Abstract
Solid cancers develop within a supportive microenvironment that promotes tumor formation and growth through the elaboration of mitogens and chemokines. Within these tumors, monocytes (macrophages and microglia) represent rich sources of these stromal factors. Leveraging a genetically engineered mouse model of neurofibromatosis type 1 (NF1) low-grade brain tumor (optic glioma), we have previously demonstrated that microglia are essential for glioma formation and maintenance. To identify potential tumor-associated microglial factors that support glioma growth (gliomagens), we initiated a comprehensive large-scale discovery effort using optimized RNA-sequencing methods focused specifically on glioma-associated microglia. Candidate microglial gliomagens were prioritized to identify potential secreted or membrane-bound proteins, which were next validated by quantitative real-time polymerase chain reaction as well as by RNA fluorescence in situ hybridization following minocycline-mediated microglial inactivation in vivo. Using these selection criteria, chemokine (C-C motif) ligand 5 (Ccl5) was identified as a chemokine highly expressed in genetically engineered Nf1 mouse optic gliomas relative to nonneoplastic optic nerves. As a candidate gliomagen, recombinant Ccl5 increased Nf1-deficient optic nerve astrocyte growth in vitro. Importantly, consistent with its critical role in maintaining tumor growth, treatment with Ccl5 neutralizing antibodies reduced Nf1 mouse optic glioma growth and improved retinal dysfunction in vivo. Collectively, these findings establish Ccl5 as an important microglial growth factor for low-grade glioma maintenance relevant to the development of future stroma-targeted brain tumor therapies.
Introduction
Studies in various experimental model systems have demonstrated that cancers develop within complex tissue environments that dramatically influence tumor cell growth, transformation, and metastasis. Within the microenvironment of most solid tumors are a variety of nonneoplastic cell types, including fibroblasts, immune system cells, and endothelial cells. Each of these stromal cell types has the capacity to produce growth/survival factors, chemokines, extracellular matrix, and angiogenic molecules that can change the local milieu in which neoplastic cells grow and infiltrate. Although the importance of the cancer microenvironment was initially explored in non–nervous system tumors [1], it is now clearly appreciated to be a fundamental determinant of brain cancer biology. Similar to cancers in other organs, the brain tumor microenvironment contains endothelial cells and monocytes (macrophages and microglia). As such, pioneering studies by Judah Folkman and colleagues revealed a critical role for endothelial cells in the tumor milieu [2], leading to brain tumor therapies that focus on inhibiting vascular endothelial growth factor activity [3], [4].
In addition to endothelial cells, brain tumor macrophages and microglia represent other logical targets for stroma-directed therapies. Analyses of human gliomas have revealed that 30% to 50% of the cells in these central nervous system tumors are microglia or macrophages [5], [6], [7], [8], where the monocyte content has been associated with increasing glioma malignancy grade [9]. Moreover, numerous studies have revealed critical roles for microglia in high-grade glioma growth and progression. In these studies, microglia produce factors (gliomagens) that increase the growth and migration of glioma cells [10], [11], [12]. Importantly, pharmacological or genetic disruption of microglia function in mouse high-grade glioma models results in attenuated tumor growth and progression [13], [14], [15].
In contrast to their high-grade counterparts, less is known about the role of microglia in low-grade gliomas. The most common inherited cause of low-grade glioma is the neurofibromatosis type 1 (NF1) cancer predisposition syndrome, in which 15% to 20% of children develop pilocytic astrocytomas (PAs) involving the optic pathway [16]. Children with NF1 are born with one mutated copy of the NF1 gene and develop tumors following somatic inactivation of the remaining normal NF1 gene in astroglial progenitors [5], [17]. Similar to their human counterparts, nearly all Nf1+/− mice with somatic Nf1 gene inactivation in neuroglial progenitors develop low-grade gliomas of the optic nerve and chiasm [18]. These resulting low-grade tumors are composed of neoplastic cells with low proliferative indices embedded within a microenvironment containing microglia and endothelial cells [18], [19], [20]. Moreover, pharmacological (minocycline treatment, JNK inhibition) or genetic (CD11b-thymidine kinase–mediated monocyte reduction or the use of Cx3cr1 knockout mice) inhibition of microglial function is sufficient to attenuate optic glioma formation and maintenance [8], [21], [22], [23].
In an effort to define the molecular mechanism(s) underlying stromal maintenance of glioma growth in vivo, we sought to identify candidate gliomagens uniquely expressed in tumor-associated microglia. Whereas our previous studies employed Nf1+/− microglia expanded in vitro [22], we now specifically focus on Nf1+/− microglia present in the glioma as a means to discover critical glioma-maintaining factors. Building on recent advances in RNA sequencing and the analysis of low-abundance and low-quality RNA [24], [25], [26], [27], Nf1+/− microglia were isolated from control and tumor-bearing optic nerves for this large-scale discovery effort. Following secondary validation and analysis, chemokine (C-C motif) ligand 5 (Ccl5) was identified as a candidate gliomagen elaborated by tumor-associated microglia in genetically engineered Nf1 mouse optic gliomas, which is also overexpressed in human PAs. Importantly, minocycline-mediated microglia inactivation decreased Ccl5 expression in vivo, whereas exogenous Ccl5 treatment increased the proliferation of Nf1-deficient optic nerve astrocytes in vitro. In addition, neutralizing Ccl5 antibody administration reduced glioma growth and optic glioma–associated retinal defects in vivo. Collectively, these experimental results establish a critical role for stromal Ccl5 in the pathobiology of low-grade brain tumors.
Materials and Methods
Mice
Three independent Nf1 optic glioma GEM models were used based on the timing of Nf1 inactivation or the presence of additional genetic changes. The first model (Nf1flox/mut; GFAP-Cre (FMC) [18]) was generated by successive breeding of Nf1 +/− mice with Nf1flox/flox (WT) mice [28] and GFAP-Cre mice [29]. In this model, Nf1 inactivation occurs in neuroglial progenitors at E14.5, whereas in the second model (FMC* [30], [31]), Nf1 loss occurs in neuroglial progenitors at E11.5. The third Nf1 optic glioma GEM model harbors Pten reduction and Nf1 loss in astroglial cells [32]. Nf1flox/mut; Ptenflox/wt; GFAP-Cre (FMPC) mice [33] were generated by intercrossing Ptenflox/flox mice [34] with Nf1flox/flox; GFAP-Cre mice [29]. The resulting Ptenflox/wt; Nf1flox/flox; GFAP-Cre mice were then mated with Nf1flox/mut (FM) mice to generate FMPC mice. In this model, Nf1 inactivation and Pten reduction occur in neuroglial progenitors at E14.5. WT and FM (Nf1+/−) littermates were used as non-glioma controls (Table 1). All mice were maintained on a C57BL/6 background and used in accordance with approved animal studies protocols at the Washington University School of Medicine. Mice were euthanized at 3 months of age, and optic nerves were collected from anesthetized and Ringer’s solution–perfused mice for histological analyses, RNA expression, and fluorescence-activated cell sorting (FACS). For all in vivo experiments, mice were randomly assigned to the treatment group, and the analyses were conducted in a blinded fashion.
Table 1.
Genetically Engineered Nf1 Mouse Models.
| Short Form | Genotype | Cell Type | Description | Time Point | CNS Abnormality | References |
|---|---|---|---|---|---|---|
| WT | Nf1flox/flox | Every cell | Exons 31 and 32 of the Nf1 gene flanked by loxP sites | None | Zhu et al., 2001 | |
| FM | Nf1flox/mut | Every cell | Reduced Nf1 gene expression | No glioma | ||
| FMC | Nf1flox/mut; GFAP-Cre | Neuroglial progenitors Surrounding cells |
Complete Nf1 loss Reduced Nf1 gene expression |
E14.5 | Optic glioma | Bajenaru et al., 2003 |
| FMC* | Nf1flox/mut; GFAP-Cre | Neuroglial progenitors | Complete Nf1 loss | E11.5 | Optic glioma | Hegedus et al., 2008 |
| Surrounding cells | Reduced Nf1 gene expression | |||||
| FMPC | Nf1flox/mut; Pten flox/wt; GFAP-Cre | Neuroglial progenitors | Complete Nf1 loss and Pten +/− | E14.5 | Optic glioma | Kaul et al., 2014 |
| Surrounding cells | Reduced Nf1 gene expression |
Minocycline Treatment
Minocycline hydrochloride (Sigma-Aldrich, St. Louis, MO) was dissolved in PBS, and 50 mg/kg was administered 5 days/week for 2 weeks. FMC mice were divided into two groups: one received intraperitoneal (i.p.) injections of minocycline, whereas the other received injections of vehicle alone (sterile PBS). Each cohort contained at least four mice. After the last injection, mice were euthanized and the optic nerves collected and processed for sectioning.
Anti-Ccl5 Antibody Treatment
FMC mice were treated by i.p. injection with 250 μg of either anti-Ccl5 antibody (clone 53405; R&D Systems, Minneapolis, MN) or an IgG2A isotype-matched control antibody (R&D Systems) suspended in sterile PBS. Mice received treatment every day for 2 weeks. Each cohort contained at least five mice. After the last injection, mice were euthanized and the optic nerves collected and processed for sectioning.
Human Tissue Samples
Pathologically normal optic nerve (n = 4) and optic glioma (n = 5) tissues were obtained at autopsy from female and male patients between 3 days and 17 years of age. Tissue was embedded in paraffin, and 6-μm–thick sections were cut and processed for immunohistochemical staining. These autopsy specimens were obtained in accordance with an active and approved Human Studies Institutional Review Board protocol at the Washington University School of Medicine.
Primary Astrocyte Cultures
Primary astrocyte cultures were established from the optic nerves of postnatal day 1 to 2 Nf1flox/flox pups [35]. Nf1-deficient (Nf1 −/−) cultures were generated following infection with adenovirus type 5 containing Cre recombinase (University of Iowa Gene Transfer Vector Core, Iowa City, IA). To measure cell proliferation, 5 × 104 astroglial cells were plated in 24-well dishes, allowed to adhere, and maintained in astrocyte growth media for 16 hours. Astrocyte cultures were then treated with either murine recombinant Ccl5 (250 ng/ml; R&D Systems) or PBS alone for up to 16 hours. The optimal cytokine concentration for the proliferation assay was predetermined using dose escalation experiments (data not shown).
Immunocytochemistry
Astrocytes were fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. Following overnight incubation with Ki67 antibodies (Abcam, Cambridge, MA), visualization was performed following incubation with Alexa Fluor 488 IgG secondary antibodies (Invitrogen, Carlsbad, CA). Cells were counterstained with DAPI. For each independent culture, at least five distinct microscopic fields were analyzed on a Nikon Eclipse TE300 fluorescence inverted microscope (Nikon, Tokyo, Japan) equipped with an optical camera (Optronics, Goleta, CA) and MetaMorph image analysis software (Molecular Devices, Dowingtown, PA).
Immunohistochemistry
Optic nerves were prepared for sectioning and immunostaining as previously described [36]. For paraffin section immunohistochemistry, HRP-conjugated secondary antibodies (Vector Laboratories, Burlingame, CA) were used in combination with Vectastain Elite ABC development and hematoxylin counterstaining. In the immunofluorescence detection experiments, appropriate Alexa Fluor–tagged secondary antibodies (Invitrogen) were used, followed by DAPI counterstaining. Amplification of the Brn3a and Ccl5 antibody signal was performed using a biotinylated secondary antibody, followed by HRP conjugation using Vectastain Elite ABC kit, and the fluorescent signal was amplified with Tyramide Signal Amplification Plus Cyanine 3 system (Perkin-Elmer, Billerica, MA) according to the manufacturer's instructions. Terminal deoxynucleotide transferase-mediated dUTP nick-end labeling (TUNEL) staining was performed using the ApopTag Plus in situ apoptosis fluorescein detection kit (Millipore, Billerica, MA) according to the manufacturer's recommendations. Images were subsequently acquired on a Nikon Eclipse TE300 fluorescence inverted microscope or a Nikon Eclipse E600 microscope equipped with an optical camera (Leica, Buffalo Grove, IL) and Leica LAS EZ image analysis software (Leica).
Alternatively, optic nerves were processed for O.C.T. (Tissue-Tek, Miles, Elkhart, IN) embedding (frozen sections). Immunofluorescence labeling was performed after blocking in PBS containing 3% normal donkey serum (Jackson Immunoresearch Labs, Westgrove, PA), 1% cold water fish gelatin (Sigma-Aldrich), and 0.1% Triton X-100 for 1 hour at room temperature before incubation with appropriate antibodies (Supplementary Table 1) in 10% normal donkey serum for 16 hours at 4°C. Fluorescence-conjugated secondary antibodies (1:100 dilution) were applied for 4 hours at 4°C. Images were acquired on a FluoView 1000 confocal microscope (Olympus, Tokyo, Japan).
The number of lineage antibody-positive, Ki67+, or TUNEL+ cells was quantitated as a percentage of total cells (DAPI+ cells or nuclei).
Microglial morphology was analyzed using ImageJ (version 1.48, NIH) software. The length of the microglial processes was defined as the distance between the nucleus and the tip of an extended process as identified by Iba1 immunostaining (Supplementary Figure 1).
Supplementary Figure 1.
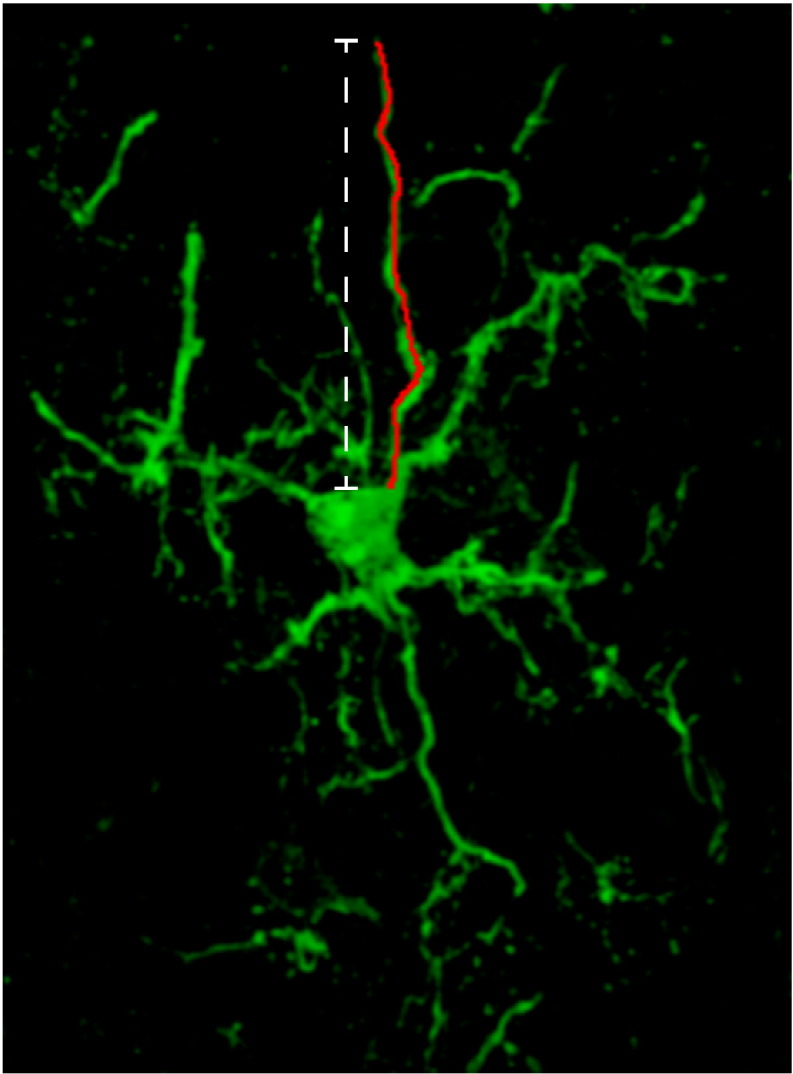
Image of a representative single Iba1+ microglia used for analysis. Microglial morphology was determined using ImageJ (version 1.48, NIH) software. The red line represents the process traced through the section. The length of the microglial processes was defined as the distance between the nucleus and the tip of an extended process as identified by Iba1 immunostaining.
In vivo BrdU Labeling and Immunohistological Analysis
IgG and anti-Ccl5–treated animals were injected with 50 mg/kg of BrdU (Sigma-Aldrich). Three hours after BrdU injection, animals were transcardially perfused with ice-cold Ringer’s solution and fixed with 4% paraformaldehyde. The optic nerves were dissected, fixed, and processed for paraffin sectioning, and BrdU immunostaining was performed as described previously [22].
RNA Fluorescence In Situ Hybridization (FISH)
FISH was performed using the QuantiGene ViewRNA kit (Affymetrix Inc., Frederick, MD) according to the manufacturer’s instructions. Conditions were optimized to include 10-minute boiling and 10-minute protease treatments. The oligonucleotide probes were commercially designed using murine Ccl5 (accession number NM_013653.3) and Cxcl13 (NM_ 018866.2) sequences. Images were obtained on a Nikon Eclipse TE300 fluorescence inverted microscope (Nikon) and analyzed using MetaMorph image analysis software (Molecular Devices). Individual mRNA punctae were manually counted, and the number of mRNA molecules per DAPI+ cell was calculated.
Fluorescence-Activated Cell Sorting
CD11b+/CD45low microglia from pools of 9 to 10 optic nerves/set (FM and FMC) were collected and processed for antibody-mediated flow sorting (Supplementary Table 1) using appropriate controls for gating, as previously described [8], [37]. FACS samples were sorted directly into TRIzol (Life Technologies Corporation, Carlsbad, CA) for total RNA extraction. Sorting was performed at the High-Speed Cell Sorter Core Facility at the Siteman Cancer Center, Washington University, and data were subsequently analyzed using FlowJo (Tree Star, Inc., Ashland, OR).
RNA Extraction
TRIzol-chloroform extraction was used to isolate total RNA from flow-sorted microglia. Extracted RNA samples were resuspended in Ambion Nuclease-free water (Life Technologies), snap frozen, and stored at − 80°C. Before cDNA library construction and quantitative real-time polymerase chain reaction (qRT-PCR), residual DNA was eliminated with the TURBO DNA-free kit (Invitrogen). RNA quality was assayed using the Agilent Eukaryotic Total RNA 6000 and quantified using the Quant-iT RNA assay kit on a Qubit Fluorometer (Life Technologies).
RNA Sequencing
The Ovation RNA-Seq method was employed for cDNA synthesis according to the manufacturer’s instructions (NuGen, San Carlos, CA). cDNA was then concentrated and suspended in 10 mM Tris-HCl (pH 8) using MinElute spin columns (Qiagen, Valencia, CA). cDNA was quantified using the Quant-iT dsDNA HS Assay (Life Technologies Corporation), whereas the molecular weight distribution was determined using the BioAnalyzer 2100 and the Agilent DNA 7500 Chip Assay (Agilent Technologies, Santa Clara, CA). A total of 500 ng of cDNA (10ng/μl) was used for Illumina library construction with the Illumina paired-end LT indexing protocol as previously published [38], [39]. Briefly, 500 ng of each library was hybridized with the Agilent mouse exome reagent [40]. Eight mouse sample sets were sequenced from independently generated biological replicates that included four samples of FM microglia and four samples of tumor-associated microglia. Corresponding RNA-Seq paired-end reads were processed using the TopHat suite [41] with Cufflinks [42], [43], [44]. All RNA-sequencing data will be uploaded into Gene Expression Omnibus (GEO) database.
Quantitative Real-Time Polymerase Chain Reaction
The Ovation Pico WTA System V2 was employed for cDNA synthesis according to the manufacturer’s instructions (NuGen). cDNA was then purified using the MinElute Reaction Cleanup kit (Qiagen), followed by assessment of the concentration using a NanoDrop2000 spectrophotometer. qRT-PCR was performed using the Bio-Rad CFX96 Real-Time System (Bio-Rad Laboratories Inc., Hercules, CA) with SYBR Green detection (Life Technologies Corporation). Primer sequences were designed with Primer-BLAST (NCBI http://www.ncbi.nlm.nih.gov/tools/primer-blast/) to span exon-exon junctions and target known splice variants (Suppl. Table 2). The ∆∆CT method was used to calculate fold expression changes.
Human CCL5 Expression Analysis
CCL5 expression in human PAs was analyzed from the GEO dataset accession GSE44971 and GSE42656.
Statistical Analyses
All in vitro experiments were performed on independent litters, repeated at least three times, and analyzed in blinded fashion. Statistical analysis was performed using GraphPad Prism 5.0 software (GraphPad, La Jolla, CA). Data were presented as mean values with SEM. Data between two groups were compared using unpaired two-tailed Student’s t tests. Data among multiple groups were compared using Kruskal-Wallis test followed by Dunn’s multiple comparison testing, with a significance level set at P < .05. Grubbs outlier test was used to determine statistical outliers.
Results and Discussion
Optic Glioma–Associated Microglia Exhibit Morphologic Changes Suggestive of Activation
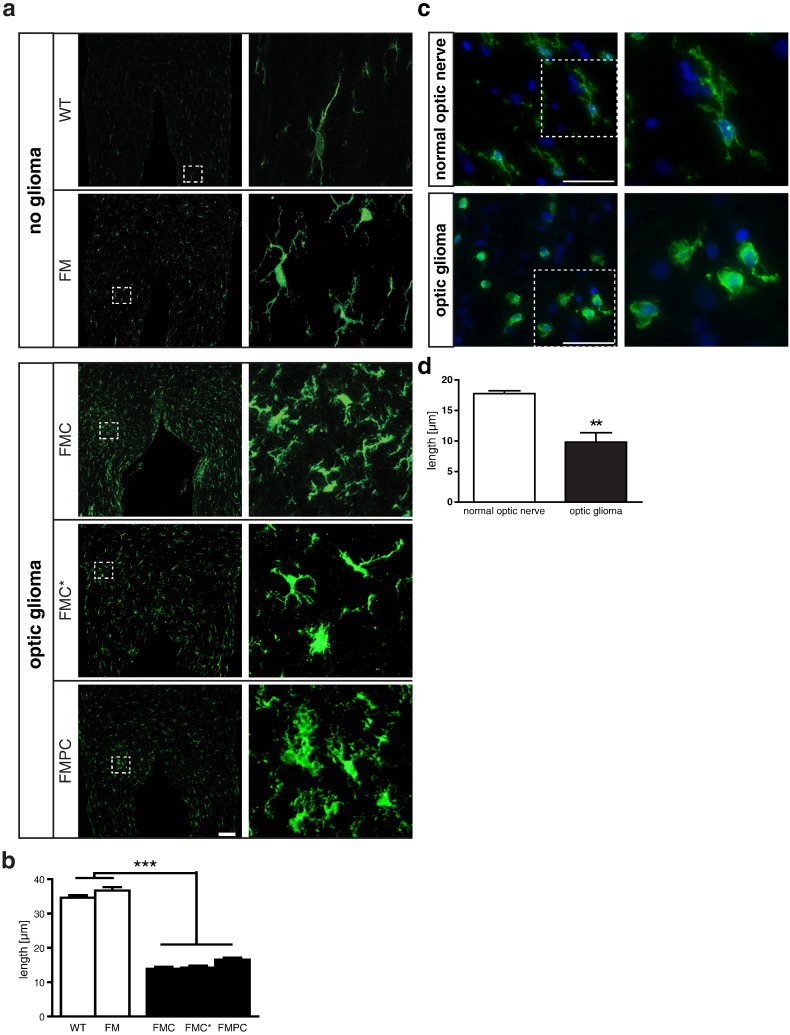
To examine microglia associated with mouse low-grade glioma, we employed three distinct models of Nf1 optic glioma. These three Nf1 murine optic glioma models differ by the timing of somatic Nf1 gene inactivation in neuroglial cells (FMC and FMC* mice; [18], [30]) or the presence of additional neoplastic cell genetic changes (heterozygous Pten mutation; FMPC mice; [45]) reported in rare NF1-associated optic pathway gliomas (OPGs) [46]. Previous studies from our laboratory have demonstrated that the tissue monocytes in these tumors represent resident microglia (CD11b+; CD45low) with robust Cx3cr1 expression rather than bone marrow–derived macrophages (CD11b+; CD45high) [8], [23]. Whereas prior studies of murine and human NF1 optic nerve gliomas demonstrated an increased percentage of microglia in these tumors [8], [18], [19], confocal image analysis of these microglia within the mouse optic gliomas also revealed striking morphological changes: Nonglioma (resident) microglia in the optic nerve from either WT or Nf1 +/− (FM) mice exhibited morphologies typical of ramified microglia, with elongated, fine processes that extended into their surroundings, whereas tumor-associated Nf1 +/− microglia (FMC, FMC*, and FMPC mice) harbored shorter processes that were slightly thickened or lacked processes completely (Figure 1, a and b). Similar changes in morphology were also observed in human OPG specimens relative to their normal tissue (optic nerve) counterparts (Figure 1, c and d), including one specimen from a child with NF1.
Figure 1.
Human and murine optic glioma–associated microglia exhibit morphological changes. (a) Whereas microglia from normal (WT and FM; n = 5/genotype) mouse optic nerves were ramified cells with long Iba1+ processes, glioma-associated (FMC, FMC*, and FMPC; n = 5/genotype) microglia were amoeboid-shaped monocytic cells with shorter processes. Scale bar, 100 μm. The differences in microglial process lengths were quantified in panel b. Similar to the mouse tumors, human optic glioma–-associated microglia also harbored short or no processes (c). The differences in microglial process lengths were quantified in panel d. DAPI (blue) was used as a counterstain to identify all cells in the sections. Scale bar, 50 μm. The dotted areas denote the regions from which the high-power images in the right panels derived. Each error bar represents the mean ± SEM. Asterisks denote statistically significant differences.
These pronounced changes in microglial morphology have previously been interpreted as microglia “activation” [47]. As such, under normal physiological conditions, microglia have surveillance functions, with an extensive array of fine processes that constantly scan their surroundings [48]. However, in the setting of brain pathology, like Alzheimer’s or Parkinson’s disease [49], microglia undergo morphological changes that include thickening and shortening, or even retraction, of their cellular processes to assume an “amoeboid” morphology. The presence of these morphologic changes in tumor-associated Nf1 +/− microglia prompted us to specifically examine these tissue macrophages as the source of potential glioma-maintaining stroma-derived growth factors [50], distinct from those made by resting Nf1 +/− microglia within the nonneoplastic optic nerve.
Ccl5 and Cxcl13 Are Highly Expressed Glioma-Associated Microglia Transcripts
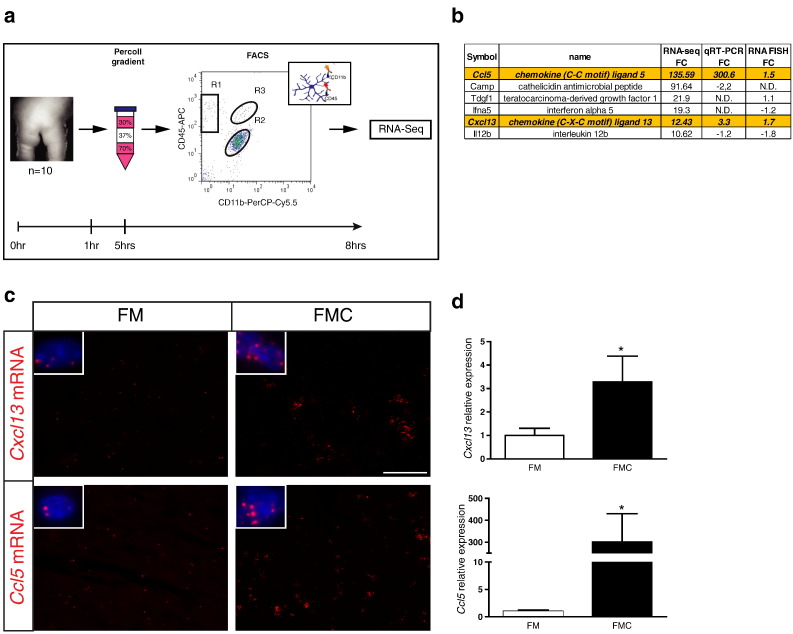
To identify potential gliomagens produced by tumor-associated microglia, we initiated a large-scale RNA-sequencing discovery effort. We were specifically interested in transcripts unique to Nf1+/− microglia within optic gliomas relative to those found in Nf1+/− microglia within the normal (non-neoplastic) optic nerve. Because the mouse optic nerve is composed of fewer than 10% microglia [23] and only ~ 2500 total cells per nerve can be recovered by FACS after processing, we pooled at least nine 3-month-old mice per genotype to obtain sufficient numbers of cells for FACS and RNA isolation. Because of the extended processing times (~ 8 hours) required, the small amounts of starting material, and the inherent cell loss associated with FACS separation, RNA isolated from microglia under these conditions is in low abundance and frequently of low quality (Figure 2a), necessitating optimization of RNA isolation and analysis methods [24], [25], [26], [27]. To circumvent these issues, we employed a combination of exome capture enrichment and Illumina RNA sequencing (cDNA-Capture sequencing) on microglia isolated from optic glioma–bearing FMC mice and Nf1 +/− non–tumor-bearing (FM) mice. Samples were analyzed using the Cufflinks platform [42], [43], [44] to calculate the differential expression of genes from each pool (FMC and FM microglia). We further narrowed the candidate list to transcripts whose predicted protein products were either secreted or cell surface associated and also exhibited at least a 10-fold change in expression (six genes). Of these six candidates, only Ccl5 and Cxcl13 were independently validated by RNA FISH and qRT-PCR as potential optic glioma–associated microglia gliomagens (Figure 2b). Other transcripts previously identified in studies using either high-grade glioma-associated or non–tumor-associated Nf1 +/− brain microglia maintained in vitro, including interleukin-6, interleukin-10 [51], [52], stem cell factor [53], transforming growth factor beta [54], hyaluronidase, hepatocyte growth factor [55], pleiotrophin, jagged-1, and insulin-like growth factor–1 [22], were not uniquely expressed by optic glioma–associated microglia relative to their nonneoplastic Nf1 +/− optic nerve microglia counterparts.
Figure 2.
Cxcl13 and Ccl5 expression are increased in murine optic glioma. (a) CD11b+ CD45low microglia from pools of 9 to 10 optic nerves/set (FM and FMC) were collected and processed for antibody-mediated flow sorting. (b) Six transcripts were identified that represented predicted secreted or cell surface–associated proteins with at least a 10-fold overexpression in FMC relative to FM microglia (six genes). FC, fold change (FMC/FM). N.D., not done. (c) RNA FISH demonstrated increased Cxcl13 and Ccl5 transcripts in optic glioma (FMC; n = 3) compared with non–tumor-bearing (FM; n = 6) optic nerves. Representative images are shown with insets of cells containing mRNA molecules (red). DAPI (blue) was used as a counterstain to identify all cells in the sections. Scale bar, 50 μm. (d) SYBR-Green qRT-PCR of independently generated FACS-isolated FMC microglia revealed increased Cxcl13 and Ccl5 expression relative to FM microglia. Each error bar represents the mean ± SEM. Asterisks denote statistically significant differences: (*) P = .0312.
Specifically, RNA FISH demonstrated increased Cxcl13 and Ccl5 expression in murine Nf1 optic glioma (FMC) nerves compared with FM control optic nerves (Figure 2c). In addition, microglia samples from the optic nerves of 3-month-old FM and FMC mice were independently isolated by Percoll density gradient centrifugation [56] and FACS. qRT-PCR analysis demonstrated that tumor-associated Nf1 +/− microglia had a 3.3-fold increase in Cxcl13 RNA expression and a 300-fold increase in Ccl5 RNA expression relative to FM (Nf1 +/−) microglia (Figure 2d). Collectively, these results implicate Ccl5 and Cxcl13 as potential tumor-associated microglia gliomagens.
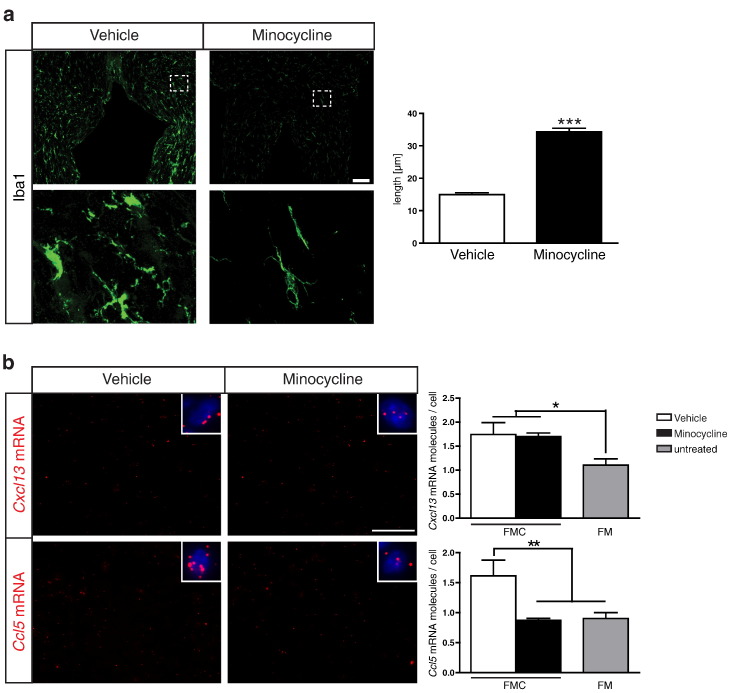
Minocycline Treatment Reduces Optic Glioma Microglial Ccl5 Expression
Previous studies have demonstrated that minocycline inactivation of microglia reduced Nf1 optic glioma proliferation in vivo [22]. To provide further support for a role of Ccl5 and Cxcl13 in mouse Nf1 optic glioma growth, FMC mice (n = 4 mice/group) were i.p. injected with either minocycline or vehicle over a 2-week period, and the optic nerves were analyzed by immunohistochemistry and RNA FISH. Following minocycline treatment, the morphology of microglia (thin, long, ramified processes) was restored to that observed in WT or FM mice (Figure 3a). Importantly, whereas minocycline treatment had no effect on Cxcl13 expression, Ccl5 expression was decreased to control (FM) levels (Figure 3b). These observations suggest that Ccl5, but not Cxcl13, is particularly worthy of further exploration as a potential tumor-associated microglia gliomagen.
Figure 3.
Minocycline treatment reduces Ccl5 mRNA expression. (a) Following minocycline hydrochloride treatment (n = 4 mice/group), Iba1+ microglia harbored long processes relative to vehicle-injected mice (n = 4 mice/group), similar to WT mice (Figure 1). Boxes indicate the regions from which the high-power images in the lower panels derived. Scale bar, 100 μm. The differences in microglial process lengths were quantified (right panel). (b) Whereas minocycline treatment resulted in no change in Cxcl13 expression in FMC optic nerves, Ccl5 mRNA expression was reduced to normal (FM) levels. Representative images are shown with insets of cells containing mRNA molecules (red). DAPI (blue) was used as a counterstain to identify all cells in the sections. Scale bar, 50 μm. Each error bar represents the mean ± SEM. Asterisks denote statistically significant differences: (*) P < .0225, (**) P = .0096, (***) P < .0001.
Ccl5 Is Important for Nf1 Optic Glioma Growth
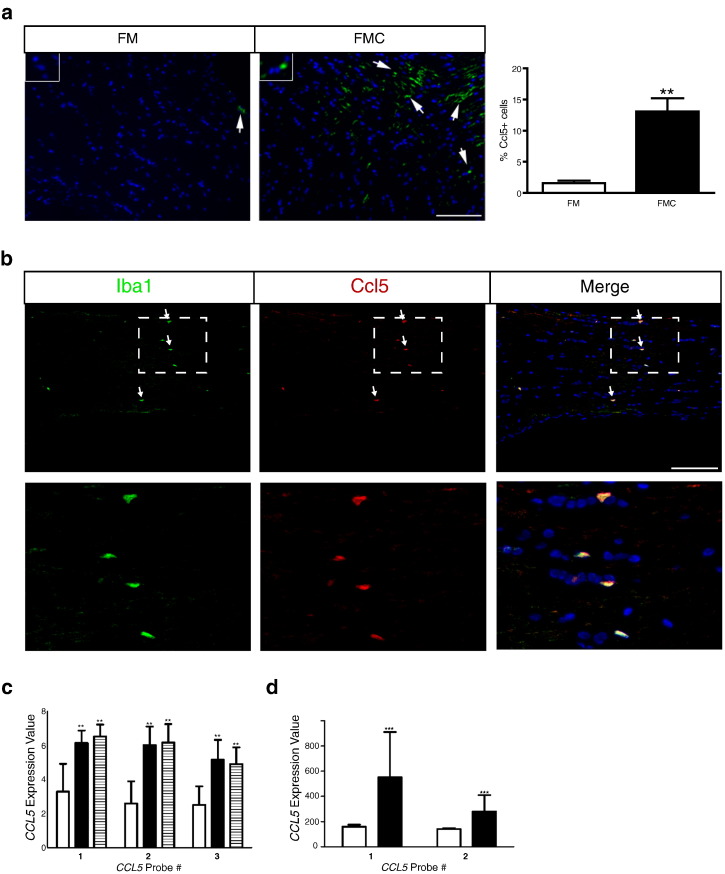
Because only Ccl5 expression was decreased following minocycline treatment, we focused subsequent experiments on this high-priority candidate. First, we examined Ccl5 expression at the protein level in FM and FMC optic nerves. Relative to their nonneoplastic counterparts, there was an 8.4-fold increase in the percentage of Ccl5+ cells in the optic gliomas (Figure 4a). Moreover, nearly all of the Ccl5+ cells were Iba1+ cells (microglia) in these tumors by double-labeling immunofluorescent microscopy (Figure 4b).
Figure 4.
Ccl5 expression is increased in murine OPGs and human PAs. (a) Immunofluorescence of murine optic nerve tissue reveals increased Ccl5 (green) protein expression in Nf1 optic glioma specimens (FMC, n = 5) relative to control optic nerve tissue (FM, n = 5). Quantification is represented in the adjacent graph. Each error bar represents the mean ± SEM. Asterisks denote statistically significant differences: (**) P = .0079. (b) Double labeling of Ccl5 and Iba1 reveals that the chemokine Ccl5 is expressed in Iba1+ cells (microglia). Scale bar, 50 μm. DAPI (blue) was used as a counterstain to identify all cells in the sections. Representative images are shown, with insets demonstrating immunopositive cells (arrows). (c) In the GSE44971 GEO dataset, CCL5 RNA expression in human PAs (n = 46 sporadic tumors and 3 from individuals with NF1) was increased for all three CCL5 probe sets relative to normal brain (n = 9). The white bars denote normal brain samples, the black bars denote the sporadic PA specimens, and the hatched bars denote the NF1-associated PAs. Each error bar represents the mean ± SD. (**) P = .0015 using a one-way analysis of variance with Bonferroni posttest correction. (d) In the GSE42656 GEO dataset, CCL5 RNA expression was also increased for both CCL5 probe sets. The white bars denote normal brain samples (n = 16), whereas the black bars denote the PA specimens (n = 14). Each error bar represents the mean ± SD. (***) P < .0001 using a two-way analysis of variance with Bonferroni post-test correction.
Second, because symptomatic NF1-associated optic gliomas are commonly treated without a prior tissue diagnosis (biopsy), we leveraged available transcriptomal human PA data sets (GSE42556 [57] and GSE44971 [58]). Analysis of the GSE42556 specimens revealed increased CCL5 expression (all three independent CCL5 probe sets) in sporadic PAs (n = 46) relative to normal brain controls (n = 9), including three additional tumors from individuals with NF1 (Figure 4c). Similarly, analysis of the GSE44971 specimens revealed increased CCL5 expression (both of the two independent CCL5 probe sets) in the sporadic PAs (n = 14) relative to normal brain controls (n = 16) (Figure 4d). In contrast, no increase in CXCL13 expression was observed in the PA tumors from either data set (data not shown), further underscoring the importance of CCL5 in low-grade glioma.
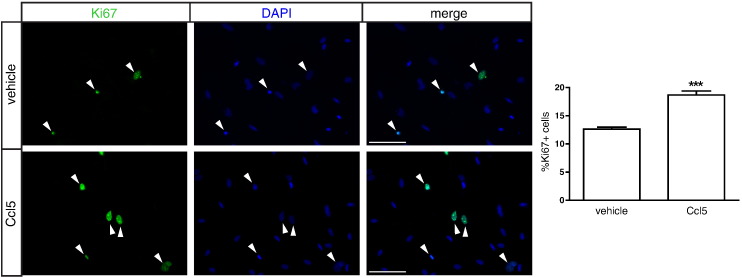
Third, to determine whether Ccl5 has the capacity to increase astrocyte growth in vitro, we employed Nf1-deficient optic nerve glia cultures following acute Nf1 gene inactivation (adenovirus-mediated Cre transduction). Because these cultures contain 70% NG2+ cells and 30% GFAP+ cells, in which only the GFAP+ astrocytes hyperproliferate following Nf1 gene inactivation [59], we analyzed the proliferation of optic nerve astroglial cells using Ki67 labeling in vitro. Following treatment with murine recombinant Ccl5 (250 ng/ml), there was a 1.5-fold increase in astrocyte proliferation (Figure 5), demonstrating that Ccl5 is sufficient to increase Nf1-deficient astrocyte growth.
Figure 5.
Ccl5 increases optic nerve astrocyte proliferation in vitro. Following incubation with 250 ng of murine recombinant Ccl5, there was an increase in Nf1-deficient astrocyte proliferation (Ki67 labeling) relative to vehicle treatment in vitro. Representative Ki67+ proliferating cells (green) and DAPI+ nuclei (blue) are shown. Arrowheads indicate Ki67-positive cells. Scale bar, 100 μm. Each error bar represents mean ± SEM. Asterisks denote statistically significant differences.
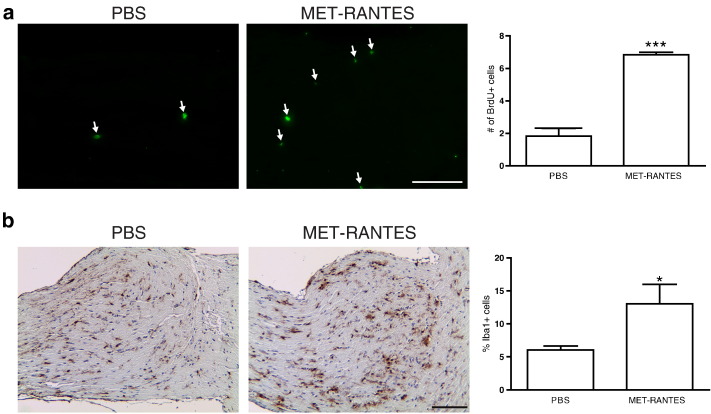
Fourth, to establish that increased microglial Ccl5 expression is necessary for Nf1 optic glioma growth, we sought to inhibit Ccl5 function in vivo. Because there are few selective small molecule inhibitors of Ccl5 that cross the blood-brain barrier, we initially employed MET-RANTES, an amino-terminal–modified methionylated Ccl5 derivative [60], [61] originally identified to competitively bind to Ccr1 and Ccr5 and inhibit the signaling pathways activated by Ccl5 [61]. In other experimental models of nervous system disease, mice treated with MET-RANTES had less severe clinical symptoms and reduced inflammation [62], [63], [64]. However, MET-RANTES–treated mice exhibited increased Nf1 optic glioma growth and more Iba1+ microglia compared with PBS-treated mice (Supplementary Figure 2). The paradoxical effect of MET-RANTES treatment is likely attributable to its function as a partial agonist [65], which prompted us to explore other methods to target Ccl5.
Supplementary Figure 2.
MET-RANTES treatment increases Nf1 optic glioma growth. MET-RANTES or vehicle (PBS) was injected into FMC mice, and 14 days after the injections, optic nerves were dissected. MET-RANTES–treated animals displayed an increase in (A) proliferation, as measured by BrdU incorporation, and (B) the percentage of Iba1+ microglia relative to animals treated with vehicle alone. Representative images are shown. Arrows denote immunopositive cells. Scale bar, 100 μm. Each error bar represents mean ± SEM. Asterisks denote statistically significant differences: (*) P = .0132, (***) P < .0006.
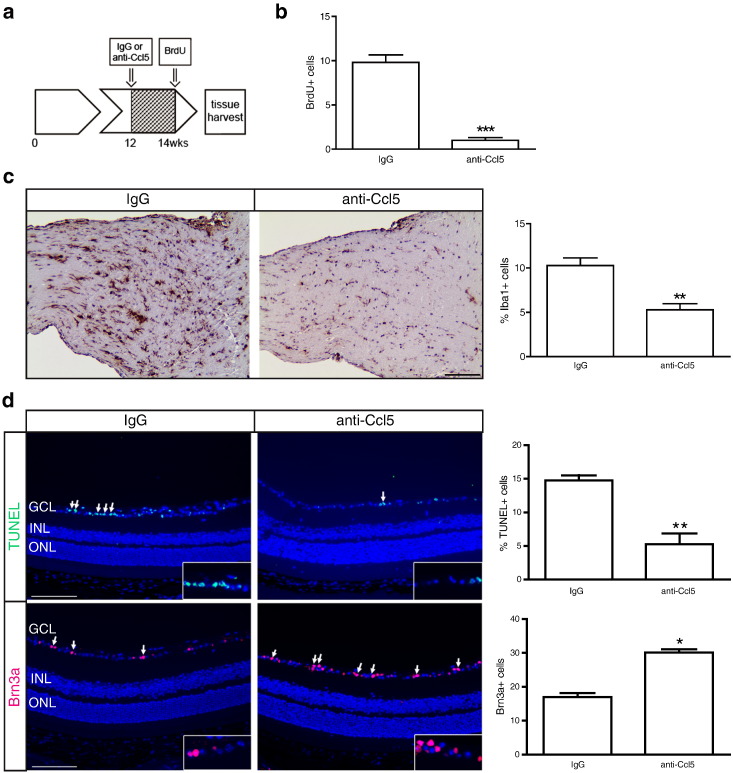
Previous preclinical studies have used anti-Ccl5 neutralizing antibodies to interfere with Ccl5 function and attenuate tumor growth in vivo [66], [67]. Specifically, treatment with Ccl5 antibody blocked leukocyte adhesion and infiltration in experimental mouse models of multiple sclerosis [68], [69]. Based on these studies, 3-month-old FMC mice were treated with either anti-Ccl5 (250 μg) or IgG isotype control antibodies i.p. daily for 2 weeks (n = 5 mice/group). Following the completion of treatment, mice were injected with BrdU (50 mg/kg), and the optic nerves were removed 3 hours later for analysis (Figure 6a). Anti-Ccl5 treatment resulted in a 9.8-fold reduction in Nf1 optic glioma proliferation (BrdU+ cells) relative to IgG control–treated mice in vivo (P < .0001; Figure 6b). Interestingly, consistent with the known chemoattractant properties of Ccl5 [70], [71], [72], anti-Ccl5 treatment also reduced the number of Iba1+ microglia into these tumors in vivo (Figure 6c).
Figure 6.
Ccl5 is necessary for murine Nf1 optic glioma growth in vivo. (a) At 3 months of age, FMC mice received i.p. injections of either Ccl5 neutralizing antibodies (n = 5 mice) or an IgG2A isotype-matched control antibody (n = 5 mice) every day for 2 weeks. Three hours before euthanasia, mice were i.p. injected with 50 mg/kg of BrdU. Ccl5 treatment decreased (b) murine Nf1 optic glioma proliferation as measured by BrdU incorporation and (c) the percentage of Iba1+ microglia. Scale bar, 100 μm. (d) Following anti-Ccl5 treatment, TUNEL staining (n = 5 mice/group; top panels) and Brn3a immunofluorescence (n = 5 mice/group; bottom panels) revealed decreased retinal cell apoptosis (%TUNEL+ cells) and increased RGC numbers (Brn3a+ cells) relative to IgG-treated controls. Representative individual TUNEL+ and Brn3a+ cells within the ganglion cell layer are shown in the insets. Quantification is represented in the adjacent graphs. Scale bar, 50 μm. Each error bar represents the mean ± SEM. Asterisks denote statistically significant differences. INL, inner nuclear layer; GCL, ganglion cell layer; ONL, outer nuclear layer.
Because optic gliomas cause clinical morbidity in children with NF1 as a result of reduced visual function [73], the impact of anti-Ccl5 treatment on optic glioma–induced retinal dysfunction was assessed. In Nf1 optic glioma mice, visual impairment occurs following retinal ganglion cell (RGC) apoptosis and loss [33]. Following anti-Ccl5 treatment, there were a 2.8-fold decrease in the percent of apoptotic (%TUNEL+) cells and a 1.78-fold increase in RGCs (Brn3a+ cells; Figure 6d) relative to IgG-treated controls. Collectively, these findings demonstrate that microglia-produced Ccl5 is both necessary and sufficient to increase Nf1-deficient astrocyte proliferation relevant to Nf1 optic glioma maintenance and that inhibiting Ccl5 function reduces retinal pathology in the setting of murine Nf1 optic glioma.
The identification of Ccl5 as a candidate gliomagen in Nf1 murine optic glioma suggests a new stromal target for therapeutic drug design. Ccl5 is also known as RANTES (regulated upon activation, normal T-cell–expressed and secreted), where it was originally identified as an inducer of leukocyte recruitment to sites of inflammation [74]. Ccl5 induces leukocyte migration by binding to three distinct seven-transmembrane G-protein–coupled receptors [75], [76], [77]. In the setting of brain pathology, increased CCL5 expression in astrocytes and microglia has been reported following viral infection [78], [79], [80], [81], where its inhibition results in decreased leukocyte adhesion within the microcirculation of infected mice [82]. Importantly, mice lacking Ccr5 expression exhibit reduced neuronal injury in responses to HIV-1 infection [83], supporting a general role for CCL5 signaling in neurologic disease.
One previous study examined CCL5 in malignant glioma, demonstrating increased expression of CCL5 and its receptors (CCR1, CCR5) in the murine high-grade GL261 astrocytoma cell line [84]. In contrast, CCL5 function in low-grade gliomas has not been explored, and the impact of CCL5 function on glioma biology remains unclear. Whereas little is known about CCL5 in brain tumors, increased CCL5 expression has been reported in lung, prostate, melanoma, colorectal, and breast cancer [67], [85], [86], [87], [88], where a positive correlation between increased CCL5 expression and disease progression was identified [89], [90]. In addition, CCL5-expressing melanoma cells form increasingly aggressive tumors in a concentration-dependent fashion [91], and Ccl5 increases the growth of breast cancer cells in vitro [92]. Finally, blocking Ccl5 receptor binding using neutralizing antibodies or siRNA knockdown reduces tumor growth in experimental models of pancreatic adenocarcinoma [93], colon cancer [67], and gastric cancer [94].
In the current study, we provide experimental evidence for a critical role for Ccl5 in the maintenance of murine Nf1 optic glioma. Following inhibition of Ccl5 with neutralizing antibodies, there were reduced tumor proliferation and attenuated retinal pathology. In addition, Ccl5 inhibition resulted in decreased microglia within the optic glioma, suggesting that Ccl5 may be a key stromal determinant of tumor growth and associated optic nerve damage as well as monocyte recruitment to the glioma. Although we specifically chose to directly inhibit Ccl5 function, additional preclinical investigation could involve the use of Ccl5 receptor antagonists. It should be recognized that Ccl5 can bind to one of three receptors (Ccr1, Ccr3, and Ccr5) to promote cell growth [95]. However, only Ccr1 and Ccr5 are expressed by optic nerve astrocytes (data not shown), complicating the decision regarding which Ccl5 receptor to inhibit. In this regard, Ccr5 antagonism using Maraviroc [96] had no effect on Nf1 optic glioma growth (data not shown), whereas MET-RANTES increased Nf1 optic glioma growth. Based on these findings, future preclinical studies will need to carefully consider the specific effects of each Ccl5 inhibitor and the engagement of its specific cognate receptors.
In summary, the observations in this report not only underscore the importance of microglia in the glioma microenvironment in maintaining tumor growth but also expand our understanding of the role of stroma-derived chemokines in glioma biology. In this regard, CXCL12 is produced by both microglia [97] and endothelial cells [98], [99], where it can dually act to further attract additional microglia as well as independently stimulate tumor growth. In addition, chemokines can also be produced by glioma cells to direct microglia migration and recruitment. For example, CX3CL1 acting on the microglial CX3CR1 receptor is critical for microglia infiltration and tumor formation in mouse Nf1 optic glioma strains [23]. Similarly, colony-stimulating factor 1 (CSF-1) produced by mouse glioblastoma cells attracts microglia through the CSF-1 receptor to further increase glioma growth [100], such that attenuation of this CSF-1/CSF-1 receptor axis reduced murine glioblastoma growth in vivo [15]. Collectively, these studies suggest that future brain tumor therapies might target the unique interplay between stromal cells and cancer cells important for maintaining this specialized ecosystem.
The following are the supplementary data related to this article.
Supplementary material.
Conflict of Interest
The authors declare no conflicts of interest.
Authors’ Contributions
W. W. P., V. M., M. H. E., E. R. M., and D. H. G. jointly supervised research. A. C. S., W. W. P., E. R. M., and D. H. G. conceived and designed the experiments. J. W., T. W., V. M., and D. H. contributed to experimental design. A. C. S., W. W. P., P. J. C., K. Y. K., and J. A. T. performed the experiments. A. C. S., W. W. P., J. A. T., J. W., T. W., V. M., M. G., and O. L. G. analyzed the data. V. M., M. H. E., E. R. M., and D. H. G. contributed reagents/materials/analysis tools. A. C. S., W. W. P., K. Y. K., J. A. T., J. W., T. W., V. M., M. G., and O. L. G. contributed to the preparation of the manuscript. A. C. S., W. W. P., and D. H. G. wrote the manuscript. All authors edited and approved the manuscript.
Acknowledgements
We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, MO, for the use of the Siteman Flow Cytometry Core, which provided FACS services, and the Washington University Siteman Cancer Center Tumor Tissue Repository. The Siteman Cancer Center is supported in part by the NCI Cancer Center Support Grant #P30 CA91842. We additionally thank Vanessa Chu for technical assistance. This work was supported by grants from the National Cancer Institute (U01-CA160882 and U01-CA141549 to D. H. G.) and the National Institutes of Health (RC4 NS072916 to D. H. G.). W. W. P. was partly supported by a grant from the W.M. Keck Foundation. J.A.T. was supported by funding from the Research Training Program in the Vision Sciences (5-T32-EY13360) and in Neurology (5-T32-NS007205-33). M. H. E. and D. H. G. were supported by a grant from the James S. McDonnell Foundation.
Footnotes
This work was supported by grants from the National Cancer Institute (U01-CA160882 and U01-CA141549 to D. H. G.) and the National Institutes of Health (RC4 NS072916 to D. H. G.). W. W. P. was partly supported by a grant from the W.M. Keck Foundation. J.A.T. was supported by funding from the Research Training Program in the Vision Sciences (5-T32-EY13360) and in Neurology (5-T32-NS007205-33). M. H. E. and D. H. G. were supported by a grant from the James S. McDonnell Foundation.
References
- 1.Tremblay G. Stromal aspects of breast carcinoma. Exp Mol Pathol. 1979;31(1):248–260. doi: 10.1016/0014-4800(79)90026-1. [DOI] [PubMed] [Google Scholar]
- 2.Folkman J. Anti-angiogenesis: new concept for therapy of solid tumors. Ann Surg. 1972;175(3):409–416. doi: 10.1097/00000658-197203000-00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willett C.G., Boucher Y., di Tomaso E., Duda D.G., Munn L.L., Tong R.T. Direct evidence that the VEGF-specific antibody bevacizumab has antivascular effects in human rectal cancer. Nat Med. 2004;10(2):145–147. doi: 10.1038/nm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pechman K.R., Donohoe D.L., Bedekar D.P., Kurpad S.N., Schmainda K.M. Evaluation of combined bevacizumab plus irinotecan therapy in brain tumors using magnetic resonance imaging measures of relative cerebral blood volume. Magn Reson Med. 2012;68(4):1266–1272. doi: 10.1002/mrm.23315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gutmann D.H., McLellan M.D., Hussain I., Wallis J.W., Fulton L.L., Fulton R.S. Somatic neurofibromatosis type 1 (NF1) inactivation characterizes NF1-associated pilocytic astrocytoma. Genome Res. 2013;23(3):431–439. doi: 10.1101/gr.142604.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morantz R.A., Wood G.W., Foster M., Clark M., Gollahon K. Macrophages in experimental and human brain tumors. Part 2: studies of the macrophage content of human brain tumors. J Neurosurg. 1979;50(3):305–311. doi: 10.3171/jns.1979.50.3.0305. [DOI] [PubMed] [Google Scholar]
- 7.Rossi M.L., Hughes J.T., Esiri M.M., Coakham H.B., Brownell D.B. Immunohistological study of mononuclear cell infiltrate in malignant gliomas. Acta Neuropathol. 1987;74(3):269–277. doi: 10.1007/BF00688191. [DOI] [PubMed] [Google Scholar]
- 8.Simmons G.W., Pong W.W., Emnett R.J., White C.R., Gianino S.M., Rodriguez F.J. Neurofibromatosis-1 heterozygosity increases microglia in a spatially and temporally restricted pattern relevant to mouse optic glioma formation and growth. J Neuropathol Exp Neurol. 2011;70(1):51–62. doi: 10.1097/NEN.0b013e3182032d37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roggendorf W., Strupp S., Paulus W. Distribution and characterization of microglia/macrophages in human brain tumors. Acta Neuropathol. 1996;92(3):288–293. doi: 10.1007/s004010050520. [DOI] [PubMed] [Google Scholar]
- 10.Bettinger I., Thanos S., Paulus W. Microglia promote glioma migration. Acta Neuropathol. 2002;103(4):351–355. doi: 10.1007/s00401-001-0472-x. [DOI] [PubMed] [Google Scholar]
- 11.Yongjun Y., Shuyun H., Lei C., Xiangrong C., Zhilin Y., Yiquan K. Atorvastatin suppresses glioma invasion and migration by reducing microglial MT1-MMP expression. J Neuroimmunol. 2013;260(1–2):1–8. doi: 10.1016/j.jneuroim.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 12.Fonseca A.C., Romao L., Amaral R.F., Assad Kahn S., Lobo D., Martins S. Microglial stress inducible protein 1 promotes proliferation and migration in human glioblastoma cells. Neuroscience. 2012;200:130–141. doi: 10.1016/j.neuroscience.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 13.Markovic D.S., Vinnakota K., Chirasani S., Synowitz M., Raguet H., Stock K. Gliomas induce and exploit microglial MT1-MMP expression for tumor expansion. Proc Natl Acad Sci U S A. 2009;106(30):12530–12535. doi: 10.1073/pnas.0804273106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blacher E., Ben Baruch B., Levy A., Geva N., Green K.D., Garneau-Tsodikova S. Inhibition of glioma progression by a newly discovered CD38 inhibitor. Int J Cancer. 2015;136(6):1422–1433. doi: 10.1002/ijc.29095. [DOI] [PubMed] [Google Scholar]
- 15.Pyonteck S.M., Akkari L., Schuhmacher A.J., Bowman R.L., Sevenich L., Quail D.F. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19(10):1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guillamo J.S., Creange A., Kalifa C., Grill J., Rodriguez D., Doz F. Prognostic factors of CNS tumours in neurofibromatosis 1 (NF1): a retrospective study of 104 patients. Brain. 2003;126(Pt 1):152–160. doi: 10.1093/brain/awg016. [DOI] [PubMed] [Google Scholar]
- 17.Legius E., Marchuk D.A., Collins F.S., Glover T.W. Somatic deletion of the neurofibromatosis type 1 gene in a neurofibrosarcoma supports a tumour suppressor gene hypothesis. Nat Genet. 1993;3(2):122–126. doi: 10.1038/ng0293-122. [DOI] [PubMed] [Google Scholar]
- 18.Bajenaru M.L., Hernandez M.R., Perry A., Zhu Y., Parada L.F., Garbow J.R. Optic nerve glioma in mice requires astrocyte Nf1 gene inactivation and Nf1 brain heterozygosity. Cancer Res. 2003;63(24):8573–8577. [PubMed] [Google Scholar]
- 19.Kim K.Y., Ju W.K., Hegedus B., Gutmann D.H., Ellisman M.H. Ultrastructural characterization of the optic pathway in a mouse model of neurofibromatosis-1 optic glioma. Neuroscience. 2010;170(1):178–188. doi: 10.1016/j.neuroscience.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y.H., McGowan L.D., Cimino P.J., Dahiya S., Leonard J.R., Lee da Y. Mouse low-grade gliomas contain cancer stem cells with unique molecular and functional properties. Cell Rep. 2015;10(11):1899–1912. doi: 10.1016/j.celrep.2015.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daginakatte G.C., Gianino S.M., Zhao N.W., Parsadanian A.S., Gutmann D.H. Increased c-Jun-NH2-kinase signaling in neurofibromatosis-1 heterozygous microglia drives microglia activation and promotes optic glioma proliferation. Cancer Res. 2008;68(24):10358–10366. doi: 10.1158/0008-5472.CAN-08-2506. [DOI] [PubMed] [Google Scholar]
- 22.Daginakatte G.C., Gutmann D.H. Neurofibromatosis-1 (Nf1) heterozygous brain microglia elaborate paracrine factors that promote Nf1-deficient astrocyte and glioma growth. Hum Mol Genet. 2007;16(9):1098–1112. doi: 10.1093/hmg/ddm059. [DOI] [PubMed] [Google Scholar]
- 23.Pong W.W., Higer S.B., Gianino S.M., Emnett R.J., Gutmann D.H. Reduced microglial CX3CR1 expression delays neurofibromatosis-1 glioma formation. Ann Neurol. 2013;73(2):303–308. doi: 10.1002/ana.23813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pong W.W., Walker J., Wylie T., Magrini V., Luo J., Emnett R.J. F11R is a novel monocyte prognostic biomarker for malignant glioma. PLoS One. 2013;8(10):e77571. doi: 10.1371/journal.pone.0077571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tariq M.A., Kim H.J., Jejelowo O., Pourmand N. Whole-transcriptome RNAseq analysis from minute amount of total RNA. Nucleic Acids Res. 2011;39(18):e120. doi: 10.1093/nar/gkr547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cabanski C.R., Magrini V., Griffith M., Griffith O.L., McGrath S., Zhang J. cDNA hybrid capture improves transcriptome analysis on low-input and archived samples. J Mol Diagn. 2014;16(4):440–451. doi: 10.1016/j.jmoldx.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solga A.C., Pong W.W., Walker J., Wylie T., Magrini V., Apicelli A.J. RNA-sequencing reveals oligodendrocyte and neuronal transcripts in microglia relevant to central nervous system disease. Glia. 2014;63(4):531–548. doi: 10.1002/glia.22754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Y., Romero M.I., Ghosh P., Ye Z., Charnay P., Rushing E.J. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15(7):859–876. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bajenaru M.L., Zhu Y., Hedrick N.M., Donahoe J., Parada L.F., Gutmann D.H. Astrocyte-specific inactivation of the neurofibromatosis 1 gene (NF1) is insufficient for astrocytoma formation. Mol Cell Biol. 2002;22(14):5100–5113. doi: 10.1128/MCB.22.14.5100-5113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Y., Harada T., Liu L., Lush M.E., Guignard F., Harada C. Inactivation of NF1 in CNS causes increased glial progenitor proliferation and optic glioma formation. Development. 2005;132(24):5577–5588. doi: 10.1242/dev.02162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hegedus B., Banerjee D., Yeh T.H., Rothermich S., Perry A., Rubin J.B. Preclinical cancer therapy in a mouse model of neurofibromatosis-1 optic glioma. Cancer Res. 2008;68(5):1520–1528. doi: 10.1158/0008-5472.CAN-07-5916. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez F.J., Ligon A.H., Horkayne-Szakaly I., Rushing E.J., Ligon K.L., Vena N. BRAF duplications and MAPK pathway activation are frequent in gliomas of the optic nerve proper. J Neuropathol Exp Neurol. 2012;71(9):789–794. doi: 10.1097/NEN.0b013e3182656ef8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaul A., Toonen J.A., Gianino S.M., Gutmann D.H. The impact of coexisting genetic mutations on murine optic glioma biology. Neuro-Oncology. 2014;17(5):670–677. doi: 10.1093/neuonc/nou287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lesche R., Groszer M., Gao J., Wang Y., Messing A., Sun H. Cre/loxP-mediated inactivation of the murine Pten tumor suppressor gene. Genesis. 2002;32(2):148–149. doi: 10.1002/gene.10036. [DOI] [PubMed] [Google Scholar]
- 35.Yeh T.H., Lee da Y., Gianino S.M., Gutmann D.H. Microarray analyses reveal regional astrocyte heterogeneity with implications for neurofibromatosis type 1 (NF1)–regulated glial proliferation. Glia. 2009;57(11):1239–1249. doi: 10.1002/glia.20845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hegedus B., Dasgupta B., Shin J.E., Emnett R.J., Hart-Mahon E.K., Elghazi L. Neurofibromatosis-1 regulates neuronal and glial cell differentiation from neuroglial progenitors in vivo by both cAMP- and Ras-dependent mechanisms. Cell Stem Cell. 2007;1(4):443–457. doi: 10.1016/j.stem.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 37.Garcia J.A., Cardona S.M., Cardona A.E. Isolation and analysis of mouse microglial cells. Curr Protoc Immunol. 2014;104 doi: 10.1002/0471142735.im1435s104. [Unit 14 35] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Govindan R., Ding L., Griffith M., Subramanian J., Dees N.D., Kanchi K.L. Genomic landscape of non–small cell lung cancer in smokers and never-smokers. Cell. 2012;150(6):1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mardis E.R., Ding L., Dooling D.J., Larson D.E., McLellan M.D., Chen K. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsushita H., Vesely M.D., Koboldt D.C., Rickert C.G., Uppaluri R., Magrini V.J. Cancer exome analysis reveals a T-cell–dependent mechanism of cancer immunoediting. Nature. 2012;482(7385):400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts A., Pimentel H., Trapnell C., Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27(17):2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 43.Roberts A., Trapnell C., Donaghey J., Rinn J.L., Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12(3):R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaul A., Toonen J.A., Gianino S.M., Gutmann D.H. The impact of coexisting genetic mutations on murine optic glioma biology. Neuro Oncol. 2014;17(6):843–853. doi: 10.1093/neuonc/nou287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez E.F., Scheithauer B.W., Giannini C., Rynearson A., Cen L., Hoesley B. PI3K/AKT pathway alterations are associated with clinically aggressive and histologically anaplastic subsets of pilocytic astrocytoma. Acta Neuropathol. 2011;121(3):407–420. doi: 10.1007/s00401-010-0784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker F.R., Beynon S.B., Jones K.A., Zhao Z., Kongsui R., Cairns M. Dynamic structural remodelling of microglia in health and disease: a review of the models, the signals and the mechanisms. Brain Behav Immun. 2014;37:1–14. doi: 10.1016/j.bbi.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 48.Nimmerjahn A., Kirchhoff F., Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308(5726):1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 49.Doorn K.J., Goudriaan A., Blits-Huizinga C., Bol J.G., Rozemuller A.J., Hoogland P.V. Increased amoeboid microglial density in the olfactory bulb of Parkinson's and Alzheimer's patients. Brain Pathol. 2014;24(2):152–165. doi: 10.1111/bpa.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kremlev S.G., Roberts R.L., Palmer C. Differential expression of chemokines and chemokine receptors during microglial activation and inhibition. J Neuroimmunol. 2004;149(1–2):1–9. doi: 10.1016/j.jneuroim.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 51.Huettner C., Czub S., Kerkau S., Roggendorf W., Tonn J.C. Interleukin 10 is expressed in human gliomas in vivo and increases glioma cell proliferation and motility in vitro. Anticancer Res. 1997;17(5A):3217–3224. [PubMed] [Google Scholar]
- 52.Goswami S., Gupta A., Sharma S.K. Interleukin-6–mediated autocrine growth promotion in human glioblastoma multiforme cell line U87MG. J Neurochem. 1998;71(5):1837–1845. doi: 10.1046/j.1471-4159.1998.71051837.x. [DOI] [PubMed] [Google Scholar]
- 53.Berdel W.E., de Vos S., Maurer J., Oberberg D., von Marschall Z., Schroeder J.K. Recombinant human stem cell factor stimulates growth of a human glioblastoma cell line expressing c-kit protooncogene. Cancer Res. 1992;52(12):3498–3502. [PubMed] [Google Scholar]
- 54.Li W., Graeber M.B. The molecular profile of microglia under the influence of glioma. Neuro Oncol. 2012;14(8):958–978. doi: 10.1093/neuonc/nos116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koochekpour S., Jeffers M., Rulong S., Taylor G., Klineberg E., Hudson E.A. Met and hepatocyte growth factor/scatter factor expression in human gliomas. Cancer Res. 1997;57(23):5391–5398. [PubMed] [Google Scholar]
- 56.Campanella M., Sciorati C., Tarozzo G., Beltramo M. Flow cytometric analysis of inflammatory cells in ischemic rat brain. Stroke. 2002;33(2):586–592. doi: 10.1161/hs0202.103399. [DOI] [PubMed] [Google Scholar]
- 57.Henriquez N.V., Forshew T., Tatevossian R., Ellis M., Richard-Loendt A., Rogers H. Comparative expression analysis reveals lineage relationships between human and murine gliomas and a dominance of glial signatures during tumor propagation in vitro. Cancer Res. 2013;73(18):5834–5844. doi: 10.1158/0008-5472.CAN-13-1299. [DOI] [PubMed] [Google Scholar]
- 58.Lambert S.R., Witt H., Hovestadt V., Zucknick M., Kool M., Pearson D.M. Differential expression and methylation of brain developmental genes define location-specific subsets of pilocytic astrocytoma. Acta Neuropathol. 2013;126(2):291–301. doi: 10.1007/s00401-013-1124-7. [DOI] [PubMed] [Google Scholar]
- 59.Solga A.C., Gianino S.M., Gutmann D.H. NG2-cells are not the cell of origin for murine neurofibromatosis-1 (Nf1) optic glioma. Oncogene. 2014;33(3):289–299. doi: 10.1038/onc.2012.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Proudfoot A.E., Power C.A., Hoogewerf A.J., Montjovent M.O., Borlat F., Offord R.E. Extension of recombinant human RANTES by the retention of the initiating methionine produces a potent antagonist. J Biol Chem. 1996;271(5):2599–2603. doi: 10.1074/jbc.271.5.2599. [DOI] [PubMed] [Google Scholar]
- 61.Proudfoot A.E., Buser R., Borlat F., Alouani S., Soler D., Offord R.E. Amino-terminally modified RANTES analogues demonstrate differential effects on RANTES receptors. J Biol Chem. 1999;274(45):32478–32485. doi: 10.1074/jbc.274.45.32478. [DOI] [PubMed] [Google Scholar]
- 62.Huang Y., Jiao S., Tao X., Tang Q., Jiao W., Xiao J. Met-CCL5 represents an immunotherapy strategy to ameliorate rabies virus infection. J Neuroinflammation. 2014;11(1):146. doi: 10.1186/s12974-014-0146-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Plater-Zyberk C., Hoogewerf A.J., Proudfoot A.E., Power C.A., Wells T.N. Effect of a CC chemokine receptor antagonist on collagen induced arthritis in DBA/1 mice. Immunol Lett. 1997;57(1–3):117–120. doi: 10.1016/s0165-2478(97)00075-8. [DOI] [PubMed] [Google Scholar]
- 64.Shahrara S., Proudfoot A.E., Woods J.M., Ruth J.H., Amin M.A., Park C.C. Amelioration of rat adjuvant-induced arthritis by Met-RANTES. Arthritis Rheum. 2005;52(6):1907–1919. doi: 10.1002/art.21033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong M., Uddin S., Majchrzak B., Huynh T., Proudfoot A.E., Platanias L.C. Rantes activates Jak2 and Jak3 to regulate engagement of multiple signaling pathways in T cells. J Biol Chem. 2001;276(14):11427–11431. doi: 10.1074/jbc.M010750200. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Y., Lv D., Kim H.J., Kurt R.A., Bu W., Li Y. A novel role of hematopoietic CCL5 in promoting triple-negative mammary tumor progression by regulating generation of myeloid-derived suppressor cells. Cell Res. 2013;23(3):394–408. doi: 10.1038/cr.2012.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cambien B., Richard-Fiardo P., Karimdjee B.F., Martini V., Ferrua B., Pitard B. CCL5 neutralization restricts cancer growth and potentiates the targeting of PDGFRbeta in colorectal carcinoma. PLoS One. 2011;6(12):e28842. doi: 10.1371/journal.pone.0028842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Glass W.G., Hickey M.J., Hardison J.L., Liu M.T., Manning J.E., Lane T.E. Antibody targeting of the CC chemokine ligand 5 results in diminished leukocyte infiltration into the central nervous system and reduced neurologic disease in a viral model of multiple sclerosis. J Immunol. 2004;172(7):4018–4025. doi: 10.4049/jimmunol.172.7.4018. [DOI] [PubMed] [Google Scholar]
- 69.dos Santos A.C., Barsante M.M., Arantes R.M., Bernard C.C., Teixeira M.M., Carvalho-Tavares J. CCL2 and CCL5 mediate leukocyte adhesion in experimental autoimmune encephalomyelitis—an intravital microscopy study. J Neuroimmunol. 2005;162(1–2):122–129. doi: 10.1016/j.jneuroim.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 70.Lee J.K., Schuchman E.H., Jin H.K., Bae J.S. Soluble CCL5 derived from bone marrow–derived mesenchymal stem cells and activated by amyloid beta ameliorates Alzheimer's disease in mice by recruiting bone marrow–induced microglia immune responses. Stem Cells. 2012;30(7):1544–1555. doi: 10.1002/stem.1125. [DOI] [PubMed] [Google Scholar]
- 71.Schall T.J., Bacon K., Toy K.J., Goeddel D.V. Selective attraction of monocytes and T lymphocytes of the memory phenotype by cytokine RANTES. Nature. 1990;347(6294):669–671. doi: 10.1038/347669a0. [DOI] [PubMed] [Google Scholar]
- 72.Skuljec J., Sun H., Pul R., Benardais K., Ragancokova D., Moharregh-Khiabani D. CCL5 induces a pro-inflammatory profile in microglia in vitro. Cell Immunol. 2011;270(2):164–171. doi: 10.1016/j.cellimm.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 73.Listernick R., Ferner R.E., Liu G.T., Gutmann D.H. Optic pathway gliomas in neurofibromatosis-1: controversies and recommendations. Ann Neurol. 2007;61(3):189–198. doi: 10.1002/ana.21107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tillie-Leblond I., Hammad H., Desurmont S., Pugin J., Wallaert B., Tonnel A.B. CC chemokines and interleukin-5 in bronchial lavage fluid from patients with status asthmaticus. Potential implication in eosinophil recruitment. Am J Respir Crit Care Med. 2000;162(2 Pt 1):586–592. doi: 10.1164/ajrccm.162.2.9907014. [DOI] [PubMed] [Google Scholar]
- 75.Raport C.J., Gosling J., Schweickart V.L., Gray P.W., Charo I.F. Molecular cloning and functional characterization of a novel human CC chemokine receptor (CCR5) for RANTES, MIP-1beta, and MIP-1alpha. J Biol Chem. 1996;271(29):17161–17166. doi: 10.1074/jbc.271.29.17161. [DOI] [PubMed] [Google Scholar]
- 76.Heath H., Qin S., Rao P., Wu L., LaRosa G., Kassam N. Chemokine receptor usage by human eosinophils. The importance of CCR3 demonstrated using an antagonistic monoclonal antibody. J Clin Invest. 1997;99(2):178–184. doi: 10.1172/JCI119145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pakianathan D.R., Kuta E.G., Artis D.R., Skelton N.J., Hebert C.A. Distinct but overlapping epitopes for the interaction of a CC-chemokine with CCR1, CCR3 and CCR5. Biochemistry. 1997;36(32):9642–9648. doi: 10.1021/bi970593z. [DOI] [PubMed] [Google Scholar]
- 78.Vago L., Nebuloni M., Bonetto S., Pellegrinelli A., Zerbi P., Ferri A. Rantes distribution and cellular localization in the brain of HIV-infected patients. Clin Neuropathol. 2001;20(4):139–145. [Research Support, Non-U.S. Gov't] [PubMed] [Google Scholar]
- 79.Avdoshina V., Biggio F., Palchik G., Campbell L.A., Mocchetti I. Morphine induces the release of CCL5 from astrocytes: potential neuroprotective mechanism against the HIV protein gp120. Glia. 2010;58(13):1630–1639. doi: 10.1002/glia.21035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ovanesov M.V., Ayhan Y., Wolbert C., Moldovan K., Sauder C., Pletnikov M.V. Astrocytes play a key role in activation of microglia by persistent Borna disease virus infection. J Neuroinflammation. 2008;5:50. doi: 10.1186/1742-2094-5-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lokensgard J.R., Hu S., Sheng W., vanOijen M., Cox D., Cheeran M.C. Robust expression of TNF-alpha, IL-1beta, RANTES, and IP-10 by human microglial cells during nonproductive infection with herpes simplex virus. J Neurovirol. 2001;7(3):208–219. doi: 10.1080/13550280152403254. [DOI] [PubMed] [Google Scholar]
- 82.Vilela M.C., Mansur D.S., Lacerda-Queiroz N., Rodrigues D.H., Lima G.K., Arantes R.M. The chemokine CCL5 is essential for leukocyte recruitment in a model of severe Herpes simplex encephalitis. Ann N Y Acad Sci. 2009;1153:256–263. doi: 10.1111/j.1749-6632.2008.03959.x. [DOI] [PubMed] [Google Scholar]
- 83.Maung R., Hoefer M.M., Sanchez A.B., Sejbuk N.E., Medders K.E., Desai M.K. CCR5 knockout prevents neuronal injury and behavioral impairment induced in a transgenic mouse model by a CXCR4-using HIV-1 glycoprotein 120. J Immunol. 2014;193(4):1895–1910. doi: 10.4049/jimmunol.1302915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pham K., Luo D., Liu C., Harrison J.K. CCL5, CCR1 and CCR5 in murine glioblastoma: immune cell infiltration and survival rates are not dependent on individual expression of either CCR1 or CCR5. J Neuroimmunol. 2012;246(1–2):10–17. doi: 10.1016/j.jneuroim.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yun H.M., Park K.R., Park M.H., Kim D.H., Jo M.R., Kim J.Y. PRDX6 promotes tumor development via the JAK2/STAT3 pathway in a urethane-induced lung tumor model. Free Radic Biol Med. 2015;10 doi: 10.1016/j.freeradbiomed.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 86.Zemskova M.Y., Song J.H., Cen B., Cerda-Infante J., Montecinos V.P., Kraft A.S. Regulation of prostate stromal fibroblasts by the PIM1 protein kinase. Cell Signal. 2015;27(1):135–146. doi: 10.1016/j.cellsig.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jehs T., Faber C., Juel H.B., Bronkhorst I.H., Jager M.J., Nissen M.H. Inflammation-induced chemokine expression in uveal melanoma cell lines stimulates monocyte chemotaxis. Invest Ophthalmol Vis Sci. 2014;55(8):5169–5175. doi: 10.1167/iovs.14-14394. [DOI] [PubMed] [Google Scholar]
- 88.Zhang Y., Liao S., Fan W., Wei W., Wang C., Sun S. Tunicamycin-induced ER stress regulates chemokine CCL5 expression and secretion via STAT3 followed by decreased transmigration of MCF-7 breast cancer cells. Oncol Rep. 2014;32(6):2769–2776. doi: 10.3892/or.2014.3479. [DOI] [PubMed] [Google Scholar]
- 89.Niwa Y., Akamatsu H., Niwa H., Sumi H., Ozaki Y., Abe A. Correlation of tissue and plasma RANTES levels with disease course in patients with breast or cervical cancer. Clin Cancer Res. 2001;7(2):285–289. [PubMed] [Google Scholar]
- 90.Luboshits G., Shina S., Kaplan O., Engelberg S., Nass D., Lifshitz-Mercer B. Elevated expression of the CC chemokine regulated on activation, normal T cell expressed and secreted (RANTES) in advanced breast carcinoma. Cancer Res. 1999;59(18):4681–4687. [PubMed] [Google Scholar]
- 91.Mrowietz U., Schwenk U., Maune S., Bartels J., Kupper M., Fichtner I. The chemokine RANTES is secreted by human melanoma cells and is associated with enhanced tumour formation in nude mice. Br J Cancer. 1999;79(7–8):1025–1031. doi: 10.1038/sj.bjc.6690164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Murooka T.T., Rahbar R., Fish E.N. CCL5 promotes proliferation of MCF-7 cells through mTOR-dependent mRNA translation. Biochem Biophys Res Commun. 2009;387(2):381–386. doi: 10.1016/j.bbrc.2009.07.035. [DOI] [PubMed] [Google Scholar]
- 93.Tan M.C., Goedegebuure P.S., Belt B.A., Flaherty B., Sankpal N., Gillanders W.E. Disruption of CCR5-dependent homing of regulatory T cells inhibits tumor growth in a murine model of pancreatic cancer. J Immunol. 2009;182(3):1746–1755. doi: 10.4049/jimmunol.182.3.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sugasawa H., Ichikura T., Kinoshita M., Ono S., Majima T., Tsujimoto H. Gastric cancer cells exploit CD4 + cell-derived CCL5 for their growth and prevention of CD8 + cell-involved tumor elimination. Int J Cancer. 2008;122(11):2535–2541. doi: 10.1002/ijc.23401. [DOI] [PubMed] [Google Scholar]
- 95.Vaday G.G., Peehl D.M., Kadam P.A., Lawrence D.M. Expression of CCL5 (RANTES) and CCR5 in prostate cancer. Prostate. 2006;66(2):124–134. doi: 10.1002/pros.20306. [DOI] [PubMed] [Google Scholar]
- 96.Mencarelli A., Graziosi L., Renga B., Cipriani S., D'Amore C., Francisci D. CCR5 antagonism by maraviroc reduces the potential for gastric cancer cell dissemination. Transl Oncol. 2013;6(6):784–793. doi: 10.1593/tlo.13499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hattermann K., Sebens S., Helm O., Schmitt A.D., Mentlein R., Mehdorn H.M. Chemokine expression profile of freshly isolated human glioblastoma-associated macrophages/microglia. Oncol Rep. 2014;32(1):270–276. doi: 10.3892/or.2014.3214. [DOI] [PubMed] [Google Scholar]
- 98.Salmaggi A., Gelati M., Pollo B., Frigerio S., Eoli M., Silvani A. CXCL12 in malignant glial tumors: a possible role in angiogenesis and cross-talk between endothelial and tumoral cells. J Neurooncol. 2004;67(3):305–317. doi: 10.1023/b:neon.0000024241.05346.24. [DOI] [PubMed] [Google Scholar]
- 99.Kenig S., Alonso M.B., Mueller M.M., Lah T.T. Glioblastoma and endothelial cells cross-talk, mediated by SDF-1, enhances tumour invasion and endothelial proliferation by increasing expression of cathepsins B, S, and MMP-9. Cancer Lett. 2010;289(1):53–61. doi: 10.1016/j.canlet.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 100.Coniglio S.J., Eugenin E., Dobrenis K., Stanley E.R., West B.L., Symons M.H. Microglial stimulation of glioblastoma invasion involves epidermal growth factor receptor (EGFR) and colony stimulating factor 1 receptor (CSF-1R) signaling. Mol Med. 2012;18:519–527. doi: 10.2119/molmed.2011.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.