Abstract
The long non-coding RNA (LncRNA) H19 is one of the most highly abundant and conserved transcripts involved in the mammalian development and tumorigenesis. H19 is expressed in both embryonic cells and tumor cells, but its physical and pathological functions still need to be further studied. Our results showed that microRNA-675, a microRNA in the first exon of H19, expressed in glioma. Over-expression of microRNA-675 in a range of glioma cell lines resulted in their immoderate proliferation and migration. In addition, H19 derived microRNA-675 was down-regulated in the glioma, and CDK6, a pivotal regulator in cell cycle, was a target of microRNA-675. The survival of glioma patients with low CDK6 expression significantly increased as compared to patients with high CDK6 expression. Moreover, the CDK6 expression was inversely correlated with microRNA-675 expression in the glioma. Our results suggest that H19 derived microRNA-675 may regulate giloma cell proliferation and migration through CDK6, and predict a poor prognosis of glioma patients.
Keywords: H19, microRNA-675, glioma
Introduction
Malignant gliomas are the most common tumors of the center nervous system, and characterized by rapid cell proliferation, high invasiveness and reduced cell apoptosis [1,2]. Despite multimodal treatments (such as surgery, chemotherapy and radiotherapy) have been employed, the overall survival of most glioma patients remains poor, particularly in case of glioblastoma [3,4]. Therefore, to better understand the molecules and signal pathways involved in the glioma cell proliferation, invasion and migration is of particular relevance to the development of novel targets and individual therapies.
Long non-coding RNAs (LncRNAs) are non-protein coding transcripts longer than 200 nucleotides and have been considered as one type of gene expression regulator for decades. The first imprinting lncRNA identified, H19, which is expressed in the maternal allele rather than paternal, is transcribed from the H19/IGF2 gene cluster located on human chromosome 11p15.5 [5-7]. H19 has been implicated in tumor suppression, but its physiological and pathological functions are still poorly understood. Expressed from opposite parental alleles but co-regulated, H19 and IGF2 share a common imprinting mechanism and are found to be dysregulated in many cancers and fetal overgrowth syndromes in humans [8,9]. The H19 RNA itself does not have a role in the imprinting mechanism, consistent with its cytoplasmic localization. Instead, the LncRNA has a tumor suppressive effect both in vitro and in vivo, and is also able to regulate a network of imprinted genes at transcription level [10-13]. How the H19 exerts effects and what its physiological and pathological roles remain unknown. One way by which LncRNAs may acquire functionality is to act as a precursor of small-non-coding RNAs (such as microRNAs) with regulatory functions. Indeed, the exon 1 of H19 gene harbors a microRNA-containing hairpin and has been found to serve as the template for two distinct microRNAs, microRNA-675,3p and microRNA-675,5p. It has been suggested that these microRNAs may function on H19 [10,14-17]. Moreover, the microRNA-675 stem loop is shown to be one of the most highly conserved features of the H19 RNA during the mammalian evolution, indicating that microRNA-675 may be an important mediator through which H19 functions [18-22]. In addition, emerging evidence indicates that, although H19 gene has a critical role in the cancer progression as an oncogene in some types of cancers, it may also act as a tumor suppressor gene depending on the cancer type and cells [7]. To date, the expression and role of H19 have not been confirmed in glioma.
MicroRNAs, small non-coding RNAs of 20-22 nucleotides, have been found to be involved in multiple biological processes, such as cell differentiation, proliferation, oncogenesis, angiogenesis, tumor invasion and tumor metastasis [6,23]. It has been demonstrated that microRNAs play pivotal roles in the human cancer cell growth, invasion and migration. MicroRNAs recognize and bind to the 3’ untranslated region (3’UTR) of mRNAs in a sequence-specific manner and negatively regulate their target mRNAs [24-26]. The post-transnational gene regulation by microRNAs provides a novel tool for the inhibition of a specific gene in cancers [27,28]. Further, due to the close relationship between microRNAs and multiple biological aspects of cancer progression, microRNAs are considered as potential targets for the anti-cancer therapies [24,26,28,29]. As described above, microRNA-675 is derived from LncRNAH19, and H19 can generate two mature microRNAs, microRNA-675,3p and microRNA-675,5p in a Drosha and Dicer splicing dependent manner [6,30]. MicroRNA-675 represses the expression of retinoblastoma tumor suppressor in a classical way and promotes the proliferation of colon cancer cells [19,31]. However, the inverse relationship between microRNA-675 and placenta growth indicates microRNA-675 acts as a growth restrictor in the embryonic development [26,29]. Furthermore, the strict regulation of the excision of microRNA-675 from H19 by some RNA binding factors (such as HuR) indicates the complicated roles of H19 and microRNA-675 in different physiological and pathological conditions [19,26,29,31].
Mammalian CDKs are widely recognized proteins with well-established roles in orchestrating the steps of cell-cycle progression [32-34]. CDK4 and CDK6 form a complex with cyclin D to promote the G1 to S phase progression through phosphorylating the retinoblastoma (Rb) protein and transcription factors with roles in the proliferation, differentiation, tumor invasion and metastasis [35-40]. CDK6 has been found aberrantly regulated in many tumors including glioma, suggesting that CDK6 may serve as a therapeutic target [41-46]. However, the mechanism underlying the CDK6 dysregulation in distinct cancers is still unclear [41-49]. Alternatively, this kinase may have additional functions unrelated to cell cycle progression that are of importance only in certain cell types [23,41,50].
Materials and methods
Cell culture
The human U251 and T98G glioblastoma cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Tianhang Biotechnology, Zhejiang, China) and 1% penicillin/streptomycin (Gibco) at 37°C in an environment with 5% carbon dioxide.
Human glioma samples
Human glioma samples were obtained from adult patients with pathologically proven glioma who received therapy in the Affiliated Sun-Yat-Sen Memorial Hospital of Sun-Yat-Sen University. Informed consent was obtained before study. Samples were collected during surgery and immediately frozen in liquid nitrogen for subsequent total RNA extraction or fixed in formalin for subsequent immunohistochemistry.
RNA extraction and quantitative real-time polymerase chain reaction
Total RNA was isolated from cultured cells, normal brain tissues and glioma tissues with Trizol Reagent (Invitrogen), according to the manufacturer’s instructions. Then, 100 ng of total RNA was reverse transcribed into cDNA using the Master Mix kit (TAKARA Biotechnology, Dalian, China). Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) was performed in the Bio-rad systems and the mRNA expression of target genes was normalized to that of U6 and glyceraldehyde 3-phosphate dehydrogenase (GADPH). Total RNA from normal brain tissues served as a control in the measurement of microRNA-675 expression in the glioma tissues. microRNA-675 expression in the glioma tissues was measured using the SYBR Green PCR Master Mix kit in accordance with the manufacturer’s instructions (Takara Biotechnology, Dalian, China). The mRNA expressions of microRNA-675 and CDK6 in cultured cells were also determined with the SYBR Green PCR Master Mix kit (Takara Biotechnology, Dalian, China). The reaction was done with 10 μL of mixture including 5×SYBR green master mix, gene-specific primers and cDNA. Fold changes in the gene expression were calculated using the 2-ΔΔCt method.
Oligonucleotide synthesis and transfection
microRNA-675 mimic is the extrinsic mature double-stranded microRNA and can mimic the endogenous microRNA-675. microRNA-675 inhibitor is the extrinsic mature complementary single-stranded microRNA, and can inhibit the endogenous microRNA-675. The microRNA-675 mimic, microRNA-675 inhibitor, microRNA-675 mimic negative control and microRNA-675 inhibitor negative control were synthesized in the GenePharma in China. The sequences were as follows: H19 siRNA: CUUUCUGUCACAUUGACCACACCUG or UCUGAUUGCAGCAUCUUCUUGAUUC (H19 negative control was designed by GenePharm [Suzhou, China]); microRNA-675 mimic: CTGTATGCCCTCACCGCTCA; microRNA-675 inhibitor: TGAGCGGTGAGGGCATACAG (microRNA-675 mimic negative control, microRNA-675 inhibitor negative control were designed by RiboBio [Guangzhou, China]); miRNA quantification: Bulge-loop TM miRNA qRT-PCR Primer Sets (one RT primer and a pair of qPCR primers for each set) specific for hsa-mir-675-3p and hsa-mir-675-5p were designed by RiboBio (Guangzhou, China).
All the sequences above were transfected into cultured cells at 100 nM by using the Lipofectamine 2000 reagent (Life) according to the manufacturer’s instructions.
Protein isolation and western blot assay
Cells were washed thrice with ice-cold phosphate buffer saline (PBS), and lysed in RIPA buffer (cwbiotechnology, Beijing, China). Western blot assay was performed according to standard protocols with a primary antibody against CDK6 (1:500; F-7, Santa Cruz). Anti-α-tubulin was used as a loading control (1:5000, CST). Chemiluminescence was detected using the ECL detection solution (cwbiotechnology, Beijing, China), and western blot assay was done at least thrice, and averages were obtained.
Luciferase assay
The microRNA-675 target sites within the CDK6 were PCR amplified using primers containing artificial XhoI and EcoRI restriction sites and cloned into the psiCheck2 vector (Promega) downstream of a luciferase reporter gene. miRNA-binding sites were mutated by the amplification of psiCheck2-CDK6 vector with primers containing the required point mutations and verified by sequencing (IGE Biotechnology, Guangzhou, China). Plasmids (200 μg) were co-transfected into U251 and T98G cells with microRNA-675 mimic, microRNA-675 mimic negative control, microRNA-675 inhibitor or microRNA-675 inhibitor negative control (RiboBio, Guangzhou, China), and the luciferase activity was measured at 48 h after transfection using the Dual Luciferase Reporter Assay Kit (Beyotime, Shanghai, China), according to the manufacturer’s instructions.
MTT assay
Glioblastoma cells were seeded into 96-well plates at 20000 cells/well. After transfection as described previously, 20 μL of 3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide (MTT) (0.5 mg/mL) was added to each well at 0, 24, 36, 48, 72 and 96 h after treatment. After incubation for 4 h, and the supernatant was removed, and 200 μL of DMSO was added to each well. The optical density (OD) was measured at 570 nm. At least 3 independent experiments were performed.
Colony formation assay
Cells were seeded in 6-well plates (400 cells/well) and cultured for 10 days. The colonies were stained with 0.1% crystal violet for 15 min after fixation in 4% formaldehyde for 30 min. Viable colonies that contained more than 50 cells were counted. The experiment was performed at least thrice for each cell line.
Wound healing assay
One day before transfection, glioma cells (2×105) were seeded into 6-well plates. When cells confluence reached about 90% at 24 h after transfection, a wound was made on the monolayer with the tip of a sterile 100-μL micropipette. Then, the debris was removed by washing with PBS. At different time points, cells migrating into the wounded area or cells with extended protrusions from the wound border were counted at 100× under a light microscope.
Immunohistochemistry
Immunohistochemistry on formalin-fixed and paraffin-embedded tissues was performed using a standard protocol. In brief, the above tissues were incubated with the anti-CDK6 primary antibody (Santa Cruz), washed in PBST (3 times, 5 min for each) and then incubated with secondary antibody (DAKO, polymer HRP-labelled anti-mouse, Envision) for 1 h at room temperature. The tissues were washed in PBST (3 times, 5 min for each). DAB (3,3’-Diaminobenzidine) was added for visualization. Signals were observed under a light microscope. Sections were washed in Milli-Q water for 5 min, stained with Mayer’s haematoxylin solution for 10 min and washed with flowing water for 30 min. Sections were finally mounted by Eukitt quick-hardening mounting medium (ASGB-BIO) and covered with coverslips. The CDK6 protein expression was independent evaluated by two experienced pathologists as follows: the percentage of CDK6-positive cells was 0% (score=0), 1-30% (score=1), 30-60% (score=2), and >60% (score=3). CDK6 staining intensity was graded as follows: no staining (score=0), weak (score=1), moderate (score=2) and strong (score=3). The products of both scores were obtained as the final CDK6 score.
Bioinformatics
The candidate targets of microRNA-675 were generated using the publicly available algorithms TargetScan (http://targetscan.org/), miRanda (http://microrna.org/), Pic Tar (pictar.mdc-berlin.de) and miRDB (http://mirdb.org).
Statistical analysis
Data were analyzed with GraphPad Prism 5 software (GraphPad Software, Inc, La Jolla, CA, USA). Quantitative data are presented as mean ± standard deviation (S.D.). Comparisons between two groups were done with t-test, and Chi-square test and Fisher’s exact test were employed for the comparisons of remaining data. Survival curves were plotted with the Kaplan-Meier method and log-rank test was employed for the survival analysis. Bivariate correlations between variables were evaluated by the Spearman’s rank correlation coefficient. A value of P<0.05 was considered statistically significant.
Results
microRNA-675 is down-regulated in human glioma and directly related to H19
To explore the relationship between microRNA-675 expression and pathological grade of glioma, 3 normal brain tissues, 4 glioma tissues at low grade (grade I, II) and 4 glioma at high grade (grade III and IV) were processed for the detection of miRNA-675. The 8 glioma tissues were categorized as tumor core and tumor marginal (adjacent normal) tissues by resecting the core and marginal tissues from the same patient during surgery. Total RNA was extracted from above tissues and stem-loop quantitative real time PCR (qRT-PCR) was performed. Results showed that the microRNA-675 expression in glioma tissues was down-regulated when compared with normal brain tissues (Figure 1A). The microRNA-675 expression was also related to the glioma grade: the lower the glioma grade, the higher the microRNA-675 expression. In addition, the microRNA-675 expression was also related to the different biological features of glioma, and the microRNA-675 expression in tumor core tissues was lower than in tumor marginal tissues in the same patient. These findings provided evidence that microRNA-675 expression is downregulated in glioma, and inversely related to the glioma grade.
Figure 1.

microRNA-675 expression was down-regulated in glioma tissues and glioma cell lines (U251 cells and T98G cells). A. Average expression microRNA-675 in human normal brain (n=3), low grade glioma marginal tissues (n=4), low grade glioma core tissues (n=4), high grade glioma marginal tissues (n=4), high grade glioma core tissues (n=4). Average expression of microRNA-675 in human low grade glioma was higher than in human high grade glioma, and average expression of microRNA-675 in glioma marginal tissues was also higher than in glioma core tissues. The microRNA-675 expression was normalized to that of U6. B. microRNA-675 expression in U251 cells and T98G cells significantly decreased as compared to negative control (NC) (P<0.001).
To investigate whether microRNA-675 is partly derived from LncRNAH19, H19 expression was down-regulated with H19siRNA in U251 and T98G cell lines, and the microRNA-675 expression was measured with stem-loop qRT-PCR. Results showed that the microRNA-675 expression in U251 cells and T98G cells significantly decreased as compared to negative control (NC) (Figure 1B).
microRNA-675 directly suppresses CDK6 in glioma cells
To identify the mRNA targets of microRNA-675, miRNA target prediction databases including TargetScan, miRanda, miRDB and PicTar were searched. Using a multiplicity of these prediction algorithms, several common targets could be predicted. Among the targets validated in cell lines, CDK6 was of particular interest because it is a key molecule in the regulation of cell cycle and has pivotal function in the tumor cell proliferation. CDK6 was a predicted target of microRNA-675 and contains a mer-7 seed match in its 3’UTR (Figure 2E). In addition, CDK6 was found to be highly expressed in the glioma and CDK6 expression correlated with the glioma grade. To confirm that CDK6 was a target gene of microRNA-675 in the glioma, the protein and mRNA expressions of CDK6 were examined in cells independently transfected with microRNA-675 mimic, microRNA-675 mimic negative control, microRNA-675 inhibitor and microRNA-675 inhibitor negative control by western blot assay and qRT-PCR, respectively (Figure 2C and 2D). To validate that H19 derived microRNA-675 can directly target CDK6 3’UTR, qRT-PCR and Western blot assay were employed to examine the expression of CDK6 in T98G cells and U251 cells which were transfected with H19siRNA and negative control (NC). Results showed that the CDK6 expression was significantly down-regulated after transfection with H19siRNA when compared with negative control (Figure 2A and 2B). Collectively, these findings strongly indicate that CDK6 is a direct target of microRNA-675 in the glioma. Luciferase assay was also performed to further confirm that CDK6 is a direct target for H19 derived microRNA-675. The wildtype CDK6 3’UTR and mutant CDK6 3’UTR containing the seed sequences were cloned to psiCheck2 vector containing a luciferase reporter construct which was then co-transfected into cells with microRNA-675 mimic, microRNA-675 mimic negative control (MN), microRNA-675 inhibitor, microRNA-675 inhibitor negative control (IN) or transfection reagent (condition). Results showed that luciferase activity reduced by more than 30% in the presence of microRNA-675 mimic when compared with condition group, and the luciferase activity of cells transfected with microRNA-675 inhibitor increased by nearly 30% when compared with condition group (Figure 2G and 2H). This was not observed when the microRNA-675-binding sites were mutated (Figure 2I and 2J). Thus, CDK6 is a potential target of microRNA-675.
Figure 2.
microRNA-675 targets CDK6 by binding its 3’UTR. A, B. Total cell lysates were used to determine CDK6 expression by qRT-PCR and Western blot assay in T98G cells and U251 cells after transfection of H19siRNA and NC (negative control), and GADPH served as an internal control for qRT-PCR and α-Tubulin as a loading control for Western blot assay. C, D. Cell lysate was used to determine the CDK6 expression by qRT-PCR and Western blot assay in T98G cells and U251 cells after transfection of microRNA-675 mimic, microRNA-675 mimic negative control, microRNA-675 inhibitor, or microRNA-675 inhibitor negative control. GADPH served as an internal control for qRT-PCR and α-Tubulin as a loading control for Western blot assay. CDK6 is a direct, primary target of microRNA-675. E. CDK6 possesses binding sites for microRNA-675 in its 3’UTR. F. Wildtype and mutant CDK6 3’UTR were constructed to psiCheck2 vector. G, H. Effect of microRNA-675 mimic, microRNA-675 mimic negative control, microRNA-675 inhibitor, microRNA-675 inhibitor negative control and transfection reagent (condition) transfection on T98G cells and U251 cells transfected with pisCheck2 vector fused to the wildtype CDK6 3’UTR. I, J. Effect of microRNA-675 mimic, microRNA-675 inhibitor and transfection reagent (condition) transfection on T98G cells and U251 cells transfected with psiCheck2 vector fused to either wildtype or mutant CDK6 3’UTR.
Reduction in H19 derived microRNA-675 in glioma cells induces tumor cell proliferation
To explore the biological effects of microRNA-675 on the glioma cell growth, glioma cells were transfected with microRNA-675 mimic, microRNA-675 mimic negative control, micorRNA-675 inhibitor or microRNA-675 inhibitor negative control, and then the cell viability was measured by MTT assay. Results showed that the viability of T98G cells transfected with microRNA-675 inhibitor increased by 29.91% when compared with T98G cells transfected with microRNA-675 inhibitor negative control, the viability of T98G cells transfected with microRNA-675 mimic reduced by 22.87% when compared with T98G cells transfected with microRNA-675 mimic negative control. In addition, the viability of U251 cells transfected with microRNA-675 mimic reduced by 25.07% when compared with U251 cells transfected with microRNA-675 mimic negative control, the viability of U251 cells transfected with microRNA-675 inhibitor increased by 50.36% when compared with U251 cells transfected with microRNA-675 inhibitor negative control (Figure 3A). Moreover, colony formation assay was also employed to evaluate the cell proliferation. Results showed T98G cells transfected with microRNA-675 mimic displayed much fewer and smaller colonies when compared with T98G cells transfected with microRNA-675 mimic negative control, and T98G cells transfected with microRNA-675 inhibitor showed more and bigger colonies when compared with cells transfected with microRNA-675 inhibitor negative control. Similar findings were also observed in U251 cells (Figure 3A). In order to further demonstrate the microRNA-675 is derived from H19 and H19 can indirectly regulate glioma cell proliferation, T98G cells and U251 cells were independently transfected with H19siRNA and negative control (NC), and MTT assay and colony formation assay were performed. Similar findings were observed (Figure 3B).
Figure 3.
microRNA-675 promotes the proliferation of glioma cells. A. MTT assay of T98G cells and U251 cells transfected with microRNA-675 mimic, microRNA-675 mimic negative control, microRNA-675 inhibitor, or microRNA-675 inhibitor negative control. B. MTT assay of T98G cells and U251 cells transfected with H19siRNA or negative control. *P<0.05 mimic vs condition in T98G, **P<0.01 inhibitor vs condition T98G or mimic vs condition in U251, ***P<0.001 mimic vs inhibitor in T98G or inhibitor vs condition in U251. C. Representative photographs (Left) and quantification (Right) of colonies formed by T98G cells and U251 cells at 10 days after transfection with microRNA-675 mimic, microRNA-675 inhibitor or condition (cells with transfection). D. Representative photographs (Left) and quantification (Right) of colonies formed by T98G cells at 10 days after transfection with H19siRNA or negative control.
To confirm our findings, the growth of T98G cells and U251 cells was evaluated by colony formation assay. T98G cells were transiently transfected with microRNA-675 mimic, microRNA-675 inhibitor, and cells transfected with transfection buffer alone were treated as control (condition). When compared with condition group, the cell growth was inhibited by 49.19% in T98G cells after transfection with microRNA-675 mimic (P<0.05), and that was increased by 178.22% in T98G cells after transfection with microRNA-675 inhibitor (P<0.01) (Figure 3C). In U251 cells, the cell growth was inhibited by 55.43% after transfection with microRNA-675 mimic when compared with condition group (cells transfected with transfection buffer) (P<0.01), and that was increased by 219.56% after transfection with microRNA-675 inhibitor when compared with condition group (P<0.001) (Figure 3C).
To further elucidate that H19 derived microRNA-675 is able to regulate cell proliferation, the cell growth was evaluated in T98G cells after transfection with H19siRNA or negative control (NG). Results showed that the cell growth was increased by 120.62% in T98G cells after transfection with H19siRNA when compared with cells transfected with negative control (P<0.01) (Figure 3D).
Reduction in H19 derived microRNA-675 in glioma cells enhances tumor cell migration
To examine the effect of microRNA-675 on the glioma cell migration, microRNA-675 mimic, microRNA-675 mimic negative control, microRNA-675 inhibitor, and microRNA-675 inhibitor negative control were transiently transfected into U251 cells and T98G cells. Tumor cell migration capability was determined by wound healing assay and transwell assay. Results of wound healing assay in T98G cells showed that, at 24 and 48 h, 21.1% and 33.4% of the wound was closed after transfection with microRNA-675, respectively, 39.5% and 61.8% of the wound was closed after transfection with microRNA-675 inhibitor, respectively, and 32.4% and 43.5% of the wound was closed after transfection with transfection buffer, respectively, in T98G cells (Figure 4A and 4B). In U251 cells, at 24 and 48 h, 19.8% and 37.8% of the wound was closed after transfection with microRNA-675, respectively, 41.5% and 63.5% of the wound was closed after transfection with microRNA-675 inhibitor, respectively, and 32.4% and 49.2% of the wound was closed after transfection with transfection condition, respectively, in U251 cells (Figure 4C and 4D). Furthermore, results of transwell assay displayed that the number of cells transfected with microRNA-675 mimic, microRNA-675 mimic negative control (MN), microRNA-675 inhibitor and microRNA-675 inhibitor negative control (IN) migrating through the membrane was 37±7, 117±31, 212±52, and 108±29, respectively, in T98G cells (Figure 4E and 4F); the number of cells transfected with microRNA-675 mimic, microRNA-675 mimic negative control (MN), microRNA-675 inhibitor and microRNA-675 inhibitor negative control (IN) migrating through the membrane was 22±4, 45±7, 72±11, and 47±6 respectively, in U251 cells (Figure 4G and 4I). Above findings indicate that H19 derived microRNA-675 is able to suppress the migration of glioma cells.
Figure 4.
Over-expression of microRNA-675 promotes glioma cell migration. A, C. Representative photographs of wound healing assay were obtained at different time points (Original Magnifiction: 100×). B, D. Quantification of cell migration was conducted by measuring the wound width. The amount of migrating cells was displayed as the percent of migrating cells at a specific time point. The migration abilities of T98G cells and U251 cells transfected with microRNA-675 mimic, microRNA-675 mimic negative control (MN), microRNA-675 inhibitor, or microRNA-675 inhibitor negative control (IN) (transwell assay). E, G. Representative photographs are shown (Original Magnification: 100×). F, H. Quantification of migrating cells from three independent experiments.
CDK6 is over-expressed in glioma and CDK6 expression is associated with tumor aggressiveness
The CDK6 protein expression was detected in 20 glioma tissues at low grade (I, II), 20 glioma tissues at high grade (III, IV), their corresponding adjacent normal tissues and 2 normal brain tissues by immunohistochemistry. Strong CDK6-positive staining was observed in the nucleus and cytoplasm in high grade glioma, but weak CDK6-positive staining in low grade glioma. The proportion of positive cells and staining intensity were compared between tumor core tissues and tumor marginal tissues from the same patient. Results showed that the staining intensity of tumor marginal tissues was significantly lower than that in tumor core tissues. The normal brain tissues had no CDK6 expression (Figure 5A and 5B).
Figure 5.
H19 derived microRNA-675 targets CDK6 and CDK6 detected in glioma tissues. A. The CDK6 expression in normal brain tissues. B. The CDK6 expression in glioma at different grades and glioma tissues at different sites. C. The average immunohistochemical scores of 40 clinical samples. *P<0.01 vs low grade tumor core and tumor marginal tissues; #P<0.01 vs tumor marginal tissues, &P<0.05 vs tumor marginal tissues.
The relationship between pathological features of glioma and CDK6 expression in glioma was further evaluated. Results showed CDK6 expression was positively correlated with the disease stage (Figure 5B). Moreover, the low grade glioma and high grade glioma were independent divided into 2 subgroups according to the tumor sites. Results showed that the CDK6 expression was significantly different between low grade tumor and high grade tumor (Figure 5B). Furthermore, the CDK6 expression in the tumor core tissues was markedly higher than in corresponding tumor marginal tissues (Figure 5B and 5C). These findings indicate that CDK6 plays a critical role in the glioma development and progression and may serve as a valuable biomarker for the glioma.
CDK6 expression is inversely associated with microRNA-675 expression in glioma tissues
In this study, low grade glioma tissues (n=4) and high grade glioblastoma tissues (n=4) were collected and divided into tumor marginal and tumor core tissues. The microRNA-675 expression was detected in 16 samples (Figure 6A) and the CDK6 expression was also determined by immunohistochemistry and scored in the same samples (Figure 6B and 6C). Our results showed a significant inverse association between microRNA-675 expression and CDK6 expression was observed in these samples (r=-0.8996, Figure 6D).
Figure 6.

CDK6 expression is up-regulated in glioma tissues and inversely correlated with microRNA-675 expression. A. microRNA-675 expression in human low grade tumor marginal tissues, low grade tumor core tissues, high grade tumor marginal tissues and high grade tumor core tissues. B, C. CDK6 expression in human low grade tumor marginal tissues, low grade tumor core tissues, high grade tumor marginal tissues, and high grade tumor core tissues. D. An inverse association between microRNA-675 and CDK6 expression was observed in human glioma tissues (r=-0.8996). E. Overall survival (OS) curves of Kaplan-Meier analysis. OS was analyzed according to the CDK6 expression. Low CDK6 expression was related to long OS (P<0.05).
Furthermore, the CDK6 expression was detected in glioma tissues by immunohistochemistry and the relationship between CDK6 expression and survival of glioma patients was evaluated. Results revealed that the high CDK6 expression was directly correlated with a poor survival (Figure 6E). Taken together, these findings indicate that the regulation of CDK6 expression by H19 derived microRNA-675 may affect the clinical prognosis of glioma patients.
microRNA-675 expression reduces in glioma cells and tissues and is correlated with survival
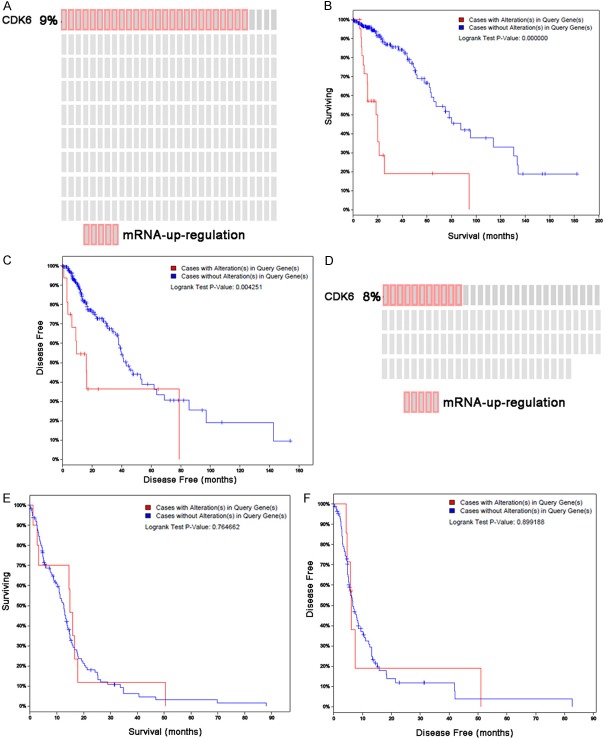
As shown above, microRNA-675 significantly decreased in glioma tissues (Figure 1) and CDK6 was the downstream target of microRNA-675. To confirm these results and explore the relationship between CDK6 expression and patients’ prognosis, glioma tissues at low and high grade were collected, and CDK6 expression was detected. On the basis of TCGA data, the association between CDK6 expression and OS or disease free survival (DFS) was investigated in low grade glioma patients and high grade glioma patients using the Kaplan-Meier analysis. Our results showed CDK6 expression was significantly up-regulated in 9% of low grade glioma patients (n=286) and highly up-regulated in 8% in high grade glioblastoma patients (n=136) (Figure 7A and 7B) and low CDK6 expression was significantly correlated with general survival and DFS as compared to high CDK6 expression in low grade glioma patients. In particular, high CDK6 expression apparently shortened both OS and DFS as compared to low CDK6 expression (median OS: 18.45 vs 78.15 months; P<0.001; median DFS: 15.97 vs 42.9 months; P<0.01) (Figure 7C and 7D). Moreover, the relationship between CDK6 expression and OS/DFS was also evaluated in high grade glioma patients (median OS: 14.91 vs 12.55 months; P>0.05; median DFS: 6.11 vs 6.67 months; P>0.05), but there was no marked correlation between CDK6 expression and OS/DFS (Figure 7E and 7F). These findings indicated that CDK6 expression may serve as a predictor of low grade glioma.
Figure 7.
CDK6 expression is up-regulated in glioma and glioblastoma and related to OS and DSF of glioma and glioblastoma patients. A. CDK6 expression was up-regulated in glioma patients by TCGA dataset. B, C. OS and DSF curves of Kaplan-Meier analysis. The OS and DFS were analyzed according to the CDK6 expression. Low CDK6 expression was related to long OS and DFS in the TCGA dataset at the indicated cut-off. D. CDK6 expression was up-regulated in glioblastoma patients in TCGA dataset. E, F. OS and DFS curves of Kaplan-Meier analysis. The OS and DFS were analyzed according to CDK6 expression. The OS and DFS of patients with low CDK6 expression were comparable to those of patients with high CDK6 expression in the TCGA dataset at the indicated cut-off.
Discussion
Increasing evidence demonstrates that microRNAs play essential roles in both physiological and pathological processes, including cancer initiation, development, invasion and migration [6,23]. In this study, our results revealed that LncRNAH19 and microRNA-675 significantly decreased in the glioma, and their expressions were inversely correlated with OS and DFS of glioma patients. Inhibition of microRNA-675 remarkably alleviated the malignancy of glioma characterized by the inhibition of glioma cell proliferation, invasion and metastasis. Inve-stigation of molecular mechanism showed that microRNA-675 dramatically inhibited the expression of CDK6, a crucial molecule regulating cell cycle [35-40]. Moreover, microRNA-675 inhibited the glioma cell growth, invasion and migration by directly targeting CDK6 [10,14-17]. Our findings highlight the significance of microRNA-675 down-regulation in the glioma development and progression and implicate microRNA-675 as a candidate prognostic biomarker and target for the glioma therapy.
MicroRNA-675 has been involved in the physiological development and differentiation, and microRNA-675 embedded in the first exon of H19 was expressed exclusively in the placenta since the gestational stage when the placenta growth normally ceased, placenta lacking of H19 or microRNA-675 continued to grow, overexpression of microRNA-675 in a range of embryonic and extra-embryonic cell lines resulted in their reduced proliferation, and the controlled release of microRNA-675 from H19 also allowed rapid inhibition of cell proliferation in response to cellular stress and oncogenic signals [10,14-17]. However, some investigators found that, in some types of tumors, microRNA-675 expression was significantly downregulated, which lead to uncontrolled tumor cell proliferation. Our results showed H19 derived microRNA-675 expression significantly decreased in the glioma tissues and cell lines, and closely correlated with the OS and DFS of glioma patients.
Thousands of human genes are predicted to be targets of microRNAs, which may regulate gene expression post-transcriptionally by binding to their complementary sequences in the target microRNAs [35-40]. The accepted mechanism of microRNA targeting involves an interaction between 5’-end of the microRNA and 3’-UTR of the mRNA, which promotes mRNA degradation and inhibitors’ translation [35-40]. In our study, luciferase assay showed that the transfection of microRNA-675 into cells containing the 3’-UTR of CDK6 wt increased the luciferase activity by more than 30% as compared to cells containing the 3’-UTR of CDK4 mut, suggesting that microRNA-675 binds directly to the 3’-UTR of CDK6. A dose-dependent decrease or increase in CDK6 expression was induced after transfection of a microRNA-675 mimic or inhibitor, respectively, further confirming this interaction. Real-Time PCR of 3 normal brain tissues, 4 low grade glioma tissues and 4 high grade glioma tissues and immunohistochemistry of tissues from 40 patients also suggested CDK6 served as a target of microRNA-675, which was consistent with the results from the analysis of samples from TCGA dataset.
CDK6 is well-known as an essential kinase that can phosphate Rb, a pivotal transcriptional factor, and regulate the cell cycle, proliferation and migration [35-40]. Growing studies have suggested that aberrant CDK6 expression in tumors mainly contributes to enhanced tumor invasion and migration [35-40]. Functional inhibition or knock-down of CDK6 expression significantly promotes G1 to S phase progression in cell cycle, leads to uncontrolled cell growth and also directly or indirectly influences the tumor invasion and migration [35-40]. Recent studies suggest that microRNAs are involved in the regulation of CDK6 expression and may affect its function [35-40].
CDK6 is related to cell proliferation, invasion and migration and was identified by a bioinformatics approach using various algorithms as a target of microRNA675. The CDK6 expression was detected after transfection of microRNA-675 mimic, microRNA-675 mimic negative control, microRNA-675 inhibitor, microRNA-675 inhibitor control. Results showed that the microRNA-675 expression was inversely correlated with CDK6 expression. Then, this was further confirmed by luciferase assay. Thus, we speculate that microRNA-675 may directly target CDK6 3’-UTR by its seed sequences. High grade gliomas with low microRNA-765 expression showed CDK6 over-expression, indicating that high CDK6 mRNA expression is a prognostic factor for glioma. Our studies demonstrated CDK6 served as a target for microRNA-675 and specified its roles in glioma and as a potential drug target. There are a few studies indirectly proposing microRNA-675 as an important regulator of CDK6 in glioma, wherein microRNA-675 expression is inversely correlated with CDK6 expression in glioma [35-40], thereby inducing characteristic changes, including cell growth, invasion and migration. Importantly, microRNA-675 mimic, microRNA-675 mimic negative control, microRNA-675 inhibitor, microRNA-675 inhibitor negative control were transfected into glioma cells to induce and knockdown microRNA-675 expression, and results showed the over-expression and suppression of microRNA-675 inhibited and promoted the glioma cell proliferation, invasion and migration, respectively, suggesting that microRNA-675 acts as a key meditator in the regulation of cell growth, tumor invasion and migration via attenuating the influence of CDK6.
The growth-suppressive effects of microRNA-675 possibly stem from its potential to inhibit cell cycle progression because the expression of proteins such as CDK6, Rb, as well as other CDK6 related proteins important to invasion and migration decreased in microRNA-675-overexpressing glioma cells [35-40]. These findings point to a regulatory framework centered on microRNA-675 in glioma cells. MicroRNA-675 coordinately downregulates the expression of oncogenes,including CDK6, IGFR1, and CDH11 [35-40]. The roles of CDK6 in the proliferation, survival, invasion and migration in the glioma have been well documented [35-40]. Other reports show that the expression of molecules like Rb and its downstream targets will be down-regulated when CDK6 expression is suppressed [35-40]. Although the expression of these downstream genes was not detected in the present study, we hypothesize that microRNA-675 may execute its tumor-suppressor function by inhibiting CDK6, causing down-regulated expression of these pro-survival genes. In other words, microRNA-675 down-regulates CDK6 expression directly and down-regulate the expression of Rb and its downstream genes indirectly through CDK6. CDK6 is an important regulator in the cell cycle progression and closely related to the uncontrolled glioma cell proliferation, invasion and migration. Hence, our study demonstrates that the interaction of microRNA-675 with its target CDK6 plays crucial roles in the progression, invasion and migration of glioma cells. Our studies demonstrate the role of microRNA-675 as a tumor suppressor and it may serve as an important target for the glioblastoma therapy.
Acknowledgements
This study was supported a grant from the Key Laboratory of Malignant Tumor Molecular Mechanism and Translational Medicine of Guangzhou Bureau of Science and Information (No. [2013] 163).
Disclosure of conflict of interest
None.
References
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Chen R, Nishimura MC, Bumbaca SM, Kharbanda S, Forrest WF, Kasman IM, Greve JM, Soriano RH, Gilmour LL, Rivers CS, Modrusan Z, Nacu S, Guerrero S, Edgar KA, Wallin JJ, Lamszus K, Westphal M, Heim S, James CD, VandenBerg SR, Costello JF, Moorefield S, Cowdrey CJ, Prados M, Phillips HS. A hierarchy of self-renewing tumorinitiating cell types in glioblastoma. Cancer Cell. 2010;17:362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 3.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 4.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, Min W, Bennett AM, Gregory RI, Ding Y, Huang Y. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okamura E, Matsuzaki H, Sakaguchi R, Takahashi T, Fukamizu A, Tanimoto K. The H19 imprinting control region mediates preimplantation imprinted methylation of nearby sequences in yeast artificial chromosome transgenic mice. Mol Cell Biol. 2013;33:858–871. doi: 10.1128/MCB.01003-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan L, Zhou J, Gao Y, Ghazal S, Lu L, Bellone S, Yang Y, Liu N, Zhao X, Santin AD, Taylor H, Huang Y. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene. 2015;34:3076–3084. doi: 10.1038/onc.2014.236. [DOI] [PubMed] [Google Scholar]
- 9.Shoshani O, Massalha H, Shani N, Kagan S, Ravid O, Madar S, Trakhtenbrot L, Leshkowitz D, Rechavi G, Zipori D. Polyploidization of murine mesenchymal cells is associated with suppression of the long noncoding RNA H19 and reduced tumorigenicity. Cancer Res. 2012;72:6403–6413. doi: 10.1158/0008-5472.CAN-12-1155. [DOI] [PubMed] [Google Scholar]
- 10.Venkatraman A, He XC, Thorvaldsen JL, Sugimura R, Perry JM, Tao F, Zhao M, Christenson MK, Sanchez R, Yu JY, Peng L, Haug JS, Paulson A, Li H, Zhong XB, Clemens TL, Bartolomei MS, Li L. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature. 2013;500:345–349. doi: 10.1038/nature12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medrzycki M, Zhang Y, Zhang W, Cao K, Pan C, Lailler N, McDonald JF, Bouhassira EE, Fan Y. Histone h1.3 suppresses h19 noncoding RNA expression and cell growth of ovarian cancer cells. Cancer Res. 2014;74:6463–6473. doi: 10.1158/0008-5472.CAN-13-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan LH, Wang W, Yeung W, Deng Y, Yuan P, Mak KK. Hedgehog signaling induces osteosarcoma development through Yap1 and H19 overexpression. Oncogene. 2014;33:4857–4866. doi: 10.1038/onc.2013.433. [DOI] [PubMed] [Google Scholar]
- 13.Oh SH, Jung YH, Gupta MK, Uhm SJ, Lee HT. H19 gene is epigenetically stable in mouse multipotent germline stem cells. Mol Cells. 2009;27:635–640. doi: 10.1007/s10059-009-0084-1. [DOI] [PubMed] [Google Scholar]
- 14.Pidsley R, Dempster E, Troakes C, Al-Sarraj S, Mill J. Epigenetic and genetic variation at the IGF2/H19 imprinting control region on 11p15.5 is associated with cerebellum weight. Epigenetics. 2012;7:155–163. doi: 10.4161/epi.7.2.18910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawahara M, Morita S, Takahashi N, Kono T. Defining contributions of paternally methylated imprinted genes at the Igf2-H19 and Dlk1-Gtl2 domains to mouse placentation by transcriptomic analysis. J Biol Chem. 2009;284:17751–17765. doi: 10.1074/jbc.M109.000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guibert S, Zhao Z, Sjolinder M, Gondor A, Fernandez A, Pant V, Ohlsson R. CTCFbinding sites within the H19 ICR differentially regulate local chromatin structures and cisacting functions. Epigenetics. 2012;7:361–369. doi: 10.4161/epi.19487. [DOI] [PubMed] [Google Scholar]
- 17.Matsuzaki H, Okamura E, Shimotsuma M, Fukamizu A, Tanimoto K. A randomly integrated transgenic H19 imprinting control region acquires methylation imprinting independently of its establishment in germ cells. Mol Cell Biol. 2009;29:4595–4603. doi: 10.1128/MCB.00275-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding GL, Wang FF, Shu J, Tian S, Jiang Y, Zhang D, Wang N, Luo Q, Zhang Y, Jin F, Leung PC, Sheng JZ, Huang HF. Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes. 2012;61:1133–1142. doi: 10.2337/db11-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim NH, Choi SH, Kim CH, Lee CH, Lee TR, Lee AY. Reduced MiR-675 in exosome in H19 RNA-related melanogenesis via MITF as a direct target. J Invest Dermatol. 2014;134:1075–1082. doi: 10.1038/jid.2013.478. [DOI] [PubMed] [Google Scholar]
- 20.Goodell MA. Parental permissions: H19 and keeping the stem cell progeny under control. Cell Stem Cell. 2013;13:137–138. doi: 10.1016/j.stem.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 21.Loke YJ, Galati JC, Morley R, Joo EJ, Novakovic B, Li X, Weinrich B, Carson N, Ollikainen M, Ng HK, Andronikos R, Aziz NK, Saffery R, Craig JM. Association of maternal and nutrient supply line factors with DNA methylation at the imprinted IGF2/H19 locus in multiple tissues of newborn twins. Epigenetics. 2013;8:1069–1079. doi: 10.4161/epi.25908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrell DT. Aberrant methylation of the H19 imprinting control region may increase the risk of spontaneous abortion. Epigenomics. 2013;5:23–24. [PubMed] [Google Scholar]
- 23.Xia B, Yang S, Liu T, Lou G. miR-211 suppresses epithelial ovarian cancer proliferation and cell-cycle progression by targeting Cyclin D1 and CDK6. Mol Cancer. 2015;14:57. doi: 10.1186/s12943-015-0322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K, Wang X, Zou J, Zhang A, Wan Y, Pu P, Song Z, Qian C, Chen Y, Yang S, Wang Y. miR-92b controls glioma proliferation and invasion through regulating Wnt/beta-catenin signaling via Nemo-like kinase. Neuro Oncol. 2013;15:578–588. doi: 10.1093/neuonc/not004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He D, Wang J, Zhang C, Shan B, Deng X, Li B, Zhou Y, Chen W, Hong J, Gao Y, Chen Z, Duan C. Down-regulation of miR-675-5p contributes to tumor progression and development by targeting pro-tumorigenic GPR55 in non-small cell lung cancer. Mol Cancer. 2015;14:73. doi: 10.1186/s12943-015-0342-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie C, Lu H, Nomura A, Hanse EA, Forster CL, Parker JB, Linden MA, Karasch C, Hallstrom TC. Co-deleting Pten with Rb in retinal progenitor cells in mice results in fully penetrant bilateral retinoblastomas. Mol Cancer. 2015;14:93. doi: 10.1186/s12943-015-0360-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang JF, He ML, Fu WM, Wang H, Chen LZ, Zhu X, Chen Y, Xie D, Lai P, Chen G, Lu G, Lin MC, Kung HF. Primate-specific microRNA-637 inhibits tumorigenesis in hepatocellular carcinoma by disrupting signal transducer and activator of transcription 3 signaling. Hepatology. 2011;54:2137–2148. doi: 10.1002/hep.24595. [DOI] [PubMed] [Google Scholar]
- 28.Huang D, Qiu S, Ge R, He L, Li M, Li Y, Peng Y. miR-340 suppresses glioblastoma multiforme. Oncotarget. 2015;6:9257–9270. doi: 10.18632/oncotarget.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dudek KA, Lafont JE, Martinez-Sanchez A, Murphy CL. Type II collagen expression is regulated by tissue-specific miR-675 in human articular chondrocytes. J Biol Chem. 2010;285:24381–24387. doi: 10.1074/jbc.M110.111328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim NH, Choi SH, Lee TR, Lee CH, Lee AY. Cadherin 11, a miR-675 target, induces N-cadherin expression and epithelial-mesenchymal transition in melasma. J Invest Dermatol. 2014;134:2967–2976. doi: 10.1038/jid.2014.257. [DOI] [PubMed] [Google Scholar]
- 32.Douet-Guilbert N, Tous C, Le Flahec G, Bovo C, Le Bris MJ, Basinko A, Morel F, De Braekeleer M. Translocation t(2;7)(p11;q21) associated with the CDK6/IGK rearrangement is a rare but recurrent abnormality in B-cell lymphoproliferative malignancies. Cancer Genet. 2014;207:83–86. doi: 10.1016/j.cancergen.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 33.Otto T, Sicinski P. The kinase-independent, second life of CDK6 in transcription. Cancer Cell. 2013;24:141–143. doi: 10.1016/j.ccr.2013.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paternot S, Colleoni B, Bisteau X, Roger PP. The CDK4/CDK6 inhibitor PD0332991 paradoxically stabilizes activated cyclin D3-CDK4/6 complexes. Cell Cycle. 2014;13:2879–2888. doi: 10.4161/15384101.2014.946841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bellail AC, Olson JJ, Hao C. SUMO1 modification stabilizes CDK6 protein and drives the cell cycle and glioblastoma progression. Nat Commun. 2014;5:4234. doi: 10.1038/ncomms5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Placke T, Faber K, Nonami A, Putwain SL, Salih HR, Heidel FH, Kramer A, Root DE, Barbie DA, Krivtsov AV, Armstrong SA, Hahn WC, Huntly BJ, Sykes SM, Milsom MD, Scholl C, Frohling S. Requirement for CDK6 in MLLrearranged acute myeloid leukemia. Blood. 2014;124:13–23. doi: 10.1182/blood-2014-02-558114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komori T. Regulation of Rb family proteins by Cdk6/Ccnd1 in growth plates. Cell Cycle. 2013;12:2161–2162. doi: 10.4161/cc.25515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh NK, Kundumani-Sridharan V, Kumar S, Verma SK, Kotla S, Mukai H, Heckle MR, Rao GN. Protein kinase N1 is a novel substrate of NFATc1-mediated cyclin D1-CDK6 activity and modulates vascular smooth muscle cell division and migration leading to inward blood vessel wall remodeling. J Biol Chem. 2012;287:36291–36304. doi: 10.1074/jbc.M112.361220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Linden MH, Willekes M, van Roon E, Seslija L, Schneider P, Pieters R, Stam RW. MLL fusion-driven activation of CDK6 potentiates proliferation in MLL-rearranged infant ALL. Cell Cycle. 2014;13:834–844. doi: 10.4161/cc.27757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shao Y, Qu Y, Dang S, Yao B, Ji M. MiR-145 inhibits oral squamous cell carcinoma (OSCC) cell growth by targeting c-Myc and Cdk6. Cancer Cell Int. 2013;13:51. doi: 10.1186/1475-2867-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baba Y, Watanabe M, Murata A, Shigaki H, Miyake K, Ishimoto T, Iwatsuki M, Iwagami S, Yoshida N, Oki E, Sakamaki K, Nakao M, Baba H. LINE-1 hypomethylation, DNA copy number alterations, and CDK6 amplification in esophageal squamous cell carcinoma. Clin Cancer Res. 2014;20:1114–1124. doi: 10.1158/1078-0432.CCR-13-1645. [DOI] [PubMed] [Google Scholar]
- 42.Pekkonen P, Jarviluoma A, Zinovkina N, Cvrljevic A, Prakash S, Westermarck J, Evan GI, Cesarman E, Verschuren EW, Ojala PM. KSHV viral cyclin interferes with T-cell development and induces lymphoma through Cdk6 and Notch activation in vivo. Cell Cycle. 2014;13:3670–3684. doi: 10.4161/15384101.2014.964118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rader J, Russell MR, Hart LS, Nakazawa MS, Belcastro LT, Martinez D, Li Y, Carpenter EL, Attiyeh EF, Diskin SJ, Kim S, Parasuraman S, Caponigro G, Schnepp RW, Wood AC, Pawel B, Cole KA, Maris JM. Dual CDK4/CDK6 inhibition induces cell-cycle arrest and senescence in neuroblastoma. Clin Cancer Res. 2013;19:6173–6182. doi: 10.1158/1078-0432.CCR-13-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsu YH, Yao J, Chan LC, Wu TJ, Hsu JL, Fang YF, Wei Y, Wu Y, Huang WC, Liu CL, Chang YC, Wang MY, Li CW, Shen J, Chen MK, Sahin AA, Sood A, Mills GB, Yu D, Hortobagyi GN, Hung MC. Definition of PKC-alpha, CDK6, and MET as therapeutic targets in triple-negative breast cancer. Cancer Res. 2014;74:4822–4835. doi: 10.1158/0008-5472.CAN-14-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pauls E, Ruiz A, Badia R, Permanyer M, Gubern A, Riveira-Munoz E, Torres-Torronteras J, Alvarez M, Mothe B, Brander C, Crespo M, Menendez-Arias L, Clotet B, Keppler OT, Marti R, Posas F, Ballana E, Este JA. Cell cycle control and HIV-1 susceptibility are linked by CDK6-dependent CDK2 phosphorylation of SAMHD1 in myeloid and lymphoid cells. J Immunol. 2014;193:1988–1997. doi: 10.4049/jimmunol.1400873. [DOI] [PubMed] [Google Scholar]
- 46.Kollmann K, Heller G, Schneckenleithner C, Warsch W, Scheicher R, Ott RG, Schafer M, Fajmann S, Schlederer M, Schiefer AI, Reichart U, Mayerhofer M, Hoeller C, Zochbauer-Muller S, Kerjaschki D, Bock C, Kenner L, Hoefler G, Freissmuth M, Green AR, Moriggl R, Busslinger M, Malumbres M, Sexl V. A kinase-independent function of CDK6 links the cell cycle to tumor angiogenesis. Cancer Cell. 2013;24:167–181. doi: 10.1016/j.ccr.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grossel MJ, Baker GL, Hinds PW. cdk6 can shorten G(1) phase dependent upon the N-terminal INK4 interaction domain. J Biol Chem. 1999;274:29960–29967. doi: 10.1074/jbc.274.42.29960. [DOI] [PubMed] [Google Scholar]
- 48.Scheicher R, Hoelbl-Kovacic A, Bellutti F, Tigan AS, Prchal-Murphy M, Heller G, Schneckenleithner C, Salazar-Roa M, Zochbauer-Muller S, Zuber J, Malumbres M, Kollmann K, Sexl V. CDK6 as a key regulator of hematopoietic and leukemic stem cell activation. Blood. 2015;125:90–101. doi: 10.1182/blood-2014-06-584417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang LH, Contractor T, Clausen R, Klimstra DS, Du YC, Allen PJ, Brennan MF, Levine AJ, Harris CR. Attenuation of the retinoblastoma pathway in pancreatic neuroendocrine tumors due to increased cdk4/cdk6. Clin Cancer Res. 2012;18:4612–4620. doi: 10.1158/1078-0432.CCR-11-3264. [DOI] [PubMed] [Google Scholar]
- 50.Kollmann K, Sexl V. CDK6 and p16INK4A in lymphoid malignancies. Oncotarget. 2013;4:1858–1859. doi: 10.18632/oncotarget.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]