Abstract
Here we perform the first genome wide association study (GWAS) of multiple myeloma (MM) survival. In a meta-analysis of 306 MM patients treated at UCSF and 239 patients treated at the Mayo clinic, we find a significant association between SNPs near the gene FOPNL on chromosome 16p13 and survival (rs72773978; p=6 × 10−10). Patients with the minor allele are at increased risk for mortality (HR 2.65; 95% CI: 1.94 – 3.58) relative to patients homozygous for the major allele. We replicate the association in the IMMEnSE cohort including 772 patients, and a University of Utah cohort including 318 patients (rs72773978 p=0.044). Using publically available data, we find that the minor allele was associated with increased expression of FOPNL and increased expression of FOPNL was associated with higher expression of centrosomal genes and with shorter survival.. Polymorphisms at the FOPNL locus are associated with survival among MM patients.
INTRODUCTION
Multiple myeloma (MM) is an incurable hematological malignancy of plasma cells. Approximately 22,000 new cases are diagnosed each year in the United States and over 10,000 deaths occur annually1. Family history is a strong risk factor for MM2. Recent genome-wide association studies (GWAS) reported 8 loci associated with susceptibility to MM3–5.
A variety of clinical features and biomarkers are associated with MM prognosis 6. Chromosomal abnormalities are also associated with prognosis; deletions at 17p, 13q, amplifications at 1q and translocations t(4;14) and t(14;16) have been associated with a poor prognosis, while hyperdiploidy is associated with a favorable prognosis7–9. Gene expression signatures of the myeloma cells also predict survival10–14.
Germline genetic variants are associated with survival among patients with esophageal15, breast16,17,pancreatic18,19 and small cell lung cancer20. We performed the first GWAS of MM survival, by conducting a meta-analysis of two studies from the University of California San Francisco (UCSF) and the Mayo Clinic. We found a locus on chromosome 16 associated with survival. We replicated the findings in the IMMEnSE consortium and the University of Utah cohort. The top SNPs were at the FOPNL locus. Using publicly available data of gene expression from peripheral blood of normal individuals, we found that the risk alleles at the top SNPs were associated with increased expression of FOPNL We also found that increased expression of FOPNL was associated with higher “centrosome index”, a gene expression correlate of centrosome amplification in multiple myeloma cells which has been associated with poor survival.
RESULTS
Identifying a Locus for Survival
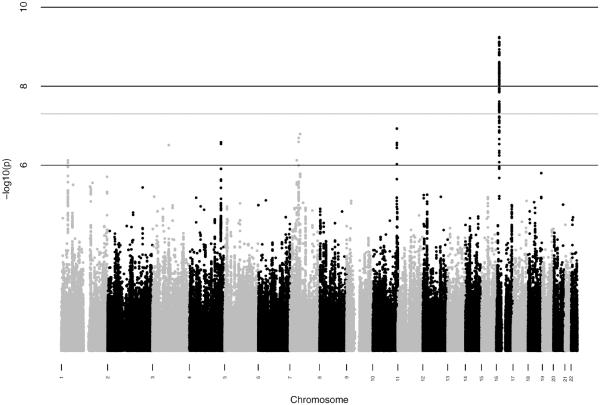
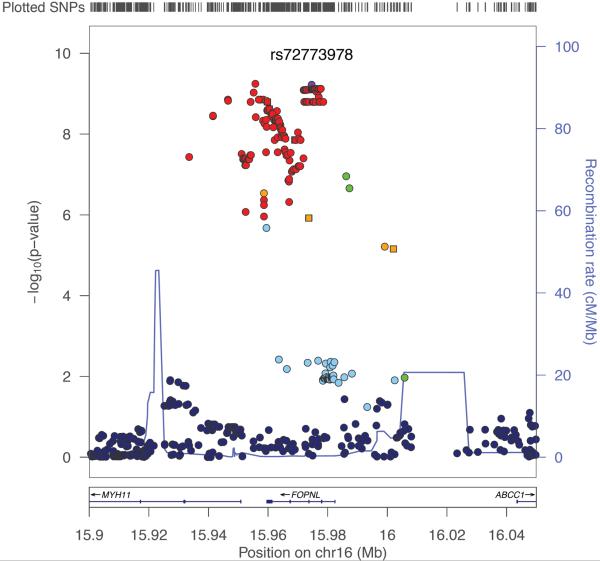
We performed a GWAS of overall survival among MM patients in cohorts (Table 1) from UCSF (n=306) and Mayo Clinic (n=239) separately. One locus mapping to chromosome 16p13.11 (Hg19) showed a suggestive association in both the UCSF (p=8.4 × 10−7; proportional hazards model) and the Mayo Clinic studies (p= 1.1 × 10−4; proportional hazards model). In a meta-analysis of these GWAS, the locus was genome-wide significant (Figure 1a) with the strongest evidence at 2 SNPs in perfect linkage disequilibrium rs72773978 and rs117863986 (p=6.0 × 10−10 for both; meta-analysis p value is calculated using inverse variance based weighting). We found no significant deviation from the proportional hazards assumption for the top SNP in either the UCSF (p=0.74; p values calculated by testing whether scaled Schoenfeld residuals vary with time) or Mayo clinic studies (p=0.95). We identified 131 SNPs at this locus associated with survival at p<5×10−8 (Supplementary Data 1); these SNPs had approximately 5–7% minor allele frequency and were in tight linkage disequilibrium (r2>0.8) with the top SNPs (figure 1b). Of the 131 top SNPs, 17 were genotyped in the UCSF dataset and 1 was genotyped in the Mayo Clinic dataset (SupplementaryData 1). The remaining SNPs were imputed, but had very high imputation quality scores (Information>0.9 or r2>0.9). We directly genotyped 8 additional SNPs in the Mayo clinic dataset, including one of the top 2 SNPs, rs117863986, and found consistently strong levels of association with the genotyped SNPs (rs117863986 HR: 2.26; 95% CI: 1.46 – 3.40; p=0.00021; proportional hazards model) and other SNPs (Supplementary Table 1).
Table 1.
Clinical characteristics of patients in discovery and replication studies
| UCSF (N=306) | Mayo Clinic (N=239) | IMMENSE Cohorts & University of Utah (N=1090) | |
|---|---|---|---|
| Age at diagnosis Mean (SD) | 55.9 ± 9.0 | 62.2 ± 11.4 | 54.4 ± 10.1 |
| Average follow up time Mean years (SD) | 3.8 ± 2.5 | 5.3 ± 3.5 | 4.8 ± 3.3 |
| Mortality N (%) | 103 (33.7) | 174 (72.8) | 423 (38.5%) |
| Type of disease (N with data) | 292 | 239 | 649 |
| IgG Kappa N (%) | 127 (43.5) | 86 (36.0) | 145 (22.3) |
| IgG Lambda N (%) | 44 (15.1) | 49 (20.5) | 64 (9.9) |
| IgG light chain not specified N (%) | 11 (3.8) | - | 173 (26.7) |
| IgA Kappa N (%) | 31 (10.6) | 35 (15.0) | 35 (5.4) |
| IgA Lambda N (%) | 21 (7.2) | 22 (9.1) | 33 (5.1) |
| IgA light chain not specified N (%) | - | - | 75 (11.6) |
| Light chain only N (%) | 51 (17.5) | 31 (13.0) | 86 (13.3) |
| Other/non-secretory N (%) | 7 (2.4) | 16 (6.7) | 38 (5.9) |
| ISS Stage (N with data) | 140 | 221 | 513 |
| 1 N (%) | 49 (35.0) | 86 (38.9) | 162 (31.6) |
| 2 N (%) | 55 (39.3) | 74 (33.5) | 165 (32.2) |
| 3 N (%) | 36 (25.7) | 61 (27.6) | 186 (36.3) |
| Durie-Salmon Stage (N with data) | 256 | 187 | 633 |
| IA or IB N (%) | 39 (15.2) | 14 (7.5) | 69 (10.9) |
| IIA or IIB N (%) | 40 (15.6) | 30 (16.4) | 149 (23.5) |
| IIIA or IIIIB N (%) | 177 (69.2) | 143 (76.5) | 415 (65.6) |
Figure 1.
GWAS results for survival among MM patients (a) Manhattan plot of results of genome-wide association analysis for survival in MM patients. Each point represents the negative log p value of the meta-analysis for association with survival using the UCSF and Mayo Clinic data (b) Locuszoom plot for association statistics at the 16p13 region in the meta-analysis of UCSF and Mayo. Each dot represents the negative log p value for the association statistics from the meta-analysis. The top associated SNP (rs72773978) is colored in purple and the remaining SNPs are colored according to linkage disequilibrium values (r2) with the top SNP.
Analysis of the genome wide distribution of association statistics (Supplementary Figure 1a) revealed minimal deviation from the expectation under the null (lambda=1.002). After removing SNPs from a 200KB region around the top locus on chromosome 16, we found no evidence for additional signal genome wide (Supplementary Figure 1b) although some other loci had some suggestive signals with p values 5×10−7 – 1×10−7 (Supplementary Data 1).
The UCSF study had a median time of 7.6 months (interquartile range 5.7–8.9 months) between date of diagnosis and date of ascertainment. Therefore, we considered whether this delay affected our results. First, we adjusted for the time difference between date of diagnosis and ascertainment in the proportional hazards models and found no attenuation (Supplementary Table 2). We also considered models stratified by the delay between date of diagnosis and ascertainment (Supplementary Table 2). We found that the effect was consistent among patients enrolled between 0 to 5.9 months after diagnosis (HR 3.23; 95% CI: 1.28 – 8.17; p=0.013; proportional hazards model) those enrolled between 6 and 11.9 months after diagnosis (HR 3.04; 95% CI: 1.67 –5.55; p=0.00028; proportional hazards model) and those enrolled from 12 to 23.9 months (HR 5.06; 95%CI: 1.18 – 21.79; p=0.029; proportional hazards model). Thus, we concluded that the delay between diagnosis and ascertainment within the first two years was unlikely to affect the association between the SNP and overall survival. We also considered models that adjusted for the difference between date of diagnosis and ascertainment in the Mayo study and found no change in the association as expected (Supplementary Table 2), since nearly all of the participants were ascertained within 1 month of diagnosis.
We searched for additional SNPs that were associated independently of the top SNP by performing conditional analyses including rs72773978 and other SNPs within 100 KB of that SNP. We performed survival analyses for all SNPs that were either in no LD (R2<0.1) or in modest LD (R2 0.1–0.5) with RS72773978, adjusting for the effect of PC's and for the effect of RS72773978. We found no other significant associations in the locus after adjusting for multiple hypothesis testing.
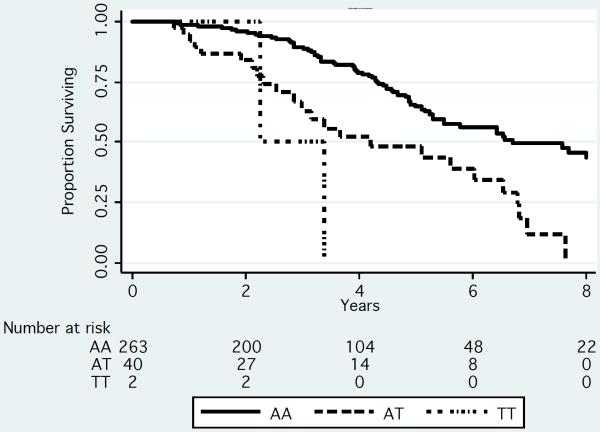
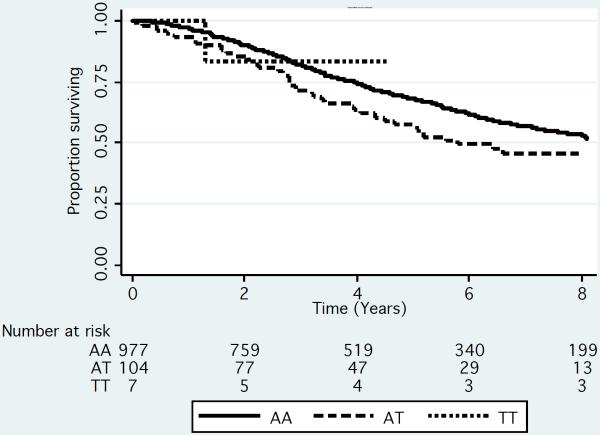
Median survival was decreased by approximately 2.7 years among patients who were either heterozygous or homozygous for the rare variant (T allele) of rs72773978 compared to patients homozygous for the common variant in both the UCSF and Mayo Clinic cohorts (Figure 2). In models that adjusted for age, gender and genetic ancestry, approximately 10–14% of patients had an increased risk of death (hazard ratio ~2.6; table 2) in the meta-analysis of these 2 datasets.
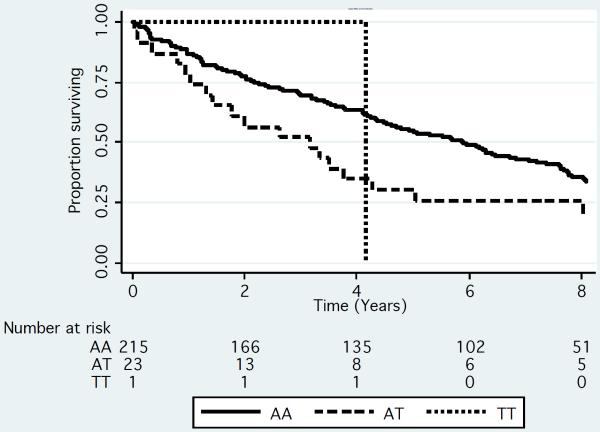
Figure 2.
Kaplan-Meier Survivorship plot by genotype for rs72773978 in the UCSF (N=306) (A) Mayo (N=239) (B) and IMMENSE/Utah (N=1080) (C) studies. We plotted the results by genotype. To generate the plots for the discovery datasets, we rounded the imputation results to the nearest whole numbers to infer genotypes of individuals.
Table 2.
Association of rs72773978 and MM survival
| Age, Sex adjusted* | ||||
|---|---|---|---|---|
| Study | Allele frequency | HR | 95% CI | P- value*** |
| UCSF (n=306) | 0.075 | 3.03 | 1.95 – 4.73 | 8.0 × 10−7 |
| Mayo (n=239) | 0.054 | 2.31 | 1.51 – 3.53 | 1.1 × 10−4 |
| Discovery Meta-analysis (UCSF & Mayo) | 2.65 | 1.94 – 3.58 | 6.0 × 10−10 | |
| Replication** (1,090): IMMENSE n=772, Utah n=315 | 0.051 | 1.34 | 1.01 – 1.78 | 0.044 |
| Discovery & Replication Meta-analysis | 1.93 | 1.54 – 2.41 | 6.7 × 10−9 | |
| Age, Sex, Stage adjusted * | |||
|---|---|---|---|
| ISS adjusted | |||
| UCSF (n=140) | 4.06 | 2.04 – 8.05 | 6.3 × 10−5 |
| Mayo (n=221) | 2.41 | 1.51 – 3.83 | 2.2 × 10−4 |
| Replication ** (n=513) | 1.70 | 1.19 – 2.44 | 0.004 |
| Salmon-Durie adjusted | |||
| UCSF (n=256) | 2.81 | 1.73 – 4.57 | 3.2 × 10−5 |
| Mayo (n=184) | 2.75 | 1.68 – 4.49 | 5.3 × 10−5 |
| Replication ** (n=625) | 1.46 | 1.04– 2.05 | 0.029 |
The UCSF and Mayo Clinic study results are also adjusted by PCA. The IMMENSE data is not due to the lack of availability of GWAS data.
The IMMENSE results are from a meta-analysis of the individual regions (see supplementary table 3).
P values are calculated from proportional hazards models.
We also performed a separate analysis of individuals who genetically clustered with Caucasians in the UCSF dataset (Supplementary Figure 2). We repeated the meta-analysis of the Mayo Clinic data with the UCSF Caucasian only sample (Supplementary Table 3), and found the association with rs72773978 remained significant (p=2.4×10−9; proportional hazards model). The UCSF cohort included African American patients (N=25 and patients of Latino (N=27) or other patients who clustered with those of mixed ancestry (N=24). In an analysis of these patients, we also found a nominally significant association between shorter survival and the minor allele (HR 2.43; 95% CI: 1.09 – 5.39; p=0.029 proportional hazards model).
We also examined whether the top SNPs that we identified were associated with susceptibility to MM in our two studies. We found no significant difference in genotype frequencies between cases and controls (Supplementary Table 4).
Relationship to Stage at Diagnosis and Treatment
We evaluated the effect of the genotype on survival after adjustment for clinical stage. In analyses that adjusted for stage using either the ISS definition or the Durie-Salmon staging system, rs72773978 genotype remained a strong predictor of survival (Table 2). Among the participants in the Mayo Clinic on whom LDH levels were available (N=154), we saw a consistent level of association (HR: 2.25; 95% CI: 1.26 – 4.03; p=0.006; proportional hazards model).
Since MM treatment has improved significantly in the last decade, we used data available from both cohorts to determine whether the SNP effect varied by initial treatment (Supplementary Table 5). We found a consistent effect of the SNP regardless of the type of treatment initiated (Table 3) and no evidence of interaction between treatment and the SNP in either the UCSF (p=0.9; p for interaction derived using proportional hazards model) or Mayo Clinic (p=0.52; p for interaction derived using proportional hazards model) cohorts.
Table 3.
Effect of SNP by initial treatment among patients in the UCSF and Mayo Clinic cohorts
| HR* | 95% CI | P value*** | |
|---|---|---|---|
| Mayo Clinic | |||
| Old treatments** | N=136, 102 deaths | ||
| RS72773978 | 1.90 | 0.98 – 3.83 | 0.057 |
| New treatments*** | N=93, 64 deaths | ||
| RS72773978 | 2.71 | 1.56 – 4.70 | 0.00045 |
| Entire sample adjusted for treatment | N=229, 166 deaths | ||
| RS72773978 | 2.18 | 1.43 – 3.32 | 0.00028 |
| UCSF | |||
| Old treatments** | N= 109 60 deaths | ||
| RS72773978 | 3.35 | 1.74 – 6.44 | 0.00028 |
| New treatments*** | N=187, 30 deaths | ||
| RS72773978 | 3.57 | 1.71 – 7.43 | 0.0007 |
| Entire Sample Adjusted for treatment | N=296, 90 deaths | ||
| RS72773978 | 3.35 | 2.07 – 5.41 | 8.2 × 10−7 |
All models are adjusted for age, gender and principal components 1–10.
Regimens including vincristine/Adriamycin/Dexamethasone or melphalan/prednisone
Treatments containing at least on of the following agents: thalidomide, botezomib or lenalidomide
P values are calculated from proportional hazards models.
Among patients in the Mayo Clinic study, 134 (56%) were treated by both high dose chemotherapy (HDC) followed by autologous stem-cell rescue. We adjusted for HDC in the proportional hazards models and found that although HDC was a strong predictor of longer survival in the cohort, there was only mild attenuation of the SNP association with survival (Supplementary Table 6). Since nearly all (97%) of the UCSF patients received HDC, the same analysis could not be done within the UCSF study.
Replication
We replicated the association of top SNPs from the UCSF and Mayo Clinic meta-analysis in a replication meta-analysis of 1,090 MM cases, including 772 European MM patients from the IMMEnSE consortium and 318 from the Utah cohort (Supplementary Table 7). We selected 2 SNPs for replication including rs72773978, one of the top associated SNPs from the meta-analysis and rs12598966, a SNP resulting in an amino acid substitution in the FOPNL gene. We performed a meta-analysis of all of the replication studies and found a significant association between rs72773978 and survival in the replication cohorts adjusted by age and sex (HR 1.34; 95% CI: 1.01 – 1.74; p=0.044; proportional hazards model) with survival shorter by approximately 1.2 years among carriers of the minor allele (figure 2c). There was no evidence of heterogeneity of effect within the replication studies (p=0.14; chi-squared test for heterogeneity). The other SNP, rs12598966, was not significantly associated with survival in the replication.
We noted a slightly stronger effect size in analyses that adjusted for stage. However, the change in effect size was not due to negative confounding between stage and the SNP, but rather to the fact that the cohorts with missing data on stage were the ones with an inconsistent point estimate (Supplementary Table 3). In a meta-analysis that did not adjust for ISS stage but just included participants without missing data on ISS stage, we saw approximately the same effect size (HR 1.71; 95% CI: 1.18 – 2.47; p=0.005; proportional hazards model) as in the meta-analysis that adjusted for ISS stage (supplementary table 7.)
Analysis of Function
The top SNPs were in a region that overlapped the entire FOPNL gene and a portion of the MYH11 gene (figure 1b). In addition, known drug transporters, ABCC1 and ABCC6, are located about 50kb and 300kb away, respectively. One of the top SNPs, rs12598966, is located in the coding sequence of FOPNL, and leads to a non-synonymous amino acid substitution: E->K at amino acid 156; however, this SNP was not significantly associated with survival in the replication and was not predicted to have a deleterious effect on protein function (SIFT score =0.89 and Polyphen 2 score=0.275). Next, we investigated the top 145 SNPs (all in tight LD (r2>0.8) with rs72773978) for an effect on gene expression using GENEVAR21. The top 2 SNPs, rs72773978 and rs117863986, were not included in the database, but six other SNPs in strong LD (r2=1) with rs72773978 are present in the database and are associated with expression of FOPNL (Supplementary Table 8). The minor allele of these SNPs predicted higher expression of FOPNL. There was no significant association between these SNPs and expression of other genes within 1 MB of the locus.
We identified 13 SNPs in LD with rs72773978 as being potentially functional (Supplementary Table 9). Six of these SNPs are in the 3' UTR of the FOPNL gene and, therefore, may be involved in transcript stability. Seven SNPs were identified as being in sites of open chromatin and thus may be involved in transcriptional regulation.
Since the top SNPs were associated with gene expression, we hypothesized that expression of FOPNL may be associated with survival among MM patients. In particular, higher expression of FOPNL is associated with the minor allele of the top SNPs from the GWAS and should also be associated with shorter survival. We used publically available data on gene expression (GSE2658) and survival from 414 MM cases to test this hypothesis13. As predicted, we found a significant association between higher expression of FOPNL and worse survival (Supplementary Table 10). FOPNL is known to localize to the centrosome and the pericentriolar satellites. Since centrosome amplification is known to be a predictor of poor prognosis, we evaluated the association between FOPNL expression and the centrosome index (CI), a previously validated gene expression signature of centrosome amplification. We found a very strong correlation (supplementary table 11) between higher FOPNL expression and increased CI in the study we analyzed for survival (GSE2658) and in two additional studies (GSE19784 and GSE26760).
DISCUSSION
We performed a GWAS for survival among MM patients and identified SNPs at chromosome 16p13 that were strongly associated with mortality. The SNPs were in the region of the FOPNL gene and a subset of the SNPs were associated with FOPNL expression levels, with the minor allele predicting higher expression22. We also found that FOPNL expression was associated with poorer survival using data from a previous study13. Thus, our results strongly suggest that FOPNL is a gene involved in myeloma progression.
FOPNL is known to be associated with centrosome function23,24. Centrosome amplification is common in MM and is associated with poor prognosis25,26. Furthermore, inhibition of centrosomal clustering may be effective in treatment of MM27,28. We found that germline variation that affects a gene involved in centrosomal function may also contribute to disease progression. Furthermore, we found an association between FOPNL expression and centrosome index in 3 datasets of gene expression from myeloma samples. However, our results implicating FOPNL as the causal gene at this locus rely on the synthesis of several datasets. We were not able to directly correlate the SNPs with gene expression, centrosome index and survival in the same dataset. It is possible that another gene/s at this locus may be responsible for the effect we observe, or that the effect is mediated by FOPNL, but that it is not through a mechanism related to centrosome amplification. Additional studies of this gene and centrosomal function will help to further define the mechanism underlying the association that we identified.
Our results imply that germline genetic variation is associated with survival among patients with MM. Other GWAS have identified loci that affect survival in other cancer types15–18,20. At least one of the known loci for MM susceptibility is associated with the risk for a particular subtype of MM4 which may also have an effect on prognosis.
MM is a heterogeneous disease with substantial variation in prognosis among different patients. Identifying patients who are at higher risk of progression may be of importance in treating these patients more aggressively earlier in their disease. Our results identify FOPNL genotype as a predictor of survival, and we found that the association remains significant after adjustment for stage at diagnosis. However, the effect size we observed in the replication cohorts was substantially attenuated compared to the effect size in the discovery cohorts. This difference is most likely due to “winner's curse” – a tendency for the initial study to over-estimate the true effect size29. Thus, the replication cohorts in our study are more likely to represent the true effect size in future studies.
Our study has several important limitations. We could not examine the association between SNPs and MM survival by cytogenetic abnormalities since the majority of our patients were diagnosed prior to the common use of cytogenetic and fluorescent in-situ hybridization (FISH) analysis in clinical practice. Therefore, it will be important to examine the utility of this genotype in the setting of cytogenetic and FISH analysis and gene expression signatures. Furthermore, we could not adjust for gene expression patterns which are also known to be associated with survival.
We found no difference between patients who were initially treated with modern vs. older therapies. However, our ability to analyze the SNP by different therapies was limited to the UCSF dataset and we had inadequate power to detect interactions between the SNPs and particular drugs. Furthermore, we only adjusted for the association between initial treatment and survival, and it is likely that many of the patients who were initially treated with older regimens received newer regimens if they survived to the era when these became available. It is possible that the effect that we saw is modified by one regimen or by one class of medications. Additional studies should be done in the context of clinical trials or other cohorts where treatment regimens are more uniform to investigate whether the effect of the locus we identified is modified by particular treatments.
In summary, we found a strong association between a locus on chromosome 16p and MM survival that is likely due to an effect on expression of the FOPNL gene. The SNPs we identified may become important clinical predictors of outcome among MM patients.
METHODS
UCSF Study
The UCSF Institutional Review Board (IRB) approved ascertainment of cases and use of existing biospecimens for genetic analysis. All participants gave informed consent. The study included 370 patients treated for MM at UCSF between 1989 and 2010. We excluded 10 samples due to insufficient clinical data. We also excluded 42 participants whose blood was collected ≥2 years after diagnosis from the survival analysis due to the potential bias towards long-term survivors among these participants. The median delay between diagnosis and ascertainment among the 42 participants excluded was 2.9 years (interquartile range 2.4 to 4.7 years). The median delay among the participants included was 7.6 months (interquartile range 5.7–8.9 months).
We used white blood cells harvested after mobilization of stem cells with granulocyte colony stimulating factor (GCSF) as a source of DNA. Mobilization of stem cells is performed prior to high dose chemotherapy followed by autologous stem cell transplantation. The patient receives GCSF and then undergoes harvesting of peripheral white blood cells via apheresis several days later30,31. Bone marrow stem cell fraction is monitored via CD34 antibody and apheresis is continued until an adequate number of CD34+ cells have been collected for stem cell rescue. The cells are then stored in liquid nitrogen.
We ascertained date of death using chart reviews and death registry data. Survival time was determined as the date of diagnosis until date of death or last clinic visit for patients who were not known to have died. Clinical stage and initial chemotherapy regimen was determined by chart review. For analysis of treatment, we dichotomized treatments into either newer regimens (including an Imid and/or proteasome Inhibitor) or older regimens (including neither an Imid nor proteasome Inhibitor). Nearly all of the participants in the UCSF study (>97%) received high dose chemotherapy with stem cell rescue (autologous bone marrow transplant).
Mayo Clinic Study
Ascertainment of MM cases and genotyping was approved by the Mayo Clinic Institutional Review Board. The study included incident MM cases seen in the regional practice between 1998 and 2007 and recruited within 6 months of initial diagnosis. Nearly all participants (96%) were recruited within 1 month of diagnosis and the remainder were recruited between 1 and 5.5 months after diagnosis. Eligible cases provided consent and a blood sample for research studies of MM. A total of 243 MM cases were used for analyses. DNA was extracted from stored peripheral blood samples. We ascertained date of death and calculated survival time as described for the UCSF study. Clinical stage was determined by chart review. Clinical data on survival could be ascertained on 239 patients. Initial chemotherapy regimen and use of high dose chemotherapy was determined by chart review. For analysis of treatment, we dichotomized treatments into either newer regimens (including an Imid and/or proteasome Inhibitor) or older regimens (including neither an Imid nor proteasome Inhibitor). Approximately 56% of participants were treated with autologous stem cell transplant.
IMMENSE Study
The International Multiple Myeloma rESEarch (IMMEnSE) consortium is a case-control study recruited from 7 different European and North American countries32. MM cases are defined by a confirmed diagnosis of MM, according to the International Myeloma Working Group (IMWG) criteria33. For each patient, demographic and clinical parameters at diagnosis and survival were collected by the responsible clinicians in each of the IMMEnSE centers. The data collected are standardized in a central database kept at the German Cancer Research Center (DKFZ, Heidelberg, Germany). For each subject, a sample of peripheral blood or extracted DNA has been collected and sent to DKFZ. A total of 772 MM cases with survival information available in the IMMEnSE consortium were included in this study.
Utah study
Sampling and genetic analysis was approved by the University of Utah Institutional Review Board. The study included prevalent MM cases in the state of Utah, ascertained up to 2012. Eligible cases provided consent and a blood or saliva sample from which DNA was extracted. Date of diagnosis was confirmed from chart review and Utah Cancer Registry data. Date of death was confirmed from chart review and death registry data. Survival time was determined as the date of diagnosis until date of death, last contact with the study, or last known event in Utah (determined from statewide vital records, driver's license renewals, and voter registrations in the Utah Population Database) for patients who were not known to have died. A total of 318 MM cases with DNA and survival information were available for this study.
All SNP positions were annotated using the Genome Reference Consortium GRCh37 (Hg19) version of the human genome.
UCSF
A pilot study of 81 MM samples used an Illumina 660 array genotyped at the UCSF Genomics Core Facility. In a second phase, we genotyped 289 MM samples using an Illumina Omni5 array at Expression Analysis (Durham, NC). Of the 370 participants in the GWAS, 52 participants were excluded from the survival analysis as noted above due to either insufficient clinical data (N=10) or due to >2 year time difference between diagnosis and ascertainment (N=42). Of the remaining 318 samples we dropped 12 since they did not pass quality control for genotyping. Eleven were dropped due to high missing genotype values (>5% missing genotypes per sample) and 1 sample was dropped due to potential contamination, leaving 306 patients. We dropped SNPs that had >5% missing values, or were monomorphic. Imputation was performed using IMPUTE234 (https://mathgen.stats.ox.ac.uk/impute/impute_v2.html#home) with all samples from 1000 Genomes dataset (Version 2, May 2011 release35) as a reference. Imputed SNPs with Information<0.5 or minor allele frequency (MAF)<0.025 were excluded, leaving 8,036,255 SNPs for analysis.
Mayo Clinic
Cases were genotyped using the Affymetrix 6.0 array. Monomorphic SNPs and those with a call rate < 95% were excluded, leaving 786,950 observed SNPs. Four samples with call rates < 95% and one sample with non-European ancestry according to principal components analysis were excluded, leaving 243 MM cases and 239 with follow-up past date of diagnosis. Imputation was performed with BEAGLE36 (http://faculty.washington.edu/browning/beagle/beagle.html), using all samples from version 2 of the 1000 Genomes data (May 2011 release) as reference. Imputed SNPs with an r2 < 0.3 or MAF < 0.025 were excluded, leaving 7,276,170 SNPs for further analyses.
We selected 2 SNPs for replication in IMMEnSE and Utah samples including one of the top two SNPs from the meta-analysis (rs72773978) and a SNP in high linkage disequilibrium with the top SNP, encoding a non-synonymous amino acid substitution in FOPNL (rs12598966). These SNPs were typed using 5' exonuclease (TaqMan) assays (ABI) at the German Cancer Research Center (DKFZ) in Heidelberg (IMMEnSE samples) and at the Genomics Core at the University of Utah (Utah samples). Duplicates of 12% of the samples were interspersed throughout the plates and concordance rate among duplicates was >99.9%.
We performed genome-wide analyses for association with survival using proportional hazards models in the UCSF and Mayo datasets separately. We inferred genetic ancestry using principal components analysis (PCA) in each cohort using SmartPCA37. Each SNP was entered into the model under an assumption of log-additive increased risk, and adjusting for PC's 1–3, age and gender. Imputed SNPs were modeled using the probability of genotypes. We tested the proportional hazards assumption for the top SNPs by calculating the scaled Schoenfeld residuals and testing whether they are significantly associated with time38.
We also performed a subset analysis of Caucasians only in the UCSF dataset. We identified Caucasians based on genetic ancestry (see supplementary figure 2). Individuals who clustered with self-described Caucasians (PC1>0, PC2<0) were included in this subset analysis (N=229).
All analyses were performed in R. For graphing survival results, we used the Kaplan-Meier estimates of the survival function and graphed the results using Stata (Version 10). We graphed the association statistics for all SNPs near the top locus using LocusZoom39.
We used Cox regression models adjusted for age and gender to test the association between SNPs and survival in the IMMEnSE consortium and the Utah cohort.
We performed a meta-analysis of the UCSF and Mayo Clinic results on a total of 6,026,834 SNPs in common from both GWAS that met the allele frequency and imputation quality thresholds. We also conducted a meta-analysis of data on two top SNPs from 7 centers within the IMMEnSE consortium and the Utah study in our replication study. We calculated a fixed effects model for each SNP using METAL 40. We used Cochran's Q statistic to test for heterogeneity.
To examine the association of SNPs and risk of MM, we compared the genotype frequencies of cases vs. ethnically matched controls from the UCSF (N=298) and the Mayo Clinic (N=295) sites respectively. We used logistic regression models, adjusting for PC1-3 age and gender.
We used the dataset from Grundberg et al22 for eQTL analyses, which consists of 856 Caucasian individuals including 154 monozygotic twin pairs, 232 dizygotic twin pairs and 84 singletons. We focused on expression in lymphocytes in this dataset. We used GENEVAR21 to query the top 145 SNPs from the GWAS and identified 6 SNPs, that were also in the Grundberg et al dataset. We queried GENEVAR for beta coefficients and p values for associations between 6 SNPs and the genes within a 1 Mb windowincluding: FOPNL, MYH11, ABCC1, NDE1, KIAA0430, ABCC6, RRN3, NTAN1 KIAA0250, KIAA0251.
We downloaded gene expression data from Zhan et al13, Broyl et al41, and Chapman et al42 from the National Institutes of Health Gene Expression Omnibus (accession number: GSE2658, GSE19784, and GSE26760, respectively). Zhan et al consisted of gene expression data from 559 MM samples assayed on Affymetrix U133 arrays; Broyl et al consisted of gene expression data from purified CD138+ plasma cells of 320 newly diagnosed myeloma patients using Affymetrix GeneChip U133 plus 2.0 arrays; Chapman et al consisted of 304 CD138-purified bone marrow samples from patients with multiple myeloma were analyzed on Affymetrix U133 Plus 2.0 microarrays.
Gene expression and MM outcome Of the samples in the Zhan et al dataset, 414 also had available clinical data and were included in the original publication13, and therefore, we used the data from these 414 samples in our analyses. We used log-transformed probe intensity values as predictors of survival, entering these as continuous variables into a proportional hazards model. We analyzed each of the two probes for FOPNL on the Affymetrix U133 array separately and also considered the average of the two probes as a predictor of overall survival in the proportional hazards model.
Analysis of potential SNP function: We used SIFT43 and Polyphen244 to determine the likelihood that a non-synonymous amino acid substitution has a deleterious effect on protein function. We used FunciSNP45 to determine whether any of the SNPs may affect gene expression, including any SNPs with r2>0.7 with rs72773928. r2 values for linkage disequilibrium were calculated in European ancestry samples from 1000 genomes.
Supplementary Material
Acknowledgements
This work was supported by the Steve and Nancy Grand Multiple Myeloma Translational Initiative and a grant from Expression Analysis and a grant from the National Cancer Institute (R21CA191896). E Ziv is supported in part by a mid-career award in patient research from the National Cancer Institute (K24169004). Collection of blood samples from Polish patients and controls from Łodz area and DNA extraction was supported by a grant from Polish Ministry of Science and Higher Education (No. NN402178334). DNA extraction from Danish healthy controls was supported by The Research Fund at Region Sjælland, Denmark. The Utah study was supported by LLS 6067-09, CA152336 and CA134674, with data collection made possible, in part, by the Utah Population Database (UPDB) and the Utah Cancer Registry (UCR). Partial support for all datasets within the UPDB is provided by the Huntsman Cancer Institute (HCI) and the HCI Cancer Center Support grant, P30 CA42014. The UCR is supported in part by NIH contract HHSN261201000026C from the NCI SEER Program with additional support from the Utah State Department of Health and the University of Utah. In Utah, Jathine Wong is thanked for support in formatting files and advice on R programming. Several authors (EZ, NC, FC, CV) are members of the International Multiple Myeloma Consortium. The authors are grateful to the leadership and other members of the International Multiple Myeloma Consortium for facilitating this collaborative study.
Footnotes
Author Contributions: Study conception: EZ, ED, CV; collection of the data: EZ, ED, KC, DC, PB, JC, CD, MD, AG, KJ, AJ, SK, DA, HM, TM, JML, VR, JS, AJV, MW, JW, NC, FC, CV; performed genotyping:, AM, RD, AL BJ; analysis of the data:, DH, DS, DC, BA, YZ; drafting of the manuscript: EZ, ED; critical revision of the manuscript: EZ, ED, DH, AM, DS, KC, DC, BA, PB, GB, YZ, JC, RD, CD, MD, LF, AG, SH, KJ, AJ, SK, DA, MG, LCA, BJ, AL, HM, TM, JML, VR, JS, AJV, MW, JW, SS, NC, FC, CV
Competing Financial Interests The authors declare no conflicts of interest.
References
- 1.Howlader N, et al. SEER Cancer Statistics Review, 1975–2010. National Cancer Institute; Bethesda, MD: 2010. [Google Scholar]
- 2.Greenberg AJ, Rajkumar SV, Vachon CM. Familial monoclonal gammopathy of undetermined significance and multiple myeloma: epidemiology, risk factors, and biological characteristics. Blood. 2012;119:5359–66. doi: 10.1182/blood-2011-11-387324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chubb D, et al. Common variation at 3q26.2, 6p21.33, 17p11.2 and 22q13.1 influences multiple myeloma risk. Nature genetics. 2013 doi: 10.1038/ng.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinhold N, et al. The CCND1 c.870G>A polymorphism is a risk factor for t(11;14)(q13;q32) multiple myeloma. Nature genetics. 2013;45:522–5. doi: 10.1038/ng.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broderick P, et al. Common variation at 3p22.1 and 7p15.3 influences multiple myeloma risk. Nature genetics. 2012;44:58–61. doi: 10.1038/ng.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajkumar SV, Greipp PR. Prognostic factors in multiple myeloma. Hematology/oncology clinics of North America. 1999;13:1295–314. xi. doi: 10.1016/s0889-8588(05)70128-3. [DOI] [PubMed] [Google Scholar]
- 7.Kumar S, et al. Trisomies in multiple myeloma: impact on survival in patients with high-risk cytogenetics. Blood. 2012;119:2100–5. doi: 10.1182/blood-2011-11-390658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fonseca R, et al. Prognostic value of chromosome 1q21 gain by fluorescent in situ hybridization and increase CKS1B expression in myeloma. Leukemia. 2006;20:2034–40. doi: 10.1038/sj.leu.2404403. [DOI] [PubMed] [Google Scholar]
- 9.Fonseca R, et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood. 2003;101:4569–75. doi: 10.1182/blood-2002-10-3017. [DOI] [PubMed] [Google Scholar]
- 10.Zhan F, et al. Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood. 2007;109:1692–700. doi: 10.1182/blood-2006-07-037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhan F, Barlogie B, Mulligan G, Shaughnessy JD, Jr., Bryant B. High-risk myeloma: a gene expression based risk-stratification model for newly diagnosed multiple myeloma treated with high-dose therapy is predictive of outcome in relapsed disease treated with single-agent bortezomib or high-dose dexamethasone. Blood. 2008;111:968–9. doi: 10.1182/blood-2007-10-119321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhan F, et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood. 2002;99:1745–57. doi: 10.1182/blood.v99.5.1745. [DOI] [PubMed] [Google Scholar]
- 13.Zhan F, et al. The molecular classification of multiple myeloma. Blood. 2006;108:2020–8. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhan F, et al. Gene expression profiling of human plasma cell differentiation and classification of multiple myeloma based on similarities to distinct stages of late-stage B-cell development. Blood. 2003;101:1128–40. doi: 10.1182/blood-2002-06-1737. [DOI] [PubMed] [Google Scholar]
- 15.Wu C, et al. Genome-wide association study identifies common variants in SLC39A6 associated with length of survival in esophageal squamous-cell carcinoma. Nature genetics. 2013;45:632–8. doi: 10.1038/ng.2638. [DOI] [PubMed] [Google Scholar]
- 16.Kiyotani K, et al. A genome-wide association study identifies locus at 10q22 associated with clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients in Japanese. Human molecular genetics. 2012;21:1665–72. doi: 10.1093/hmg/ddr597. [DOI] [PubMed] [Google Scholar]
- 17.Shu XO, et al. Novel genetic markers of breast cancer survival identified by a genome-wide association study. Cancer research. 2012;72:1182–9. doi: 10.1158/0008-5472.CAN-11-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Innocenti F, et al. A genome-wide association study of overall survival in pancreatic cancer patients treated with gemcitabine in CALGB 80303. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:577–84. doi: 10.1158/1078-0432.CCR-11-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu C, et al. Genome-wide association study of survival in patients with pancreatic adenocarcinoma. Gut. 2012 doi: 10.1136/gutjnl-2012-303477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu C, et al. Genome-wide interrogation identifies YAP1 variants associated with survival of small-cell lung cancer patients. Cancer research. 2010;70:9721–9. doi: 10.1158/0008-5472.CAN-10-1493. [DOI] [PubMed] [Google Scholar]
- 21.Yang TP, et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics. 2010;26:2474–6. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grundberg E, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nature genetics. 2012;44:1084–9. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aubusson-Fleury A, et al. The conserved centrosomal protein FOR20 is required for assembly of the transition zone and basal body docking at the cell surface. Journal of cell science. 2012;125:4395–404. doi: 10.1242/jcs.108639. [DOI] [PubMed] [Google Scholar]
- 24.Sedjai F, et al. Control of ciliogenesis by FOR20, a novel centrosome and pericentriolar satellite protein. Journal of cell science. 2010;123:2391–401. doi: 10.1242/jcs.065045. [DOI] [PubMed] [Google Scholar]
- 25.Chng WJ, et al. The centrosome index is a powerful prognostic marker in myeloma and identifies a cohort of patients that might benefit from aurora kinase inhibition. Blood. 2008;111:1603–9. doi: 10.1182/blood-2007-06-097774. [DOI] [PubMed] [Google Scholar]
- 26.Chng WJ, et al. Clinical implication of centrosome amplification in plasma cell neoplasm. Blood. 2006;107:3669–75. doi: 10.1182/blood-2005-09-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raab MS, et al. GF-15, a novel inhibitor of centrosomal clustering, suppresses tumor cell growth in vitro and in vivo. Cancer research. 2012;72:5374–85. doi: 10.1158/0008-5472.CAN-12-2026. [DOI] [PubMed] [Google Scholar]
- 28.Shiheido H, et al. A phthalimide derivative that inhibits centrosomal clustering is effective on multiple myeloma. PloS one. 2012;7:e38878. doi: 10.1371/journal.pone.0038878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zollner S, Pritchard JK. Overcoming the winner's curse: estimating penetrance parameters from case-control data. Am J Hum Genet. 2007;80:605–15. doi: 10.1086/512821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart DA, et al. Factors predicting engraftment of autologous blood stem cells: CD34+ subsets inferior to the total CD34+ cell dose. Bone Marrow Transplant. 1999;23:1237–43. doi: 10.1038/sj.bmt.1701800. [DOI] [PubMed] [Google Scholar]
- 31.Stewart DA, et al. The CD34+90+ cell dose does not predict early engraftment of autologous blood stem cells as well as the total CD34+ cell dose. Bone Marrow Transplant. 2000;25:435–40. doi: 10.1038/sj.bmt.1702171. [DOI] [PubMed] [Google Scholar]
- 32.Martino A, et al. Genetics and molecular epidemiology of multiple myeloma: the rationale for the IMMEnSE consortium (review) International journal of oncology. 2012;40:625–38. doi: 10.3892/ijo.2011.1284. [DOI] [PubMed] [Google Scholar]
- 33.Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: a report of the International Myeloma Working Group British journal of haematology. 2003;121:749–57. [PubMed] [Google Scholar]
- 34.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Genomes Project, C. et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–23. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grambsch PA, Therneau T. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;81:515–526. [Google Scholar]
- 39.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Broyl A, et al. Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood. 2010;116:2543–53. doi: 10.1182/blood-2009-12-261032. [DOI] [PubMed] [Google Scholar]
- 42.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–72. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sim NL, et al. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic acids research. 2012;40:W452–7. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Current protocols in human genetics / editorial board, Jonathan L. Haines … [et al.] 2013;Chapter 7(Unit7):20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coetzee SG, Rhie SK, Berman BP, Coetzee GA, Noushmehr H. FunciSNP: an R/bioconductor tool integrating functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs. Nucleic acids research. 2012;40:e139. doi: 10.1093/nar/gks542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.