Main Text
Nucleation is a process that initiates phase transitions. Since the classical work of Gibbs on nucleation thermodynamics at the end of the nineteenth century, research into nucleation processes has been spread into a huge variety of scientific fields. To date, nucleation has been studied in disciplines ranging from biophysics to cosmology in systems spanning atomic to planetary scales and beyond (1). Proteins in solution are also known historically to form a variety of states and structures through nucleation (2, 3). In the past two decades, the assembly of proteins into highly ordered amyloid fibrils associated with numerous human diseases such as Alzheimer’s disease, Parkinson’s disease, and Type II diabetes mellitus has sparked renewed interest in theory development, which has resulted in new kinetic models of protein fibril assembly (4, 5). With these new developments, nucleation has again come to the fore in protein biophysics through the nucleated processes in which supersaturated solutions of proteins form insoluble supramolecular protein aggregates consisting of amyloid fibrils, which are defined by their highly ordered cross-β conformation (6). Thus, amyloid assembly converts soluble proteins into insoluble fractions that can be associated with human diseases, and the nucleation mechanism that initiates this phase transition represents a hitherto unresolved process of medical and fundamental biophysical importance.
In this issue of the Biophysical Journal, Kashchiev (7) presents a theoretical study on the application of nucleation theory to a simple one-dimensional polymerization model, the Oosawa-Kasai model that describes the formation of linear and helical polymers. The thermodynamics and kinetics of three versions of such one-dimensional polymerization in terms of their homogeneous nucleation properties were evaluated. With this approach, the nucleation behavior of the polymerization models, or lack thereof, was assessed rigorously. Formulae for key thermodynamic quantities such as nucleus size, polymerization work, nucleation rate, and the nucleation energy barrier were derived for the models. Here, Kashchiev sets an important example on how classical nucleation theory can be applied to protein fibril formation. By showing how the thermodynamics and kinetics of protein fibril nucleation can be evaluated in a conceptually straightforward manner, he offers a critical perspective that links new kinetic models of protein polymerization with the classic thermodynamics and kinetics results offered by the physical chemistry of nucleation.
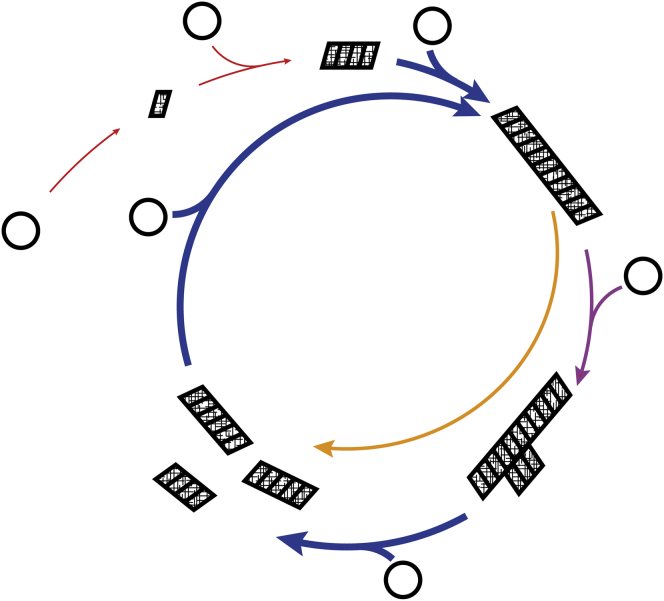
With this work, Kashchiev sets new challenges and offers new opportunities toward resolving the initial molecular steps in the mechanism of fibrous protein polymerization and amyloid formation. Firstly, this work sets a renewed challenge for researchers to design new experiments with sufficient information content and analysis approaches that can be used to validate protein nucleation models. For example, the approach used by Kashchiev predicts stationary prenuclei species (subnuclei) distribution and nucleation rates that could be observed and measured. In experimental studies of protein fibril assembly, direct or indirect observations of nucleation processes (red and purple arrows in Fig. 1) are, arguably, the most challenging to achieve compared to the other main processes involved (blue and orange arrows in Fig. 1). New experimental approaches such as microfluidics-based assays, fluorescence methods, and multimethod combinations, which allow novel detection of early monomer interactions, single nucleation events, and rare prenucleus species (4, 8, 9, 10) may resolve the molecular mechanism of protein nucleation with the help from the theoretical framework laid down here by Kashchiev. Secondly, the process of secondary nucleation (Fig. 1, purple arrow) has been implicated as a key factor that accelerates formation of disease-associated protein fibrils and produces potentially cytotoxic species (9). These secondary nucleation events are heterogeneous nucleation processes occurring on the surfaces of preformed aggregates. In addition, heterogeneous nucleation may occur at the numerous biological interfaces or artificially introduced solid-liquid/air-liquid interfaces in vivo and in vitro, which results in accelerated primary nucleation (Fig. 1, red arrows). In both cases, interfaces may catalyze heterogeneous nucleation of amyloid in a way explained by the so-called wetting phenomenon. Thereby, these key surface reactions can be described thermodynamically and kinetically by the same framework used by Kashchiev here for homogeneous nucleation (1, 7). Understanding the thermodynamic origin of these surface-catalyzed nucleation reactions will be particularly important in resolving why certain amyloid aggregates catalyze secondary nucleation on their surfaces and how potentially disease-associated heterogeneous nucleation events can be avoided or inhibited. Thirdly, Kashchiev discussed the Oosawa-Kasai model of helical polymerization, which provides a structural explanation of how small prenucleus species (subnuclei) can be less stable than postnucleus species (supernuclei), and therefore how nucleation might occur during the formation of pseudo one-dimensional protein polymers. This model provides an interesting idea in the case of amyloid polymerization and suggests that small prenucleus oligomeric species may switch to and from an amyloid conformation during the nucleation process. While this type of model is still limited in scope, the approach is flexible enough to allow expansions aimed to resolve whether the nucleation of any fibrillar protein aggregate is dominated by the increased stability that the cross-β conformation of the peptide chains confer, by the number of molecules present in the clusters, or by a combination of both.
Figure 1.
Schematic illustration of the lifecycle of amyloid. (Circles) Soluble monomeric protein. (Parallelograms) Monomeric units in the amyloid cross-β conformation. (Colored arrows) Main processes in amyloid assembly. (Red arrows) Primary nucleation, which may occur as homogeneous nucleation in solution or heterogeneous nucleation at interfaces. (Purple arrow) Secondary nucleation, which may occur as heterogeneous nucleation at surfaces presented by preformed aggregates. (Blue and orange arrows) Key nonnucleation based processes elongation growth, and breakage division, respectively. (Single arrows may represent multiple consecutive steps; arrow thickness symbolizes relative rates involved in the processes.) To see this figure in color, go online.
Ultimately, nucleated polymerization of proteins into ordered amyloid fibrils has a dimension of added complexity owning to the required structural transition from globular or intrinsically disordered states to the cross-β conformation that defines the amyloid structure (10, 11). This complexity is not yet fully captured by existing nucleation models. Consequently, how amyloid structural polymorphism, species barrier, and strains phenomenon originate in relation to primary nucleation and secondary aggregate surface mediated heterogeneous nucleation is not resolved. Kashchiev’s approach, therefore, presents an elegant generalization that has the potential to unify theories of homogeneous and heterogeneous protein nucleation, and offers a new opportunity to examine the fundamental thermodynamic basis of the birth of early amyloid species that dictates the final structures and functions of the fibril products. Recently, a hypothesis has been laid out, proposing the existence of a prebiotic amyloid world (6). “The amyloid world” hypothesis suggests that the protein assemblies with the cross-β amyloid conformation were the first self-propagating and information carrying biomolecules, which are preserved today as epigenetic information carriers. Kashchiev commented in his book on nucleation: “it should not be a surprise if it proves that even the Big Bang was a nucleation phenomenon” (1). Likewise, fundamental mechanistic understanding of nucleation underlying the birth of ordered protein fibrils may too not surprisingly suggest that even the evolution of early life started as a nucleation phenomenon.
Acknowledgments
The author thanks the members of the Xue group and the Kent Fungal Group for helpful comments and discussions.
This work was supported by the Biotechnology and Biological Sciences Research Council (UK) under grant No. BB/M02427X/1.
Editor: Rohit Pappu.
References
- 1.Kashchiev D. Butterworth-Heinemann; Oxford, UK: 2000. Nucleation: Basic Theory with Applications. [Google Scholar]
- 2.Ferrone F.A., Hofrichter J., Eaton W.A. Kinetic studies on photolysis-induced gelation of sickle cell hemoglobin suggest a new mechanism. Biophys. J. 1980;32:361–380. doi: 10.1016/S0006-3495(80)84962-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldstein R.F., Stryer L. Cooperative polymerization reactions. Analytical approximations, numerical examples, and experimental strategy. Biophys. J. 1986;50:583–599. doi: 10.1016/S0006-3495(86)83498-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xue W.-F., Homans S.W., Radford S.E. Systematic analysis of nucleation-dependent polymerization reveals new insights into the mechanism of amyloid self-assembly. Proc. Natl. Acad. Sci. USA. 2008;105:8926–8931. doi: 10.1073/pnas.0711664105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knowles T.P., Waudby C.A., Dobson C.M. An analytical solution to the kinetics of breakable filament assembly. Science. 2009;326:1533–1537. doi: 10.1126/science.1178250. [DOI] [PubMed] [Google Scholar]
- 6.Greenwald J., Riek R. Biology of amyloid: structure, function, and regulation. Structure. 2010;18:1244–1260. doi: 10.1016/j.str.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Kashchiev D. Protein polymerization into fibrils from the viewpoint of nucleation theory. Biophys. J. 2015;109:2126–2136. doi: 10.1016/j.bpj.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cremades N., Cohen S.I., Klenerman D. Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell. 2012;149:1048–1059. doi: 10.1016/j.cell.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen S.I., Linse S., Knowles T.P. Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl. Acad. Sci. USA. 2013;110:9758–9763. doi: 10.1073/pnas.1218402110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crick S.L., Ruff K.M., Pappu R.V. Unmasking the roles of N- and C-terminal flanking sequences from exon 1 of huntingtin as modulators of polyglutamine aggregation. Proc. Natl. Acad. Sci. USA. 2013;110:20075–20080. doi: 10.1073/pnas.1320626110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serio T.R., Cashikar A.G., Lindquist S.L. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]