Abstract
Signal transducers and activators of transcription (STATs) mediate essential signals for various biological processes, including immune responses, hematopoiesis, and neurogenesis. STAT3, for example, is involved in the pathogenesis of various human diseases, including cancers, autoimmune and inflammatory disorders. STAT3 activation is therefore tightly regulated at multiple levels to prevent these pathological conditions. A number of proteins have been reported to associate with STAT3 and regulate its activity. These STAT3-interacting proteins function to modulate STAT3-mediated signaling at various steps and mediate the crosstalk of STAT3 with other cellular signaling pathways. This article reviews the roles of novel STAT3 binding partners such as DAXX, zipper-interacting protein kinase, Krüppel-associated box-associated protein 1, Y14, PDZ and LIM domain 2 and signal transducing adaptor protein-2, in the regulation of STAT3-mediated signaling.
Keywords: Janus kinase/signal transducer and activator of transcription, Signal transduction, Signal transducer and activator of transcription 3, DAXX, Zipper-interacting protein kinase, Krüppel-associated box-associated protein 1, Y14, PDZ and LIM domain 2, Signal transducing adaptor protein-2, Nuclear factor-κB
Core tip: Signal transducer and activator of transcription 3 (STAT3) has been proposed its physiological and pathological significance in malignant and inflammatory diseases; therefore, the targeting of the STAT3 pathways is likely to be suitable for clinical application. In this review, we introduced novel regulatory molecules of STAT3 binding partners, such as DAXX, zipper-interacting protein kinase, Krüppel-associated box-associated protein 1, Y14, PDZ and LIM domain 2 and signal transducing adaptor protein-2. These proteins positively or negatively regulate critical steps of STAT3-mediated signals via individually unique mechanism. We hope that the information described here will help to develop a new strategy to clinically control the STAT3 activities.
INTRODUCTION
Cytokines selectively activate Janus kinases (JAKs), which in turn activate one or more signal transducers and activators of transcription (STATs) via their tyrosine phosphorylation[1-3]. STATs have cytoplasmic signaling regions, such as a Src-homology 2 (SH2) domain and tyrosine phosphorylation sites. Upon cytokine stimulation, STATs are phosphorylated and dimerize via their SH2 domains and then move into the nucleus[4]. STAT3 is a central member of the STAT protein family, and is activated by various cytokine signals, such as interleukin 6 (IL-6)[1,5-7], which plays a role in immune regulation, hematopoiesis, inflammation and oncogenesis[6,7] (Figure 1). The majority of IL-6 functions are in turn mediated by STAT3[8,9]. Of importance, abnormal expression of STAT3 has been reported in several cancer cells as well as autoimmune diseases, suggesting the involvement of STATs in a wide range of diseases[5,10-13]. Because of important physical roles, STAT3 activity is strictly regulated by multiple molecular mechanisms. For example, the protein inhibitor of activated STAT suppresses transcriptional activities of STAT3 by interfering STAT3 from DNA binding in the nucleus[14]. Suppressor of cytokine signaling (SOCS), which is induced by STAT3, participates in the negative feedback of STAT3 activities[15,16]. Cytoplasmic tyrosine phosphatases, such as SH2-containing phosphatase 1 (SHP1), SHP2 and protein-tyrosine phosphatase 1B, function to stop STAT activities[14,15]. Nuclear tyrosine phosphatases, such as TC45, also dephosphorylate nuclear STAT3, resulting in their translocation from the nucleus to the cytoplasm[14,17]. We have identified some STAT3-interacting molecules, including DAXX[18,19], zipper-interacting protein kinase (ZIPK)[20,21], Krüppel-associated box-associated protein 1 (KAP1)[22], Y14[23,24], PDZ and LIM domain 2 (PDLIM2)[25] and signal transducing adaptor protein-2 (STAP-2)[26-28]. Here, we describe functions of each of these molecules in the STAT3-mediated signaling pathway. DAXX negatively regulates STAT3-mediated transactivation and cell proliferation through the IL-6 signal transducer gp130[18,19]. ZIPK positively regulates STAT3 transactivation through STAT3 Ser727 phosphorylation[20,21]. KAP1 negatively regulates STAT3 Ser727 phosphorylation and transactivation by interacting with HDAC3 within the nucleus[22]. Y14 is a novel type of STAT3 binding partner and influences IL-6-induced STAT3 transactivation through altering its tyrosine-phosphorylation state[23,24]. PDLIM2 acts as a nuclear E3 ligase for STAT3 and terminates STAT3-mediated signaling[25]. STAP-2 is a novel adaptor protein, composed of pleckstrin homology (PH) and SH2-like domains, and a STAT3-binding (YXXQ) motif[26-28]. Taken together, STAT3 activity is positively and negatively regulated at multiple steps.
Figure 1.
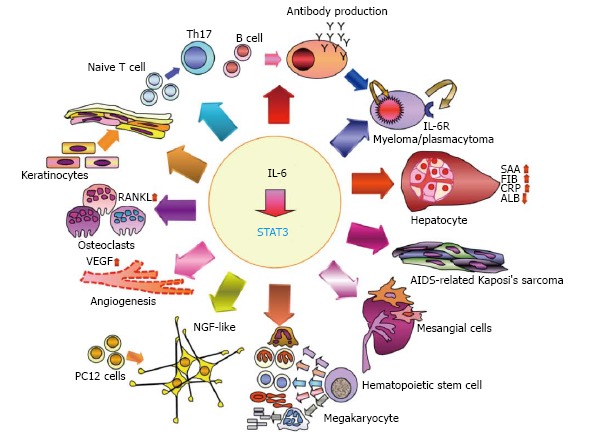
Interleukin 6 modulates a variety of physiological events, such as cell proliferation, differentiation, survival, and apoptosis, through signal transducer and activator of transcription 3. IL-6-STAT3 axis plays roles in the immune, the endocrine, the nervous and the hematopoietic systems, and on bone metabolism. IL-6 has been implicated in the pathology of different diseases including multiple myeloma, rheumatoid arthritis, Castleman’s disease, AIDS, mesangial proliferative glomerulonephritis, psoriasis, Kaposi’s sarcoma, sepsis and osteoporosis. SAA: Serum amyloid A; FIB: Fibrinogen; ALB: Albumin; CRP: C-reactive protein; NGF: Nerve growth factor; VEGF: Vascular endothelial growth factor; NF-κB: Nuclear factor-κB; RANKL: Receptor activator of NF-κB ligand; Th17: T helper type 17; IL-6: Interleukin 6; STAT3: Signal transducer and activator of transcription 3; AIDS: Acquired immunodeficiency syndrome.
NOVEL STAT3 BINDING PARTNERS
The STAT3 activities are strictly regulated, and recent reports have suggested several novel STAT3 regulators, whose characters are summarized in Table 1[25,26,29-33].
Table 1.
Novel signal transducer and activator of transcription 3-interacting proteins and their knockout mice phenotypes
| Protein | STAT3-binding site | Binding site in STAT3 | Function | Reported KO mice phenotype | Ref. |
| DAXX | N-terminal (1-240) | DNA-BD (320-493) | STAT3 suppression | Extensive apoptosis and embryonic lethality | [29] |
| ZIPK | Kinase domain (1-275) | DNA-BD (320-493) and SH2-TAD (494-750) | STAT3 activation | Not yet reported | |
| KAP1 | Not determined | Coiled-coil (138-319) and DNA-BD (320-493) | STAT3 suppression | Severe hypoproliferative anemia (hematopoietic-restricted deletion of KAP1) | [30] |
| Significant expansion of immature thymocytes, imbalances in CD4+/CD8+ cell ratios, and altered responses to TCR and TGF-β stimulation (T-cell-specific Kap1-deletion) | [31] | ||||
| Male-predominant hepatosteatosis and development of liver adenoma (Liver-specific ablation of KAP1) | [32] | ||||
| Heightened levels of anxiety-like and exploratory activity and stress-induced alterations in spatial learning and memory (Conditional Deletion of KAP1 in the Forebrain) | [33] | ||||
| Y14 | Not determined | DNA-BD (320-493) and SH2-TAD (494-750) | STAT3 activation | Not yet reported | |
| PDLIM2 | LIM domain | N-terminal (1-137), DNA-BD (320-493) and SH2-TAD (494-750) | STAT3 suppression | Enhanced Th17-cell dependent responses | [25] |
| STAP-2 | SH2 domain YXXQ motif | Coiled-coil (138-319), DNA-BD (320-493) and SH2-TAD (494-750) | STAT3 activation | Reduction in LPS-induced acute phase response | [26] |
STAT3: Signal transducer and activator of transcription 3; KO: Knock out; DNA-BD: DNA binding domain; SH2: Src homology 2 domain; TAD: Transactivation domain; TCR: T cell receptor; TGF: Transforming growth factor; Th17: T helper type 17; LPS: Lipopolysaccharide.
A nuclear STAT3 repressor, Death domain-associated protein (DAXX)
DAXX, which mainly located in the nucleus, has an ability to modulate transcription as well as cell death[34]. DAXX interacts with a number of transcription factors, including ETS1[35], PAX5[36], Glucocorticoid receptor[37], RelA[38], RelB[39], TCF4[40], SMAD4[41], C/EBP[42] and AIRE[43], and regulates their transcriptional activities. Because DAXX is also known to bind to histone deacetylases[44], DNA methyltransferases and their associated proteins[29,45,46], and the chromatin-modifying α-thalassemia syndrome protein[47,48], DAXX is likely to regulate cellular processes by regulating the transcription of specific genes via epigenetic modification. We found that DAXX regulates STAT3-transcriptional activity and that IFN-induced DAXX functionally links to IL-6/LIF/STAT3-mediated signaling[18]. Pretreatment of HeLa and Hep3B cells with IFN caused a decrease of IL-6-induced STAT3 transcriptional activities. Importantly, DAXX directly interacts with STAT3 in the nucleus, leading to the decreased STAT3-transcriptional activities. Indeed, knockdown of DAXX significantly enhanced STAT3 activation and gene expression after IL-6-stimulation.
The IL-6 family cytokines recognize gp130 membrane protein as a signal-transducing receptor component[8,9]. Dimerization of gp130 activates JAK family proteins (JAK1, JAK2 and TYK2), which then phosphorylate and activate STAT3. In lymphocytes, STAT3 is involved in IL-6- and/or IL-27-dependent cell growth[49,50]. In addition, STAT3 is also required for pro-B cell survival as well as efficient B lymphocyte production[51]. DAXX was reported to play a role in STAT3-mediated growth signals through gp130[19]. DAXX constitutively interacts with STAT3, leading to the impairment of STAT3 binding to the consensus DNA sequences of its target genes. In this regard, DAXX preferentially suppresses gp130-mediated Bcl-2 expression, which control cell survival. During lymphocyte apoptosis, an inverse correlation between DAXX and Bcl-2 expression levels is often observed. When progenitor B lymphocytes were treated with IFN-β, DAXX expression and nuclear localization were enhanced in parallel to Bcl-2 down-regulation[52].
Therefore, DAXX has an important function to control STAT3 activity and Bcl-2 expression during cytokine stimulation.
A STAT3 Ser727 kinase, ZIPK
Tyrosine and/or serine residues of STATs are phosphorylated in response to ligand stimulation[53,54]. In the case of STAT3, a single serine phosphorylation (serine residue at the position of amino acid 727; Ser727) in the transcriptional activation domain is needed for its maximal transcriptional activity. A mutant form of S727A of STAT3, in which serine 727 was replaced by alanine, was estimated to have approximately 50% of transcriptional activity when compared with wild type[54]. To analyze the meaning of Ser727 phosphorylation in vivo, SA mutant mice whose STAT3 Ser727 was substituted to alanine, were produced[55]. Embryonic fibroblasts from SA/SA homozygous mice displayed approximately 50% of the transcriptional cellular responses when compared with wild-type mice; therefore, Ser727 phosphorylation is important for maximal transcriptional activities of STAT3 even in vivo. Serine phosphorylation increases STAT3 activity via the association with some cofactors, such as p300[56]. Several kinases were implicated in serine phosphorylation of STAT3, and interactions between STAT signaling and serine kinase signaling pathways have been proposed[53].
With a yeast two-hybrid screen using the C-terminal region of STAT3 as bait, we identified ZIPK as a new STAT3-binding protein[20]. ZIPK selectively bound to STAT3, but not other STAT proteins, in mammalian cells. Furthermore, the kinase domain of ZIPK interacted with the DNA binding and C-terminal domains of STAT3 although ZIPK kinase activities were not essential for their binding. Of importance, ZIPK phosphorylates STAT3 Ser727 in the nucleus, and functionally enhances STAT3-mediated transcription after IL-6- or LIF-stimulation. siRNA-mediated knock down of endogenous ZIPK expression also proposed participation of ZIPK in STAT3-mediated transcriptional activation and target gene expression after LIF-stimulation. ZIPK, a serine/threonine-specific protein kinase, binds to ATF4, which belongs to the activating transcription factor/cyclic AMP-responsive element binding protein family[57]. ZIPK aggregates via its leucine zipper domain to become an active enzyme form. Over-expression of wild type ZIPK, but not the kinase-inactive mutant ZIPK K42A, induces apoptosis in NIH 3T3 cells, indicating that ZIPK stimulates the apoptotic process via its catalytic activity[57]. The kinase domain of ZIPK shows high sequence homology to that of death-associated protein kinase (DAPK), and these proteins establish a family with DAPK2/DRP-1, DRAK1 and DRAK2, all of which are related to apoptosis[58-60]. In collaboration with DAXX and Par-4, ZIPK induces apoptosis by way of nuclear PML oncogenic domains (PODs)[61]. We previously reported that activated STAT3 enhanced ZIPK activity after IL-6- or LIF-stimulation[20,21]. In this regard, IL-6/LIF/STAT3 signaling is likely to mediate apoptotic activity via inducing the translocation of ZIPK into PODs, together with PML and DAXX. Conversely, ZIPK induces STAT3 Ser727 phosphorylation, and enhances STAT3-mediated transcription. However, ZIPK K42A expression decreased STAT3 Ser727 phosphorylation in early but not late phase of IL-6-stimulation, suggesting that other kinases may be involved in the late phase of STAT3 Ser727 activation after IL-6-stimulation. Phosphorylation of Ser727 can increase STAT3 activity via associations with some co-activators, such as p300[56]. Of importance, ZIPK also interacts with p300 and forms a complex with STAT3.
Therefore, the binding of STAT3 to ZIPK in the nucleus may contribute to the stabilization of coactivator-transcription factor complexes.
A nuclear STAT3 binder, KAP1
KAP1, also known as transcriptional intermediary factor 1β and Tripartite motif-containing 28 (TRIM28), is a co-repressor of Krüppel-associated box-domain-containing zinc finger proteins[62-64]. KAP1 has an ability to coordinate various components involving in gene silencing; therefore, it can control the histone deacetylase (HDAC) complex[65-67] and a histone methyltransferase[68]. In other words, KAP1 inhibits the transcription of its target genes via orchestrating functions of the co-repressor complexes.
We isolated KAP1 as a STAT3-interacting protein using a yeast two-hybrid screening of a mouse embryo cDNA library[22]. Co-immunoprecipitation experiments confirmed that KAP1 binds to STAT3 in Hep3B cells. Endogenous KAP1 was present within the nucleus even in the absence of stimulation. After IL-6 stimulation, STAT3 was predominantly found in the nucleus, where it overlapped with KAP1, demonstrating that activated STAT3 translocates into the nucleus and interacts with KAP1. In Hep3B cells, KAP1 knockdown by specific siRNA significantly enhanced STAT3 activation as well as mRNA expression of SOCS3 and C/EBPδ in response to IL-6. Thus, KAP1 negatively regulates STAT3-mediated transcriptional activation and gene expression after IL-6-stimulation. Importantly, phosphorylation of STAT3 Ser727, but not STAT3 Tyr705, increased in parallel to reduction of KAP1 expression. Coincident with these data, reduction of KAP1 expression showed enhanced nuclear accumulation of STAT3 phosphorylated at Ser727. This may be in part related to the association with some cofactors, such as p300. Therefore, KAP1 is likely to recruit protein phosphatases to dephosphorylate STAT3 Ser727 in the nucleus. Alternatively, the direct interaction of KAP1 with HDACs may also be another mechanism for KAP1-mediated transcriptional repression because STAT3 has an ability to associate with HDAC3[69].
Therefore, KAP1 has a potential to suppress transcriptional activities of STAT3 in multiple ways.
A novel type of STAT3 binder, Y14
We identified Y14 as a novel associating protein with STAT3[23,24]. Y14, an RNA-binding protein, forms an exon-junction complex (EJC) with MAGOH. This complex selectively recognizes spliced forms of mRNAs immediately upstream of exon-exon junctions, and the binding is kept even after nuclear export[70,71]. In general, mRNAs produced by splicing are translated more efficiently than those from similar intronless precursors[72,73]. The EJC is in part involved in this translational enhancement because both Y14 and MAGOH recognize spliced form of mRNAs in the cytoplasm until mRNAs are translated. Human Y14 is known to shuttle mRNAs to interact with MAGOH[74]. However, only limited information is available regarding an mRNA shuttling protein involved in the regulation of transcription factors, such as STAT3.
We found that endogenous Y14 directly binds to STAT3 in Hep3B cells and affects STAT3 transactivation activity at several steps of IL-6-mediated signaling, including the tyrosine-phosphorylation, the nuclear accumulation and the DNA-binding of STAT3[23,24]. Furthermore, MAGOH inhibits complex formation between STAT3 and Y14, and MAGOH knockdown by specific siRNA enhances IL-6-induced gene expression.
Therefore, Y14 positively regulates IL-6-induced STAT3 activation, and MAGOH interferes with this effect by displacing Y14 from STAT3.
PDLIM2, a nuclear E3 ligase for STAT3
STAT3 activation is tightly regulated at multiple levels, including the ubiquitin/proteasome-dependent degradation of STAT3[75,76]. We found that a nuclear ubiquitin E3 ligase, PDLIM2 (also known as SLIM or mystique) binds to and degrades STAT3. PDLIM2 is a nuclear protein, composed of PDZ (postsynaptic density 65-discs large-zonula occludens 1) and LIM (abnormal cell lineage 11-isket 1-mechanosensory abnormal 3) domains[77,78]. PDLIM2 promoted to polyubiquitinate and degrade STAT3 in a proteasome-dependent manner by means of its LIM domain[25]. Consistently, PDLIM2-deficiency, as well as targeted gene disruption or knockdown of PDLIM2, caused insufficient STAT3 degradation, leading to nuclear accumulation of STAT3 and enhanced STAT3-mediated gene expression.
The LIM domain of PDLIM2 is needed for the recognition of STAT3. Ubiquitination reactions require three types of enzymes: An ubiquitin-activating enzyme (E1), an ubiquitin-conjugating enzyme (E2) and an ubiquitin ligase (E3). RING-type E3 ligases provide the polyubiquitin chain from E2 to their substrate by binding to E2 via their RING-finger domain, as well as by interacting with substrate proteins via the other domain[79]. The LIM domain forms a zinc finger structure and PHD domains. Proteins containing these domains generally possess ubiquitin E3 ligase activity and polyubiquitinate their target proteins[80]. Thus, the LIM domain of PDLIM2 is thought to be enough to bind to both E2 and its substrate STAT3. This possibility was consistent with the finding that the LIM domain of PDLIM2 could individually polyubiquitinate STAT3 in vitro. Interestingly, PDLIM2 binds to both phosphorylated and unphosphorylated STAT3, but PDLIM2 was shown to bind to phosphorylated but not unphosphorylated STAT4.
Therefore, PDLIM2 may regulate STAT3 activation via a different mechanism from that used on STAT4.
STAP-2 as a novel adaptor protein for STAT3
STAP-2, which we isolated as a c-fms-interacting protein, is composed of an N-terminal PH domain and an SH2-like domain[26]. A proline-rich region as well as a STAT3-binding YXXQ motif are also present in its C-terminal region[26]. STAP-2 is a murine homologue of an adaptor molecule BKS, which is a substrate of BRK tyrosine kinase[81]. Upon stimulation with epidermal growth factor, STAP-2 is tyrosine-phosphorylated and moves to the plasma membrane in STAP-2-overexpressing fibroblasts. IL-6 strongly induced STAP-2 mRNA in cultured hepatocytes; in addition, lipopolysaccharide-injection also induced STAP-2 mRNA in mice liver. In STAP-2-deficient hepatocytes, mRNA expression of acute-phase proteins and the tyrosine-phosphorylation of STAT3 are specifically impaired at the late phase of IL-6 stimulation. Thus, STAP-2 regulates IL-6/STAT3-mediated acute-phase protein responses during systemic inflammation. Furthermore, transient overexpression of STAP-2 mutant constructs revealed that STAP-2 enhances STAT3 activation through the YXXQ motif[26]. STAP-2 tyrosine-250 (Tyr250), a major tyrosine phosphorylation site by v-Src, JAK2 and LIF, is also required for the enhancement of STAT3 activity[27]. Indeed, the Y250F mutant, in which Tyr250 is substituted with phenylalanine, does not enhance STAT3 transcriptional activity.
As announced first, STAP-2 is a substrate of BRK[81]. BRK, also known as PTK6, is a non-receptor tyrosine kinase, composed of an SH3 domain, an SH2 domain and a tyrosine kinase catalytic domain, lacking an N-terminal myristoylation site for membrane targeting[82]. BRK is expressed by several malignant cells, such as metastatic melanomas and colon and prostate tumors as well as breast cancers[83-87]. In mammary gland, a large proportion of breast cancer cells express BRK, while normal mammary cells do not[88]. Notably, growth of breast cancer cells was impaired by siRNA-mediated down-regulation of BRK expression[89]. Our manipulation of STAP-2 expression indicates that STAP-2 plays an essential role in STAT3 activation by BRK. Indeed, STAP-2 bound to both BRK and STAT3, and STAP-2 knockdown by specific siRNA greatly decreased STAT3 activation induced by BRK in a breast cancer line T47D. Notably, an artificial STAP-2-BRK fusion protein had robust kinase activity and strongly induced activation and tyrosine phosphorylation of STAT3[28].
Therefore, STAP-2 is involved in BRK-mediated STAT3 activation and tumor cell growth.
Possible clinical utility of targeting STAT3-related molecules in future
Of note, most of these STAT3 binding proteins also directly interact with nuclear factor-κB (NF-κB) (p65/RelA) or NF-κB signaling molecules[38,78,90-92]. NF-κB, as well as STAT3, is a central signaling hub in inflammation and oncogenesis. NF-κB is also a transcription factor, which regulates gene expression of antiapoptosis as well as proinflammatory cytokines and chemokines[93,94]. Like STAT3, constitutively active NF-κB is found in many types of cancers[95]. Both STAT3 and NF-κB are also involved in the expression of target genes relating to tumor cell growth, migration and invasion[93,94,96]. Furthermore, target genes regulated by positive or negative crosstalk between STAT3 and NF-κB are gradually increasing[96,97]. In normal immune cells, activated STAT3 promotes serine-phosphorylation and subsequent proteasome-mediated degradation of IκBα, resulting in the activation of IκB kinase[98]. In cardiomyocytes as well as non-small cell lung cancer cells, the activation of NF-κB up-regulates STAT3 expression[99,100]. Importantly, STAT3 is known to directly bind to the transactivation domain of NF-κB through its DNA-binding domain[101,102]. Furthermore, it has been shown that, besides nuclear translocation after cytokine-stimulation, STAT3 continuously shuttles between the cytoplasm and the nucleus, independently of its tyrosine phosphorylation. Unphosphorylated STAT3 can interact with transcription factors, such as NF-κB, bind to DNA and drive gene expression in a distinct manner from phosphorylated STAT3[103]. Thus, direct interactions between STAT3 and NF-κB can regulate gene expression in several forms of NF-κB-dependent transcription. Therefore, STAT3-regulating molecules as well as STAT3 are likely to be key players during oncogenesis or inflammation, proposing that STAT3 could be a suitable target for malignant and/or inflammatory diseases. Although many manuscripts have showed that STAT3 has physiological and/or pathological significance, clinical meanings of the interactions with STAT3 and its binding partners should be clarified in future. STAT3 binding proteins described here are likely to have a potential to regulate STAT3 activity under some malignant or inflammatory circumstance; therefore, further experiments, including the establishment of low molecular compounds to inhibit their interaction with STAT3 could help for us to gather information about their clinical utility as well as physiological and/or pathological significance. Because STAT3 deficient mice are embryonic lethal[104], the targeting of STAT3 binding proteins may have fewer adverse effects than that of STAT 3 itself.
CONCLUSION
In this review, we summarized the functions of newly identified STAT3-interacting proteins. DAXX negatively regulates STAT3-mediated transactivation and cell proliferation through an IL-6 signal transducer, gp130[18,19]. ZIPK positively regulates STAT3 transactivation through STAT3 Ser727 phosphorylation[20,21]. KAP1 negatively regulates STAT3 Ser727 phosphorylation and transactivation by interacting with HDAC3 inside the nucleus[22]. Y14 regulates STAT3 transactivation via influencing tyrosine-phosphorylation after IL-6-stimulation[23,24]. PDLIM2 acts as a nuclear E3 ligase for STAT3 and terminates STAT3-signals[25]. STAP-2, a new adaptor protein, recognizes STAT3 through its YXXQ motif and stimulates STAT3 transactivation[26]. Although constitutive STAT3 activation is frequently observed in malignancies, few mutations in the STAT3 gene have yet been described. Therefore, it is very informative to clarify the mechanism how STAT3 is activated in malignant cells. Although direct proof is lacking, STAT3-associated proteins described here may be involved in this malignant process.
Footnotes
P- Reviewer: Fang Y, Park JH, Shao R S- Editor: Gong XM L- Editor: A E- Editor: Lu YJ
Conflict-of-interest statement: The authors declare no conflict of interest.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: January 20, 2015
First decision: April 10, 2015
Article in press: September 2, 2015
References
- 1.Darnell JE, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 2.Ihle JN. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 3.O'Shea JJ. Jaks, STATs, cytokine signal transduction, and immunoregulation: are we there yet? Immunity. 1997;7:1–11. doi: 10.1016/s1074-7613(00)80505-1. [DOI] [PubMed] [Google Scholar]
- 4.Darnell JE. STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 5.Hirano T, Ishihara K, Hibi M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene. 2000;19:2548–2556. doi: 10.1038/sj.onc.1203551. [DOI] [PubMed] [Google Scholar]
- 6.Kishimoto T, Akira S, Taga T. Interleukin-6 and its receptor: a paradigm for cytokines. Science. 1992;258:593–597. doi: 10.1126/science.1411569. [DOI] [PubMed] [Google Scholar]
- 7.Matsuda T, Suematsu S, Kawano M, Yoshizaki K, Tang B, Tanabe O, Nakajima T, Akira S, Hirano T, Kishimoto T. IL-6/BSF2 in normal and abnormal regulation of immune responses. Ann N Y Acad Sci. 1989;557:466–476; discussion 476-477. doi: 10.1111/j.1749-6632.1989.tb24039.x. [DOI] [PubMed] [Google Scholar]
- 8.Hirano T, Nakajima K, Hibi M. Signaling mechanisms through gp130: a model of the cytokine system. Cytokine Growth Factor Rev. 1997;8:241–252. doi: 10.1016/s1359-6101(98)80005-1. [DOI] [PubMed] [Google Scholar]
- 9.Kishimoto T, Taga T, Akira S. Cytokine signal transduction. Cell. 1994;76:253–262. doi: 10.1016/0092-8674(94)90333-6. [DOI] [PubMed] [Google Scholar]
- 10.Jove R. Preface: STAT signaling. Oncogene. 2000;19:2466–2467. doi: 10.1038/sj.onc.1203549. [DOI] [PubMed] [Google Scholar]
- 11.Levy DE, Darnell JE. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 12.Bromberg J, Darnell JE. The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19:2468–2473. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 13.Levy DE, Lee CK. What does Stat3 do? J Clin Invest. 2002;109:1143–1148. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 15.Yasukawa H, Sasaki A, Yoshimura A. Negative regulation of cytokine signaling pathways. Annu Rev Immunol. 2000;18:143–164. doi: 10.1146/annurev.immunol.18.1.143. [DOI] [PubMed] [Google Scholar]
- 16.Krebs DL, Hilton DJ. SOCS proteins: negative regulators of cytokine signaling. Stem Cells. 2001;19:378–387. doi: 10.1634/stemcells.19-5-378. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto T, Sekine Y, Kashima K, Kubota A, Sato N, Aoki N, Matsuda T. The nuclear isoform of protein-tyrosine phosphatase TC-PTP regulates interleukin-6-mediated signaling pathway through STAT3 dephosphorylation. Biochem Biophys Res Commun. 2002;297:811–817. doi: 10.1016/s0006-291x(02)02291-x. [DOI] [PubMed] [Google Scholar]
- 18.Muromoto R, Nakao K, Watanabe T, Sato N, Sekine Y, Sugiyama K, Oritani K, Shimoda K, Matsuda T. Physical and functional interactions between Daxx and STAT3. Oncogene. 2006;25:2131–2136. doi: 10.1038/sj.onc.1209235. [DOI] [PubMed] [Google Scholar]
- 19.Muromoto R, Kuroda M, Togi S, Sekine Y, Nanbo A, Shimoda K, Oritani K, Matsuda T. Functional involvement of Daxx in gp130-mediated cell growth and survival in BaF3 cells. Eur J Immunol. 2010;40:3570–3580. doi: 10.1002/eji.201040688. [DOI] [PubMed] [Google Scholar]
- 20.Sato N, Kawai T, Sugiyama K, Muromoto R, Imoto S, Sekine Y, Ishida M, Akira S, Matsuda T. Physical and functional interactions between STAT3 and ZIP kinase. Int Immunol. 2005;17:1543–1552. doi: 10.1093/intimm/dxh331. [DOI] [PubMed] [Google Scholar]
- 21.Sato N, Kamada N, Muromoto R, Kawai T, Sugiyama K, Watanabe T, Imoto S, Sekine Y, Ohbayashi N, Ishida M, et al. Phosphorylation of threonine-265 in Zipper-interacting protein kinase plays an important role in its activity and is induced by IL-6 family cytokines. Immunol Lett. 2006;103:127–134. doi: 10.1016/j.imlet.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 22.Tsuruma R, Ohbayashi N, Kamitani S, Ikeda O, Sato N, Muromoto R, Sekine Y, Oritani K, Matsuda T. Physical and functional interactions between STAT3 and KAP1. Oncogene. 2008;27:3054–3059. doi: 10.1038/sj.onc.1210952. [DOI] [PubMed] [Google Scholar]
- 23.Ohbayashi N, Taira N, Kawakami S, Togi S, Sato N, Ikeda O, Kamitani S, Muromoto R, Sekine Y, Matsuda T. An RNA biding protein, Y14 interacts with and modulates STAT3 activation. Biochem Biophys Res Commun. 2008;372:475–479. doi: 10.1016/j.bbrc.2008.05.073. [DOI] [PubMed] [Google Scholar]
- 24.Muromoto R, Taira N, Ikeda O, Shiga K, Kamitani S, Togi S, Kawakami S, Sekine Y, Nanbo A, Oritani K, et al. The exon-junction complex proteins, Y14 and MAGOH regulate STAT3 activation. Biochem Biophys Res Commun. 2009;382:63–68. doi: 10.1016/j.bbrc.2009.02.127. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka T, Yamamoto Y, Muromoto R, Ikeda O, Sekine Y, Grusby MJ, Kaisho T, Matsuda T. PDLIM2 inhibits T helper 17 cell development and granulomatous inflammation through degradation of STAT3. Sci Signal. 2011;4:ra85. doi: 10.1126/scisignal.2001637. [DOI] [PubMed] [Google Scholar]
- 26.Minoguchi M, Minoguchi S, Aki D, Joo A, Yamamoto T, Yumioka T, Matsuda T, Yoshimura A. STAP-2/BKS, an adaptor/docking protein, modulates STAT3 activation in acute-phase response through its YXXQ motif. J Biol Chem. 2003;278:11182–11189. doi: 10.1074/jbc.M211230200. [DOI] [PubMed] [Google Scholar]
- 27.Ikeda O, Miyasaka Y, Sekine Y, Mizushima A, Muromoto R, Nanbo A, Yoshimura A, Matsuda T. STAP-2 is phosphorylated at tyrosine-250 by Brk and modulates Brk-mediated STAT3 activation. Biochem Biophys Res Commun. 2009;384:71–75. doi: 10.1016/j.bbrc.2009.04.076. [DOI] [PubMed] [Google Scholar]
- 28.Ikeda O, Sekine Y, Mizushima A, Nakasuji M, Miyasaka Y, Yamamoto C, Muromoto R, Nanbo A, Oritani K, Yoshimura A, et al. Interactions of STAP-2 with Brk and STAT3 participate in cell growth of human breast cancer cells. J Biol Chem. 2010;285:38093–38103. doi: 10.1074/jbc.M110.162388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michaelson JS, Bader D, Kuo F, Kozak C, Leder P. Loss of Daxx, a promiscuously interacting protein, results in extensive apoptosis in early mouse development. Genes Dev. 1999;13:1918–1923. doi: 10.1101/gad.13.15.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barde I, Rauwel B, Marin-Florez RM, Corsinotti A, Laurenti E, Verp S, Offner S, Marquis J, Kapopoulou A, Vanicek J, et al. A KRAB/KAP1-miRNA cascade regulates erythropoiesis through stage-specific control of mitophagy. Science. 2013;340:350–353. doi: 10.1126/science.1232398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santoni de Sio FR, Barde I, Offner S, Kapopoulou A, Corsinotti A, Bojkowska K, Genolet R, Thomas JH, Luescher IF, Pinschewer D, et al. KAP1 regulates gene networks controlling T-cell development and responsiveness. FASEB J. 2012;26:4561–4575. doi: 10.1096/fj.12-206177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bojkowska K, Aloisio F, Cassano M, Kapopoulou A, Santoni de Sio F, Zangger N, Offner S, Cartoni C, Thomas C, Quenneville S, et al. Liver-specific ablation of Krüppel-associated box-associated protein 1 in mice leads to male-predominant hepatosteatosis and development of liver adenoma. Hepatology. 2012;56:1279–1290. doi: 10.1002/hep.25767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakobsson J, Cordero MI, Bisaz R, Groner AC, Busskamp V, Bensadoun JC, Cammas F, Losson R, Mansuy IM, Sandi C, et al. KAP1-mediated epigenetic repression in the forebrain modulates behavioral vulnerability to stress. Neuron. 2008;60:818–831. doi: 10.1016/j.neuron.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 34.Salomoni P, Khelifi AF. Daxx: death or survival protein? Trends Cell Biol. 2006;16:97–104. doi: 10.1016/j.tcb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Li R, Pei H, Watson DK, Papas TS. EAP1/Daxx interacts with ETS1 and represses transcriptional activation of ETS1 target genes. Oncogene. 2000;19:745–753. doi: 10.1038/sj.onc.1203385. [DOI] [PubMed] [Google Scholar]
- 36.Emelyanov AV, Kovac CR, Sepulveda MA, Birshtein BK. The interaction of Pax5 (BSAP) with Daxx can result in transcriptional activation in B cells. J Biol Chem. 2002;277:11156–11164. doi: 10.1074/jbc.M111763200. [DOI] [PubMed] [Google Scholar]
- 37.Lin DY, Lai MZ, Ann DK, Shih HM. Promyelocytic leukemia protein (PML) functions as a glucocorticoid receptor co-activator by sequestering Daxx to the PML oncogenic domains (PODs) to enhance its transactivation potential. J Biol Chem. 2003;278:15958–15965. doi: 10.1074/jbc.M300387200. [DOI] [PubMed] [Google Scholar]
- 38.Park J, Lee JH, La M, Jang MJ, Chae GW, Kim SB, Tak H, Jung Y, Byun B, Ahn JK, et al. Inhibition of NF-kappaB acetylation and its transcriptional activity by Daxx. J Mol Biol. 2007;368:388–397. doi: 10.1016/j.jmb.2007.02.047. [DOI] [PubMed] [Google Scholar]
- 39.Croxton R, Puto LA, de Belle I, Thomas M, Torii S, Hanaii F, Cuddy M, Reed JC. Daxx represses expression of a subset of antiapoptotic genes regulated by nuclear factor-kappaB. Cancer Res. 2006;66:9026–9035. doi: 10.1158/0008-5472.CAN-06-1047. [DOI] [PubMed] [Google Scholar]
- 40.Tzeng SL, Cheng YW, Li CH, Lin YS, Hsu HC, Kang JJ. Physiological and functional interactions between Tcf4 and Daxx in colon cancer cells. J Biol Chem. 2006;281:15405–15411. doi: 10.1074/jbc.M601807200. [DOI] [PubMed] [Google Scholar]
- 41.Chang CC, Lin DY, Fang HI, Chen RH, Shih HM. Daxx mediates the small ubiquitin-like modifier-dependent transcriptional repression of Smad4. J Biol Chem. 2005;280:10164–10173. doi: 10.1074/jbc.M409161200. [DOI] [PubMed] [Google Scholar]
- 42.Wethkamp N, Klempnauer KH. Daxx is a transcriptional repressor of CCAAT/enhancer-binding protein beta. J Biol Chem. 2009;284:28783–28794. doi: 10.1074/jbc.M109.041186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meloni A, Fiorillo E, Corda D, Incani F, Serra ML, Contini A, Cao A, Rosatelli MC. DAXX is a new AIRE-interacting protein. J Biol Chem. 2010;285:13012–13021. doi: 10.1074/jbc.M109.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hollenbach AD, McPherson CJ, Mientjes EJ, Iyengar R, Grosveld G. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J Cell Sci. 2002;115:3319–3330. doi: 10.1242/jcs.115.16.3319. [DOI] [PubMed] [Google Scholar]
- 45.Puto LA, Reed JC. Daxx represses RelB target promoters via DNA methyltransferase recruitment and DNA hypermethylation. Genes Dev. 2008;22:998–1010. doi: 10.1101/gad.1632208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muromoto R, Sugiyama K, Takachi A, Imoto S, Sato N, Yamamoto T, Oritani K, Shimoda K, Matsuda T. Physical and functional interactions between Daxx and DNA methyltransferase 1-associated protein, DMAP1. J Immunol. 2004;172:2985–2993. doi: 10.4049/jimmunol.172.5.2985. [DOI] [PubMed] [Google Scholar]
- 47.Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, Qin J, Zhou S, Higgs D, Wang W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc Natl Acad Sci USA. 2003;100:10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang J, Wu S, Liu H, Stratt R, Barak OG, Shiekhattar R, Picketts DJ, Yang X. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J Biol Chem. 2004;279:20369–20377. doi: 10.1074/jbc.M401321200. [DOI] [PubMed] [Google Scholar]
- 49.Takeda K, Kaisho T, Yoshida N, Takeda J, Kishimoto T, Akira S. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J Immunol. 1998;161:4652–4660. [PubMed] [Google Scholar]
- 50.Owaki T, Asakawa M, Morishima N, Mizoguchi I, Fukai F, Takeda K, Mizuguchi J, Yoshimoto T. STAT3 is indispensable to IL-27-mediated cell proliferation but not to IL-27-induced Th1 differentiation and suppression of proinflammatory cytokine production. J Immunol. 2008;180:2903–2911. doi: 10.4049/jimmunol.180.5.2903. [DOI] [PubMed] [Google Scholar]
- 51.Chou WC, Levy DE, Lee CK. STAT3 positively regulates an early step in B-cell development. Blood. 2006;108:3005–3011. doi: 10.1182/blood-2006-05-024430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gongora R, Stephan RP, Zhang Z, Cooper MD. An essential role for Daxx in the inhibition of B lymphopoiesis by type I interferons. Immunity. 2001;14:727–737. doi: 10.1016/s1074-7613(01)00152-2. [DOI] [PubMed] [Google Scholar]
- 53.Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19:2628–2637. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- 54.Wen Z, Zhong Z, Darnell JE. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 55.Shen Y, Schlessinger K, Zhu X, Meffre E, Quimby F, Levy DE, Darnell JE. Essential role of STAT3 in postnatal survival and growth revealed by mice lacking STAT3 serine 727 phosphorylation. Mol Cell Biol. 2004;24:407–419. doi: 10.1128/MCB.24.1.407-419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schuringa JJ, Schepers H, Vellenga E, Kruijer W. Ser727-dependent transcriptional activation by association of p300 with STAT3 upon IL-6 stimulation. FEBS Lett. 2001;495:71–76. doi: 10.1016/s0014-5793(01)02354-7. [DOI] [PubMed] [Google Scholar]
- 57.Kawai T, Matsumoto M, Takeda K, Sanjo H, Akira S. ZIP kinase, a novel serine/threonine kinase which mediates apoptosis. Mol Cell Biol. 1998;18:1642–1651. doi: 10.1128/mcb.18.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kögel D, Prehn JH, Scheidtmann KH. The DAP kinase family of pro-apoptotic proteins: novel players in the apoptotic game. Bioessays. 2001;23:352–358. doi: 10.1002/bies.1050. [DOI] [PubMed] [Google Scholar]
- 59.Shohat G, Shani G, Eisenstein M, Kimchi A. The DAP-kinase family of proteins: study of a novel group of calcium-regulated death-promoting kinases. Biochim Biophys Acta. 2002;1600:45–50. doi: 10.1016/s1570-9639(02)00443-0. [DOI] [PubMed] [Google Scholar]
- 60.Inbal B, Cohen O, Polak-Charcon S, Kopolovic J, Vadai E, Eisenbach L, Kimchi A. DAP kinase links the control of apoptosis to metastasis. Nature. 1997;390:180–184. doi: 10.1038/36599. [DOI] [PubMed] [Google Scholar]
- 61.Kawai T, Akira S, Reed JC. ZIP kinase triggers apoptosis from nuclear PML oncogenic domains. Mol Cell Biol. 2003;23:6174–6186. doi: 10.1128/MCB.23.17.6174-6186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim SS, Chen YM, O’Leary E, Witzgall R, Vidal M, Bonventre JV. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc Natl Acad Sci USA. 1996;93:15299–15304. doi: 10.1073/pnas.93.26.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 64.Agata Y, Matsuda E, Shimizu A. Two novel Krüppel-associated box-containing zinc-finger proteins, KRAZ1 and KRAZ2, repress transcription through functional interaction with the corepressor KAP-1 (TIF1beta/KRIP-1) J Biol Chem. 1999;274:16412–16422. doi: 10.1074/jbc.274.23.16412. [DOI] [PubMed] [Google Scholar]
- 65.Underhill C, Qutob MS, Yee SP, Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem. 2000;275:40463–40470. doi: 10.1074/jbc.M007864200. [DOI] [PubMed] [Google Scholar]
- 66.Schultz DC, Friedman JR, Rauscher FJ. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Satou A, Taira T, Iguchi-Ariga SM, Ariga H. A novel transrepression pathway of c-Myc. Recruitment of a transcriptional corepressor complex to c-Myc by MM-1, a c-Myc-binding protein. J Biol Chem. 2001;276:46562–46567. doi: 10.1074/jbc.M104937200. [DOI] [PubMed] [Google Scholar]
- 68.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 70.Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 71.Kim VN, Yong J, Kataoka N, Abel L, Diem MD, Dreyfuss G. The Y14 protein communicates to the cytoplasm the position of exon-exon junctions. EMBO J. 2001;20:2062–2068. doi: 10.1093/emboj/20.8.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Palacios IM. RNA processing: splicing and the cytoplasmic localisation of mRNA. Curr Biol. 2002;12:R50–R52. doi: 10.1016/s0960-9822(01)00671-6. [DOI] [PubMed] [Google Scholar]
- 73.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 74.Kataoka N, Diem MD, Kim VN, Yong J, Dreyfuss G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. EMBO J. 2001;20:6424–6433. doi: 10.1093/emboj/20.22.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ulane CM, Rodriguez JJ, Parisien JP, Horvath CM. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J Virol. 2003;77:6385–6393. doi: 10.1128/JVI.77.11.6385-6393.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Perry E, Tsruya R, Levitsky P, Pomp O, Taller M, Weisberg S, Parris W, Kulkarni S, Malovani H, Pawson T, et al. TMF/ARA160 is a BC-box-containing protein that mediates the degradation of Stat3. Oncogene. 2004;23:8908–8919. doi: 10.1038/sj.onc.1208149. [DOI] [PubMed] [Google Scholar]
- 77.Tanaka T, Soriano MA, Grusby MJ. SLIM is a nuclear ubiquitin E3 ligase that negatively regulates STAT signaling. Immunity. 2005;22:729–736. doi: 10.1016/j.immuni.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 78.Tanaka T, Grusby MJ, Kaisho T. PDLIM2-mediated termination of transcription factor NF-kappaB activation by intranuclear sequestration and degradation of the p65 subunit. Nat Immunol. 2007;8:584–591. doi: 10.1038/ni1464. [DOI] [PubMed] [Google Scholar]
- 79.Liu YC. Ubiquitin ligases and the immune response. Annu Rev Immunol. 2004;22:81–127. doi: 10.1146/annurev.immunol.22.012703.104813. [DOI] [PubMed] [Google Scholar]
- 80.Capili AD, Schultz DC, RauscherIII FJ, Borden KL. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 2001;20:165–177. doi: 10.1093/emboj/20.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mitchell PJ, Sara EA, Crompton MR. A novel adaptor-like protein which is a substrate for the non-receptor tyrosine kinase, BRK. Oncogene. 2000;19:4273–4282. doi: 10.1038/sj.onc.1203775. [DOI] [PubMed] [Google Scholar]
- 82.Mitchell PJ, Barker KT, Martindale JE, Kamalati T, Lowe PN, Page MJ, Gusterson BA, Crompton MR. Cloning and characterisation of cDNAs encoding a novel non-receptor tyrosine kinase, brk, expressed in human breast tumours. Oncogene. 1994;9:2383–2390. [PubMed] [Google Scholar]
- 83.Lee ST, Strunk KM, Spritz RA. A survey of protein tyrosine kinase mRNAs expressed in normal human melanocytes. Oncogene. 1993;8:3403–3410. [PubMed] [Google Scholar]
- 84.Derry JJ, Prins GS, Ray V, Tyner AL. Altered localization and activity of the intracellular tyrosine kinase BRK/Sik in prostate tumor cells. Oncogene. 2003;22:4212–4220. doi: 10.1038/sj.onc.1206465. [DOI] [PubMed] [Google Scholar]
- 85.Easty DJ, Mitchell PJ, Patel K, Flørenes VA, Spritz RA, Bennett DC. Loss of expression of receptor tyrosine kinase family genes PTK7 and SEK in metastatic melanoma. Int J Cancer. 1997;71:1061–1065. doi: 10.1002/(sici)1097-0215(19970611)71:6<1061::aid-ijc24>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 86.Llor X, Serfas MS, Bie W, Vasioukhin V, Polonskaia M, Derry J, Abbott CM, Tyner AL. BRK/Sik expression in the gastrointestinal tract and in colon tumors. Clin Cancer Res. 1999;5:1767–1777. [PubMed] [Google Scholar]
- 87.Schmandt RE, Bennett M, Clifford S, Thornton A, Jiang F, Broaddus RR, Sun CC, Lu KH, Sood AK, Gershenson DM. The BRK tyrosine kinase is expressed in high-grade serous carcinoma of the ovary. Cancer Biol Ther. 2006;5:1136–1141. doi: 10.4161/cbt.5.9.2953. [DOI] [PubMed] [Google Scholar]
- 88.Barker KT, Jackson LE, Crompton MR. BRK tyrosine kinase expression in a high proportion of human breast carcinomas. Oncogene. 1997;15:799–805. doi: 10.1038/sj.onc.1201241. [DOI] [PubMed] [Google Scholar]
- 89.Harvey AJ, Crompton MR. Use of RNA interference to validate Brk as a novel therapeutic target in breast cancer: Brk promotes breast carcinoma cell proliferation. Oncogene. 2003;22:5006–5010. doi: 10.1038/sj.onc.1206577. [DOI] [PubMed] [Google Scholar]
- 90.Kamitani S, Togi S, Ikeda O, Nakasuji M, Sakauchi A, Sekine Y, Muromoto R, Oritani K, Matsuda T. Krüppel-associated box-associated protein 1 negatively regulates TNF-α-induced NF-κB transcriptional activity by influencing the interactions among STAT3, p300, and NF-κB/p65. J Immunol. 2011;187:2476–2483. doi: 10.4049/jimmunol.1003243. [DOI] [PubMed] [Google Scholar]
- 91.Togi S, Shiga K, Muromoto R, Kato M, Souma Y, Sekine Y, Kon S, Oritani K, Matsuda T. Y14 positively regulates TNF-α-induced NF-κB transcriptional activity via interacting RIP1 and TRADD beyond an exon junction complex protein. J Immunol. 2013;191:1436–1444. doi: 10.4049/jimmunol.1300501. [DOI] [PubMed] [Google Scholar]
- 92.Sekine Y, Yumioka T, Yamamoto T, Muromoto R, Imoto S, Sugiyma K, Oritani K, Shimoda K, Minoguchi M, Akira S, et al. Modulation of TLR4 signaling by a novel adaptor protein signal-transducing adaptor protein-2 in macrophages. J Immunol. 2006;176:380–389. doi: 10.4049/jimmunol.176.1.380. [DOI] [PubMed] [Google Scholar]
- 93.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 94.Chen LF, Greene WC. Shaping the nuclear action of NF-kappaB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 95.Darnell JE. Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 96.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 97.Lee H, Herrmann A, Deng JH, Kujawski M, Niu G, Li Z, Forman S, Jove R, Pardoll DM, Yu H. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell. 2009;15:283–293. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Welte T, Zhang SS, Wang T, Zhang Z, Hesslein DG, Yin Z, Kano A, Iwamoto Y, Li E, Craft JE, et al. STAT3 deletion during hematopoiesis causes Crohn’s disease-like pathogenesis and lethality: a critical role of STAT3 in innate immunity. Proc Natl Acad Sci USA. 2003;100:1879–1884. doi: 10.1073/pnas.0237137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lu Y, Zhou J, Xu C, Lin H, Xiao J, Wang Z, Yang B. JAK/STAT and PI3K/AKT pathways form a mutual transactivation loop and afford resistance to oxidative stress-induced apoptosis in cardiomyocytes. Cell Physiol Biochem. 2008;21:305–314. doi: 10.1159/000129389. [DOI] [PubMed] [Google Scholar]
- 100.Dalwadi H, Krysan K, Heuze-Vourc’h N, Dohadwala M, Elashoff D, Sharma S, Cacalano N, Lichtenstein A, Dubinett S. Cyclooxygenase-2-dependent activation of signal transducer and activator of transcription 3 by interleukin-6 in non-small cell lung cancer. Clin Cancer Res. 2005;11:7674–7682. doi: 10.1158/1078-0432.CCR-05-1205. [DOI] [PubMed] [Google Scholar]
- 101.Yu Z, Kone BC. The STAT3 DNA-binding domain mediates interaction with NF-kappaB p65 and inducible nitric oxide synthase transrepression in mesangial cells. J Am Soc Nephrol. 2004;15:585–591. doi: 10.1097/01.asn.0000114556.19556.f9. [DOI] [PubMed] [Google Scholar]
- 102.Kesanakurti D, Chetty C, Rajasekhar Maddirela D, Gujrati M, Rao JS. Essential role of cooperative NF-κB and Stat3 recruitment to ICAM-1 intronic consensus elements in the regulation of radiation-induced invasion and migration in glioma. Oncogene. 2013;32:5144–5155. doi: 10.1038/onc.2012.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang J, Stark GR. Roles of unphosphorylated STATs in signaling. Cell Res. 2008;18:443–451. doi: 10.1038/cr.2008.41. [DOI] [PubMed] [Google Scholar]
- 104.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci USA. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]


