were assessed using the Cox proportional hazards model. We found one CDKN1B haplotype CCT/ACT (constructed by rs36228499 C>A, rs34330 C>T and rs2066827 T>G) significantly associated with decreased OS of HCC patients, compared to the common haplotype ACT/CTT both in univariate analysis (P=0.013, HR=1.198, 95% CI: 1.039-1.381) and multivariate analysis (P=0.006, HR=1.224, 95% CI: 1.059-1.413). We also find two SNPs (rs560191 G>C and rs2602141 T>G) in TP53BP1 shown to be marginally significantly associated with decreased OS of HCC patients. However, none of the other SNPs or haplotypes were significantly associated with HCC OS. Our results illustrated the potential use of CDKN1B haplotype as a prognostic marker for HCC patients with surgical resection of tumor.
Keywords: Hepatocellular carcinoma, survival, apoptosis, CDKN1B, genetic polymorphisms
Introduction
Hepatocellular carcinoma (HCC) is diagnosed in more than half a million people worldwide every year, and it is one of the leading causes of cancer-related deaths worldwide [1]. China alone accounts for about 50% of the total number of HCC cases and deaths [2]. In 2012, estimated 782,500 new HCC cases and 745,500 cancer-related deaths occurred worldwide [1], making the incidence and mortality rates almost equal. Although multiple clinical factors of HCC, such as large tumor size, vascular invasion, positive portal vein thrombosis, increased serum α-fetoprotein (AFP) and advanced tumor nodes metastasis (TNM) stage have been indicated to be useful to evaluate HCC patients’ prognosis [3], they cannot meet clinical requirements for precise prediction of HCC course. Therefore, it is of great significance to identify potential biomarkers for improving the efficiency of prognosis prediction, thus establishing more appropriate cancer management strategies and improving better clinical outcomes of HCC.
Apoptosis is a genetically controlled cell suicide mechanism, which enables multicellular organisms to regulate cell number in tissues and to eliminate unnecessary or damaged cells [4]. Defects in apoptosis are implicated in tumor progression and metastasis through maintaining survival of tumor cells, leading to clonal expansion within tumor and further invading surrounding tissues [5]. It is assumed that a decreased ability to eliminate cells with DNA damage may facilitate the accumulation of somatic mutations, and thereby contribute to tumor initiation, progression, and metastasis [6-9]. There are considerable inter-individual variations in apoptotic capacity, which are largely attributed to an individual’s genetic constitution [10,11]. Many studies have demonstrated that several polymorphisms in apoptosis-related genes affect either the expression or the activities of enzymes, and thus associated with the risk of various human cancers, including HCC [12-15]. Accordingly, it is reasonable to suggest that alterations in apoptotic capacity related polymorphisms of apoptosis-related genes could affect prognosis of patients with HCC. However, evidence is still limited to the demonstration of the effects of apoptotic gene-related polymorphisms on the prognosis of HCC.
In this study, we systematically selected 16 potentially functional single nucleotide polymorphisms (SNPs) from 10 genes in the apoptotic pathway, including TP53, TP53INP1, TP53BP1, CDKN2A, CDKN1A, CDKN1B, MDM2, BAX, CCDN1 and BCL2 to assess their prognostic significance for HCC in a Chinese cohort of 362 HCC patients.
Materials and methods
Patients and samples collection
A total of 362 newly diagnosed HCC patients receiving surgical resection of HCC tumor were recruited by the Qidong Liver Cancer Institute in Qidong, Jiangsu province, China from April 1996 to September 2009. All of the HCC patients were Han Chinese. The clinical outcomes of HCC were recorded until October 2014, with a median follow-up time of 53.0 months (range 2-110 months).
The diagnosis of HCC was based on histopathological examination and the National Comprehensive Cancer Network (NCCN) clinical practice guidelines in oncology. All tumors were proven to be HCC by two pathologists. All patients had no other cancers as determined by initial screening examination and were followed up prospectively every 3 months from the time of enrollment by personal or family contacts until death or last time of follow-up.
There were no recruitment restrictions on age, gender and tumor stage. 5 ml whole blood for each subject was extracted. Clinical information was collected at the time the blood specimens were collected from medical records with patients’ consent. The histologic grade of tumor differentiation was assigned by the Edmondson grading system. The clinical typing of tumors was determined according to the TNM classification system of International Union against Cancer (edition 6). The study endpoint was OS, which was calculated from the date of pathologic diagnosis/recruitment to death or the end of available follow-up.
The methods were carried out in accordance with the approved guidelines and in accordance with the Helsinki Declaration as revised in 2000. This study was approved by the Department of Scientific Research of Fudan University and the Qidong Liver Cancer Institute, and a written informed consent with a signature was obtained from each patient before enrollment.
SNP selection
To select the potential functional SNPs of apoptosis-related genes, we utilized the International HapMap Project database (http://hapmap.ncbi.nlm.nih.gov/), and dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/) to search for candidate variants in the promoter region, all exons including intron-exon boundaries and the 3’-untranslated region (3’-UTR). We also selected SNPs previously reported to be associated with the outcome of cancers. Finally, a total of 16 potentially functional SNPs were selected for genotyping (Table 1).
Table 1.
The selected functional SNPs in 10 apoptosis-related genes and their allele frequencies
| Gene | Chromosome | Location | Position | SNP | Allele* | MAF (CHB)† | MAF (observed)‡ |
|---|---|---|---|---|---|---|---|
| TP53 | 17p13.1 | exon 4 | 7579472 | rs1042522 | G/C | 0.489 | 0.413 |
| TP53INP1 | 8q22 | 3’UTR (miR-330-5p target site) | 95938422 | rs7760 | G/T | 0.109 | 0.113 |
| TP53BP1 | 15q15-q21 | promoter | 43803621 | rs1869258 | T/G | 0.489 | 0.452 |
| exon 9 | 43767774 | rs560191 | C/G | 0.444 | 0.424 | ||
| exon 17 | 43724646 | rs2602141 | G/T | 0.478 | 0.425 | ||
| CDKN2A | 9p21 | exon 3 | 21968199 | rs11515 | C/G | 0.011 | 0.022 |
| exon 3 | 21968159 | rs3088440 | T/C | 0.107 | 0.123 | ||
| CDKN1A | 6p21.2 | exon 2 | 36651971 | rs1801270 | A/C | 0.449 | 0.488 |
| 3’UTR | 36653597 | rs1059234 | T/C | 0.442 | 0.496 | ||
| CDKN1B | 12p13.1-p12 | promoter | 12869936 | rs36228499 | A/C | 0.408 | 0.414 |
| promoter | 12870695 | rs34330 | T/C | 0.470 | 0.461 | ||
| exon 1 | 12871099 | rs2066827 | G/T | 0.044 | 0.022 | ||
| MDM2 | 12q14.3-q15 | promoter | 69202580 | rs2279744 | T/G | 0.358 | 0.465 |
| BAX | 19q13.3-q13.4 | promoter | 49457938 | rs4645878 | A/G | 0.051 | 0.068 |
| CCND1 | 11q13 | exon 4 | 69462910 | rs603965 (rs9344) | G/A | 0.438 | 0.422 |
| BCL2 | 18q21.3 | P2 promoter (5’flanking) | 60986837 | rs2279115 | A/C | 0.433 | 0.411 |
Abbreviations: MAF, minor allele frequency; CHB, Chinese Han in Beijing.
Minor allele/major allele.
Obtained from the International Hap Map Project database (http://hapmap.ncbi.nlm.nih.gov/).
Estimated from 362 HCC patients receiving surgical resection of the tumor.
DNA extraction, genotyping, and haplotypes reconstruction
Genomic DNA was extracted from blood samples using the QIAamp DNA Mini Kit (QIAGEN GmbH, Hilden, Germany). Genotyping was performed with Sequenom MassARRAY iPLEX platform by use of allele-specific MALDI-TOF mass spectrometry assay. Polymerase chain reaction (PCR) and extension primers for these 16 SNPs were designed using the MassARRAY Assay Design 3.0 software (Sequenom). PCR and extension reactions were performed according to the manufacturer’s instructions, and extension product sizes were determined by mass spectrometry using the Sequenom iPLEX system. Duplicate test samples and two water samples (PCR negative controls) that were blinded to the technician were included in each 96-well plate. Genotyping quality was examined by a detailed QC procedure consisting of >95% successful call rate, duplicate calling of genotypes, internal positive control samples.
The linkage disequilibrium (LD) status among SNPs was measured with Lewontin D and r2 by using the Haploview software package (http:// www.broad.mit.edu/mpg/haploview). LD blocks were inferred from the definition proposed by Gabriel and colleagues [16]. Probable haplotypes were calculated on the basis of a Bayesian algorithm [17] using PHASE software (ver 2.1.1, Seattle, WA, USA).
Statistical analysis
The effects of the study variables including clinical variables, single SNP and haplotype on HCC OS were assessed using the Cox proportional hazards model. For single SNP analysis, the major homozygote genotype was regarded as reference, the heterozygote and minor homozygote genotypes as well as the combination of heterozygote and minor homozygote genotype were compared to the major homozygote genotype. While for haplotype analysis, the most popular haplotype was considered as reference and other haplotypes were compared to it. HRs and 95% CIs were estimated for each analysis. In multivariate analysis of single SNP and haplotype, only clinical variables found to be significant in univariate analysis were considered in the Cox model. Survival curves were estimated according to the Kaplan-Meier method, and the statistical differences in the survival curves of different subgroups of subjects were analyzed using the log-rank test. Data analysis, with the exception of haplotype construction, was performed with SPSS software version 22 (SPSS, Chicago, IL). All tests were two-sided and a P<0.05 was considered statistically significant.
Results
Patient characteristics and clinical outcomes
This study included 362 HCC patients with an overall median survival time (MST) of 34.0 months and median follow-up time of 53.0 months. At the time of analysis, 225 (62.2%) of the patients had died. The clinical pathologic characteristics and their association with OS are summarized in Table 2. By univariate analysis, tumor size and venous invasion were significantly associated with overall survival (OS) (P<0.05). Therefore, we calculated hazard ratio (HR) and its corresponding P-value for each single SNP and haplotype in multivariate analysis using Cox proportional hazard models, adjusted for tumor size and venous invasion.
Table 2.
Clinical characteristics and their prediction of overall survival in 362 HCC patients receiving surgical resection for the tumor
| Characteristics | No of patients | No of events | 5-y-survival (%) | Overall survival | ||
|---|---|---|---|---|---|---|
|
| ||||||
| MST (95% CI) | Hazard ratio (95% CI) | P | ||||
| Number | 362 | 225 | 30 | 34.0 (27.4-40.6) | ||
| Age (year) | ||||||
| ≤50 | 186 | 113 | 30 | 35.0 (23.2-46.7) | 1.000 | |
| >50 | 176 | 112 | 29 | 33.0 (24.2-41.8) | 1.098 (0.845-1.426) | 0.483 |
| Sex | ||||||
| Female | 63 | 41 | 27 | 31.0 (24.2-37.8) | 1.000 | |
| Male | 299 | 184 | 30 | 37.0 (27.5-46.5) | 0.885 (0.631-1.242) | 0.481 |
| Smoking | ||||||
| Never | 224 | 144 | 26 | 31.0 (23.2-38.8) | 1.000 | |
| Ever | 138 | 81 | 37 | 39.0 (27.3-50.7) | 0.858 (0.653-1.127) | 0.270 |
| Drinking | ||||||
| Never | 142 | 86 | 28 | 35.0 (19.6-50.4) | 1.000 | |
| Ever | 220 | 139 | 31 | 33.0 (25.0-41.0) | 1.071 (0.818-1.401) | 0.619 |
| Family history | ||||||
| Absent | 263 | 158 | 31 | 37.0 (27.3-46.7) | 1.000 | |
| Present | 81 | 55 | 25 | 29.0 (17.4-40.6) | 1.192 (0.877-1.620) | 0.262 |
| Unknow | 18 | 12 | ||||
| HbsAg | ||||||
| Negative | 59 | 40 | 35 | 22.0 (6.7-37.3) | 1.000 | |
| Positive | 303 | 185 | 28 | 37.0 (30.2-43.8) | 0.913 (0.648-1.287) | 0.603 |
| AFP | ||||||
| Negative | 142 | 95 | 26 | 33.0 (25.7-40.3) | 1.000 | |
| Positive | 214 | 127 | 32 | 35.0 (26.1-43.9) | 0.892 (0.684-1.164) | 0.400 |
| Unknow | 6 | 3 | ||||
| Tumor size (cm) | ||||||
| ≤5 | 183 | 107 | 35 | 39.0 (28.1-49.9) | 1.000 | |
| >5 | 179 | 118 | 24 | 30.0 (21.0-39.0) | 1.343 (1.033-1.747) | 0.028† |
| Differentiation | ||||||
| I+II | 196 | 122 | 28 | 37.0 (27.5-46.5) | 1.000 | |
| III+IV | 155 | 96 | 32 | 34.0 (26.0-42.0) | 0.926 (0.708-1.120) | 0.572 |
| Unknow | 11 | 7 | ||||
| Tumor capsule | ||||||
| Absent | 177 | 113 | 28 | 31.0 (22.7-39.3) | 1.000 | |
| Present | 181 | 110 | 31 | 37.0 (26.1-47.9) | 0.913 (0.702-1.188) | 0.499 |
| Unknow | 4 | 2 | ||||
| Venous invasion | ||||||
| Absent | 257 | 150 | 33 | 39.0 (29.3-48.7) | 1.000 | |
| Present | 102 | 73 | 22 | 26.0 (20.1-31.9) | 1.368 (1.033-1.811) | 0.029† |
| Unknow | 3 | 2 | ||||
| Cirrhosis | ||||||
| Absent | 121 | 79 | 30 | 27.0 (13.6-40.4) | 1.000 | |
| Present | 239 | 145 | 30 | 36.0 (29.6-42.4) | 0.949 (0.721-1.249) | 0.708 |
| Unknow | 2 | 1 | ||||
| Tumor number | ||||||
| Solitary | 279 | 172 | 30 | 34.0 (26.0-42.0) | 1.000 | |
| Multiple | 83 | 53 | 27 | 35.0 (24.5-45.5) | 1.061 (0.780-1.445) | 0.704 |
| pTNM stage | ||||||
| I+II | 309 | 188 | 31 | 37.0 (30.7-43.3) | 1.000 | |
| III+IV | 39 | 27 | 24 | 22.0 (13.0-31.0) | 1.280 (0.855-1.916) | 0.231 |
| Unknow | 14 | 10 | ||||
Abbreviations: MST, median survival time; CI, confidence interval; AFP, α-fetoprotein.
P<0.05.
Association analysis of individual SNPs with OS of HCC patients
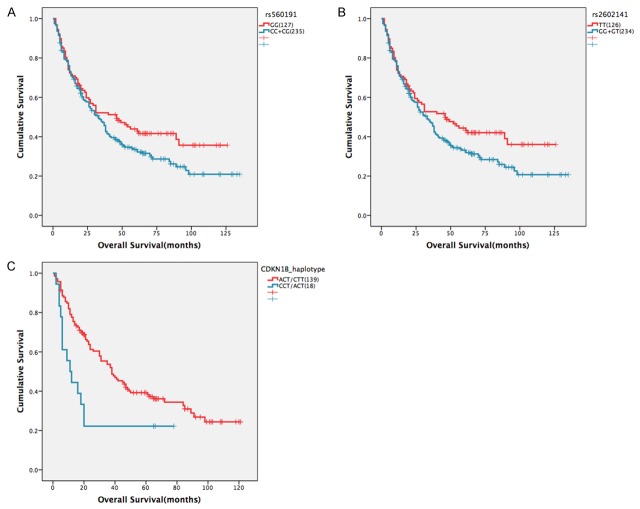
Table 3 shows the data for all the 16 SNPs among 10 genes (TP53, TP53INP1, TP53BP1, CDKN2A, CDKN1A, CDKN1B, MDM2, BAX, CCDN1 and BCL2) analyzed for OS of HCC patients. In the univariate analysis, of all the 16 SNPs, only two SNPs (rs560191 and rs2602141), which are resided in TP53BP1 gene, showed suggestive evidence of an association with OS of HCC patients (Table 3). We observed rs560191 CC+CG genotype has a marginally significant association with decreased OS (P=0.080; HR=1.288, 95% confident interval [CI]: 0.971-1.708), compared with the GG genotypes (Table 3, Figure 1A). Similar result was found for rs2602141 GG+GT genotype, which was marginally significantly associated with decreased OS (P=0.065; HR=1.306, 95% CI: 0.983-1.736), compared with the TT genotypes (Table 3; Figure 1B). However, none of the other 14 SNPs examined were significantly associated with OS (Table 3).
Table 3.
Univariate and multivariate Cox regression analysis of genotype of all selected SNPs in 362 HCC patients with surgical resection for the tumor
| Genotype | No of patients | No of events | 5-y-survival (%) | MST (95% CI) | Overall survival | |||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Univariate analysis | Multivariate analysis | |||||||
|
| ||||||||
| Hazard ratio (95% CI) | P | Hazard ratio (95% CI) | P | |||||
| TP53_rs1042522 | ||||||||
| GG | 114 | 73 | 30 | 36.0 (25.8-46.2) | 1.000 | 1.000 | ||
| GC | 191 | 114 | 32 | 30.0 (22.1-37.9) | 0.960 (0.715-1.288) | 0.785 | 0.967 (0.720-1.300) | 0.825 |
| CC | 52 | 33 | 26 | 51.0 (38.5-63.5) | 0.910 (0.603-1.374) | 0.654 | 0.899 (0.590-1.371) | 0.622 |
| CC+GC | 243 | 147 | 30 | 35.0 (23.2-46.8) | 0.950 (0.717-1.258) | 0.719 | 0.946 (0.713-1.255) | 0.699 |
| TP53INP1_rs7760 | ||||||||
| TT | 279 | 178 | 28 | 31.0 (23.5-38.5) | 1.000 | 1.000 | ||
| GT | 75 | 44 | 32 | 41.0 (25.0-57.0) | 0.838 (0.602-1.166) | 0.294 | 0.875 (0.628-1.218) | 0.428 |
| GG | 3 | 2 | 33 | 16.0 (0.0-33.6) | 1.244 (0.309-5.018) | 0.759 | 1.467 (0.361-5.956) | 0.592 |
| GG+GT | 78 | 46 | 32 | 38.0 (21.7-54.3) | 0.850 (0.614-1.176) | 0.326 | 0.891 (0.643-1.233) | 0.485 |
| TP53BP1_rs1869258 | ||||||||
| TT | 112 | 62 | 38 | 46.0 (23.0-69.0) | 1.000 | 1.000 | ||
| GT | 172 | 112 | 26 | 35.0 (25.4-44.6) | 1.223 (0.897-1.669) | 0.204 | 1.214 (0.887-1.659) | 0.225 |
| GG | 77 | 50 | 27 | 30.0 (20.9-39.1) | 1.263 (0.870-1.834) | 0.220 | 1.307 (0.899-1.902) | 0.161 |
| GG+GT | 249 | 162 | 26 | 33.0 (26.7-39.3) | 1.239 (0.924-1.660) | 0.152 | 1.249 (0.930-1.677) | 0.140 |
| TP53BP1_rs560191 | ||||||||
| GG | 127 | 70 | 38 | 46.0 (25.5-66.5) | 1.000 | 1.000 | ||
| CG | 163 | 107 | 24 | 36.0 (26.1-45.9) | 1.260 (0.932-1.704) | 0.133 | 1.264 (0.933-1.713) | 0.130 |
| CC | 72 | 48 | 26 | 30.0 (19.2-40.8) | 1.161 (0.966-1.396) | 0.112 | 1.176 (0.978-1.415) | 0.085 |
| CC+CG | 235 | 155 | 25 | 33.0 (26.7-39.3) | 1.288 (0.971-1.708) | 0.080 | 1.303 (0.980-1.732) | 0.068 |
| TP53BP1_rs2602141 | ||||||||
| TT | 126 | 69 | 39 | 46.0 (25.7-66.3) | 1.000 | 1.000 | ||
| GT | 162 | 107 | 24 | 35.0 (25.3-44.7) | 1.282 (0.947-1.736) | 0.108 | 1.284 (0.946-1.742) | 0.108 |
| GG | 72 | 48 | 26 | 30.0 (19.2-40.8) | 1.357 (0.938-1.963) | 0.105 | 1.393 (0.961-2.019) | 0.080 |
| GG+GT | 234 | 155 | 25 | 33.0 (26.7-39.3) | 1.306 (0.983-1.736) | 0.065 | 1.320 (0.992-1.757) | 0.057 |
| CDKN2A_rs11515 | ||||||||
| CC | 344 | 213 | 30 | 35.0 (29.0-41.0) | 1.000 | 1.000 | ||
| GC | 16 | 11 | 31 | 12.0 (0.0-27.7) | 1.312 (0.715-2.406) | 0.381 | 1.382 (0.752-2.540) | 0.298 |
| GG | 0 | 0 | ||||||
| CDKN2A_rs3088440 | ||||||||
| CC | 277 | 173 | 29 | 33.0 (26.1-39.9) | 1.000 | 1.000 | ||
| CT | 79 | 48 | 32 | 36.0 (21.8-50.2) | 0.922 (0.660-1.269) | 0.617 | 0.964 (0.699-1.330) | 0.825 |
| TT | 5 | 4 | 12 | 61.0 (35.2-86.8) | 0.844 (0.313-2.277) | 0.738 | 0.860 (0.318-2.325) | 0.767 |
| TT+CT | 84 | 52 | 30 | 39.0 (25.3-52.7) | 0.915 (0.671-1.247) | 0.574 | 0.955 (0.699-1.304) | 0.771 |
| CDKN1A_rs1801270 | ||||||||
| AA | 97 | 55 | 36 | 36.0 (12.0-60.0) | 1.000 | 1.000 | ||
| CA | 177 | 115 | 27 | 35.0 (29.1-40.9) | 1.215 (0.881-1.676) | 0.236 | 1.170 (0.846-1.617) | 0.342 |
| CC | 88 | 55 | 28 | 28.0 (11.1-44.9) | 1.270 (0.873-1.848) | 0.211 | 1.193 (0.817-1.742) | 0.361 |
| CC+CA | 265 | 170 | 27 | 33.0 (26.0-40.0) | 1.230 (0.907-1.667) | 0.183 | 1.186 (0.873-1.610) | 0.275 |
| CDKN1A_rs1059234 | ||||||||
| CC | 93 | 59 | 28 | 27.0 (11.4-42.6) | 1.000 | 1.000 | ||
| TC | 179 | 115 | 27 | 37.0 (31.1-42.9) | 0.937 (0.684-1.283) | 0.685 | 0.924 (0.673-1.267) | 0.623 |
| TT | 90 | 51 | 37 | 35.0 (11.4-58.6) | 0.783 (0.538-1.140) | 0.202 | 0.842 (0.575-1.233) | 0.377 |
| TT+TC | 269 | 166 | 30 | 36.0 (29.5-42.5) | 0.883 (0.656-1.188) | 0.411 | 0.893 (0.662-1.204) | 0.457 |
| CDKN1B_rs36228499 | ||||||||
| CC | 127 | 74 | 34 | 33.0 (21.1-44.9) | 1.000 | 1.000 | ||
| CA | 170 | 109 | 28 | 37.0 (28.1-45.9) | 1.080 (0.804-1.451) | 0.609 | 1.045 (0.776-1.406) | 0.773 |
| AA | 65 | 42 | 26 | 30.0 (16.5-43.5) | 1.147 (0.785-1.675) | 0.479 | 1.082 (0.739-1.585) | 0.685 |
| AA+CA | 235 | 151 | 27 | 35.0 (28.1-41.9) | 1.101 (0.834-1.455) | 0.497 | 1.062 (0.803-1.405) | 0.672 |
| CDKN1B_rs34330 | ||||||||
| TT | 110 | 66 | 33 | 31.0 (20.7-41.2) | 1.000 | 1.000 | ||
| CT | 169 | 105 | 29 | 38.0 (28.8-47.2) | 0.946 (0.695-1.287) | 0.723 | 0.907 (0.666-1.237) | 0.539 |
| CC | 82 | 54 | 27 | 24.0 (15.5-32.5) | 1.176 (0.820-1.685) | 0.378 | 1.108 (0.771-1.593) | 0.578 |
| CC+CT | 251 | 159 | 28 | 37.0 (29.3-44.7) | 1.019 (0.764-1.357) | 0.900 | 0.975 (0.731-1.300) | 0.862 |
| CDKN1B_rs2066827 | ||||||||
| TT | 346 | 215 | 30 | 35.0 (28.2-41.8) | 1.000 | 1.000 | ||
| GT | 16 | 10 | 26 | 33.0 (12.9-53.1) | 1.093 (0.579-2.060) | 0.784 | 0.992 (0.524-1.877) | 0.981 |
| GG | 0 | 0 | ||||||
| MDM2_rs2279744 | ||||||||
| GG | 103 | 60 | 32 | 39.0 (23.6-54.4) | 1.000 | 1.000 | ||
| GT | 179 | 112 | 28 | 35.0 (26.2-43.8) | 1.082 (0.791-1.481) | 0.622 | 1.070 (0.779-1.468) | 0.677 |
| TT | 78 | 51 | 32 | 28.0 (15.2-40.8) | 1.118 (0.769-1.626) | 0.558 | 1.157 (0.790-1.694) | 0.454 |
| TT+GT | 257 | 163 | 29 | 31.0 (24.6-37.4) | 1.092 (0.812-1.468) | 0.561 | 1.094 (0.811-1.474) | 0.577 |
| BAX_rs4645878 | ||||||||
| GG | 313 | 190 | 32 | 34.0 (26.2-41.8) | 1.000 | 1.000 | ||
| GA | 49 | 35 | 14 | 37.0 (28.3-45.7) | 1.197 (0.834-1.717) | 0.329 | 1.200 (0.834-1.725) | 0.326 |
| AA | 0 | 0 | ||||||
| CCND1_ rs9344 | ||||||||
| AA | 121 | 80 | 23 | 35.0 (26.2-43.8) | 1.000 | 1.000 | ||
| GA | 175 | 101 | 36 | 39.0 (25.5-52.5) | 0.852 (0.635-1.144) | 0.287 | 0.893 (0.663-1.202) | 0.455 |
| GG | 65 | 44 | 23 | 24.0 (16.3-31.7) | 1.110 (0.923-1.335) | 0.266 | 1.085 (0.900-1.308) | 0.392 |
| GG+GA | 240 | 145 | 32 | 34.0 (25.7-42.3) | 0.941 (0.716-1.237) | 0.664 | 0.965 (0.733-1.271) | 0.802 |
| BCL2_rs2279115 | ||||||||
| CC | 126 | 75 | 31 | 37.0 (27.7-46.3) | 1.000 | 1.000 | ||
| CA | 173 | 110 | 30 | 31.0 (17.3-44.7) | 1.067 (0.795-1.431) | 0.666 | 1.043 (0.777-1.401) | 0.778 |
| AA | 62 | 39 | 28 | 31.0 (23.6-38.4) | 1.109 (0.752-1.634) | 0.602 | 1.124 (0.759-1.665) | 0.559 |
| AA+CA | 235 | 149 | 30 | 31.0 (23.4-38.6) | 1.076 (0.815-1.420) | 0.606 | 1.058 (0.801-1.398) | 0.691 |
Abbreviations: MST, median survival time; CI, confidence interval.
Figure 1.
Kaplan-Meier survival curves of overall survival in 362 HCC patients receiving surgical resection for the tumor are shown for polymorphisms of (A) rs560191, (B) rs2602141, and (C) CDKN1B_haplotype.
A multivariate analysis of genotype effects on OS of HCC patients was conducted using Cox proportional hazards models adjusted for tumor size and venous invasion, and similar results were found as the univariate analysis. As shown in Table 3, only two SNPs (rs560191 and rs2602141) in TP53BP1 were confirmed to be marginally significantly associated with clinical outcomes of HCC patients, with the CC+CG genotype of rs560191 presenting a suggestively negative effect on OS (P=0.068, HR=1.303, 95% CI: 0.980-1.732), compared to the common GG genotype, and with the GG+GT genotype of rs2602141 presenting a suggestively negative effect on OS (P=0.057, HR=1.320, 95% CI: 0.992-1.757), compared to the common TT genotype (Table 3).
Association analysis of haplotypes with OS of HCC patients
Furthermore, we examined the associations of the haplotypes with OS of HCC patients. When examining combinations of SNPs for the TP53BP1 (rs1869258 G>T, rs560191 G>C and rs2602141 T>G), CDKN2A (rs11515 G>C and rs3088440 C>T), CDKN1A (rs1801270 C>A and rs1059234 C>T), CDKN1B (rs36228499 C>A, rs34330 C>T and rs2066827 T>G), which has at least two tested SNPs in this study, we attained 4 haplotypes of TP53BP1, 3 haplotypes of CDKN2A, 4 haplotypes of CDKN1A, and 6 haplotypes of CDKN1B. The inferred haplotypes and their associations with OS are shown in Table 4. It shows that only one CDKN1B haplotype CCT/ACT was significantly related with OS (P=0.013, HR=1.198, 95% CI: 1.039-1.381), compared to the common haplotype ACT/CTT in univariate analysis (Table 4; Figure 1C).
Table 4.
Univariate and multivariate Cox regression analysis of haplotype of the apoptosis-related genes in 362 HCC patients with surgical resection for the tumor
| Haplotype | No of patients | No of events | 5-y-survival (%) | MST (95% CI) | Overall survival | |||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Univariate analysis | Multivariate analysis | |||||||
|
| ||||||||
| Hazard ratio (95% CI) | P | Hazard ratio (95% CI) | P | |||||
| TP53BP1_haplotype | ||||||||
| GCG/TGT | 157 | 104 | 24 | 35.0 (25.4-44.6) | 1.000 | 1.000 | ||
| TGT/TGT | 112 | 62 | 38 | 47.0 (24.3-69.7) | 0.793 (0.579-1.087) | 0.150 | 0.799 (0.582-1.098) | 0.166 |
| GCG/GCG | 71 | 47 | 27 | 30.0 (22.0-38.0) | 1.054 (0.746-1.489) | 0.765 | 1.099 (0.777-1.553) | 0.593 |
| Rare* | 22 | 12 | 38 | 46.0 (1.5-90.5) | 0.786 (0.432-1.429) | 0.430 | 0.812 (0.446-1.478) | 0.495 |
| CDKN2A_haplotype | ||||||||
| CC/CC | 263 | 163 | 29 | 35.0 (27.5-42.5) | 1.000 | 1.000 | ||
| CC/CT | 77 | 47 | 31 | 36.0 (22.0-50.0) | 0.945 (0.683-1.307) | 0.732 | 1.002 (0.723-1.390) | 0.989 |
| Rare* | 20 | 14 | 27 | 26.0 (8.6-43.4) | 1.1639 (0.673-2.008) | 0.588 | 1.203 (0.695-2.082) | 0.510 |
| CDKN1A_haplotype | ||||||||
| AT/CC | 172 | 111 | 27 | 37.0 (31.2-42.8) | 1.000 | 1.000 | ||
| AT/AT | 90 | 51 | 37 | 35.0 (11.4-58.6) | 0.840 (0.603-1.171) | 0.304 | 0.880 (0.630-1.230) | 0.455 |
| CC/CC | 88 | 55 | 28 | 28.0 (11.1-44.9) | 1.016 (0.864-1.195) | 0.845 | 1.021 (0.868-1.201) | 0.803 |
| Rare* | 12 | 8 | 25 | 30.0 (0.0-69.8) | 1.002 (0.788-1.272) | 0.990 | 1.030 (0.808-1.313) | 0.814 |
| CDKN1B_haplotype | ||||||||
| ACT/CTT | 139 | 87 | 29 | 38.0 (26.8-49.2) | 1.000 | 1.000 | ||
| CTT/CTT | 101 | 61 | 32 | 28.0 (20.3-35.7) | 1.078 (0.777-1.496) | 0.654 | 1.132 (0.814-1.575) | 0.460 |
| ACT/ACT | 56 | 34 | 31 | 31.0 (8.5-53.5) | 1.034 (0.848-1.261) | 0.739 | 1.017 (0.832-1.242) | 0.871 |
| CTT/CCT | 21 | 10 | 42 | 61.0 (/-/) | 0.875 (0.703-1.088) | 0.875 | 0.876 (0.702-1.092) | 0.239 |
| CCT/ACT | 18 | 14 | 22 | 11.0 (4.8-17.2) | 1.198 (1.039-1.381) | 0.013† | 1.224 (1.059-1.413) | 0.006† |
| Rare* | 27 | 19 | 21 | 34.0 (13.2054.8) | 1.054 (0.954-1.165) | 0.299 | 1.044 (0.945-1.155) | 0.398 |
Abbreviations: MST, median survival time; CI, confidence interval.
All that genotypes with number less than 18 (5% of 362) were combined as rare genotypes.
P<0.05.
Similar result was found in multivariate analysis adjusted for tumor size and venous invasion, the CDKN1B CCT/ACT haplotype (P=0.006, HR=1.224, 95% CI: 1.059-1.413) present an independent negative effect on OS, compared to the common haplotype ACT/CTT in CDKN1B (Table 4). None of the haplotypes carrying variant alleles from TP53BP1, CDKN2A and CDKN1A showed any significant association with OS.
Discussion
HCC has a highly variable clinical courses and includes several subgroups with distinct pathways of hepatocarcinogenesis [18]. These processes share common mechanisms with embryogenesis and can be considered as an aberrant form of organogenesis [19]. However, the critical steps both with respect to molecular genetics and phenotypic characteristics in the prognosis of HCC are still not well characterized. While some germline genetic factors have been suspected of playing an important role in prognosis, none have been firmly established [20,21]. Most investigations into SNPs in apoptosis-related genes have just focused on their effects on risk rather than prognosis of HCC [12-15,22,23]. The aim of our study was to evaluate the role of genetic variants of apoptosis-related genes in determining the clinical outcomes of HCC patients. To the best of our knowledge, this is the first evidence showing the relationship between genetic variants of apoptosis-related genes and the prognosis of HCC patients.
In the present study, we found that one haplotype in CDKN1B gene was significantly associated with OS in 362 HCC patients. The haplotype GCT/TCT (constructed by rs36228499 C>A, rs34330 C>T and rs2066827 T>G) in CDKN1B gene was significantly associated with decreased OS, compared with the common TCT/GTT haplotype both in univariate analysis and in multivariate analysis adjusted for tumor size and venous invasion. This haplotype presents an independent negative effect on OS and could be used to predict which HCC patients are at risk for poor clinical outcomes in the future.
CDKN1B (p27Kip1), encoded by CDKN1B gene, is an enzyme inhibitor in humans and belongs to the cip/kip family of CDKI [24]. CDKN1B shares significant homology with its other family members (p21 and p57), specifically in the amino terminal domain [25]. The protein was firstly identified as an inhibitor of CDK2 containing complexes in G1 arrested lung epithelial cells under contact inhibition or when treated with transformation growth factor beta (TGF-β) [26]. Subsequently, the gene encoding CDKN1B was cloned and also identified in a yeast tri-hybrid screen as a cyclin D-CDK4 interacting protein [25,27]. Since then CDKN1B has not only emerged as a prime regulator of cell cycle progression but has also been implicated in numerous malignancies including HCC [28]. In cancer cells, CDKN1B can also be mislocalized to the cytoplasm in order to facilitate metastasis. The mechanisms by which it acts on motility differ between cancers. In HCC cells, CDKN1B co-localizes with actin fibers to act on GTPase Rac and induce cell migration [29], and CDKN1B promotes cell migration in metastatic HCC cells through the regulation of RhoA activity [30]. Moreover, studies in several tumor types indicate that CDKN1B expression levels have both prognostic and therapeutic implications [31]. To date, accumulating evidence has suggested that decreased CDKN1B expression can be considered as an adverse prognostic biomarker in HCC [32-38].
Besides CDKN1B gene, none of additional genetic polymorphisms reached significance and could be served as an independent prognostic factor for OS. One explanation is that even though we selected and investigated these SNPs in a systematical way, due to limited techniques, labor and resources, we missed some key SNPs which play a predominant role in regulating the expression of the apoptosis-related genes. For this reason, we are not capable of concluding that the SNPs and the other haplotypes of these genes are not associated with the prognosis of HCC. Instead, a more comprehensive analysis of polymorphisms in the apoptosis related-genes is imperative to illustrate the close correlation between apoptosis related-genes and HCC prognosis.
It is worth mentioning that there were some limitations in our study. Firstly, the cohort size of the present study was relatively small. Therefore, larger well-designed longitudinal follow-up studies and functional evaluation are warranted to confirm our findings. Secondly, though several clinical and pathologic characteristics showed significant associations with OS, including tumor size and venous invasion, it is regretful that we failed to collect adequate and accurate information of these factors in our study. In order to make the greatest use of the genotype polymorphisms information of the 362 HCC patients, we had to operate the multivariate analysis by adjusting all these potential prognostic factors. Future studies are essential to investigate the role of genetic polymorphisms in HCC patients with more complete and comprehensive clinical pathologic characteristics. Last but not the least, as mentioned above, all of our samples are blood from each HCC patients treated with surgery. This drawback prevented us to conduct analysis of the relationship between apoptosis related genes expression in tissues and HCC prognosis. Accordingly, analyses of tissue samples are urgent to figure out the unknown modulation of these genes in HCC prognosis.
In summary, our results demonstrated the potential use of CDKN1B gene haplotype as a prognostic marker for HCC patients. However, neither the SNPs nor the other haplotypes from apoptosis-related genes were recognized having any significant association with HCC prognosis. More comprehensive studies are needed to evaluate the association between genetic polymorphisms of apoptosis-related genes and prognosis of HCC.
Acknowledgements
We thank all the study participants, research staff and students who took part in this work. The study is supported by the National Natural Science Foundation of China for Creative Research Groups (30024001; to L.Y.), the National Key Sci-Tech Special Project of China (2008ZX10002-020 and 2013ZX10002010; to L.Y.), the Project of the Shanghai Municipal Science and Technology Commission (to L.Y.), the National Natural Science Foundation of China (31071193 to L.Y., 31100895, and 81472618 to D.-K.J.), Director Foundation of the State Key Laboratory of Genetic Engineering (to L.Y.), the Research Fund of the State Key Laboratory of Genetic Engineering, Fudan University (to L.Y.), Outstanding Young Scholar Project of Fudan University (to D.-K.J.), as well as an intramural research grant for new young teachers from Fudan University (to D.-K.J.), an intramural research grant for promotion of the scientific research ability of young teachers from Fudan University (to D.-K.J.).
Disclosure of conflict of interest
None.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Yang JD, Roberts LR. Hepatocellular carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu AX. Molecularly targeted therapy for advanced hepatocellular carcinoma in 2012: current status and future perspectives. Semin Oncol. 2012;39:493–502. doi: 10.1053/j.seminoncol.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Raff M. Cell suicide for beginners. Nature. 1998;396:119–122. doi: 10.1038/24055. [DOI] [PubMed] [Google Scholar]
- 5.Vermeulen K, Van Bockstaele DR, Berneman ZN. Apoptosis: mechanisms and relevance in cancer. Ann Hematol. 2005;84:627–639. doi: 10.1007/s00277-005-1065-x. [DOI] [PubMed] [Google Scholar]
- 6.Schulte-Hermann R, Bursch W, Low-Baselli A, Wagner A, Grasl-Kraupp B. Apoptosis in the liver and its role in hepatocarcinogenesis. Cell Biol Toxicol. 1997;13:339–348. doi: 10.1023/a:1007495626864. [DOI] [PubMed] [Google Scholar]
- 7.Lowe SW, Lin AW. Apoptosis in cancer. Carcinogenesis. 2000;21:485–495. doi: 10.1093/carcin/21.3.485. [DOI] [PubMed] [Google Scholar]
- 8.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 9.Kountouras J, Zavos C, Chatzopoulos D. Apoptosis in hepatocellular carcinoma. Hepatogastroenterology. 2003;50:242–249. [PubMed] [Google Scholar]
- 10.Schmitz A, Bayer J, Dechamps N, Thomas G. Intrinsic susceptibility to radiation-induced apoptosis of human lymphocyte subpopulations. Int J Radiat Oncol Biol Phys. 2003;57:769–778. doi: 10.1016/s0360-3016(03)00637-0. [DOI] [PubMed] [Google Scholar]
- 11.Camplejohn RS, Hodgson S, Carter N, Kato BS, Spector TD. Heritability of DNA-damageinduced apoptosis and its relationship with age in lymphocytes from female twins. Br J Cancer. 2006;95:520–524. doi: 10.1038/sj.bjc.6603257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bayram S, Akkiz H, Bekar A, Akgollu E. The association between the survivin -31G/C promoter polymorphism and hepatocellular carcinoma risk in a Turkish population. Cancer Epidemiol. 2011;35:555–559. doi: 10.1016/j.canep.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Xu Y, Liu L, Liu J, Zhang Y, Zhu J, Chen J, Liu S, Liu Z, Shi H, Shen H, Hu Z. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma. Int J Cancer. 2011;128:412–417. doi: 10.1002/ijc.25342. [DOI] [PubMed] [Google Scholar]
- 14.Deng B, Liu F, Wei Y, Luo L, Chen X, Yan L, Li B. Association of a p73 exon 2 G4C14-to-A4T14 polymorphism with risk of hepatocellular carcinoma in a Chinese population. Tumour Biol. 2013;34:293–299. doi: 10.1007/s13277-012-0550-9. [DOI] [PubMed] [Google Scholar]
- 15.Zha Y, Gan P, Liu Q, Tan J. Relationship between polymorphism of angiotensin-converting enzyme gene insertion/deletion and risk of hepatocellular carcinoma in a Chinese Dai population. J Renin Angiotensin Aldosterone Syst. 2015;16:695–9. doi: 10.1177/1470320314539829. [DOI] [PubMed] [Google Scholar]
- 16.International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 17.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sha L, Dong L, Lv L, Bai L, Ji X. HOXB9 promotes epithelial-to-mesenchymal transition via transforming growth factor-beta1 pathway in hepatocellular carcinoma cells. Clin Exp Med. 2015;15:55–64. doi: 10.1007/s10238-014-0276-7. [DOI] [PubMed] [Google Scholar]
- 19.Zaret KS. Regulatory phases of early liver development: paradigms of organogenesis. Nat Rev Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 20.Jin F, Xiong WJ, Jing JC, Feng Z, Qu LS, Shen XZ. Evaluation of the association studies of single nucleotide polymorphisms and hepatocellular carcinoma: a systematic review. J Cancer Res Clin Oncol. 2011;137:1095–1104. doi: 10.1007/s00432-010-0970-0. [DOI] [PubMed] [Google Scholar]
- 21.Nishida N, Kudo M. Recent advancements in comprehensive genetic analyses for human hepatocellular carcinoma. Oncology. 2013;84(Suppl 1):93–97. doi: 10.1159/000345897. [DOI] [PubMed] [Google Scholar]
- 22.Chen TC, Ng KF, Lien JM, Jeng LB, Chen MF, Hsieh LL. Mutational analysis of the p27(kip1) gene in hepatocellular carcinoma. Cancer Lett. 2000;153:169–173. doi: 10.1016/s0304-3835(00)00366-9. [DOI] [PubMed] [Google Scholar]
- 23.Liu F, Wei YG, Luo LM, Wang WT, Yan LN, Wen TF, Xu MQ, Yang JY, Li B. Genetic variants of p21 and p27 and hepatocellular cancer risk in a Chinese Han population: a case-control study. Int J Cancer. 2013;132:2056–2064. doi: 10.1002/ijc.27885. [DOI] [PubMed] [Google Scholar]
- 24.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 25.Polyak K, Lee MH, Erdjument-Bromage H, Koff A, Roberts JM, Tempst P, Massague J. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell. 1994;78:59–66. doi: 10.1016/0092-8674(94)90572-x. [DOI] [PubMed] [Google Scholar]
- 26.Polyak K, Kato JY, Solomon MJ, Sherr CJ, Massague J, Roberts JM, Koff A. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev. 1994;8:9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- 27.Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell. 1994;78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 28.Huang AM, Ding Y, Liu JF, Gao LY, Zang SB, Chen SP. [Expression of survivin, p27 and PTEN in hepatocellular carcinoma and their clinical significances] . Zhonghua Gan Zang Bing Za Zhi. 2008;16:17–20. [PubMed] [Google Scholar]
- 29.McAllister SS, Becker-Hapak M, Pintucci G, Pagano M, Dowdy SF. Novel p27(kip1) C-terminal scatter domain mediates Rac-dependent cell migration independent of cell cycle arrest functions. Mol Cell Biol. 2003;23:216–228. doi: 10.1128/MCB.23.1.216-228.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang XQ, Lui EL, Cai Q, Ching WY, Liu KS, Poon RT, Fan ST. p27Kip1 promotes migration of metastatic hepatocellular carcinoma cells. Tumour Biol. 2008;29:217–223. doi: 10.1159/000152939. [DOI] [PubMed] [Google Scholar]
- 31.Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat Rev Cancer. 2008;8:253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- 32.Fiorentino M, Altimari A, D’Errico A, Cukor B, Barozzi C, Loda M, Grigioni WF. Acquired expression of p27 is a favorable prognostic indicator in patients with hepatocellular carcinoma. Clin Cancer Res. 2000;6:3966–3972. [PubMed] [Google Scholar]
- 33.Tannapfel A, Grund D, Katalinic A, Uhlmann D, Kockerling F, Haugwitz U, Wasner M, Hauss J, Engeland K, Wittekind C. Decreased expression of p27 protein is associated with advanced tumor stage in hepatocellular carcinoma. Int J Cancer. 2000;89:350–355. doi: 10.1002/1097-0215(20000720)89:4<350::aid-ijc6>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 34.Qin LF, Ng IO. Expression of p27(KIP1) and p21(WAF1/CIP1) in primary hepatocellular carcinoma: clinicopathologic correlation and survival analysis. Hum Pathol. 2001;32:778–784. doi: 10.1053/hupa.2001.27105. [DOI] [PubMed] [Google Scholar]
- 35.Armengol C, Boix L, Bachs O, Sole M, Fuster J, Sala M, Llovet JM, Rodes J, Bruix J. p27(Kip1) is an independent predictor of recurrence after surgical resection in patients with small hepatocellular carcinoma. J Hepatol. 2003;38:591–597. doi: 10.1016/s0168-8278(03)00025-4. [DOI] [PubMed] [Google Scholar]
- 36.Zhou Q, He Q, Liang LJ. Expression of p27, cyclin E and cyclin A in hepatocellular carcinoma and its clinical significance. World J Gastroenterol. 2003;9:2450–2454. doi: 10.3748/wjg.v9.i11.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuda Y. Molecular mechanism underlying the functional loss of cyclindependent kinase inhibitors p16 and p27 in hepatocellular carcinoma. World J Gastroenterol. 2008;14:1734–1740. doi: 10.3748/wjg.14.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuda Y, Wakai T, Kubota M, Takamura M, Yamagiwa S, Aoyagi Y, Osawa M, Fujimaki S, Sanpei A, Genda T, Ichida T. Clinical significance of cell cycle inhibitors in hepatocellular carcinoma. Med Mol Morphol. 2013;46:185–192. doi: 10.1007/s00795-013-0047-7. [DOI] [PubMed] [Google Scholar]