Abstract
Background: MicroRNAs (miRNA) have been documented playing critical roles in cancer progression. Aberrant expression of miR-205 has been reported in several types of cancer. However, the role and mechanism of miR-205 in colorectal cancer (CRC) remains unclear. Methods: In this study, the expression levels of miR-205 in CRC tissues and cell lines were detected by quantitative real-time PCR (qRT-PCR). MTT assay and transwell assays were applied to assess the effect of miR-205 on CRC cell proliferation, migration and invasion abilities in vitro. Furthermore, Dual-luciferase reporter assay was conducted to confirm the direct binding of miR-205 and target genes. Results: In the present study, we found that the expression level of miR-205 in CRC tissues and cell lines was significantly down-regulated. Functional assays showed that overexpression of miR-205 suppressed CRC cell proliferation, migration and invasion in vitro. In addition, cAMP responsive element binding protein 1 (CREB1) was identified as targets of miR-205. Silencing CREB1 had similar effects of miR-205 restoration on proliferation, migration and invasion in CRC cells. Conclusions: Our results demonstrated that miR-205 may act as a tumor suppressor by targeting the CREB1 gene and suppressing CRC cell proliferation, migration and invasion. Thus, miR-205 may represent a potential therapeutic target for CRC intervention.
Keywords: Colorectal cancer, miR-205, cAMP responsive element binding protein 1
Introduction
Colorectal cancer (CRC) is one of the most common malignancies and a leading cause of cancer-related morbidity all over the world [1]. Despite innovative therapeutic strategies applied to CRC treatment, the prognosis for CRC patients has not significantly changed in the last 20 years [2]. The poor prognosis of CRC is related mainly to frequent tumor metastasis and tumor recurrence after surgical resection [3]. Thus, understanding the molecular mechanisms which regulate the metastasis and recurrence in CRC is of crucial significance to the development of therapeutic strategies for CRC patients.
microRNAs (miRNA) are a class of are small non coding RNAs that repress protein translation through binding to the 3’-untranslated region (UTR) of their target mRNAs in a sequence-specific manner [4]. In recent years, miRNAs have been proven to play important roles in the cell viability, proliferation, invasion and metastasis of human cancer, and it is believed that miRNAs function both as tumor suppressors and oncogenes in cancer progression [5,6]. For example, Yang et al reported that miR-506 was downregulated in clear cell renal cell carcinoma and could inhibit renal cancer cell growth and metastasis via suppressing FLOT1 expression [7]. Sun et al showed that miR-646 was downregulated in osteosarcoma and inhibited osteosarcoma cell migration and invasion by downregulating FGF2 expression [8]. Xu et al demonstrated that miR-221/222 was overexpressed in pancreatic cancer and promoted pancreatic cancer cell proliferation and invasion via regulating TIMP-2 expression [9]. However, to our knowledge the underlying mechanism of miR-205 in CRC remains unclear.
In this study, we showed that miR-205 was downregulated in CRC tissues and cell lines. Overexpression of miR-205 inhibited CRC cell proliferation, migration and invasion in vitro. Furthermore, we identified cAMP responsive element binding protein 1 (CREB1) as a direct and functional target of miR-205. Knockdown of CREB1 exerted similar effects of miR-205 restoration on proliferation, migration and invasion of CRC cells. Thus, our data suggested that miR-205 play a critical role in CRC cell proliferation, migration and invasion through suppression of CREB1, indicating that miR-205 might serves as a potential therapeutic target of CRC.
Materials and methods
Tissue specimens and cell cultures
Human CRC tissues and matched adjacent non-tumor tissues were collected from 20 CRC patients who underwent surgical resection at the Department of Surgery, Affiliated Hospital of Nantong University. This study was approved by the Ethics Committee of Nantong University and informed consent was obtained from each patient. Normal colon epithelium cell line FHC and CRC cell lines SW480, HT29 and HCT116 were purchased from the American Type Culture Collection (ATCC). All cells were maintained in RPMI-1640 culture medium supplemented with 10% fetal bovine serum (FBS) in a humidified cell incubator with an atmosphere of 5% CO2 at 37°C.
Transfection experiments
miR-205 mimics and negative control (NC) were purchased from GenePharma (China). Small interfering RNA against CREB1 (si-CREB1) and negative control (si-NC) were synthesized by RiboBio (China). The sequences of the si-CREB1 were: sense 5’-AUUGUUAGCCAGCUGUAUUG-3’, antisense 5’-CAAUACAGCUGGCUAACAAU-3’ Transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Transfection efficiency was monitored by qRT-PCR.
MTT assay
Cell proliferation was measured by MTT assay. Cells were seeded in 96-well culture plates at a density of 5 × 103 cells/well. Transfection was performed the next day at the concentration described above. Cell proliferation was analyzed at 24, 48, 72 and 96 h, and the optical absorbance was read at 490 nm by EnSpire Multimode Plate Reader (PerkinElmer).
Transwell assays
For transwell migration assay, 5 × 104 cells were plated in the top chamber of each insert (BD Biosciences) with a non-coated membrane. For invasion assay, 1 × 105 cells were added to the upper chamber with 100 µg Matrigel (BD Biosciences). For both assay types, 800 µl of medium supplemented with 10% FBS was injected into the lower chambers. After harvest, the inserts were fixed and stained in a dye solution containing 0.1% crystal violet and 20% methanol. Cells adhering to the lower membrane of the inserts were imaged with microscope (Olympus).
Luciferase reporter assay
The 3’-untranslated region (3’UTR) sequence of CREB1 predicted to interact with miR-205 or a mutated sequence within the predicted target sites was synthesized and inserted into the XbaI and FseI sites of the pGL3 control vector (Promega). These constructs were named as Wt CREB1-3’UTR or Mut CREB1-3’UTR, respectively. For the reporter assay, HEK293 cells were seeded into 24-well plates and transfected with the above constructs and miR-205 mimics or negative control. After 48 h, the cells were harvested and Renilla luciferase activity was measured using the Dual luciferase reporter assay (Promega) according to the manufacturer’s instructions.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from CRC tissues and cell lines using an mirVana™ miRNA Isolation Kit (Applied Biosystems) according to the manufacturer’s instructions. The expression level of miR-205 in CRC tissues and cell lines was measured by qRT-PCR according to the Taqman MicroRNA Assays protocol (Applied Biosystems) and normalized using U6 small nuclear RNA. The expression level of CREB1 mRNA was measured by qRT-PCR according to the SYBR Green real-time PCR Assay protocol (Applied Biosystems) and GAPDH was used for normalization. All experiments were performed using the 2-ΔΔCt method. Each experiment was performed in triplicate.
Western blot
Cells were lysed using the mammalian protein extraction reagent RIPA (Beyotime) supplemented with a protease inhibitor cocktail (Roche) and phenylmethylsulfonylfluoride (Roche). Equal amount proteins were separated by 10% SDS-PAGE gel and transferred to PVDF membranes (Bio-Rad). Membranes were probed with primary antibodies against CREB1 or GAPDH (Abcam) at 4°C overnight followed by incubation with HRP conjugated secondary antibodies. Protein was detected with Image Acquisition using Image Quant™ LAS 4000 (GE Healthcare Life Sciences).
Statistical analysis
Statistical analyses were performed using SPSS version 18.0. All data are presented as the mean ± SD from at least three independent experiments. Differences between groups were analyzed using Student’s t test or one-way ANOVA analysis. P < 0.05 was considered statistically significant.
Results
miR-205 was decreased in CRC tissues and cell lines
To determine whether miR-205 was involved in the tumorigenesis of CRC, we firstly examined the expression of miR-205 in CRC tissues and cell lines. Among 20 paired CRC tissue samples, miR-205 was significantly downregulated in CRC tissues compared to the adjacent non-tumor tissues (Figure 1A). The expression of miR-205 was further evaluated by qRT-PCR in CRC cell lines and normal colon epithelium cell line FHC. Remarkable downregulation of miR-205 could also be observed in CRC cell lines than that of FHC cell line (Figure 1B). These results indicated that miR-205 might play an important role in CRC progression.
Figure 1.
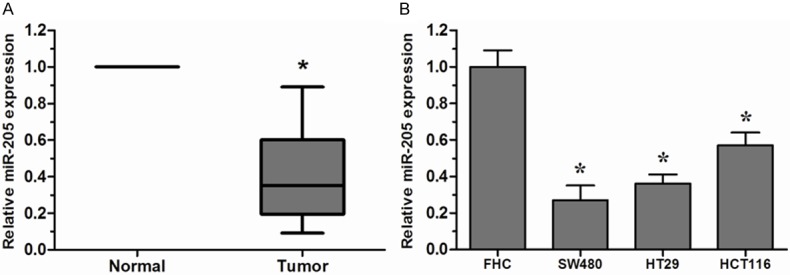
miR-205 was downregulated in CRC tissues and cell lines. A. Expression of miR-205 in 20 paired CRC tissues and their corresponding non-tumor tissues was detected by qRT-PCR. U6 was used as an internal control. B. Expression of miR-205 in three CRC cell lines (SW480, HT29 and HCT116) and normal colon epithelium cell line FHC was detected by qRT-PCR. *P < 0.05.
miR-205 suppressed CRC cell proliferation, migration and invasion
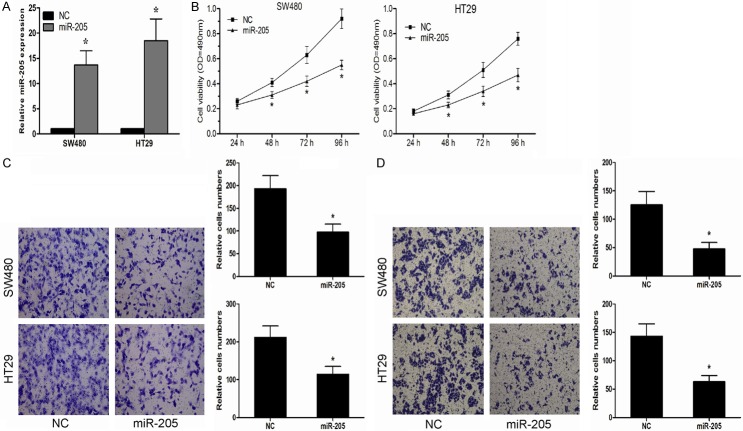
To investigate the function of miR-205 in the CRC progression, SW480 and HT29 cells were transfected with miR-205 mimics. The effect of miR-205 mimics was confirmed by qRT-PCR (Figure 2A). Ectopic overexpression of miR-205 significantly suppressed proliferation of SW480 and HT29 cells (Figure 2B). Furthermore, we studied the effects of miR-205 on CRC cell migration and invasion, our data showed that enforced expression of miR-205 significantly inhibited in vitro migration and invasion capabilities of SW480 and HT29 cells (Figure 2C, 2D).
Figure 2.
miR-205 inhibited CRC cell proliferation, migration and invasion in vitro. A. Expression of miR-205 in SW480 and HT29 cells transfected with miR-205 mimics or negative control (NC) was detected by qRT-PCR. B. miR-205 inhibited SW480 and HT29 cell proliferation ability. C. miR-205 decreased the migration ability of SW480 and HT29 cells. D. miR-205 suppressed the invasion ability of of SW480 and HT29 cells. *P < 0.05.
CREB1 is identified as a functional target of miR-205 in CRC
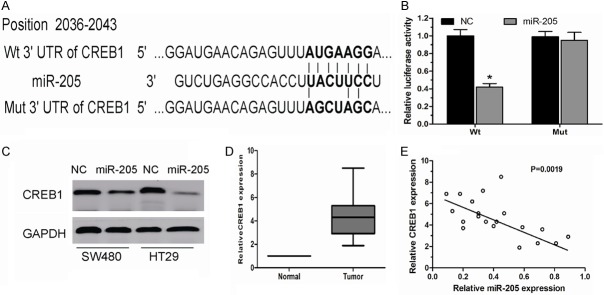
To elucidate the underlying molecular mechanisms through which miR-205 executes its function, TargetScan 6.2 was used to screen the target of miR-205. CREB1, which is known to act as an oncogene and play critical roles in carcinogenesis and cancer progression [10] was predicted as one of the targets (Figure 3A). Luciferase activity assay showed that miR-205 significantly inhibited the wild-type (Wt) but not the Mutant (Mut) 3’UTR of the CREB1 gene luciferase activity in HEK293 cells (Figure 3B). Moreover, western blot analysis further demonstrated that miR-205 overexpression significantly suppressed the expression of CREB1 at the protein levels in SW480 and HT29 cells (Figure 3C). We further examined the expression of CREB1 in the 20 cases of CRC tissues used before. Our results showed that CREB1 was significantly increased and strongly inverse correlated with miR-205 expression in CRC tissues (Figure 3D, 3E).
Figure 3.
miR-205 negatively regulated CREB1 by binding to the CREB1 3’UTR. A. The potential miR-205 binding sequence of CREB1 3’UTR and the mutant. B. A luciferase reporter assay showed the inhibitory effect of miR-205 on CREB1 3’UTR luciferase activity in HEK293 cells. C. CREB1 protein levels were analyzed by Western blot in SW480 and HT29 cells transfected with miR-205 mimics or negative control (NC). D. Relative expression levels of CREB1 in CRC tissues and adjacent non-tumor tissues. E. Correlation of CREB1 expression to miR-205 expression in 20 CRC tissues using simple linear regression analysis. P < 0.05.
CREB1 knockdown inhibited CRC cell proliferation, migration and invasion
In order to explore the biological functions of CREB1, si-CREB1 was used to knockdown the expression of CREB1 and to evaluate its function in SW480 cells. Western blot was used to determinate the expression of CREB1 (Figure 4A). MTT assay showed that CREB1 knockdown dramatically suppressed CRC cell proliferation in SW480 cells (Figure 4B). Moreover, transwell assays showed that CREB1 downregulation inhibited the migration and invasion capacities of SW480 cells (Figure 4C). These results were similar to the inhibitory effects on CRC cells observed with miR-205 mimics. Taken together, these data indicated that CREB1 was a direct target of miR-205 in CRC.
Figure 4.
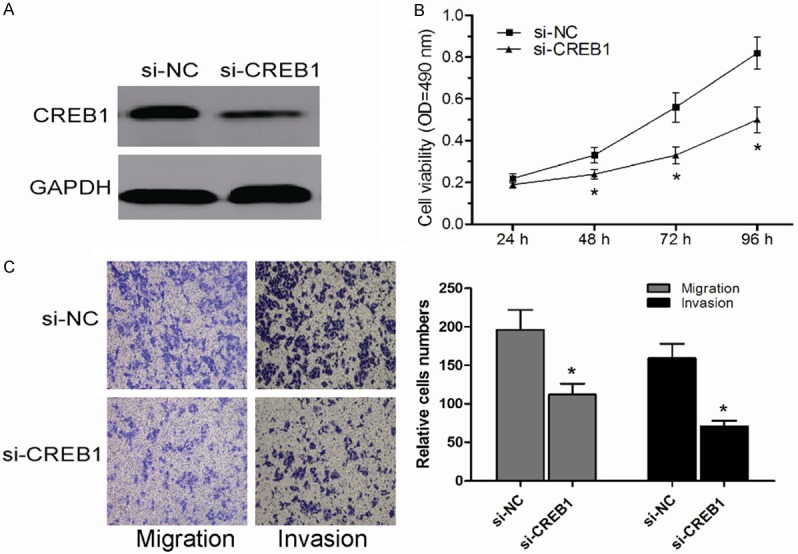
Knockdown of CREB1 inhibited CRC cell proliferation, migration and invasion in vitro. A. Expression of CREB1 in SW480 cells transfected with si-CREB1 or si-NC was detected by Western blot. B. Transfection of si-CREB1 inhibited SW480 cell proliferation ability. C. Transfection of si-CREB1 suppressed SW480 cell migration and invasion ability. *P < 0.05.
Discussion
Emerging evidence has shown that miRNAs could act as a novel class of oncogenic or tumor suppressive genes [11]. Recently, multiple of studies revealed that the aberrant expression of miRNAs in a wide range of human cancers, and the aberrant expression of miRNAs contributed to carcinogenesis by inhibiting the expression of their target genes [12]. Here, we found that miR-205 was downregulated in CRC tissues and cell lines, consistent with the report of Orang et al [13]. Ectopic expression of miR-205 significantly inhibited CRC cell proliferation, migration and invasion. Furthermore, we identified that CREB1 was a direct target of miR-205, and decreased expression of CREB1 have the similar effects of miR-205 restoration on proliferation, migration and invasion of CRC cells. These results demonstrated that miR-205 could act as a novel potential therapeutic strategy for CRC.
miR-205 has been found to be downregulated in many types of cancer. For example, Hagman et al showed that miR-205 were significantly decreased in prostate cancer patients with metastases [14]. Orang et al showed miR-205 was significantly downregulated in CRC samples and decreased miR-205 correlated with lymphatic metastasis [13]. Kim et al found that miR-205 might act as a tumor suppressor in KB human oral cancer cell via inhibiting Axin-2 expression [15]. Elgamal et al reported that in breast cancer cells miR-205 was significantly decreased, and overexpression of miR-205 inhibited breast cancer cell proliferation and invasion via inhibiting the expression of HMGB3 [16].
The cAMP responsive element binding protein 1 (CREB1) is a proto-oncogenic transcription factor and involves in oncogenesis of many organs. As a potent oncogene, through gene amplification, chromosome translocation, interaction with viral oncoproteins, and inactivation of tumor suppressor genes, CREB1 promotes tumorigenesis by significantly impacting on growth, proliferation, survival, metastasis and invasion of tumor cells [17,18]. For example, Peng et al reported that CREB1 was high expressed in glioma tissues, and ectopic expression of CREB1 attenuated the growth suppressive phenotypes of glioma cells caused by miR-200b [19]. Kong et al showed that oncogenic CREB1 was increased in gastric cancer and miR-182 targeted the CREB1 gene and suppresses gastric adenocarcinoma cell growth [20]. Yang et al showed that miR-433 inhibition of expression of the CREB1 repressed hepatocellular carcinoma cell migration [21]. In our study, we found that silencing CREB1 expression had similar effects of miR-205 restoration on proliferation, migration and invasion in CRC cells.
In conclusion, our data revealed that miR-205 was dramatically downregulated in CRC tissues and cell lines, and enforced expression of miR-205 could suppress CRC cell proliferation, migration and invasion. Moreover, we found that the tumor suppressor function of miR-205 was mediated by the downregulation of its downstream target gene CREB1. These results suggested that miR-205 deregulation might play important roles in tumor progression and miR-205 could be a potential therapeutic strategy for the treatment of CRC.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Benatti P, Gafa R, Barana D, Marino M, Scarselli A, Pedroni M, Maestri I, Guerzoni L, Roncucci L, Menigatti M, Roncari B, Maffei S, Rossi G, Ponti G, Santini A, Losi L, Di Gregorio C, Oliani C, Ponz de Leon M, Lanza G. Microsatellite instability and colorectal cancer prognosis. Clin Cancer Res. 2005;11:8332–8340. doi: 10.1158/1078-0432.CCR-05-1030. [DOI] [PubMed] [Google Scholar]
- 3.Folprecht G, Grothey A, Alberts S, Raab HR, Kohne CH. Neoadjuvant treatment of unresectable colorectal liver metastases: correlation between tumour response and resection rates. Ann Oncol. 2005;16:1311–1319. doi: 10.1093/annonc/mdi246. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 7.Yang FQ, Zhang HM, Chen SJ, Yan Y, Zheng JH. MiR-506 Is Down-Regulated in Clear Cell Renal Cell Carcinoma and Inhibits Cell Growth and Metastasis via Targeting FLOT1. PLoS One. 2015;10:e0120258. doi: 10.1371/journal.pone.0120258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun XH, Geng XL, Zhang J, Zhang C. miRNA-646 suppresses osteosarcoma cell metastasis by downregulating fibroblast growth factor 2 (FGF2) Tumour Biol. 2015;36:2127–2134. doi: 10.1007/s13277-014-2822-z. [DOI] [PubMed] [Google Scholar]
- 9.Xu Q, Li P, Chen X, Zong L, Jiang Z, Nan L, Lei J, Duan W, Zhang D, Li X, Sha H, Wu Z, Ma Q, Wang Z. miR-221/222 induces pancreatic cancer progression through the regulation of matrix metalloproteinases. Oncotarget. 2015;6:14153–14164. doi: 10.18632/oncotarget.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang YW, Chen X, Gao JW, Zhang H, Ma RR, Gao ZH, Gao P. High expression of cAMP-responsive element-binding protein 1 (CREB1) is associated with metastasis, tumor stage and poor outcome in gastric cancer. Oncotarget. 2015;6:10646–10657. doi: 10.18632/oncotarget.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 12.Zimmerman AL, Wu S. MicroRNAs, cancer and cancer stem cells. Cancer Lett. 2011;300:10–19. doi: 10.1016/j.canlet.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orang AV, Safaralizadeh R, Hosseinpour Feizi MA, Somi MH. Diagnostic and prognostic value of miR-205 in colorectal cancer. Asian Pac J Cancer Prev. 2014;15:4033–4037. doi: 10.7314/apjcp.2014.15.9.4033. [DOI] [PubMed] [Google Scholar]
- 14.Hagman Z, Haflidadottir BS, Ceder JA, Larne O, Bjartell A, Lilja H, Edsjo A, Ceder Y. miR-205 negatively regulates the androgen receptor and is associated with adverse outcome of prostate cancer patients. Br J Cancer. 2013;108:1668–1676. doi: 10.1038/bjc.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JS, Park SY, Lee SA, Park MG, Yu SK, Lee MH, Park MR, Kim SG, Oh JS, Lee SY, Kim CS, Kim HJ, Chun HS, Moon SM, Kim do K. MicroRNA-205 suppresses the oral carcinoma oncogenic activity via down-regulation of Axin-2 in KB human oral cancer cell. Mol Cell Biochem. 2014;387:71–79. doi: 10.1007/s11010-013-1872-7. [DOI] [PubMed] [Google Scholar]
- 16.Elgamal OA, Park JK, Gusev Y, Azevedo-Pouly AC, Jiang J, Roopra A, Schmittgen TD. Tumor suppressive function of mir-205 in breast cancer is linked to HMGB3 regulation. PLoS One. 2013;8:e76402. doi: 10.1371/journal.pone.0076402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conkright MD, Montminy M. CREB: the unindicted cancer co-conspirator. Trends Cell Biol. 2005;15:457–459. doi: 10.1016/j.tcb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Klemm DJ, Watson PA, Frid MG, Dempsey EC, Schaack J, Colton LA, Nesterova A, Stenmark KR, Reusch JE. cAMP response element-binding protein content is a molecular determinant of smooth muscle cell proliferation and migration. J Biol Chem. 2001;276:46132–46141. doi: 10.1074/jbc.M104769200. [DOI] [PubMed] [Google Scholar]
- 19.Peng B, Hu S, Jun Q, Luo D, Zhang X, Zhao H, Li D. MicroRNA-200b targets CREB1 and suppresses cell growth in human malignant glioma. Mol Cell Biochem. 2013;379:51–58. doi: 10.1007/s11010-013-1626-6. [DOI] [PubMed] [Google Scholar]
- 20.Kong WQ, Bai R, Liu T, Cai CL, Liu M, Li X, Tang H. MicroRNA-182 targets cAMP-responsive element-binding protein 1 and suppresses cell growth in human gastric adenocarcinoma. FEBS J. 2012;279:1252–1260. doi: 10.1111/j.1742-4658.2012.08519.x. [DOI] [PubMed] [Google Scholar]
- 21.Yang Z, Tsuchiya H, Zhang Y, Hartnett ME, Wang L. MicroRNA-433 inhibits liver cancer cell migration by repressing the protein expression and function of cAMP response element-binding protein. J Biol Chem. 2013;288:28893–28899. doi: 10.1074/jbc.M113.502682. [DOI] [PMC free article] [PubMed] [Google Scholar]