Abstract
Although microRNAs (miRNAs) play an important role in esophageal squamous cell carcinoma (ESCC), their roles in radiotherapy response remain unexplored. Our study aims to investigate whether plasma miRNAs can be used as predictors of radiotherapy outcome in ESCC patients. We selected nine miRNAs, which were reported to be associated with carcinogenesis or radiobiology of ESCC, as the targets of our study. Plasma miRNA expression was investigated with 24 subjects (pre-, at the first week and post-radiotherapy). Tumor radiographic response and 3-year overall survival were used to evaluate the response. The results showed that the level of miR-16 in the patients with good outcome was significantly higher than that in the patients with poor outcome (P < 0.05, AUC: 0.762) at the post-radiotherapy. We also found that the variation tendency of miRNA expression were related to its radiotherapy response. Moreover, miR-16 levels increased by more than 2-fold following treatment, which were shown to be associated with longer OS (log-rank P = 0.009). In conclusion, miR-16 has considerable clinical value in predicting radiotherapy outcomes for ESCC patients.
Keywords: Esophageal squamous cell carcinoma, microRNAs, plasma, radiotherapy
Introduction
Esophageal carcinoma is the seventh most common malignant tumor in the world, the mortality of which occupies the sixth position among the malignant tumors worldwide [1], and its incidence rate has increased significantly in recent years [2], especially in China. The incidence and treatment rates of esophageal cancer will continue to increase substantially with the development of an aging population and the social economic development. As the dominant type of esophageal cancer in China, esophageal squamous cell carcinoma (ESCC) has a generally poor prognosis due to the lack of effective clinical methods for its response detection.
At present, radiotherapy is the main treatment for esophageal cancer and it is important for local tumor control. Radiotherapy combined with chemotherapy is the standard treatment model for locally advanced disease. A complete pathological response in patients with neoadjuvant chemoradiation treatment is superior to chemotherapy treatment alone [3]. Although the remaining treated patients can gain potential benefits from radiotherapy, they are subject to the risk of toxicity, complications after radiotherapy, and being delayed to other treatment options [4]. Consequently, the identification of biomarkers and molecular mechanisms of radioresistance may be critical to the stratification of treatment, which can enhance the efficacy of radiotherapy.
Radiotherapy induces a complex cellular response involving multiple pathways [5]. Apparently, microRNA (miRNA) must be involved in the regulation of radiation biology, such as cell cycle and DNA repair [6,7]. It has been demonstrated in a number of cancers that miRNA plays a role in modulating the cellular response to radiation [8]. Although numerous studies have confirmed a role for miRNA in tumorigenesis, progression, and abnormal apoptosis of esophageal cancer [9,10], the role of miRNA in the response to radiotherapy in ESCC is unknown.
MiRNAs are short, non-coding, endogenously expressed RNAs that play a key role in gene expression regulation by binding to target messenger RNAs and either degrading or blocking their translation. Their aberrant expression is common in malignancy and may be involved in disease initiation and progression [11]. As the key regulators of post-transcriptional regulation, miRNAs are associated with the cellular response to radiotherapy [12,13]. In addition, recent studies have shown the existence of a large amount of highly stable miRNAs in human serum/plasma and it was found that circulating miRNA levels are altered in cancer and other diseases, although the origin and function of miRNAs in the blood remains poorly defined [14,15].
Our study investigated plasma miRNAs to predict the outcome of radiotherapy in ESCC patients. In this paper, we presented the successful discovery of a circulating miRNA in discriminating the good outcome from the poor outcome in ESCC patients receiving radiotherapy, and the correlation between the dynamic changes of miRNA and the patient’ survival rate. This miRNA has potential clinical value on evaluating radiotherapy responses in ESCC patients.
Materials and methods
Clinical specimens and radiation treatment plan
The study population consisted of 24 ESCC patients who were recruited at Shanghai Sixth Hospitals from August 2009 to June 2013. The eligibility criteria for the selection subjects were as follows: 1. Age ≥ 18 years and ≤ 85 years; 2. Has ability to give informed consent; 3. Diagnosis must be supported by tissue and two image reports (ultrasound B, computed tomography or magnetic resonance imaging). For selection of the samples in the ESCC patients, we used routine histological classification according to the World Health Organization Classification of tumor system [16]. All the lesions were diagnosed by two experienced pathologists and staged using TNM classification [17]. The 7th edition of American Joint Committee on Cancer (AJCC) staging system for esophageal cancer was used to evaluate the clinical staging of the diseases. The blood samples of the cancer patients were obtained from the three different radiation treatment periods. This study was approved by the local institutional review board and the written informed consent was obtained from all study participants.
The therapeutic protocol for all the patients was about 32 sessions of radiotherapy, respectively. The patients were irradiated with a 6 MV X-ray linear accelerator (Artiste™ Solutions; Siemens, Germany). The mean dose of radiation was (60.2 ± 9.1) Gy, at 2 Gy per day administered 5 days per week. From the included patients, we collected blood from 24 patients in various stages of therapeutic intervention: before radiotherapy (ESCC-0), radiotherapy for a week (ESCC-1), and one day after the end of radiotherapy (ESCC-2).
Follow-up and evaluation of radiotherapy response
The patients were followed every 3 months after the radiotherapy. The clinical end point of this study was death or the end of the study period with a median follow-up period of 23 months (range: 4-40 months). Overall survival (OS) was defined as the period from the end of radiotherapy to death. All data including physical examination and computed tomography findings were collected from hospital records or by patient interviews.
Tumor radiographic responses were assessed from computed tomography scans both in the beginning and at the end of radiotherapy. The best response rate was defined as the greatest reduction in tumor size (%) in comparison with the intial size; otherwise, it would be based on tumor size stability or the least progression (%). Complete response (CR) was defined as the disappearance of lesions. Partial response (PR) was defined as a 30% or greater reduction of the overall total sum of lesion diameter. Progressive disease (PD) was defined as an increase of at least 20% in the sum of the lesion diameter. Stable disease (SD) was defined as a tumor response not fulfilling the criteria for CR, PR and PD. When considering the radiologic responses based on the RECIST criteria version 1.1 and clinical response: no patient had a PD, 12.5% of patients (3/24) had a CR, 45.8% (11/24) had a PR, and 41.7% (10/24) had a SD.
The determination of candidate miRNAs
A large number of studies have confirmed that miRNA plays an important role in tumor incidence, outcome, and biological behavior of tumor radiation. Therefore, we selected nine miRNAs from the literature published for this research (miR-16, miR-21, miR-22, miR-126, miR-148b, miR-185, miR-221, miR-223 and miR-375). Functions of the above mentioned candidate miRNAs are summarized in Table 1. The ultimately selected miRNAs were then further analyzed by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR). By comparing the differences between the miRNA expression and the outcome of radiotherapy, we hoped to find biomarkers which can be used in radiotherapy response evaluation.
Table 1.
The functions and clinical value of nine candidate miRNAs. According to the published literature, we summarized the nine candidate miRNAs including the functions, source of study, tumor types, and the clinical value generated from the regulation of miRNA expression
| Candidates | Functions | Source | Types | Up-/down regulation | Effect | References |
|---|---|---|---|---|---|---|
| miR-21 | Functions as a phospho-Akt protein, downstream target of PTEN; Involved in cellular proliferation and invasion. | cell | ESCC | ↑ | radioresistant | [18] |
| serum | ESCC | ↑ | prognosis | [19] | ||
| tissue | ESCC | ↑ | diagnosis | [20,21] | ||
| plasma | ESCC | ↑ | diagnosis | [22] | ||
| miR-16 | Apoptosis; NF-κB subunit IκB, Radio-related signal transduction pathways. | cell | HepG2 | ↑ | radiosensitivity | [23,24] |
| miR-126 | PI3K-Akt pathway. | cell tissue | NSCLC | ↑ | radiosensitivity | [25] |
| miR-223 | Cell cycle; Metastasis. | tissue | ESCC | ↑ | survival | [26] |
| tissue/cell | ESCC | ↑ | diagnostic | [27] | ||
| serum | ESCC | ↑ | diagnostic | [28] | ||
| miR-375 | Inhibits tumour growth and metastasis. | tissue | EAC | ↓ | diagnosis and prognosis | [29] |
| tissue | ESCC | ↓ | prognosis | [30] | ||
| plasma | ESCC | ↓ | diagnosis | [22] | ||
| miR-22 | Migration and invasion; Regulates hypoxia signaling; Activation of PTEN. | serum | ESCC | ↑ | diagnostic | [28] |
| cell/tissue | NSCLC | ↓ | radiosensitivity | [25] | ||
| miR-148b | Inhibition of migration and invasion; Promoting radiation-induced apoptosis. | serum | ESCC | ↑ | diagnostic | [28] |
| cell | NHL | ↑ | radiosensitivity | [31] | ||
| miR-185 | HIF-1 may bind to HRE2 of miR185 and initiate its transcription. | cell | PC | ↑ | diagnostic | [32] |
| miR-221 | Involved in PTEN/AKT pathway; Radio-related signal transduction pathways. | cell | HEK 293 | ↓ | radiosensitivity | [33] |
| cell | GC | ↓ | radiosensitivity | [34] |
Abbreviations: ESCC: esophageal squamous cell carcinoma; HepG2: liver hepatocellular cells; NSCLC: non-small cell carcinoma; EAC: esophageal adenocarcinoma; NHL: non-Hodgkin’s Lymphoma; PC: pancreatic cancer; HEK 293: human embryonic kidney cell line; GC: gastric carcinoma.
Plasma preparation and RNA isolation
For plasma preparation, 4 ml peripheral blood was drawn into EDTA tubes. Within 1 hour, the tubes were centrifuged at 820 g for 10 min. Next, 1 ml aliquots of the plasma were transferred to 1.5 ml tubes and centrifuged at 16,000 g for 10 min to pellet any remaining cellular debris. Subsequently, the supernatant was transferred to fresh tubes and stored at -80°C. Total RNA of the plasma samples was extracted by mirVana PARIS miRNA Isolation kit according to the instructions from the manufacturer (Ambion, Austin, TX). The concentration was quantified by NanoDrop 1000 Spectrophotometer (NanoDrop Technologies, Waltham, MA). The mean concentration of triplicate measurements for each of the RNA samples was used to calculate the input total RNA for qRT-PCR [35].
qRT-PCR
For testing of candidate miRNA, qRT-PCR was performed using Taqman MicroRNA Assays (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. qRT-PCR was performed in a total reaction volume of 10 µl for relative quantification with an ABI 7900HT fast system (Applied Biosystems, Foster City, CA). The cycle threshold (Ct) was calculated using the second derivative method in the ABI software. The Taqman MicroRNA Assays were performed on 72 plasma samples for nine candidate miRNAs. The expression level of miR-1228 was used as an endogenous control. All assays were carried out in triplicate. A RNA sample was discarded for further analysis if the endogenous control miR-1228 or target miRNA showed Ct values above 35 cycles.
Statistical analysis
For qRT-PCR, ΔCt value was used to represent individual miRNA expression levels. The ΔCt of the miRNAs was calculated by subtracting the Ct value of the endogenous control miR-1228. Mann-Whitney unpaired test was used to determine the difference of miRNA expression level between the various groups. The predicted probability of being diagnosed with ESCC was used as a surrogate marker to construct receiver operating characteristic (ROC) curve [36]. Area under the ROC curve (AUC) was used as an accuracy index for evaluating the diagnostic performance of the selected miRNA [37]. OS was calculated as the time from the last date of radiotherapy to the date of death from any cause or to the last visit before Sep 30, 2014. Each variable, such as age, differentiation, metastasis, the dynamic changes of miRNA levels, etc., was assessed in a univariate analysis, and the variables that reached a P value < 0.05 were evaluated in a multivariate analysis. Survival curves were plotted using the Kaplan-Meier method and used the log-rank test. After testing for variable interactions, a forward stepwise elimination procedure was used to determine the best-fitting model using the proportional hazards model using Cox regression. P values < 0.05 were regarded as statistically significant. All statistical analyses were performed using MedCalc (10.4.7.0) software (Mariakerke, Belgium).
Results
Patient characteristics
The clinical characteristics of the studied subjects were presented in Table 2. Of the 24 patients, 54% (13/24) had non-metastatic disease (as evidenced by histologic analysis of surgically excised tumors), while the remaining 46% (11/24) patients had vascular metastasis, lymph node metastasis, or other distant metastasis. Tumor location and differentiation were distributed more evenly. In clinical tumor staging, I-II stage and III-IV stage accounted for 54.2% (13/24) and 45.8% (11/24) respectively.
Table 2.
Summary of clinic features of ESCC patients in the study
| Characteristics | Patients (n = 24) No. (%) |
|---|---|
| Sex | |
| Male | 18 (75%) |
| Female | 6 (25%) |
| Age -- year | |
| Mean ± SD | 68 ± 12.3 |
| Tumor location | |
| Upper | 5 (20.8%) |
| Middle | 10 (41.7%) |
| Lower | 9 (37.5%) |
| Differentiation | |
| Well | 8 (33.3%) |
| Moderate | 7 (29.2%) |
| Poor | 9 (37.5%) |
| TNM stage | |
| I | 6 (25%) |
| II | 7 (29.2%) |
| III | 6 (25%) |
| IV | 5 (20.8%) |
| Metastasis | |
| Presence | 11 (45.8%) |
| Absence | 13 (54.2%) |
| Chemo/Radiotherapy | |
| Yes | 11 (45.8%) |
| No | 13 (54.2%) |
| Surgery | |
| Yes | 6 (25%) |
| No | 18 (75%) |
Comparison of miRNA expression of ESCC in different periods of radiotherapy
Each patient’s blood samples were obtained before the start of radiotherapy, at the first week and at the end of radiotherapy, which was defined as the three subgroups (ESCC-0, ESCC-1 and ESCC-2). The expression profiles of the nine candidate miRNAs were evaluated with qRT-PCR on the 24 patients according to the three subgroups. According to the above criteria of short term tumor radiographic response evaluation, patients were divided into two groups at the end of the radiotherapy, the good outcome group and the poor outcome group. The good outcome group included CR and PR, while the poor outcome group included SD. We were surprised to find that the expression levels of all candidate miRNAs increased during the course of radiotherapy in the good outcome group (Figure 1A), while in the poor outcome group, the expression levels of the same miRNAs increased temporarily in the first week of radiotherapy but decreased quickly at the end of radiotherapy (Figure 1B). By comparing the miRNA expression levels of each subgroup in the good outcome group, we found that the expression levels of miR-16, miR-21, miR-126 and miR-185 significantly increased in the ESCC-2 subgroup, greater than that in ESCC-0 subgroup (P < 0.05), especially for miR-16 (P < 0.01) (Figure 1).
Figure 1.
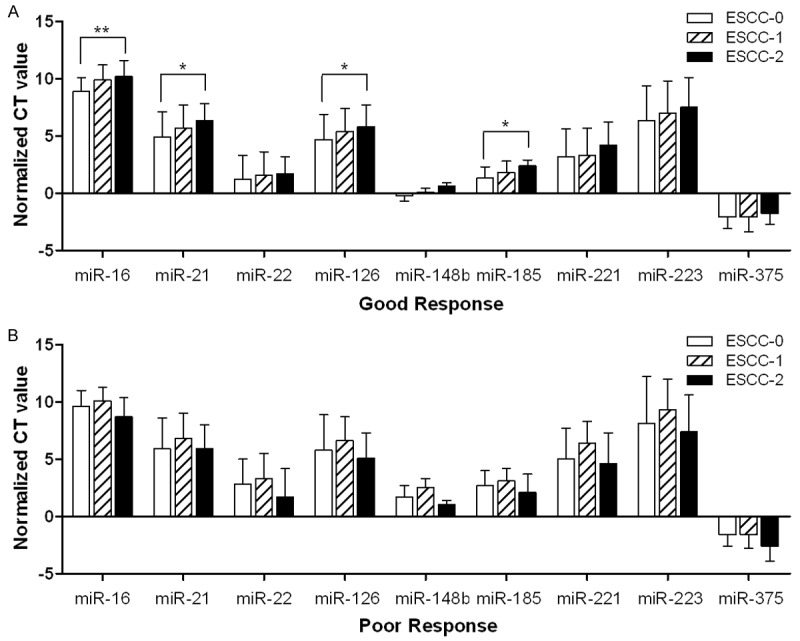
Changes tendency of candidate microRNA expression in different periods of radiotherapy treatment. A: The change tendency of miR-16, miR-21, miR-22, miR-126, miR-148b, miR-185, miR-221, miR-223 and miR-375 in the course of radiotherapy in good response group, respectively; B: The change tendency of the nine candidate targets in the course of radiotherapy in poor response group, respectively; Good response: complete response and partial response; Poor response: stable disease and progressive disease; ΔCt value was used to represent individual normalized Ct value. The ΔCt of the miRNA was calculated by subtracting the Ct value of the endogenous control miR-1228. ESCC-0, ESCC-1 and ESCC-2 represent three different periods of treatment (before, the first week, and the end of radiotherapy), respectively. Analysis was performed by Mann-Whitney unpaired test; *P < 0.05; **P < 0.01.
Differentially expression level of miRNA possibly predicting radiotherapy response
We further analyzed whether the plasma miRNA expression levels were associated with prognosis. In each subgroup, the level of miRNAs was divided into the good and poor outcome groups. The analysis showed that the expression levels of the nine candidate miRNAs had no significant differences between ESCC-0 and ESCC-1 subgroups. However, the results showed that miR-16 had a significantly higher expression level in the good outcome group than that in the poor outcome group (P = 0.038, Fold change = 2.85) in ESCC-2 subgroups. The diagnostic accuracy of miR-16, measured by AUC, was 0.762 (95% CI, 0.540 to 0.913, Sensitivity = 92.9%, Specificity = 55.6%) (Figure 2). With the exception of miR-16, other candidates did not show any significant difference in expression level between groups (Fold change = 1.43-3.00, AUC = 0.548-0.714). The results were shown in Table 3.
Figure 2.

ROC curve analysis of the candidate miRNAs in discriminating the good response patients from the poor response group. AUC estimation for the plasma miR-16 (A), miR-21 (B), miR-126 (C) and miR-185 (D) in discriminating the good response patients from the poor response group.
Table 3.
Evaluation the correlation between the miRNA levels and the treatment outcome (good response group vs. poor response group)
| P-value | Fold change | AUC | 95% CI | |
|---|---|---|---|---|
| miR-16 | 0.038* | 2.85 | 0.762 | 0.540-0.913 |
| miR-21 | 0.102 | 2.36 | 0.706 | 0.482-0.875 |
| miR-126 | 0.089 | 3.00 | 0.714 | 0.490-0.881 |
| miR-223 | 0.284 | 2.23 | 0.635 | 0.411-0.823 |
| miR-375 | 0.284 | 1.43 | 0.635 | 0.411-0.823 |
| miR-22 | 0.529 | 1.56 | 0.579 | 0.358-0.780 |
| miR-148b | 0.706 | 1.55 | 0.548 | 0.329-0.754 |
| miR-185 | 0.231 | 2.12 | 0.651 | 0.426-0.835 |
| miR-221 | 0.614 | 1.76 | 0.563 | 0.343-0.767 |
Good response: complete response and partial response; Poor response: stable disease and progressive disease; AUC: area under the receiver operating characteristic (ROC) curve; Analysis was performed by Mann-Whitney unpaired test;
P < 0.05.
Correlation between the dynamic changes of miR-16 expression and OS in ESCC patients
With a median follow-up of 40 months (range, 4-40 months), the 3-year OS of all the patients was 37%, and the median OS time was 23 months. We performed Kaplan-Meier analyses to assess the relationship between the dynamic changes of plasma miR-16 levels and clinical outcome in 24 ESCC patients. We evaluated the correlation between OS and the up-regulation of miR-16 at various points of radiotherapy. That is to say, we used the fold change of miR-16 up-regulation as the index to evaluate the effect of treatment response. The results showed that the patients whose miR-16 expression level had increased more than 2-fold (fold change: ESCC-2/ESCC-0 > 2) would experience significantly longer OS (HR = 3.875, 95% CI: 1.398-10.737, P = 0.009) (Figure 3).
Figure 3.
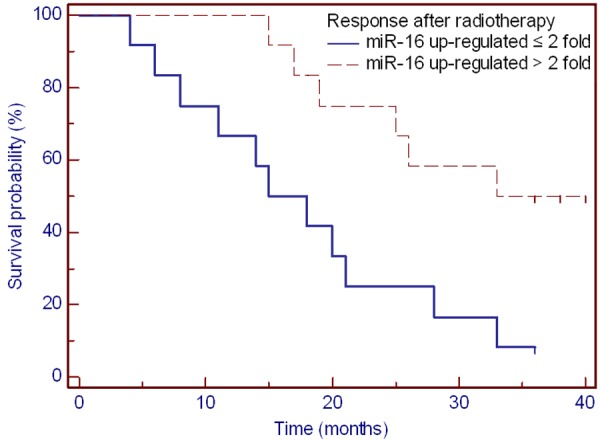
Overall survival of patients with miR-16 level up-regulated more than 2-fold after radiotherapy. (log-rank P = 0.009).
Correlation of clinical factors with patient survival
A univariate analysis of clinical factors showed that tumor radiographic response after radiotherapy had significant associations with OS (P = 0.040), while other clinical did not show a good correlation with OS (Table 4), such as age, sex, differentiation, clinical stage, lesion length, metastasis, primary tumor location and radiotherapy dose. Clinical factors that were statistically significant (P < 0.05) in univariate analysis were analyzed further in a multivariate analysis with a stepwise selection of variables. Only patients who had tumor radiographic response after radiotherapy (P = 0.040) and ESCC-2/ESCC-0 up-regulated more than 2-fold (P = 0.009) were selected by a stepwise selection as factors in the final models. A multivariate analysis of these factors showed that miR-16 ESCC2/ESCC0 up-regulated more than 2-fold and tumor radiographic response after radiotherapy maintained their significance as independent prognostic factors for OS (HR = 0.263, 95% CI: 0.093-0.745, P = 0.012; and HR = 0.344, 95% CI: 0.125-0.947, P = 0.039, respectively).
Table 4.
Univariate analysis of prognostic factors of overall survival (n = 24)
| Variable | HR† | 95% CI | P value |
|---|---|---|---|
| Age (> 60 y vs. ≤ 60 y) | 0.960 | 0.361-2.557 | 0.936 |
| Sex (male vs. female) | 1.338 | 0.422-4.241 | 0.621 |
| Differentiation (well vs. moderate vs. poor) | 1.7753 | 0.527-5.976 | 0.354 |
| Clinical stage (I-II vs. III-IV) | 1.328 | 0.502-3.513 | 0.568 |
| Lesion length (> 6 cm vs. ≤ 6 cm) | 0.928 | 0.289-2.986 | 0.901 |
| Metastasis (yes vs. no) | 1.562 | 0.583-4.187 | 0.375 |
| Primary tumor location (Upper vs. Middle vs. Lower) | 1.293 | 0.347-4.814 | 0.702 |
| RT dose (> 65 Gy vs. ≤ 65 Gy) | 2.081 | 0.767-5.642 | 0.150 |
| Response after radiotherapy (Good response vs. Poor response)‡ | 3.203 | 1.052-9.751 | 0.040* |
HR: Hazard ratios;
Radiographic assessment when the end of radiotherapy. Good response: complete response and partial response; Poor response: stable disease and progressive disease;
P < 0.05.
Discussion
ESCC is a common cancer of the digestive system. The treatment of locally advanced esophageal carcinoma remains a great challenge. Radiotherapy is a good move for those patients who are unwilling to undergo surgery [38]. However, the prognosis for such patients is very poor, with 5-year survival rates of about 20% [39]. Radiosensitivity is thought to be a key factor that affects therapeutic efficacy. However, biomarkers for radiosensitivity are not yet available in the clinic. Identification of biomarkers for predicting radiation sensitization could provide a useful indicator for individual-specific radiotherapy of patients with esophageal cancer. Some studies found evidence from the perspective of clinical pathology and imaging diagnosis such as TNM staging system, histological grading, tumor location and tumor length, as well as depth of esophageal wall invasion [40,41]. Other studies explored the possible radiotherapy biomarker in ESCC from the molecular level such as Dicer 1, DNA methyltransferase 1 (Dnmt1), and let-7b, etc. [42,43]. But the prognosis for patients with advanced disease was still poor. We acknowledge that there were several limitations to this study, and it was these limitations which prevented proper prediction of the curative effect.
In our study, we were the first to analyze the dynamic expression of the nine miRNAs in the process of radiotherapy and found that miRNA expression patterns are related to its radiotherapy response. It suggests that the nine candidate genes may be involved in radiation sensitivity and it warrants further study on its mechanism. Moreover, our study reveals that plasma miR-16 is a potential circulating marker for radiotherapy response in ESCC. In the good response group, the results show that miR-16 expression levels increased continuously during the course of therapy, and the expression level in the ESCC-2 subgroups was significantly higher than that in ESCC-0 subgroups. Simultaneously, miR-16 demonstrates high diagnostic accuracy (AUC 0.762, sensitivity 92.9% and specificity 55.6%) in distinguishing the good outcome patients from the poor outcome patients receiving radiotherapy. We have shown that the plasma levels of miR-16 can serve as a prognostic indicator of radiographic response in ESCC patients. We further queried whether the plasma level of miR-16 factored in the patients’ response to different stages of radiotherapy. We divided the patients into three subgroups according to their therapeutic stage (ESCC-0, ESCC-1 and ESCC-2) and analyzed the dynamic changes of miR-16 level for each patient in different treatment period. The data showed that the therapeutic response in patients with miR-16 plasma levels up-regulated by more than 2-fold significantly prolonged the OS. Therefore, dynamic up-regulation of miR-16 levels is able to reverse the poor prognosis of ESCC patients with radiotherapy and significantly improve the ESCC patient survival.
MiR-16 has been identified to play a role in multiple pathways by regulating genes involved in different cancer types, including ESCC [44]. Evidence suggests that miR-16 can regulate the cell cycle, proliferation, apoptosis, and tumorigenicity both in vitro and in vivo [45]. These effects can be explained by the analysis of several targets of miR-16 such as Bcl-2 [23], cyclin D1, cyclin D3, cyclin E1, CDK6 and WNT3A [6,46,47]. Moreover, recent studies have confirmed that P-glycoprotein enhances radiation-induced apoptotic cell death through the regulation of miR-16 and Bcl-2 expression in hepatocellular carcinoma cells [23]. The recent research elucidates that miR-16 can enhance radiation sensitivity by regulating the TLR1/NF-κB signaling pathway and acts as a potential therapeutic approach to overcome radioresistance for lung cancer treatment [48]. In summary, miR-16 is a tumor suppressor gene can also be used as an optimal tool for ESCC radiotherapy prognosis.
At the serum or plasma level, more than 100 circulating miRNAs had been identified in the blood of healthy individuals [15] and their profiles significantly differed from that of ESCC patients. Compared to those studies of circulating miRNAs in diagnosing ESCC, very few studies are about the prognostic biomarkers of radiotherapy outcome in ESCC patients [43,49]. Our study is unique for the following reasons: First, we retrieved a large number of correlations between plasma miRNAs and the biology of radiation by publication, which enabled us to have a better opportunity to identify potential diagnostic markers. Furthermore, because ionizing radiation has a direct effect on DNA replication, transcription and protein synthesis, so the miRNAs would be regulated widely in this process. In this study, we establish for the first time that the patterns of miRNA expression changes with the radiation process. In addition, we first confirmed that the miR-16 is a promising marker for ESCC radiotherapy prognosis, and also that dynamic miR-16 up-regulation indicates a longer OS in ESCC patients. MiR-16 levels reflect the satisfactory diagnostic performance in the detection of patients with successful treatment by radiotherapy. Using this miRNA for screening plasma in ESCC in the early stage of radiotherapy would improve the rationality of treatment programs and allow for personalized treatment plans according to the patient’s response.
At the plasma level, several miRNAs were thought to be the best candidate for normalizing reference controls. However, those studies were limited due to a limited number of screened miRNAs, small sample size, an association between the reference control and disease studied, or the lack of validation by other studies [50]. One study investigated global circulating miRNA profiles to identify a stable endogenous control for quantifying circulating miRNAs using three cohorts (n = 544). The results showed miR-1228, as a housekeeping gene, presented as the most stable endogenous control across eight cancer types including the esophagus cancer. The results explained the steady expression of miR-1228 in the blood [50]. In summary, miR-1228 was selected as the stable endogenous control for quantifying circulating miRNAs in this study.
Although it has been reported that the expression levels of circulating miRNAs in the healthy human subjects are reproducibly consistent [14], many individuals in the ESCC group might still introduce a bias in the study outcome. The number of samples was too small to allow for a reasonable stratified analysis in our study. Therefore some clinical factors, such as differentiation, clinical stage, and metastasis, had no statistic difference. In this study, we just adopted both short-term imaging index at the end of radiotherapy and long-term follow-up in order to evaluate the treatment response. In fact, there was a good correlation between the short-term imaging index and the long-term follow-up. In future research, we would increase the number of patients and validate the plasma miRNA panel in in several independent cohorts from multiple medical centers.
In conclusion, our study was the first to show the importance of the miR-16 expression profile in association with radiotherapy response. It demonstrated that miR-16 can be used as a predictive indicator of the efficacy of treatment. ESCC patients with a high expression of miR-16 would be more sensitive to radiotherapy and have better prognosis. Thus, patients who would have otherwise missed the curative treatment window can benefit from the optimal therapy.
Acknowledgements
We would like to thank Prof. Ying Wu, Department of Pathology, Shanghai Medical College, Fudan University, Shanghai, China, for experimental guidance. This article is funded by the research grant (YG2012ZD02, 2JC1407400) from Shanghai Jiao Tong University and the Science and Technology Commission of Shanghai, China and the grant (81272506, 61227017) from National Natural Science Foundation of China.
Disclosure of conflict of interest
None.
References
- 1.Kamangar F, Qiao YL, Schiller JT, Dawsey SM, Fears T, Sun XD, Abnet CC, Zhao P, Taylor PR, Mark SD. Human papillomavirus serology and the risk of esophageal and gastric cancers: results from a cohort in a high-risk region in China. Int J Cancer. 2006;119:579–584. doi: 10.1002/ijc.21871. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Reynolds JV, Muldoon C, Hollywood D, Ravi N, Rowley S, O’Byrne K, Kennedy J, Murphy TJ. Long-term outcomes following neoadjuvant chemoradiotherapy for esophageal cancer. Ann Surg. 2007;245:707–716. doi: 10.1097/01.sla.0000254367.15810.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelsen DP. Multimodality therapy of esophageal cancer: an update. Cancer J. 2000;6(Suppl 2):S177–181. [PubMed] [Google Scholar]
- 5.Yin E, Nelson DO, Coleman MA, Peterson LE, Wyrobek AJ. Gene expression changes in mouse brain after exposure to low-dose ionizing radiation. Int J Radiat Biol. 2003;79:759–775. doi: 10.1080/09553000310001610961. [DOI] [PubMed] [Google Scholar]
- 6.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, Chau N, Cleary M, Jackson AL, Carleton M, Lim L. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69:1221–1229. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Josson S, Sung SY, Lao K, Chung LW, Johnstone PA. Radiation modulation of microRNA in prostate cancer cell lines. Prostate. 2008;68:1599–1606. doi: 10.1002/pros.20827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feber A, Xi L, Luketich JD, Pennathur A, Landreneau RJ, Wu M, Swanson SJ, Godfrey TE, Litle VR. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135:255–260. doi: 10.1016/j.jtcvs.2007.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo Y, Chen Z, Zhang L, Zhou F, Shi S, Feng X, Li B, Meng X, Ma X, Luo M, Shao K, Li N, Qiu B, Mitchelson K, Cheng J, He J. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68:26–33. doi: 10.1158/0008-5472.CAN-06-4418. [DOI] [PubMed] [Google Scholar]
- 11.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 12.Cellini F, Morganti AG, Genovesi D, Silvestris N, Valentini V. Role of microRNA in response to ionizing radiations: evidences and potential impact on clinical practice for radiotherapy. Molecules. 2014;19:5379–5401. doi: 10.3390/molecules19045379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halimi M, Asghari SM, Sariri R, Moslemi D, Parsian H. Cellular Response to Ionizing Radiation: A MicroRNA Story. Int J Mol Cell Med. 2012;1:178–184. [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamilton SR, Aaltonen LA. Pathology and Genetics of Tumours of Digestive System. In: Kleihues P, Sobin LH, editors. World Health Organization Classification of Tumors. Lyon: IARC Press; 2000. pp. 9–30. [Google Scholar]
- 17.Sobin LH, Fleming ID. TNM Classification of Malignant Tumors, fifth edition (1997). Union Internationale Contre le Cancer and the American Joint Committee on Cancer. Cancer. 1997;80:1803–1804. doi: 10.1002/(sici)1097-0142(19971101)80:9<1803::aid-cncr16>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 18.Huang S, Li XQ, Chen X, Che SM, Chen W, Zhang XZ. Inhibition of microRNA-21 increases radiosensitivity of esophageal cancer cells through phosphatase and tensin homolog deleted on chromosome 10 activation. Dis Esophagus. 2013;26:823–831. doi: 10.1111/j.1442-2050.2012.01389.x. [DOI] [PubMed] [Google Scholar]
- 19.Cai EH, Gao YX, Wei ZZ, Chen WY, Yu P, Li K. Serum miR-21 expression in human esophageal squamous cell carcinomas. Asian Pac J Cancer Prev. 2012;13:1563–1567. doi: 10.7314/apjcp.2012.13.4.1563. [DOI] [PubMed] [Google Scholar]
- 20.Fassan M, Realdon S, Pizzi M, Balistreri M, Battaglia G, Zaninotto G, Ancona E, Rugge M. Programmed cell death 4 nuclear loss and miR-21 or activated Akt overexpression in esophageal squamous cell carcinogenesis. Dis Esophagus. 2012;25:263–268. doi: 10.1111/j.1442-2050.2011.01236.x. [DOI] [PubMed] [Google Scholar]
- 21.Ma WJ, Lv GD, Tuersun A, Liu Q, Liu H, Zheng ST, Huang CG, Feng JG, Wang X, Lin RY, Sheyhidin I, Lu XM. Role of microRNA-21 and effect on PTEN in Kazakh’s esophageal squamous cell carcinoma. Mol Biol Rep. 2011;38:3253–3260. doi: 10.1007/s11033-010-0480-9. [DOI] [PubMed] [Google Scholar]
- 22.Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Morimura R, Nagata H, Kosuga T, Iitaka D, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Otsuji E. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2011;105:104–111. doi: 10.1038/bjc.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsang TY, Tang WY, Chan JY, Co NN, Au Yeung CL, Yau PL, Kong SK, Fung KP, Kwok TT. P-glycoprotein enhances radiation-induced apoptotic cell death through the regulation of miR-16 and Bcl-2 expressions in hepatocellular carcinoma cells. Apoptosis. 2011;16:524–535. doi: 10.1007/s10495-011-0581-5. [DOI] [PubMed] [Google Scholar]
- 24.Glazer PM, Le QT, Bristow R, Helleday T, Pelroy R, Bernhard EJ. New translational possibilities for microenvironmental modulation of radiosensitivity. Radiat Res. 2011;176:412–414. doi: 10.1667/rrxx33.1. [DOI] [PubMed] [Google Scholar]
- 25.Wang XC, Du LQ, Tian LL, Wu HL, Jiang XY, Zhang H, Li DG, Wang YY, Wu HY, She Y, Liu QF, Fan FY, Meng AM. Expression and function of miRNA in postoperative radiotherapy sensitive and resistant patients of non-small cell lung cancer. Lung Cancer. 2011;72:92–99. doi: 10.1016/j.lungcan.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 26.Kurashige J, Watanabe M, Iwatsuki M, Kinoshita K, Saito S, Hiyoshi Y, Kamohara H, Baba Y, Mimori K, Baba H. Overexpression of microRNA-223 regulates the ubiquitin ligase FBXW7 in oesophageal squamous cell carcinoma. Br J Cancer. 2012;106:182–188. doi: 10.1038/bjc.2011.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S, Li Z, Guo F, Qin X, Liu B, Lei Z, Song Z, Sun L, Zhang HT, You J, Zhou Q. miR-223 regulates migration and invasion by targeting Artemin in human esophageal carcinoma. J Biomed Sci. 2011;18:24. doi: 10.1186/1423-0127-18-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang C, Wang C, Chen X, Yang C, Li K, Wang J, Dai J, Hu Z, Zhou X, Chen L, Zhang Y, Li Y, Qiu H, Xing J, Liang Z, Ren B, Yang C, Zen K, Zhang CY. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem. 2010;56:1871–1879. doi: 10.1373/clinchem.2010.147553. [DOI] [PubMed] [Google Scholar]
- 29.Leidner RS, Ravi L, Leahy P, Chen Y, Bednarchik B, Streppel M, Canto M, Wang JS, Maitra A, Willis J, Markowitz SD, Barnholtz-Sloan J, Adams MD, Chak A, Guda K. The microRNAs, MiR-31 and MiR-375, as candidate markers in Barrett’s esophageal carcinogenesis. Genes Chromosomes Cancer. 2012;51:473–479. doi: 10.1002/gcc.21934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong KL, Kwong DL, Chan TH, Law SY, Chen L, Li Y, Qin YR, Guan XY. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut. 2012;61:33–42. doi: 10.1136/gutjnl-2011-300178. [DOI] [PubMed] [Google Scholar]
- 31.Wu Y, Liu GL, Liu SH, Wang CX, Xu YL, Ying Y, Mao P. MicroRNA-148b enhances the radiosensitivity of non-Hodgkin’s Lymphoma cells by promoting radiation-induced apoptosis. J Radiat Res. 2012;53:516–525. doi: 10.1093/jrr/rrs002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song Z, Ren H, Gao S, Zhao T, Wang X, Zhang S, Zhao X, Jia L, Sun J, Hao J. The hypoxia-inducible factor-1 regulates the microRNA185 expression through binding to hypoxia response elements sequence 2. Med Oncol. 2013;30:756. doi: 10.1007/s12032-013-0756-8. [DOI] [PubMed] [Google Scholar]
- 33.Zhang C, Kang C, Wang P, Cao Y, Lv Z, Yu S, Wang G, Zhang A, Jia Z, Han L, Yang C, Ishiyama H, Teh BS, Xu B, Pu P. MicroRNA-221 and -222 regulate radiation sensitivity by targeting the PTEN pathway. Int J Radiat Oncol Biol Phys. 2011;80:240–248. doi: 10.1016/j.ijrobp.2010.12.049. [DOI] [PubMed] [Google Scholar]
- 34.Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F, Xiao Y, Guang-Xiu W, Zhi-Fan J, Pei-Yu P, Qing-Yu Z, Chun-Sheng K. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer. 2010;10:367. doi: 10.1186/1471-2407-10-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S, Xiang J, Li Z, Lu S, Hu J, Gao X, Yu L, Wang L, Wang J, Wu Y, Chen Z, Zhu H. A plasma microRNA panel for early detection of colorectal cancer. Int J Cancer. 2015;136:152–161. doi: 10.1002/ijc.28136. [DOI] [PubMed] [Google Scholar]
- 36.Zweig MH, Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem. 1993;39:561–577. [PubMed] [Google Scholar]
- 37.Hanley JA, McNeil BJ. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology. 1983;148:839–843. doi: 10.1148/radiology.148.3.6878708. [DOI] [PubMed] [Google Scholar]
- 38.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 39.Wu KL, Chen GY, Xu ZY, Fu XL, Qian H, Jiang GL. Three-dimensional conformal radiation therapy for squamous cell carcinoma of the esophagus: a prospective phase I/II study. Radiother Oncol. 2009;93:454–457. doi: 10.1016/j.radonc.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 40.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 41.Bolton WD, Hofstetter WL, Francis AM, Correa AM, Ajani JA, Bhutani MS, Erasmus J, Komaki R, Maru DM, Mehran RJ, Rice DC, Roth JA, Vaporciyan AA, Walsh GL, Swisher SG. Impact of tumor length on long-term survival of pT1 esophageal adenocarcinoma. J Thorac Cardiovasc Surg. 2009;138:831–836. doi: 10.1016/j.jtcvs.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 42.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous endjoining. Biochem J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu S, Wang X, Chen JX, Chen Y. Predictive factors for the sensitivity of radiotherapy and prognosis of esophageal squamous cell carcinoma. Int J Radiat Biol. 2014;90:407–413. doi: 10.3109/09553002.2014.894649. [DOI] [PubMed] [Google Scholar]
- 44.Zhu Y, Xia Y, Niu H, Chen Y. MiR-16 induced the suppression of cell apoptosis while promote proliferation in esophageal squamous cell carcinoma. Cell Physiol Biochem. 2014;33:1340–1348. doi: 10.1159/000358701. [DOI] [PubMed] [Google Scholar]
- 45.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17:215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 46.Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391–5404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D’Urso L, Pagliuca A, Biffoni M, Labbaye C, Bartucci M, Muto G, Peschle C, De Maria R. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 48.Lan F, Yue X, Ren G, Li H, Ping L, Wang Y, Xia T. miR-15a/16 enhances radiation sensitivity of non-small cell lung cancer cells by targeting the TLR1/NF-kappaB signaling pathway. Int J Radiat Oncol Biol Phys. 2015;91:73–81. doi: 10.1016/j.ijrobp.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 49.Wang XC, Zhang ZB, Wang YY, Wu HY, Li DG, Meng AM, Fan FY. Increased miRNA-22 expression sensitizes esophageal squamous cell carcinoma to irradiation. J Radiat Res. 2013;54:401–408. doi: 10.1093/jrr/rrs113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu J, Wang Z, Liao BY, Yu L, Gao X, Lu S, Wang S, Dai Z, Zhang X, Chen Q, Qiu SJ, Wu Y, Zhu H, Fan J, Zhou J, Wang J. Human miR-1228 as a stable endogenous control for the quantification of circulating microRNAs in cancer patients. Int J Cancer. 2014;135:1187–1194. doi: 10.1002/ijc.28757. [DOI] [PubMed] [Google Scholar]


