Abstract
Deregulated expression of miRNAs is related to progression and initiation of human cancers. Although miR-495 has identified in various tumors, its expression and function in renal cell carcinoma (RCC) is still unknown. In this study, we found that the expression of miR-495 was downregulated in RCC cell lines and tissues. Ectopic expression of miR-495 induced G0/G1 phase arrest and suppressed cell proliferation and migration in RCC cell lines. We further validated SATB1 was a direct target of miR-495 in RCC. In addition, re-expression of SATB1 reversed the miR-495-induced inhibition of cell proliferation and migration. These data suggest that miR-495 functions as a tumor suppressor and may be a promising therapeutic target in RCC in the future.
Keywords: Renal cell carcinoma, microRNAs, miR-495, SATB1
Introduction
Renal cell carcinoma (RCC) is the most common neoplasm of the adult kidney, which accounts for approximately 85% of all primary malignant kidney tumors as well as 3% of cancers in adults [1-4]. Surgery is often curative for localized disease; however, a lot of these patients develop metastatic or relapses diseases, which are associated with poor prognosis [5-8]. The 5-year-survival rate of patients who diagnosed at the metastatic stage was only 9% [9-11]. Therefore, there is a crucial need to found new biomarkers and targeted therapies for this aggressive malignancy.
MicroRNAs (miRNAs), approximately 19-24 nucleotides in length, are highly conserved regulatory molecules that modulate gene expression through imperfect complementary sequence pairing to the 3’ untranslated region (3’UTR) of target genes resulting in either mRNA degradation or translational repression [12-14]. Several studies have reported that miRNAs play key roles in developmental regulation, cell proliferation, differentiation, invasion and apoptosis [15-17]. Abnormal expressions of miRNAs have been found in several types of human cancers, including cervical cancer, bladder cancer, breast cancer, renal cell carcinoma, hepatocellular and ovarian cancers [12,18-23]. They may play important roles in the progression and development of cancers similar to those played by tumor suppressor genes or oncogenes [24-27].
In this study, we demonstrated that miR-495 expression level was frequently downregulated in human RCC cell lines and tissues and overexpression of miR-495 suppressed RCC cell proliferation and migration. Moreover, SATB1 was identified as a direct target of miR-495 and re-expression of SATB1 reversed the miR-495-induced inhibition of cell proliferation and migration.
Materials and methods
Ethics statement
Patients gave written informed consent. This study was approved by the ethical board of Renmin Hospital of Wuhan University and complied with the Declaration of Helsinki.
Tissues and cell lines
RCC tissues and adjacent normal tissues were collected from patients undergoing surgery at our hospital. The samples were immediately snap-frozen in liquid nitrogen until protein or RNA extraction. The human RCC cell lines, 769-P, 786-O, A498, SN12-PM6 and one normal renal cell line (HK-2) were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). These cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 20% FBS.
Transient transfection
MiR-495 mimics and scramble were purchased from GenePharma (GenePharma, China). MiR-495 mimics or scramble was transfected into cells by using Lipofectamine 2000 (Invitrogen, Canada) according to the manufacturer’s instructions.
Cell proliferation, cell cycle and colony formation
CCK-8 assay was performed to evaluate the cell proliferation according the manufacturer’s instructions.
Optical density (OD) was measured at 450 nm an enzyme immunoassay instrument (BioRad, Hercules, CA, USA). For cell cycle experiment, cells were fixed in 95% ethanol, incubated at -20°C overnight and resuspended in FACS solution. Cells were detected by using a FACS Calibur flow cytometer (BD Biosciences). For colony formation assay, cells were cultured for 7 days. Then cells were then fixed with 4% formaldehyde and stained with 1.0% crystal violet.
Real-time RT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen). SYBR-Green PCR master mix (Applied Biosystems, Inc. Foster City, CA, and USA) was performed on the 7500 Real-time PCR System (Applied Biosystems). The expression of U6 or GAPDH was used as control. The primer sequences were: SATB1 forward: 5’-GAGGAAGGCTTGGGAGTA-3’, reverse: 5’-GGGCAGCAGAGCTATGTG-3; GAPDH forward: 5’-CGGAGTCAACGGATTTGGTCGTAT-3’ and reverse: 5’-AGCCTTCTCCATGGTGGTGAAGAC-3’.
Dual-luciferase reporter assay
Luciferase assay was performed as previously reported [28]. SATB1 3’-UTR containing the putative miR-495 binding site or a mutant sequence was designed and inserted into the downstream of the firefly luciferase reporter (Promega, Madison, WI, USA). The cultures transciently cotransfected with miR-495 scramble and vector (contain either WT or MUT 3’-UTR). Luciferase activity was performed by Dual Luciferase Reporter Assay System.
Western blotting
Western blotting was performed as previously described [29]. Protein was separated by electrophoresis, transferred to membranes and were probed with specific primary antibody. The membranes were incubated with the secondary antibody conjugated to horseradish peroxidase (HRP). Protein was visualized by chemiluminescence using the ECL detection system (BeyoECL Plus, Beyotime). Anti-SATB1 and GAPDH antibodies were purchased from Abcam (Cambridge, MA, USA).
Migration assay
Cell migration was performed using a wound-healing assay. The cell monolayer was scraped using a P-20 micropipette tip. The initial gap length and the residual gap length after wounding were measured by photomicrographs.
Statistical analysis
All experiments were performed in triplicate and analyzed for significant differences between two groups were analyzed using Student’s t test; One-way ANOVA was performed to analyze the more than two groups. P < 0.05 was considered statistically significant. The data are showed as means ± SD.
Results
miR-495 was downregulated in RCC cell lines and tissues
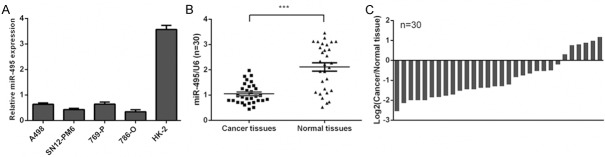
The expression of miR-495 was lower in all four human renal cell carcinoma cells lines (769-P, 786-O, A498, SN12-PM6) than in one normal renal cell line (HK-2) (Figure 1A). Moreover, qRT-PCR data demonstrated that the expression level of miR-495 was also significantly lower in clinical RCC specimens and compared with normal tissues (Figure 1B and 1C).
Figure 1.
miR-495 was downregulated in RCC cell lines and tissues (A) qRT-PCR analysis was used to detect the expression of miR-495 in four human renal cell carcinoma cells lines (769-P, 786-O, A498, SN12-PM6) and one normal renal cell line (HK-2). (B) qRT-PCR analysis was used to detect the expression of miR-495 in clinical RCC specimens and normal tissues. (C) The relative expression of miR-495 was downregulated in clinical RCC specimens compared with their corresponding nontumor tissues.
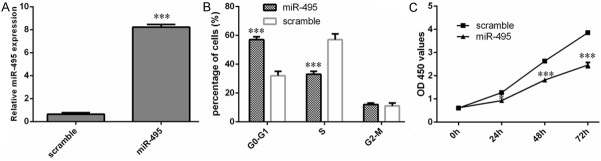
Re-expression of miR-495 suppressed RCC cell proliferation
We aimed to address the phenotypes of 786-O cells stably expressing miR-495 (Figure 2A). Cell cycle assays showed that 786-O cells transfected with miR-495 mimics had an obvious cell cycle arrest at the G0/G1 phase (Figure 2B). Re-expression of miR-495 suppressed 786-O cells proliferation using CCK-8 assay (Figure 2C).
Figure 2.
Re-expression of miR-495 suppressed RCC cell proliferation (A) qRT-PCR was performed to measure the miR-495 expression in 786-O cells at 48 hours after miR-495 mimic transfection. (B) Cell cycle assays showed that 786-O cells transfected with miR-495 mimics had an obvious cell cycle arrest at the G0/G1 phase. (C) Re-expression of miR-495 suppressed 786-O cells proliferation using CCK-8 assay.
Overexpression of miR-495 inhibited RCC cell migration
Migration assays were performed to measure the function of miR-495 in cell migration. As shown in Figure 3, cell migration was significantly repressed in the group transfected with miR-495 compared with those in the scramble group.
Figure 3.
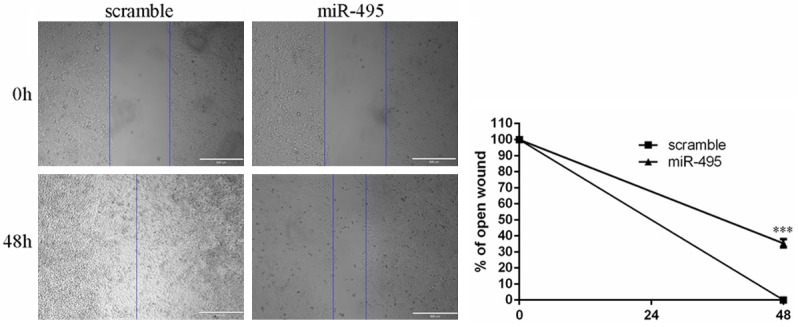
Overexpression of miR-495 inhibited RCC cell migration. Overexpression of miR-495 repressed the 786-O cells migration.
miR-495 downregulated SATB1 expression by directly targeting its 3’-UTR
The data of TargetScan showed SATB1 has the putative target of miR-495 (Figure 4A). Luciferase reporter assay showed that the luciferase reporter activity decreased approximately 67% in the 786-O cells containing the SATB1 WT 3’UTR fragment (Figure 4B). Moreover, we found that the ectopic expression of miR-495 suppressed the SATB1 mRNA and protein level in 786-O cells by using qRT-PCR and Western blot (Figure 4C and 4D).
Figure 4.
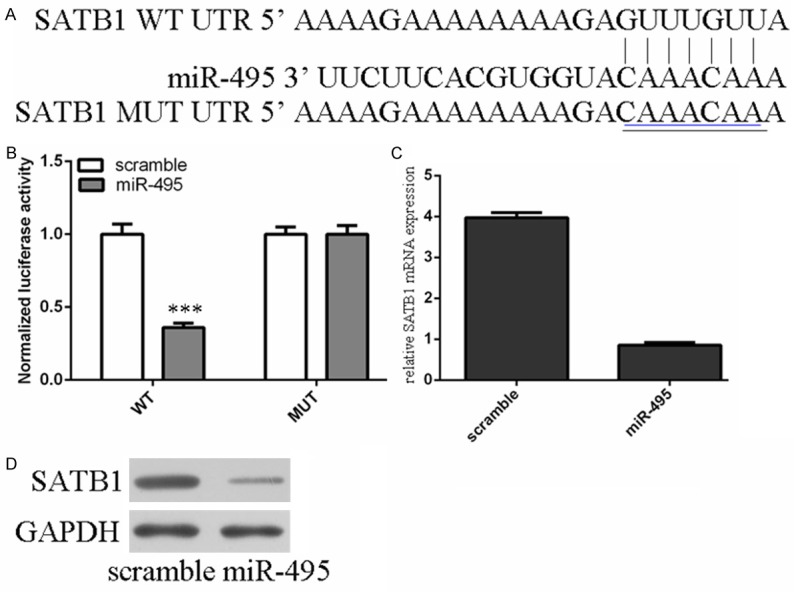
miR-495 downregulated SATB1 expression by directly targeting its 3’-UTR. A. SATB1 gene harbored a potential miR-495 binding site using a stringent bioinformatics approach. B. Luciferase reporter assay showed that the luciferase reporter activity decreased approximately 67% in the 786-O cells containing the SATB1 WT 3’UTR fragment. C. The ectopic expression of miR-495 suppressed the SATB1 mRNA by using qRT-PCR. D. Overexpression of miR-495 inhibited the SATB1 protein by using Western blot.
Re-expression of SATB1 reversed the miR-495-induced inhibition of cell proliferation and migration
Rescue experiment to confirm that miR-495 acts as a tumor suppressor in renal cell carcinoma cells by regulating SATB1 expression. The expression of SATB1 was overexpression by using SATB1 vector (Figure 5A). We rescued the expression of SATB1 in miR-495 overexpressing 786-O cells. CCK8 assay showed that re-expression of SATB1 increased the miR-495 overexpressing 786-O cells proliferation (Figure 5B). Cell cycle assays showed that miR-495 overexpressing 786-O cells transfected with SATB1 vector had an obvious cell cycle at the S phase (Figure 5C). Furthermore, the migration abilities of miR-495 overexpressing 786-O cells were increased after SATB1 transfection (Figure 5D).
Figure 5.
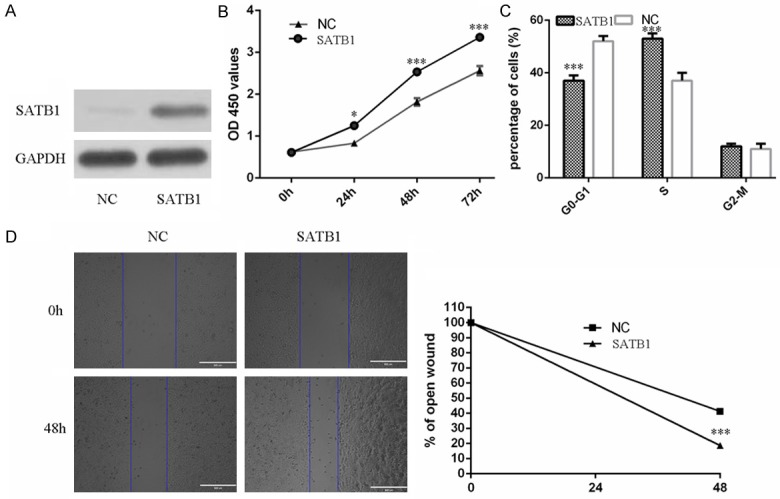
Re-expression of SATB1 reversed the miR-495-induced inhibition of cell proliferation and migration. A. The protein expression of SATB1 was measured by using Western blot. B. The data of CCK8 assay showed that re-expression of SATB1 increased the miR-495 overexpressing 786-O cells proliferation. C. Cell cycle assays showed that miR-495 overexpressing 786-O cells transfected with SATB1 vector had an obvious cell cycle at the S phase. D. The migration abilities of miR-495 overexpressing 786-O cells were increased after SATB1 transfection.
Discussion
RCC is the third most common urological cancer and is one of the most common types of cancers [3,30,31]. Despite the great improvement in cancer therapy, metastatic RCC presents a 5-year survival rate of 0-10% [32-34]. Therefore, it is important to search new treatment strategies. In this study, we demonstrated that miR-495 expression level was downregulated in human RCC cell lines and tissues and overexpression of miR-495 suppressed RCC cell proliferation and migration. Moreover, SATB1 was identified as a direct target of miR-495 and re-expression of SATB1 reversed the miR-495-induced inhibition of cell proliferation and migration. Taken together, our study suggests that miR-495 acts a tumor suppressor gene in RCC.
MiR-495 was reported to act as a tumor suppressor gene or an oncogene in a lot of cancers including non-small cell lung cancer, breast cancer, glioblastoma, gastric cancer and leukemia [35-38]. For example, miR-495 was downregulated in gastric carcinoma samples and overexpression of miR-495 inhibited gastric cancer cell migration and invasion by targeting the PRL-3 oncogene [35]. Another study showed that miR-495 likely functions as a tumor suppressor in acute myeloid leukemia (AML) with mixed lineage leukemia (MLL) rearrangements by targeting essential leukemia-related genes [37]. However, it has been demonstrated that miR-495 acts as an oncogene in breast cancer via downregulation of E-cadherin and REDD1 [38]. Cao et al. also showed that miR-495 could facilitate breast cancer progression through the repression of JAM-A [39]. In our study, our data showed that the expression of miR-495 was lower in four human renal cell carcinoma cells lines (769-P, 786-O, A498, SN12-PM6) than in one normal renal cell line (HK-2). The expression level of miR-495 was also significantly lower in clinical RCC specimens and compared with normal tissues. Moreover, re-expression of miR-495 had an obvious cell cycle arrest at the G0/G1 phase and suppressed RCC cells proliferation and invasion. These results confirmed that miR-495 acts as a tumor suppressor gene in RCC.
It is crucial to explore the molecular mechanism underlying miR-495 function in RCC. The putative target of SATB1 for miR-495 was detected by the TargetScan program. Luciferase reporter assay showed that the luciferase reporter activity decreased approximately 67% in the RCC cells containing the SATB1 WT 3’UTR fragment. To test this assumption, we detected the mRNA and protein expression of SATB1 after miR-495 overexpression. We found that ectopic expression of miR-495 suppressed the SATB1 mRNA and protein level in RCC cells. Furthermore, re-expression of SATB1 reversed the miR-495-induced inhibition of cell proliferation and migration. Our results suggest that SATB1 is a functional downstream target of miR-495 in RCC. Previous study showed that the levels of SATB1 mRNA and protein were increased in human RCC tissues and SATB1 knockdown inhibited the proliferation, migration and invasion of 786-O cells [40]. However, the detail mechanism of downregulation of SATB1 is still unknown. Our data showed that the ability of miR-495 to repress SATB1 expression may provide one such mechanism of post-transcriptional control of SATB1.
In conclusion, this study provided novel evidence that miR-495 functions as a tumor suppressor miRNA in RCC through inhibiting SATB1 expression. Our data suggested that this miRNA could be a potential target for the treatment of RCC in future.
Acknowledgements
This work was supported by Natural Science Foundation of Hainan Province (Grant Number: 20158264).
Disclosure of conflict of interest
None.
References
- 1.Liang J, Zhang Y, Jiang G, Liu Z, Xiang W, Chen X, Chen Z, Zhao J. MiR-138 induces renal carcinoma cell senescence by targeting EZH2 and is downregulated in human clear cell renal cell carcinoma. Oncol Res. 2013;21:83–91. doi: 10.3727/096504013X13775486749218. [DOI] [PubMed] [Google Scholar]
- 2.Chow TF, Youssef YM, Lianidou E, Romaschin AD, Honey RJ, Stewart R, Pace KT, Yousef GM. Differential expression profiling of microRNAs and their potential involvement in renal cell carcinoma pathogenesis. Clin Biochem. 2010;43:150–158. doi: 10.1016/j.clinbiochem.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 3.Li W, Liu M, Feng Y, Xu YF, Huang YF, Che JP, Wang GC, Yao XD, Zheng JH. Downregulated miR-646 in clear cell renal carcinoma correlated with tumour metastasis by targeting the nin one binding protein (NOB1) Br J Cancer. 2014;111:1188–1200. doi: 10.1038/bjc.2014.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez-Lago MA, Thodima VJ, Guttapalli A, Chan T, Heguy A, Molina AM, Reuter VE, Motzer RJ, Chaganti RS. Genomic deregulation during metastasis of renal cell carcinoma implements a myofibroblast-like program of gene expression. Cancer Res. 2010;70:9682–9692. doi: 10.1158/0008-5472.CAN-10-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu X, Weng L, Li X, Guo C, Pal SK, Jin JM, Li Y, Nelson RA, Mu B, Onami SH, Wu JJ, Ruel NH, Wilczynski SP, Gao H, Covarrubias M, Figlin RA, Weiss LM, Wu H. Identification of a 4-microRNA signature for clear cell renal cell carcinoma metastasis and prognosis. PLoS One. 2012;7:e35661. doi: 10.1371/journal.pone.0035661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slaby O, Redova M, Poprach A, Nekvindova J, Iliev R, Radova L, Lakomy R, Svoboda M, Vyzula R. Identification of MicroRNAs associated with early relapse after nephrectomy in renal cell carcinoma patients. Genes Chromosomes Cancer. 2012;51:707–716. doi: 10.1002/gcc.21957. [DOI] [PubMed] [Google Scholar]
- 7.Zhai Q, Zhou L, Zhao C, Wan J, Yu Z, Guo X, Qin J, Chen J, Lu R. Identification of miR-508-3p and miR-509-3p that are associated with cell invasion and migration and involved in the apoptosis of renal cell carcinoma. Biochem Biophys Res Commun. 2012;419:621–626. doi: 10.1016/j.bbrc.2012.02.060. [DOI] [PubMed] [Google Scholar]
- 8.Huang Y, Dai Y, Yang J, Chen T, Yin Y, Tang M, Hu C, Zhang L. Microarray analysis of microRNA expression in renal clear cell carcinoma. Eur J Surg Oncol. 2009;35:1119–1123. doi: 10.1016/j.ejso.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 9.Lv L, Huang F, Mao H, Li M, Li X, Yang M, Yu X. MicroRNA-21 is overexpressed in renal cell carcinoma. Int J Biol Markers. 2013;28:201–207. doi: 10.5301/JBM.2013.10831. [DOI] [PubMed] [Google Scholar]
- 10.Yu XY, Zhang Z, Liu J, Zhan B, Kong CZ. MicroRNA-141 is downregulated in human renal cell carcinoma and regulates cell survival by targeting CDC25B. Onco Targets Ther. 2013;6:349–354. doi: 10.2147/OTT.S41343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Zhao J, Zhang JW, Huang QY, Huang JZ, Chi LS, Tang HJ, Liu GQ, Zhu DJ, Ma WM. MicroRNA-217, down-regulated in clear cell renal cell carcinoma and associated with lower survival, suppresses cell proliferation and migration. Neoplasma. 2013;60:511–515. doi: 10.4149/neo_2013_066. [DOI] [PubMed] [Google Scholar]
- 12.Zhang WH, Gui JH, Wang CZ, Chang Q, Xu SP, Cai CH, Li YN, Tian YP, Yan L, Wu B. The identification of miR-375 as a potential biomarker in distal gastric adenocarcinoma. Oncol Res. 2012;20:139–147. doi: 10.3727/096504012x13522227232156. [DOI] [PubMed] [Google Scholar]
- 13.Chiang Y, Zhou X, Wang Z, Song Y, Liu Z, Zhao F, Zhu J, Xu H. Expression levels of microRNA-192 and -215 in gastric carcinoma. Pathol Oncol Res. 2012;18:585–591. doi: 10.1007/s12253-011-9480-x. [DOI] [PubMed] [Google Scholar]
- 14.Wang K, Jia Z, Zou J, Zhang A, Wang G, Hao J, Wang Y, Yang S, Pu P. Analysis of hsa-miR-30a-5p expression in human gliomas. Pathol Oncol Res. 2013;19:405–411. doi: 10.1007/s12253-012-9593-x. [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Yu X, Shen J, Chan MT, Wu WK. MicroRNA in intervertebral disc degeneration. Cell Prolif. 2015;48:278–83. doi: 10.1111/cpr.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu X, Li Z. MicroRNAs regulate vascular smooth muscle cell functions in atherosclerosis (review) Int J Mol Med. 2014;34:923–933. doi: 10.3892/ijmm.2014.1853. [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Yu X, Shen J, Wu WK, Chan MT. MicroRNA expression and its clinical implications in Ewing’s sarcoma. Cell Prolif. 2015;48:1–6. doi: 10.1111/cpr.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, Yu X, Wang Y, Shen J, Wu WK, Liang J, Feng F. By downregulating TIAM1 expression, microRNA-329 suppresses gastric cancer invasion and growth. Oncotarget. 2015;6:17559–69. doi: 10.18632/oncotarget.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo LJ, Zhang QY. Decreased serum miR-181a is a potential new tool for breast cancer screening. Int J Mol Med. 2012;30:680–686. doi: 10.3892/ijmm.2012.1021. [DOI] [PubMed] [Google Scholar]
- 20.Wu D, Tao J, Xu B, Li P, Lu Q, Zhang W. Downregulation of Dicer, a component of the microRNA machinery, in bladder cancer. Mol Med Rep. 2012;5:695–699. doi: 10.3892/mmr.2011.711. [DOI] [PubMed] [Google Scholar]
- 21.Zhao H, Bi T, Qu Z, Jiang J, Cui S, Wang Y. Expression of miR-224-5p is associated with the original cisplatin resistance of ovarian papillary serous carcinoma. Oncol Rep. 2014;32:1003–1012. doi: 10.3892/or.2014.3311. [DOI] [PubMed] [Google Scholar]
- 22.Tang T, Wong HK, Gu W, Yu MY, To KF, Wang CC, Wong YF, Cheung TH, Chung TK, Choy KW. MicroRNA-182 plays an onco-miRNA role in cervical cancer. Gynecol Oncol. 2013;129:199–208. doi: 10.1016/j.ygyno.2012.12.043. [DOI] [PubMed] [Google Scholar]
- 23.Vaira V, Faversani A, Dohi T, Montorsi M, Augello C, Gatti S, Coggi G, Altieri DC, Bosari S. miR-296 regulation of a cell polaritycell plasticity module controls tumor progression. Oncogene. 2012;31:27–38. doi: 10.1038/onc.2011.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, Yu X, Shen J, Jiang Y. MicroRNA dysregulation in uveal melanoma: a new player enters the game. Oncotarget. 2015;6:4562–8. doi: 10.18632/oncotarget.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohdaira H, Sekiguchi M, Miyata K, Yoshida K. MicroRNA-494 suppresses cell proliferation and induces senescence in A549 lung cancer cells. Cell Prolif. 2012;45:32–38. doi: 10.1111/j.1365-2184.2011.00798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo X, Dong Z, Chen Y, Yang L, Lai D. Enrichment of ovarian cancer stem-like cells is associated with epithelial to mesenchymal transition through an miRNA-activated AKT pathway. Cell Prolif. 2013;46:436–446. doi: 10.1111/cpr.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M, Yu M, Liu C, Zhu H, He X, Peng S, Hua J. miR-34c works downstream of p53 leading to dairy goat male germline stem-cell (mGSCs) apoptosis. Cell Prolif. 2013;46:223–231. doi: 10.1111/cpr.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z, Yu X, Wang Y, Shen J, Wu WK, Liang J, Feng F. By downregulating TIAM1 expression, microRNA-329 suppresses gastric cancer invasion and growth. Oncotarget. 2015;6:17559–17569. doi: 10.18632/oncotarget.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Yu X, Liang J, Wu WK, Yu J, Shen J. Leptin downregulates aggrecan through the p38-ADAMST pathway in human nucleus pulposus cells. PLoS One. 2014;9:e109595. doi: 10.1371/journal.pone.0109595. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Yuan HX, Zhang JP, Kong WT, Liu YJ, Lin ZM, Wang WP, Guo JM. Elevated microRNA-185 is associated with high vascular endothelial growth factor receptor 2 expression levels and high microvessel density in clear cell renal cell carcinoma. Tumour Biol. 2014;35:12757–12763. doi: 10.1007/s13277-014-2602-9. [DOI] [PubMed] [Google Scholar]
- 31.Yoshino H, Enokida H, Itesako T, Tatarano S, Kinoshita T, Fuse M, Kojima S, Nakagawa M, Seki N. Epithelial-mesenchymal transitionrelated microRNA-200s regulate molecular targets and pathways in renal cell carcinoma. J Hum Genet. 2013;58:508–516. doi: 10.1038/jhg.2013.31. [DOI] [PubMed] [Google Scholar]
- 32.Khella HW, White NM, Faragalla H, Gabril M, Boazak M, Dorian D, Khalil B, Antonios H, Bao TT, Pasic MD, Honey RJ, Stewart R, Pace KT, Bjarnason GA, Jewett MA, Yousef GM. Exploring the role of miRNAs in renal cell carcinoma progression and metastasis through bioinformatic and experimental analyses. Tumour Biol. 2012;33:131–140. doi: 10.1007/s13277-011-0255-5. [DOI] [PubMed] [Google Scholar]
- 33.Goto K, Oue N, Shinmei S, Sentani K, Sakamoto N, Naito Y, Hayashi T, Teishima J, Matsubara A, Yasui W. Expression of is a potential prognostic factor after nephrectomy in advanced renal cell carcinoma. Mol Clin Oncol. 2013;1:235–240. doi: 10.3892/mco.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slaby O, Jancovicova J, Lakomy R, Svoboda M, Poprach A, Fabian P, Kren L, Michalek J, Vyzula R. Expression of miRNA-106b in conventional renal cell carcinoma is a potential marker for prediction of early metastasis after nephrectomy. J Exp Clin Cancer Res. 2010;29:90. doi: 10.1186/1756-9966-29-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z, Zhang G, Li D, Jie Z, Chen H, Xiong J, Liu Y, Cao Y, Jiang M, Le Z, Tan S. Methylationassociated silencing of miR-495 inhibit the migration and invasion of human gastric cancer cells by directly targeting PRL-3. Biochem Biophys Res Commun. 2015;456:344–350. doi: 10.1016/j.bbrc.2014.11.083. [DOI] [PubMed] [Google Scholar]
- 36.Chu H, Chen X, Wang H, Du Y, Wang Y, Zang W, Li P, Li J, Chang J, Zhao G, Zhang G. MiR-495 regulates proliferation and migration in NSCLC by targeting MTA3. Tumour Biol. 2014;35:3487–3494. doi: 10.1007/s13277-013-1460-1. [DOI] [PubMed] [Google Scholar]
- 37.Jiang X, Huang H, Li Z, He C, Li Y, Chen P, Gurbuxani S, Arnovitz S, Hong GM, Price C, Ren H, Kunjamma RB, Neilly MB, Salat J, Wunderlich M, Slany RK, Zhang Y, Larson RA, Le Beau MM, Mulloy JC, Rowley JD, Chen J. MiR-495 is a tumor-suppressor microRNA down-regulated in MLL-rearranged leukemia. Proc Natl Acad Sci U S A. 2012;109:19397–19402. doi: 10.1073/pnas.1217519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang-Verslues WW, Chang PH, Wei PC, Yang CY, Huang CK, Kuo WH, Shew JY, Chang KJ, Lee EY, Lee WH. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene. 2011;30:2463–2474. doi: 10.1038/onc.2010.618. [DOI] [PubMed] [Google Scholar]
- 39.Cao M, Nie W, Li J, Zhang Y, Yan X, Guan X, Chen X, Zen K, Zhang CY, Jiang X, Hou D. MicroRNA-495 induces breast cancer cell migration by targeting JAM-A. Protein Cell. 2014;5:862–872. doi: 10.1007/s13238-014-0088-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng C, Wan F, Liu L, Zeng F, Xing S, Wu X, Chen X, Zhu Z. Overexpression of SATB1 is associated with biologic behavior in human renal cell carcinoma. PLoS One. 2014;9:e97406. doi: 10.1371/journal.pone.0097406. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]