Abstract
Leiomyosarcoma is an aggressive soft tissue sarcoma with poor patient survival. The genetic changes of leiomyosarcoma remain to be discovered. In this study, we analyzed the genetic changes of 44 cancer-related genes by using next-generation sequencing in 54 leiomyosarcomas. We identified TP53 mutations in 19 of the 54 tumors (35%) and ATRX mutations in 9 of the 54 tumors (17%). The TP53-mutated leiomyosarcomas were limited to female patients (P = 0.006). All but 2 of the TP53-mutated leiomyosarcomas were located in the uterus (n = 11) or retroperitoneum (n = 6). The ATRX mutations were associated with poorly differentiated leiomyosarcomas (P = 0.028) and the presence of tumor necrosis (P = 0.015). Kaplan-Meier survival analysis showed that patients with ATRX-mutated leiomyosarcomas had worse overall survival than did patients with ATRX-wild-type leiomyosarcomas. All of the ATRX-mutated leiomyosarcomas showed the alternative lengthening of telomere phenotype. The ATRX mutations did not correlate with ATRX protein expression, as detected using immunohistochemistry. In conclusion, we identified loss of function of the p53 and ATRX pathways being the main mechanisms for leiomyosarcomas. The molecular mechanisms may provide new opportunities to treat these aggressive neoplasms.
Keywords: Leiomyosarcoma, TP53, ATRX, alternative lengthening of telomeres
Introduction
Soft tissue sarcoma can be broadly categorised as sarcomas with simple karyotypes that frequently involve the fusion or mutation of specific genes and sarcomas with complex karyotypic abnormalities that frequently have numerical chromosomal changes, non-recurrent translocations, and deletions [1]. Sarcomas with complex karyotypes account for approximately two-thirds of sarcoma cases. They frequently harbour genetic alterations in the p53 pathway, such as inactivating TP53 mutations, homozygous deletions of CDKN2A, and MDM2 amplifications [2,3]. Other than the p53 pathway, the pathogenesis of sarcomas with complex karyotypes remains largely unknown.
Leiomyosarcoma is a prototypical example of a sarcoma with a complex karyotype. Excluding those of uterine origin, it accounts for 5%-10% of all sarcomas [1]. Leiomyosarcomas are aggressive neoplasms associated with poor patient survival. Leiomyosarcomas are tumors with complex karyotypes without recurrent chromosomal aberrations [4,5]. The current treatment modalities for leiomyosarcomas include surgical debulking, radiotherapy, and chemotherapy regimens that frequently include gemcitabine and docetaxel [6,7]. However, no clear survival benefit has been proven for chemotherapy in metastatic tumors, and most patients eventually die from the disease [8,9]. The outcome is particularly grave for deeply seated, large tumors and tumors associated with the great vessels. Further study of the pathogenesis and cell biology of these tumors is necessary for developing more effective treatment modalities.
The genetic changes of leiomyosarcoma remain to be discovered. TP53 mutation was identified in 16 of 37 extrauterine leiomyosarcomas [10]. A clinical correlation study indicated that tumors with TP53 mutations have a higher histologic grade or stage at presentation [10]. Ito et al. also reported TP53 mutations in 39% of leiomyosarcomas [11]. MED12, a gene commonly mutated in uterine leiomyoma, were found to be mutated in only 3 of 32 uterine leiomyosarcomas [12]. Whether other genes are mutated remains unknown. In this study, we analyzed the genetic changes of 44 cancer-related genes by using next-generation sequencing and identified frequent mutations of TP53 and ATRX in leiomyosarcomas. The clinicopathological significance of mutations in these 2 genes was also explored.
Materials and methods
Tumor samples
A total of 54 cases of leiomyosarcoma from various sites with available formalin-fixed and paraffin-embedded tissue blocks were retrieved from the archives of the Department of Pathology, National Taiwan University Hospital. Histological and immunohistochemical sections were reviewed to confirm the diagnoses. This study was approved by the Research Ethics Committee of National Taiwan University Hospital, and the specimens were anonymous and analyzed in a blind manner.
Immunohistochemistry and telomere-specific fluorescent in situ hybridization
The detailed methods of the immunohistochemistry and telomere-specific fluorescent in situ hybridization were described previously [13]. The immunohistochemical staining was performed as previously described using an ATRX antibody (1:500; Sigma Aldrich, St. Louis, MO, USA). An FITC-labelled PNA probe (Panagene, Daejeon, South Korea) was used for the telomere-specific fluorescent in situ hybridization.
Design of the oligonucleotide probe for capture of cancer-related genes
The capture probe was designed to enrich the open reading frame of 43 cancer-related genes and the TERT promoter (-150 to -350), with the sequence being taken from UCSC hg19, NCBI build 37. A list of the genes analyzed is shown in Table 1. The NimbleGen Sequence Capture design algorithm was used with 50-105 mer probes (Roche NimbleGen, Madison, WI, USA); a total of 2,100,000 capture probes were used for the capture reactions to ensure high-performance captures.
Table 1.
The 44 cancer-related genes included in this study
| AKT1 | CDH1 | H3F3A | MUTYH | TERT promoter |
|---|---|---|---|---|
| ALK | CDKN2A | HIST1H3B | NFE2L2 | TP53 |
| APC | CHEK2 | HRAS | NRAS | TSC2 |
| ARID1A | CTNNB1 | IDH1 | PALB2 | VHL |
| ARID2 | DAXX | IDH2 | PIK3CA | |
| ATM | DICER1 | JAK1 | PRKACA | |
| ATRX | EGFR | KIT | PRKAR1A | |
| BRAF | FGFR3 | KRAS | RNF43 | |
| BRCA1 | GNAQ | MED12 | SMAD4 | |
| BRCA2 | GNAS | MEN1 | STK11 |
Library and capture probe hybridization
Double-stranded DNA extracted from formalin-fixed, paraffin-embedded samples was quantified using a PicoGreen fluorescence assay employing the provided standard (Life Technologies, Carlsbad, CA, USA); 100-1.0 ug of dsDNA in 50 μL of TE Buffer (10 mM Tris-HCl (pH 7.6), 0.1 mM EDTA) was fragmented to 200-550 bp by using sonication. Shotgun libraries were prepared using the KAPA Library Preparation Kit (Kapa Biosystems, Wilmington, MA, USA), which contains mixes for end repair and dA addition and ligation, by performing the ‘with-bead’ protocol to maximise reproducibility and library yield. Indexed (6-bp barcodes) sequencing libraries were amplified using PCR for 9-13 cycles; size selection was performed to remove undesired DNA fragments. A total of 1.0 ug of the shotgun sequencing library, adaptor-specific blocker DNA, and Cot DNA were lyophilised in a 1.5-mL tube and suspended in a hybridization buffer, and heat denatured at 95°C for 10 min before adding the bait-set reagent. The mixture was applied to hybridization-based capture probes for 72 h at 47°C. The hybridised capture probes were then washed using a SeqCap EZ Hybridization and Wash Kits (Roche NimbleGen) at 47°C and at room temperature, respectively. The biotinylated bait was further pulled down using dynabeads M-270 Streptavidin (Life Technologies). The captured DNA was amplified through LM-PCR amplification by using the SeqCap EZ Accessory Kit v2 (Roche NimbleGen), and the PCR products were purified using the QIA Quick Purification Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions.
Quantification and qualification of captured DNA samples
Purified amplification products were eluted in 50 μL of molecular grade water, with the quantity and quality of the products being measured using a PicoGreen fluorescence assay and 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA) with a DNA 1000/7500 chip. Moreover, real-time quantitative PCR assays with 4 control loci were used to estimate the relative fold enrichment and assess the capture success. The reactions were performed using a LightCycler 480 system (Roche NimbleGen) according to the manufacturer’s instructions.
Next-generation sequencing
Post-ligation-mediated PCR products were quantified using PicoGreen dsDNA quantification reagent. The libraries were normalised to 4.0 nM and applied to Illumina MiSeq (Illumina, San Diego, CA, USA), according to the manufacturer’s protocols. On completion of the run, the data were base called and demultiplexed on the instrument (Illumina FASTQ 1.9 files with Phred+33 encoding), and the FASTQ format files were used for downstream analysis.
Bioinformatics analysis
The sequence data were mapped to the human genome (hg19) using the BWA aligner 0.77. Local-alignment optimisation was performed using the GATK 1.6. Variant calling was performed only in the genomic regions that had been targeted by the Illumina VariantStudio testing and visualisation.
Confirmatory sanger sequencing
All of the sequence variations of the TP53 and ATRX genes detected using next-generation sequencing were confirmed using Sanger sequencing. In summary, the genomic DNA was extracted using a QIAamp DNA FFPE Tissue Kit (Qiagen). The target sequences were PCR-amplified. After purification, direct sequencing was performed using an automated ABI 3770 sequencer (Applied BioSystems, Foster City, CA, USA).
Statistical analysis
The data analyses were conducted using Epi Info Version 3.3.2 software (Centres for Disease Control and Prevention) and SPSS Version 19.0 software (IBM Corp., Armonk, NY, USA). Categorical variables were compared using the χ2 test or Fisher’s exact test. Continuous variables were analyzed using the Student t test. Survival curves were generated using the Kaplan-Meier method, and differences were calculated using the log-rank test. All statistical results were considered significant at a 2-tailed P value of < 0.05.
Results
Patients and tumor samples
A total of 54 cases of leiomyosarcoma were examined. Table 2 summarises the clinical information. Forty-three of the patients were women; 24 of the cases originated in the uterus, and 30 of the cases had extrauterine origins. The most common extrauterine site was the retroperitoneum (n = 11). The remaining 19 cases originated in various other sites (intra-abdominal: n = 7; lower extremities: n = 5; upper extremity, skin, chest wall, thyroid, back, paraspinal region, thoracic cavity: each n = 1). The patients’ overall mean and median ages were 53.7 and 53.8 years, respectively (range: 28-88 years); the mean ages of the patients with tumors in the uterus and in extrauterine sites were 51 and 56 years, respectively. The tumor sizes were known in 44 cases and ranged from 1.5 to 23 cm. The overall mean and median tumor sizes in the cases were 10.5 and 10 cm, respectively.
Table 2.
Clinicopathological features of 54 leiomyosarcomas
| n = 54 | |
|---|---|
| Sex | |
| Male | 11 (20%) |
| Female | 43 (80%) |
| Age (mean ± SD) | 53.7±12.8 |
| Tumor site | |
| Uterus | 24 (44%) |
| Retroperitoneum | 11 (20%) |
| Others | 19 (35%) |
| Tumor size (cm) (mean ± SD) | 10.5±5.0 |
| Cell morphology | |
| Spindle | 36 (67%) |
| Epithelioid/pleomorphic | 18 (33%) |
| Mitotic activity | |
| 1-9/HPF | 8 (17%) |
| 10-19/HPF | 26 (55%) |
| > 19/HPF | 13 (40%) |
| Tumor differentiation | |
| Well/Moderately | 28 (60%) |
| Poorly | 19 (40%) |
| Necrosis | |
| Absent | 8 (17%) |
| Present | 39 (83%) |
A summary of the histological features is shown in Table 2. Of the tumors, 36, 12, and 6 were composed predominantly of spindle, epithelioid, and pleomorphic cells, respectively. To simplify statistical analysis, the epithelioid and pleomorphic cell morphologies were combined as a single feature. Because none of the tumors had ≥50% tumor necrosis and because there were relatively few well-differentiated tumors in our series, these parameters were also simplified to ‘absent’ or ‘present’ and ‘well/moderately differentiated’ or ‘poorly differentiated’, respectively. The tumors exhibiting predominantly epithelioid or pleomorphic cell morphologies occurred mostly in the uterus (8 and 3 cases, respectively). In 7 cases, only sections of metastatic tumors or small biopsy samples were available for histological review and were not graded.
Prevalence of mutations among the 44 genes analyzed
An adequate library was obtained from 54 formalin-fixed, paraffin-embedded tumor samples for subsequent sequencing. The mean coverage of each target region was 425×. False positive sites were removed under the following filter conditions, as determined using Integrative Genomics Viewer software: a mean coverage > 100×, each variant coverage > 20×, a variant frequency of each sample > 5%, and P < 0.01; in the coding region (except for the TERT promoter), not a synonymous mutation and not strand-biased variants, by using the Integrative Genomics Viewer (IGV) software. Subsequently, these variants were searched in the 1000 Genome database was searched for these variants to distinguish between SNPs and mutations.
In the analyzed leiomyosarcoma samples, mutations were identified in 22 of 44 genes. The genes with mutations in more than 2 tumors are shown in Figure 1. Only 3 genes-TP53, ATRX, and PALB2-had mutation frequencies higher than 10%.
Figure 1.
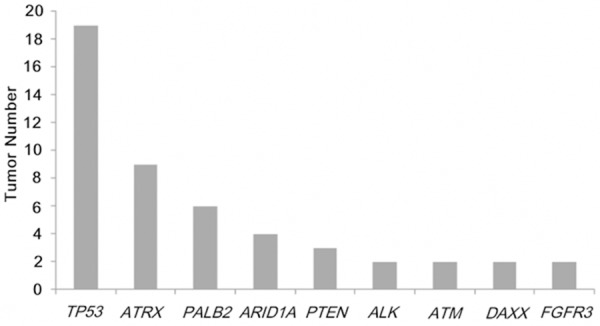
The tumour numbers of each mutated gene. Only genes mutated in 2 or more tumours were included.
In the 54 tumors, TP53 was mutated in 19 (35%), ATRX in 9 (17%), and PALB2 in 6 (11%). The 6 PALB2 mutations were p.C1127R (c.3379 A>G), p.E1018D (c.3054 C>G), p.T1012/I (c.3035 G>A), p.R825T (c.2674 C>G), p.I309V (c.925T>C), and p.D498Y (c.1492 C>A). Because only 6 tumors harboured PALB2 mutations, the following sections focus on the clinicopathological significance of the TP53 and ATRX mutations.
Clinicopathological significance of the TP53 mutation in leiomyosarcoma
Figure 2A shows the locations of the TP53 mutations in the leiomyosarcomas. All of the mutations were located in exons 4-10, mainly in the DNA-binding domains and mostly in the evolutionary-conserved regions [14]. Three leiomyosarcomas had 2 mutations. Most of the mutations were missense mutations, but one nonsense mutation and 2 frameshift mutations were also detected.
Figure 2.
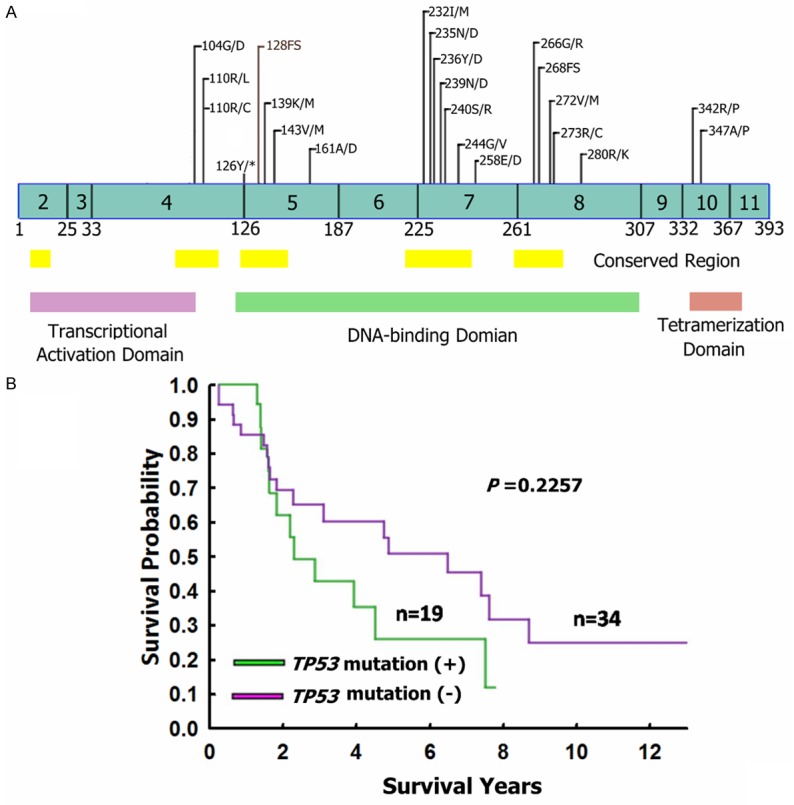
Mutations of TP53 in leiomyosarcomas. A. The distribution of TP53 mutations in leiomyosarcomas. *: stop codon; FS: Frameshift mutation. B. Kaplan-Meier survival analyses of patients with TP53-mutated and TP53-wild-type leiomyosarcomas.
The correlations of the TP53 mutation with clinicopathological features are shown in Table 3. The TP53-mutated leiomyosarcomas were limited to female patients (P = 0.006). All but 2 of the TP53-mutated leiomyosarcomas were located in the uterus (n = 11) or retroperitoneum (n = 6), whereas 49% of the TP53-wild-type leiomyosarcomas were located in body sites other than the uterus or retroperitoneum (P = 0.018). TP53 mutations were also associated with larger tumor size (12.8 ± 5.6 versus 9.1 ± 4.0, P = 0.012). The TP53 mutation did not correlate with patient age or survival, cell morphology, mitotic activity, tumor differentiation, or tumor necrosis (Table 3 and Figure 2B).
Table 3.
Relation of clinicopathological features with TP53 and ATRX mutations in leiomyosarcoma
| TP53 mutation | ATRX mutation | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| (+) | (-) | (+) | (-) | |||
|
|
|
|||||
| N (%) | N (%) | P value | N (%) | N (%) | P value | |
| Sex | 0.006 | 0.44 | ||||
| Male | 0 (0%) | 11 (31%) | 1 (11%) | 10 (22%) | ||
| Female | 19 (100%) | 24 (69%) | 8 (88%) | 35 (78%) | ||
| Tumor site | 0.018 | 0.085 | ||||
| Uterus | 11 (58%) | 13 (37%) | 7 (78%) | 17 (38%) | ||
| Retroperitoneum | 6 (32%) | 5 (14%) | 1 (11%) | 10 (22%) | ||
| Others | 2 (11%) | 17 (49%) | 1 (11%) | 18 (40%) | ||
| Age (mean ± SD) | 54.8±10.8 | 53.1±13.9 | 0.651 | 54.46±12.9 | 53.6±12.9 | 0.651 |
| Tumor size (cm) (mean ± SD) | 12.8±5.6 | 9.1±4.0 | 0.012 | 13.4±4.1 | 10.7±.5.0 | 0.097 |
| Cell morphology | 0.840 | 1.000 | ||||
| Spindle | 13 (68%) | 23 (66%) | 6 (67%) | 30 (67%) | ||
| Epithelioid/pleomorphic | 6 (32%) | 12 (34%) | 3 (33%) | 15 (30%) | ||
| Mitotic activity | 0.302 | 0.180 | ||||
| 1-9/10 HPF | 1 (6%) | 7 (12%) | 0 (0%) | 8 (21%) | ||
| 10-19/10 HPF | 11 (65%) | 15 (52%) | 4 (50%) | 22 (56%) | ||
| >19/10 HPF | 5 (29%) | 8 (36%) | 4 (50%) | 9 (23%) | ||
| Tumor differentiation | 0.937 | 0.028 | ||||
| Well/Moderately | 10 (59%) | 18 (60%) | 2 (25%) | 26 (67%) | ||
| Poorly | 7 (41%) | 12 (40%) | 6 (75%) | 13 (33%) | ||
| Necrosis | 0.126 | 0.015 | ||||
| Absent | 1 (19%) | 7 (0%) | 0 (0%) | 8 (21%) | ||
| Present | 16 (81%) | 23 (100%) | 8 (100%) | 31 (79%) | ||
| ATRX expression | 0.167 | 0.207 | ||||
| Lost | 9 (47%) | 10 (29%) | 5 (55%) | 15 (33%) | ||
| Intact | 10 (53%) | 25 (71%) | 4 (44%) | 30 (67%) | ||
| ALT | 0.163 | 0.008 | ||||
| Negative | 5 (26%) | 16 (46%) | 0 (0%) | 21 (47%) | ||
| Positive | 14 (74%) | 19 (54%) | 9 (100%) | 24 (53%) | ||
Clinicopathological significance of ATRX mutation in leiomyosarcoma
Figure 3A shows the locations of the ATRX mutations in the leiomyosarcomas. Most of the mutations were located in the helicase-like domain. Most of the mutations were missense mutations, but 2 nonsense mutations and 2 frameshift mutations were also detected. Two tumors harboured 2 mutations. One tumor harboured 5 mutations (p.S1792Y, p.I1765M, p.H1759Y, p.Q1788*, and p.P609A).
Figure 3.
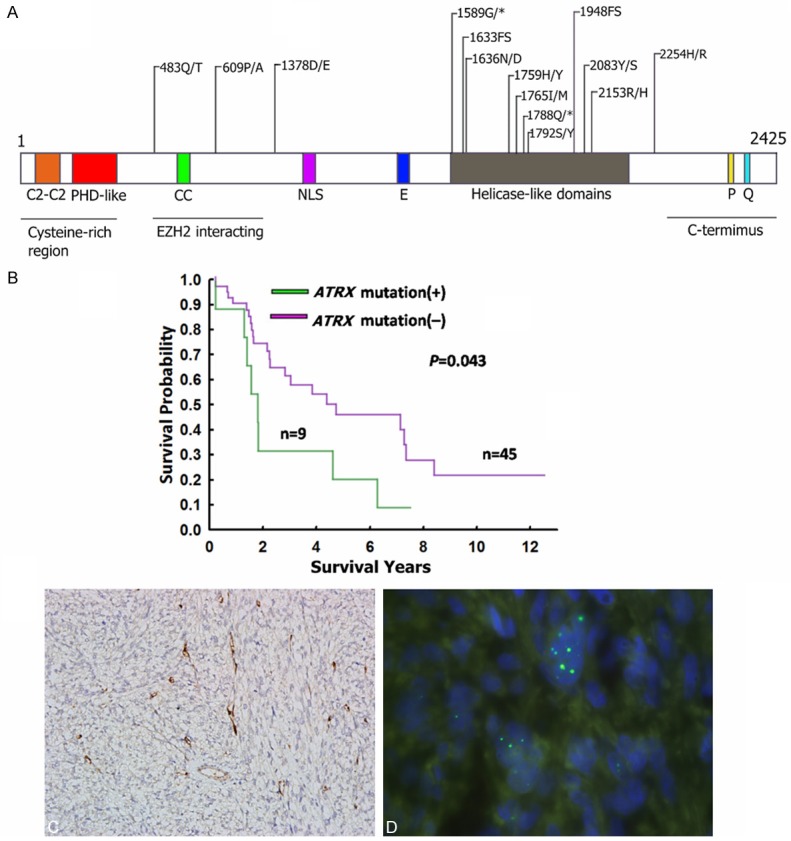
Mutations of ATRX in leiomyosarcomas. (A) The distribution of ATRX mutations in leiomyosarcomas. Two patients had the p.G1589* mutation. *: stop codon; FS: Frameshift mutation; CC: Coiled-coil domain; NLS: Nulcear localization signal; E: Negative tract; P: P domain; Q: Q domain. (B) Kaplan-Meier survival analyses of patients with ATRX-mutated and ATRX-wild-type leiomyosarcomas. (C) Immunostaining demonstrated the loss of ATRX expression in a leiomyosarcoma. (D) Telomere-specific fluorescent in situ hybridization revealed the presence of large and bright signals. (C) and (D) were taken from a case with ATRX mutation.
The correlations of the ATRX mutation with clinicopathological features are shown in Table 3. ATRX mutations were associated with poor differentiation (P = 0.028) and the presence of tumor necrosis (P = 0.015). The ATRX mutation did not correlate with patient sex or age, tumor site or size, cell morphology, or mitotic activity. The Kaplan-Meier survival analysis showed that the patients with ATRX-mutated leiomyosarcomas had worse overall survival than did patients with ATRX-wild-type leiomyosarcomas (Figure 3B).
Correlation with ATRX mutation and alternative lengthening of telomeres (ALT)
Previous studies have shown that ATRX mutations are highly correlated with the loss of ATRX expression and the ALT phenotypes in pancreatic neuroendocrine tumor and glioma [15,16]. Therefore, we performed immunostaining for ATRX protein and telomere-specific fluorescent in situ hybridization for all of the tumors. ATRX expression was lost in 19 leiomyosarcomas (35%). ALT was detected in 33 leiomyosarcomas (61%). Although the ATRX mutation did not correlate with the loss of ATRX expression, all 4 tumors with nonsense and frameshift mutations were immunonegative for ATRX. All of the ATRX-mutated leiomyosarcomas showed the ALT phenotypes (P = 0.008). Figure 3C and 3D show the results of the immunostaining and telomere-specific fluorescent in situ hybridization of an ATRX-mutated leiomyosarcoma.
Discussion
In this paper, we present a parallel sequencing analysis of 44 cancer-related genes from a collection of 54 leiomyosarcomas. Our work represents the first systematic, large-scale sequencing analysis of leiomyosarcomas. We found that TP53 and ATRX were frequently mutated in the leiomyosarcomas.
Mutations of TP53 in leiomyosarcomas have been reported previously [10,11]. Our study showed that TP53-mutated leiomyosarcomas were limited to female patients and that all but 2 of the TP53-mutated leiomyosarcomas were located in the uterus or retroperitoneum. By contrast, Konomoto et al. showed TP53 mutations in 8 of 15 superficial-type extrauterine leiomyosarcomas and 8 of 22 deep-type extrauterine leiomyosarcomas. The reason for this discrepancy is unknown. Advances in sarcoma classification since the 1998 study of Konomoto et al. may change the diagnosis of some of their cases. The TP53 mutation rate of Ito’s study (39%) was similar to that of our study. Pérot et al. showed mutations of TP53 coding regions in 15 of 35 leiomyosarcomas (42%) [17]. However, the sex distribution and tumor sites were not mentioned in these 2 studies. Our finding that TP53 mutations were highly correlated with the uterine or retroperitoneal leiomyosarcomas in female patients supports the recent hypothesis that uterine and retroperitoneal leiomyosarcomas in female patients have similar histogenesis. A cluster analysis of immunohistochemical markers showed that uterine and retroperitoneal leiomyosarcomas in female patients shared similar expression features for the estrogen and progesterone receptors and WT1 [18]. Uterine and retroperitoneal leiomyosarcomas are typically larger than their trunk and extremity counterparts [19], which may account for the size difference between TP53-mutated and TP53-wild-type leiomyosarcomas.
ALT is a major telomere-maintaining mechanism for sarcomas with complex karyotypic abnormalities [20]. ATRX and DAXX form a dimer that acts as a histone chaperone to deposit histone variant H3.3 to GC-rich regions of the genome, including the telomeres, and plays a critical role in telomere stability [21-23]. It is hypothesized that dysfunction of the dimer leads to telomere instability, increased telomere homologous recombination, and ultimately alterative lengthening of telomeres. Consistent with previous reports on pancreatic neuroendocrine tumors and glioma, all of the leiomyosarcomas with ATRX mutations in the present study were positive for the ALT phenotypes. However, 15 leiomyosarcomas in our study showed a loss of ATRX expression without somatic mutations being identified in those cases, indicating that other mechanisms may be responsible for the loss of ATRX function in these tumors. According to provisional data from The Cancer Genome Atlas (http://cancergenome.nih.gov/), 7.4% (7/56) of the sarcomas harboured homozygous ATRX deletions, which might partly account for the loss of ATRX expression in some of the cases.
In most tumors, only one mutation in the TP53 or ATRX gene was identified. The dominant negative effect of the mutant p53 over the wild-type p53 may cause the inactivation of the p53 tetramer complex [24]; consequently, monoallelic mutation may abolish the tumor suppressor activity of p53. The ATRX gene is located in the X chromosome. In female cells, one X chromosome is inactivated through methylation [25]. This Lyonization effect may have inactivated the wild-type allele of the ATRX genes in the tumors.
We detected 6 PALB2 mutations in our cohorts. PALB2 has a critical function in DNA damage repair [26]. Germline PALB2 mutations predispose individuals to a high risk of developing familial breast cancer [27]. In a nation-wide epidemiological study in Sweden, leiomyosarcoma was associated with maternal breast cancer [28]. Additional large-scale studies should be performed to determine whether the PALB2 mutation underlies the genetic mechanism for these familial breast cancer leiomyosarcoma cases.
In conclusion, we found frequent TP53 and ATRX mutations in leiomyosarcomas. The TP53 mutations were predominantly identified in the uterine and retroperitoneal leiomyosarcomas in female patients, indicating that leiomyosarcomas in those 2 sites share similar pathogenesis. The ATRX mutations were highly correlated with the ALT phenotypes. Recently, ataxia telangiectasia and Rad3-related kinase have been shown to play crucial roles in homologous recombination and ALT, with inhibitions of this kinase causing apoptosis of cancer cells with alternative telomere lengthening [29]. Identifying ATRX mutations in leiomyosarcomas may provide new opportunities for treating these aggressive neoplasms.
Acknowledgements
This study was supported by grants provided by the National Science Council, Taiwan, to CYY (MOST 104-2314-B-002-029-) and Ntaional Taiwan University Hospital (NTUH 104-P12). We thank the staff of the Department of Medical Resaerch at National Taiwan University Hospital for technical Support.
Disclosure of conflict of interest
The authors declare no conflict of interest.
Abbreviations
- ALT
alternative lengthening of telomeres
References
- 1.Antonescu CR. The role of genetic testing in soft tissue sarcoma. Histopathology. 2006;48:13–21. doi: 10.1111/j.1365-2559.2005.02285.x. [DOI] [PubMed] [Google Scholar]
- 2.Latres E, Drobnjak M, Pollack D, Oliva MR, Ramos M, Karpeh M, Woodruff JM, Cordon-Cardo C. Chromosome 17 abnormalities and TP53 mutations in adult soft tissue sarcomas. Am J Pathol. 1994;145:345–355. [PMC free article] [PubMed] [Google Scholar]
- 3.Orlow I, Drobnjak M, Zhang ZF, Lewis J, Woodruff JM, Brennan MF, Cordon-Cardo C. Alterations of INK4A and INK4B genes in adult soft tissue sarcomas: effect on survival. J Natl Cancer Inst. 1999;91:73–9. doi: 10.1093/jnci/91.1.73. [DOI] [PubMed] [Google Scholar]
- 4.Knutsen T, Gobu V, Knaus R, Padilla-Nash H, Augustus M, Strausberg RL, Kirsch IR, Sirotkin K, Ried T. The interactive online SKY/MFISH & CGH database and the Entrez cancer chromosomes search database: linkage of chromosomal aberrations with the genome sequence. Genes Chromosomes Cancer. 2005;44:52–64. doi: 10.1002/gcc.20224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang R, Lu YJ, Fisher C, Bridge JA, Shipley J. Characterization of chromosome aberrations associated with soft-tissue leiomyosarcomas by twenty-four-color karyotyping and comparative genomic hybridization analysis. Genes Chromosomes Cancer. 2001;31:54–64. doi: 10.1002/gcc.1118. [DOI] [PubMed] [Google Scholar]
- 6.Hensley ML, Maki R, Venkatraman E, Geller G, Lovegren M, Aghajanian C, Sabbatini P, Tong W, Barakat R, Spriggs DR. Gemcitabine and docetaxel in patients with unresectable leiomyosarcoma: results of a phase II trial. J. Clin. Oncol. 2002;20:2824–2831. doi: 10.1200/JCO.2002.11.050. [DOI] [PubMed] [Google Scholar]
- 7.Hensley ML, Wathen JK, Maki RG, Araujo DM, Sutton G, Priebat DA, George S, Soslow RA, Baker LH. Adjuvant therapy for high-grade, uterus-limited leiomyosarcoma: results of a phase 2 trial (SARC 005) Cancer. 2013;119:1555–1561. doi: 10.1002/cncr.27942. [DOI] [PubMed] [Google Scholar]
- 8.Giuntoli RL 2nd, Metzinger DS, DiMarco CS, Cha SS, Sloan JA, Keeney GL, Gostout BS. Retrospective review of 208 patients with leiomyosarcoma of the uterus: prognostic indicators, surgical management, and adjuvant therapy. Gynecol Oncol. 2003;89:460–469. doi: 10.1016/s0090-8258(03)00137-9. [DOI] [PubMed] [Google Scholar]
- 9.Ricci S, Giuntoli RL 2nd, Eisenhauer E, Lopez MA, Krill L, Tanner EJ 3rd, Gehrig PA, Havrilesky LJ, Secord AA, Levinson K, Frasure H, Celano P, Fader AN. Does adjuvant chemotherapy improve survival for women with early-stage uterine leiomyosarcoma? Gynecol Oncol. 2013;131:629–633. doi: 10.1016/j.ygyno.2013.08.037. [DOI] [PubMed] [Google Scholar]
- 10.Konomoto T, Fukuda T, Hayashi K, Kumazawa J, Tsuneyoshi M. Leiomyosarcoma in soft tissue: examination of p53 status and cell proliferating factors in different locations. Hum Pathol. 1998;29:74–81. doi: 10.1016/s0046-8177(98)90393-8. [DOI] [PubMed] [Google Scholar]
- 11.Ito M, Barys L, O’Reilly T, Young S, Gorbatcheva B, Monahan J, Zumstein-Mecker S, Choong PF, Dickinson I, Crowe P, Hemmings C, Desai J, Thomas DM, Lisztwan J. Comprehensive mapping of p53 pathway alterations reveals an apparent role for both SNP309 and MDM2 amplification in sarcomagenesis. Clin Cancer Res. 2011;17:416–426. doi: 10.1158/1078-0432.CCR-10-2050. [DOI] [PubMed] [Google Scholar]
- 12.Bertsch E, Qiang W, Zhang Q, Espona-Fiedler M, Druschitz S, Liu Y, Mittal K, Kong B, Kurita T, Wei JJ. MED12 and HMGA2 mutations: two independent genetic events in uterine leiomyoma and leiomyosarcoma. Mod Pathol. 2014;27:1144–1153. doi: 10.1038/modpathol.2013.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liau JY, Tsai JH, Jeng YM, Lee JC, Hsu HH, Yang CY. Leiomyosarcoma with alternative lengthening of telomeres is associated with aggressive histologic features, loss of ATRX expression, and poor clinical outcome. Am J Surg Pathol. 2015;39:236–244. doi: 10.1097/PAS.0000000000000324. [DOI] [PubMed] [Google Scholar]
- 14.Pavletich NP, Chambers KA, Pabo CO. The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev. 1993;7:2556–2564. doi: 10.1101/gad.7.12b.2556. [DOI] [PubMed] [Google Scholar]
- 15.Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, Offerhaus GJ, McLendon R, Rasheed BA, He Y, Yan H, Bigner DD, Oba-Shinjo SM, Marie SK, Riggins GJ, Kinzler KW, Vogelstein B, Hruban RH, Maitra A, Papadopoulos N, Meeker AK. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abedalthagafi M, Phillips JJ, Kim GE, Mueller S, Haas-Kogen DA, Marshall RE, Croul SE, Santi MR, Cheng J, Zhou S, Sullivan LM, Martinez-Lage M, Judkins AR, Perry A. The alternative lengthening of telomere phenotype is significantly associated with loss of ATRX expression in high-grade pediatric and adult astrocytomas: a multi-institutional study of 214 astrocytomas. Mod Pathol. 2013;26:1425–1432. doi: 10.1038/modpathol.2013.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pérot G, Chibon F, Montero A, Lagarde P, de Thé H, Terrier P, Guillou L, Ranchère D, Coindre JM, Aurias A. Constant p53 pathway inactivation in a large series of soft tissue sarcomas with complex genetics. Am J Pathol. 2010;177:2080–2090. doi: 10.2353/ajpath.2010.100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carvalho JC, Thomas DG, Lucas DR. Cluster analysis of immunohistochemical markers in leiomyosarcoma delineates specific anatomic and gender subgroups. Cancer. 2009;115:4186–4195. doi: 10.1002/cncr.24486. [DOI] [PubMed] [Google Scholar]
- 19.Worhunsky DJ, Gupta M, Gholami S, Tran TB, Ganjoo KN, van de Rijn M, Visser BC, Norton JA, Poultsides GA. Leiomyosarcoma: One disease or distinct biologic entities based on site of origin? J Surg Oncol. 2015;111:808–812. doi: 10.1002/jso.23904. [DOI] [PubMed] [Google Scholar]
- 20.Henson JD, Hannay JA, McCarthy SW, Royds JA, Yeager TR, Robinson RA, Wharton SB, Jellinek DA, Arbuckle SM, Yoo J, Robinson BG, Learoyd DL, Stalley PD, Bonar SF, Yu D, Pollock RE, Reddel RR. A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin Cancer Res. 2005;11:217–225. [PubMed] [Google Scholar]
- 21.Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee YL, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, George AJ, Morgan KA, Mann JR, Choo KH. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351–360. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willis A, Jung EJ, Wakefield T, Chen X. Mutant p53 exerts a dominant negative effect by preventing wild-type p53 from binding to the promoter of its target genes. Oncogene. 2004;23:2330–2338. doi: 10.1038/sj.onc.1207396. [DOI] [PubMed] [Google Scholar]
- 25.Chaligné R, Heard E. X-chromosome inactivation in development and cancer. FEBS Lett. 2014;588:2514–2522. doi: 10.1016/j.febslet.2014.06.023. [DOI] [PubMed] [Google Scholar]
- 26.Pauty J, Rodrigue A, Couturier A, Buisson R, Masson JY. Exploring the roles of PALB2 at the crossroads of DNA repair and cancer. Biochem J. 2014;460:331–342. doi: 10.1042/BJ20140208. [DOI] [PubMed] [Google Scholar]
- 27.Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T, Jayatilake H, McGuffog L, Hanks S, Evans DG, Eccles D Breast Cancer Susceptibility Collaboration (UK) Easton DF, Stratton MR. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji J, Eng C, Hemminki K. Familial risk for soft tissue tumors: a nation-wide epidemiological study from Sweden. J Cancer Res Clin Oncol. 2008;134:617–624. doi: 10.1007/s00432-007-0327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flynn RL, Cox KE, Jeitany M, Wakimoto H, Bryll AR, Ganem NJ, Bersani F, Pineda JR, Suvà ML, Benes CH, Haber DA, Boussin FD, Zou L. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science. 2015;347:273–277. doi: 10.1126/science.1257216. [DOI] [PMC free article] [PubMed] [Google Scholar]


