Abstract
Aims/hypothesis
Physical activity improves oxidative capacity and exerts therapeutic beneficial effects, particularly in the context of metabolic diseases. The peroxisome proliferator-activated receptor γ (PPARγ) coactivator-1α (PGC-1α) and the nuclear receptor PPARβ/δ have both been independently discovered to play a pivotal role in the regulation of oxidative metabolism in skeletal muscle, though their interdependence remain unclear. Hence, our aim was to determine the functional interaction between these two factors in mouse skeletal muscle in vivo.
Methods
Adult male control mice, PGC-1α muscle-specific transgenic (mTg) mice, PPARβ/δ muscle-specific knockout (mKO) mice and the combination PPARβ/δ mKO + PGC-1α mTg were studied under basal conditions and following PPARβ/δ agonist administration and acute exercise. Whole body metabolism was assessed by indirect calorimetry and blood analysis, while magnetic resonance was used to measure body composition. Quantitative PCR and western blot were used to determine gene expression and intracellular signaling. Proportion of oxidative muscle fiber was determined by NADH staining.
Results
Agonist-induced PPARβ/δ activation was only disrupted by PPARβ/δ knockout. We also found that the disruption of the PGC-1α-PPARβ/δ axis does not affect whole body metabolism under basal conditions. As expected, PGC-1α mTg mice exhibited higher exercise performance, peak oxygen consumption and lower blood lactate levels following exercise, though PPARβ/δ mKO+PGC-1α mTg mice showed a similar phenotype. Similarly, we found that PPARβ/δ was dispensable for PGC-1α-mediated enhancement of an oxidative phenotype in skeletal muscle.
Conclusions/interpretation
Collectively, these results indicate that PPARβ/δ is not an essential partner of PGC-1α in the control of skeletal muscle energy metabolism.
Keywords: skeletal muscle metabolism, nuclear receptors, coregulators, exercise
Introduction
The regulation of energy metabolism in skeletal muscle is highly controlled by the peroxisome proliferator-activated receptor γ (PPARγ) coactivator-1α (PGC-1α) [1]. PGC-1α drives the expression of genes involved in catabolic processes leading to aerobic ATP synthesis [1], while concomitantly promoting anabolic processes, including de novo lipogenesis [2]. Once activated, PGC-1α boosts the activity of different transcription factors to control gene programs resembling an endurance-trained phenotype in skeletal muscle [1, 3]. These adaptations are associated with an enhanced oxidative capacity, which contributes to an increased skeletal muscle fatigue resistance ex vivo and exercise performance in vivo [4-6]. Importantly, exercise is in fact one of the most efficient stimuli to induce PGC-1α in skeletal muscle [3].
Among the transcription factors regulated by PGC-1α, the nuclear receptor PPARβ/δ has been proposed to be a key partner of PGC-1α in the regulation of skeletal muscle metabolism and function, though mainly based on cell culture and pharmacological studies [7]. PGC-1α acts as a coactivator of PPARβ/δ [8-10], while PPARβ/δ can directly regulate PGC-1α expression [11, 12], indicating that this nuclear receptor acts both upstream as well as downstream of PGC-1α. Furthermore, transgenic mouse models for PPARβ/δ exhibit a similar phenotype compared to their counterparts for PGC-1α [4, 5, 13, 14]. Nevertheless, although the PGC-1α-PPARβ/δ axis appears to play a key role in the regulation of energy metabolism, the epistatic interaction between these proteins is currently unclear. We therefore aimed at directly assessing the functional interplay between PGC-1α and PPARβ/δ in the regulation of skeletal muscle oxidative metabolism in vivo.
Methods
Animals
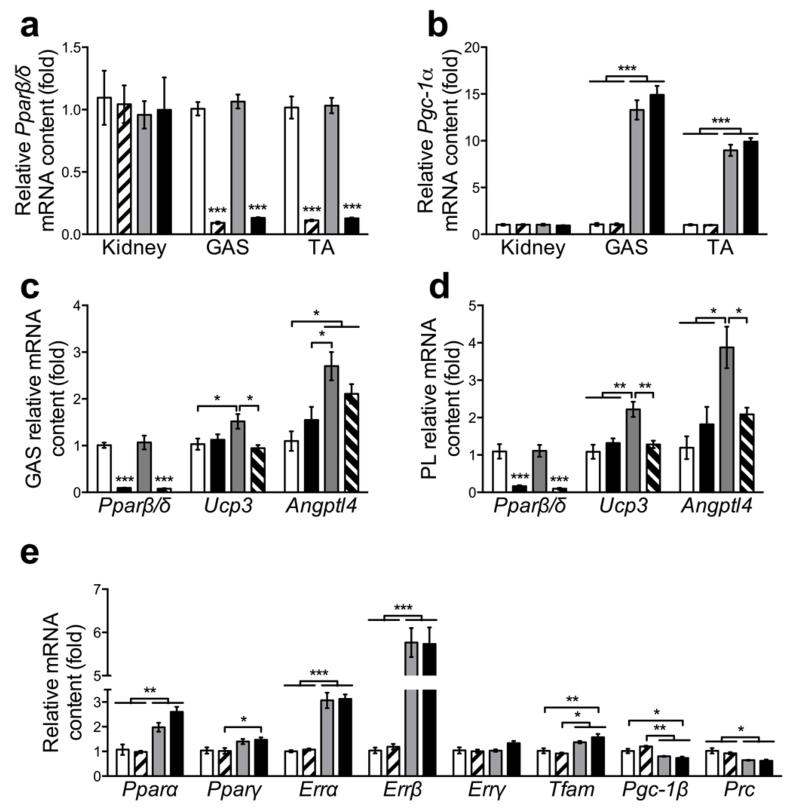
Mice were housed in a conventional facility with a 12 h night/day cycle, with free access to food/water. Experiments were performed on adult male mice with approval of the Swiss authorities. PGC-1α muscle-specific transgenic (mTg) mice have been described previously [5]. PPARβ/δ muscle-specific knockout (mKO) mice were generated by crossing PPARβ/δloxP/loxP mice with HSA-Cre transgenic mice [11]. Finally, PGC-1α mTg and HSA-Cre positive PPARβ/δloxP/loxP mKO mice were crossed to generate PPARβ/δ mKO+PGC-1α mTg mice. PPARβ/δloxP/loxP animals without Cre and PGC-1α transgene expression were used as control (CON) mice. All mice had mixed sv129 and C57BL/6 background. Genotypes were confirmed through PCR procedures (data not shown) and qPCR analysis in kidney and skeletal muscle (Fig. 1a, b).
Fig. 1.
PGC-1α and PPARβ/δ mouse models. (a and b) Pgc-1α and Pparβ/δ mRNA levels in kidney, gastrocnemius (GAS) and tibialis anterior (TA) (n = 6 per group). (c and d) Pparβ/δ, Ucp3 and Angptl4 mRNA levels in GAS and plantaris (PL) 8 h after the injection of 0.9% NaCl (as control) or 1 mg/kg of body weight of the PPARβ/δ agonist GW0742 (n = 6 per group). (e) mRNA level of different transcriptional regulators in GAS (n = 6 per group). In a, b and e CON: white, PPARβ/δ mKO: white with stripes, PGC-1α mTg: grey, PPARβ/δ mKO+ PGC-1α mTg: black. In c and d CON+NaCl: white, PPARβ/δ mKO+NaCl: black, CON+ GW0742: grey, PPARβ/δ mKO+ GW0742: black with white stripes. Values are mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. In a, c and d ***p<0.001 vs. CON and/or PGC-1α mTg for same tissue/treatment.
PPARβ/δ agonist
CON mice were subjected to an intra-peritoneal injection of either 0.9% NaCl (control) or 1 mg/kg of body weight of the PPARβ/δ agonist GW0742 (Tocris #2229, Bristol, UK), as previously described [15]. Muscles were collected 8 h following drug administration.
Body composition analysis
Lean and fat mass were measured via magnetic resonance (EchoMRI™, Houston, TX, USA).
Blood and plasma analysis
Blood samples were collected under basal conditions or immediately after maximal exercise from fed and/or overnight fasted mice, as previously described [9].
Glucose and insulin tolerance test
Intra-peritoneal glucose tolerance tests (GTT) were carried out by injecting 2 g/kg of body weight of glucose after 16 h of fasting. Insulin tolerance tests (ITT) were performed by injecting 0.8 U/kg of body weight of insulin (Novo Nordisk, Bagsvaerd, Denmark) after 6 h of fasting.
Indirect calorimetry
Animals were individually housed in a Comprehensive Lab Animal Monitoring System (Columbus Instruments, Columbus, OH, USA) during an acclimatization period of 48 h with free access to food and water. Subsequently, indirect calorimetry was performed during 48 h and data analyzed with the Oxymax software (Columbus Instruments).
Maximal exercise test
Exercise tests were performed as previously described [9]. Briefly, two days after acclimatization animals performed a maximal exercise test in a closed treadmill (Columbus Instruments), allowing the measurement of peak oxygen consumption (VO2peak) and respiratory exchange ratio (RER).
Histology
NADH staining was performed on 10 μm cross sections from tibialis anterior by exposing them to 1 mg/ml NADH (Sigma, St. Louis, MO, USA) in the presence of 1 mg/ml nitro blue tetrazolium (Sigma).
Non-esterified fatty acids (NEFA) measurement
Plasma NEFA were measured using a commercial kit (HR Series NEFA-HR(2), Wako Diagnostics, Richmond, VA, USA), according to manufacturer’s instructions. Blood samples were collected under basal conditions and following 1 h of treadmill running at 13 m min-1 with 5° slope.
Intramyocellular triacylglycerol (IMTG) extraction
Quadriceps IMTG were extracted by standard procedures using a solid-phase extraction column (UPTI-CLEAN NH2-S 100mg/1mL SPE Columns, Interchim, Montluçon, France) and quantified with a commercial kit (Triglyceride enzymatique PAP 150, Biomerieux, Marcy-l’Etoile, France), according to manufacturer’s instructions.
RNA isolation and quantitative PCR (qPCR)
Total RNA isolation from fed (ad libitum) animals and qPCR analysis was performed by standard procedures [9]. Sequences of qPCR primers are depicted in Table S1. Analysis was performed by the ΔΔCT method using TATA binding protein (TBP) as endogenous control. TBP transcript levels were not different between genotypes or experimental conditions.
Protein isolation and western blot
Protein isolation and western blot was conducted as previously described [9]. Proteins were detected with primary antibodies to Akt (Cell Signaling, Danvers, MA, USA; #9272), p-AktT308 (Cell Signaling #4056), AMPKα (Cell Signaling #2603), p-AMPKαT172 (Cell Signaling #2535, USA), total OXPHOS (Abcam #ab110413, USA) and eEF2 (Cell Signaling: #2332).
Statistical analysis
Values are expressed as mean ± standard error of the mean (SEM). Statistical significance was determined with unpaired two-tailed t-tests or one-way ANOVA with Tukey’s post-hoc test. Significance was considered with a p < 0.05.
Results
PGC-1α overexpression and PPARβ/δ deletion in mouse skeletal muscle
To elucidate the functional requirement for PPARβ/δ in the metabolic adaptations induced by PGC-1α, we crossed PPARβ/δ muscle-specific knockout (mKO) mice with PGC-1α muscle-specific transgenic (mTg) mice, referred as PPARβ/δ mKO+PGC-1α mTg animals. As expected, both PPARβ/δ mKO and PPARβ/δ mKO+PGC-1α mTg mice showed a reduction of Pparβ/δ mRNA specifically in skeletal muscle, while Pgc-1α mRNA was up-regulated by ~12 fold in skeletal muscle of PGC-1α mTg and PPARβ/δ mKO+PGC-1α mTg mice compared to control (CON) animals (Fig. 1a, b). To validate the functional consequence of PPARβ/δ deletion in skeletal muscle, we assessed the effects of the PPARβ/δ agonist GW0742 on the expression levels of PPARβ/δ target genes [7, 16]. Acute treatment with GW0742 did not affect Pparβ/δ mRNA in gastrocnemius and plantaris muscles whereas uncoupling protein 3 (Ucp3) mRNA levels were induced in CON, but not in PPARβ/δ mKO mice (Fig. 1c, d). Moreover, as previously reported [16], angiopoietin-like 4 (Angptl4) was up-regulated by GW0742 in a way that was partially dependent on PPARβ/δ (Fig. 1c, d). Importantly, PPARβ/δ deletion did not affect the transcript levels of Pparα and Pparγ (Fig. 1e). We subsequently measured the expression levels of other transcription factors and coactivators regulating metabolism, including the estrogen related receptors (Err), mitochondrial transcription factor A (Tfam), Pgc-1β and PGC-1-related coactivator (Prc). The expression of these genes was altered in skeletal muscle of PGC-1α mTg and PPARβ/δ mKO+PGC-1α mTg mice, thus independent of PPARβ/δ ablation (Fig. 1e).
Effects of skeletal muscle disruption of the PGC-1α-PPARβ/δ axis on whole body metabolism
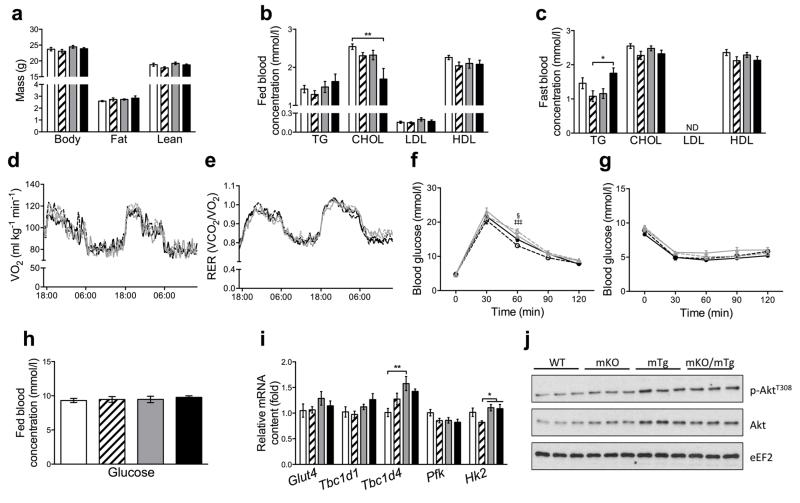
Body composition assessment revealed equal body weight, fat mass and lean mass in PPARβ/δ mKO, PGC-1α mTg, PPARβ/δ mKO+PGC-1α mTg and CON mice (Fig. 2a). Analysis of plasma triacylglycerol (TG), cholesterol (CHOL), low-density lipoproteins (LDL) and high-density lipoproteins (HDL) during fed and fasted state exhibited no differences except for a significant decrease in fed CHOL in the PPARβ/δ mKO+PGC-1α mTg mice (Fig. 2b, c). Moreover, indirect calorimetry during 48 h revealed no differences in VO2 or RER between any of the genotypes (Fig. 2d, e, electronic supplementary material (ESM) Fig. 1a, b).
Fig. 2.
Body composition, systemic parameters and glucose handling. (a) Assessment of body composition (n = 10-12 per group). (b and c) Plasma concentration of TAG, CHOL, LDL and HDL under fed and fasted conditions (n = 10-12 per group). (d and e) Measurement of VO2 (d) and RER (e) over a period of 48 h (n = 10-14 per group). (f-h) Blood glucose levels during glucose (f) and insulin tolerance tests (g). (h) Blood glucose levels in fed animals (n = 10-12 per group). (i) Gastrocnemius mRNA levels of genes involved in glucose metabolism (n = 6 per group). (j) Western blot assessment of Akt phosphorylation status in gastrocnemius (n = 6 per group). In a, b, c, h and I CON: white, PPARβ/δ mKO: white with stripes, PGC-1α mTg: grey, PPARβ/δ mKO + PGC-1α mTg: black. In d – g CON: black continuous line, PPARβ/δ mKO: black discontinuous line, PGC-1α mTg: grey continuous line, PPARβ/δ mKO + PGC-1α mTg: grey discontinuous line. Values are mean ± SEM. *p < 0.05, **p < 0.01. In f §p < 0.05 PPARβ/δ mKO vs. PGC-1α mTg; ‡‡‡p < 0.001 PPARβ/δ mKO vs. PPARβ/δ mKO + PGC-1α mTg.
Pharmacological activation of PPARβ/δ attenuates the detrimental effects of obesity and type 2 diabetes on systemic glucose homeostasis [13, 17, 18]. Compared to CON mice, neither glucose nor insulin tolerance tests were affected by PGC-1α overexpression and/or PPARβ/δ deletion in skeletal muscle in mice fed a regular chow diet (Fig. 2f, g, ESM Fig. 1c, d). Moreover, we did not find any differences in blood glucose levels in fed mice of the four different genotypes (Fig. 2h). These findings were corroborated by unchanged expression of genes involved in glucose transport and catabolism such as the glucose transporter 4 (Glut4), TBC1 domain family member 1 (Tbc1d1), phosphofructokinase (Pfk) and hexokinase 2 (Hk2) in skeletal muscle of PPARβ/δ mKO, PGC-1α mTg and PPARβ/δ mKO+PGC-1α mTg mice (Fig. 2i). In contrast, Tbc1d4 (also known as Akt substrate of 160 kDa, As160) was significantly up-regulated in PGC-1α mTg animals (Fig. 2i). Finally, we observed an increase in total Akt protein levels following PGC-1α overexpression, with no substantial effect of PPARβ/δ deletion (Fig. 2j, ESM Fig. 1e). Consistently, PGC-1α overexpression slightly decreased relative AktT308 phosphorylation levels, though this effect was not statistically significant (ESM Fig. 1f). These data hence suggest that the PGC-1α-PPARβ/δ axis is not essential for the modulation of whole body metabolism and glucose homeostasis under basal conditions in chow fed animals.
Modulation of skeletal muscle metabolism by the PGC-1α-PPARβ/δ axis
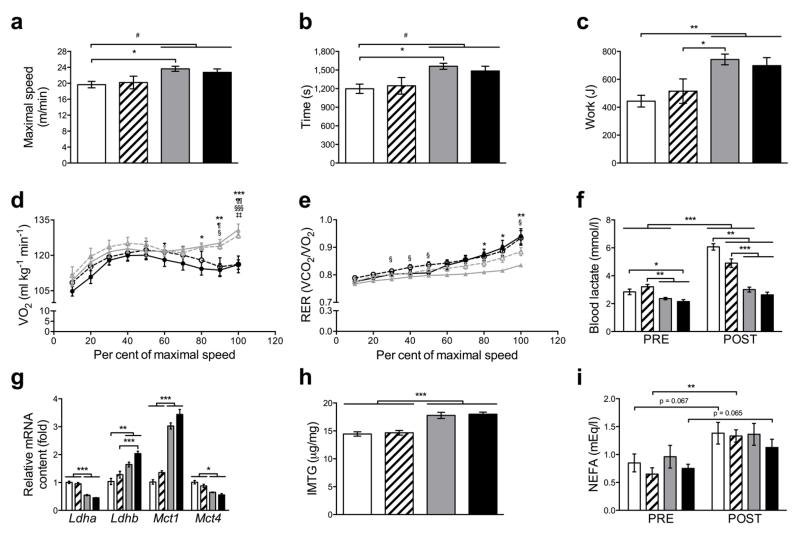
skeletal muscle PGC-1α and PPARβ/δ have been proposed to be key regulators of exercise performance and lactate metabolism [19, 20]. Consequently, we next assessed exercise performance in treadmill-based tests, which revealed a higher exercise performance in PGC-1α mTg mice as expected (Fig. 3a, b, c). Interestingly, PPARβ/δ muscle knockout did not reduce this difference when PPARβ/δ mKO+PGC-1α mTg mice were compared to CON animals (Fig. 3a, b, c). Moreover, VO2 was significantly enhanced in PGC-1α mTg and PPARβ/δ mKO+PGC-1α mTg mice during maximal exercise (Fig. 3d), thus altered by PGC-1α independent of PPARβ/δ. In contrast, the decrease in the RER in PGC-1α mTg animals was attenuated by concomitant PPARβ/δ deletion (Fig. 3e). Blood lactate concentration increased following maximal exercise in CON animals (Fig. 3f). This effect was attenuated in PPARβ/δ mKO mice and virtually abolished in both PGC-1α mTg and PPARβ/δ mKO+PGC-1α mTg mice (Fig. 3f). Similarly, pre-exercise blood lactate levels were reduced only in the mouse models with elevated skeletal muscle PGC-1α (Fig. 3f). Consistently, lactate dehydrogenase A (Ldha) and monocarboxylic acid transporters 4 (Mct4) mRNA levels were reduced only by PGC-1α overexpression in skeletal muscle, while in the same mice, Ldhb and Mct1 were up-regulated (Fig. 3g) reflecting an attenuated lactate production as well as higher catabolism. In order to assess substrate availability, we measured IMTG content and consistent with the function of PGC-1α in de novo lipogenesis [2] both PGC-1α mTg and PPARβ/δ mKO+PGC-1α mTg mice showed elevated IMTG levels, though PPARβ/δ knockout had no effect (Fig. 3h). Finally, we measured plasma levels of NEFA pre- and post-exercise. Exercise significantly increased plasma NEFA levels in PPARβ/δ mKO mice, while CON and PPARβ/δ mKO+PGC-1α mTg mice showed a trend toward an increase (Fig. 3i). These data show that, in response to maximal exercise, skeletal muscle PGC-1α is a pivotal regulator of whole body metabolism, mainly in a PPARβ/δ independent manner.
Fig. 3.
Skeletal muscle PGC-1α modulates whole body metabolism during maximal exercise. (a, b, c) Maximal speed, time and work achieved during exercise tests to exhaustion (n = 10-12 per group). (d and e) Measurement of VO2 and RER during the maximal exercise test (n = 10-12 per group). (f) Blood lactate levels before (PRE) and after (POST) maximal exercise (n = 10-12 per group). (g) mRNA levels of key genes of lactate metabolism in gastrocnemius (n = 6 per group). (h) Quadriceps IMTG content (n = 5 per group). (i) Plasma levels of NEFA before (PRE) and after (POST) exercise (n = 4-6 per group). In a, b, c, f, g, h and I CON: white, PPARβ/δ mKO: white with stripes, PGC-1α mTg: grey, PPARβ/δ mKO + PGC-1α mTg: black. In d and e CON: black continuous line, PPARβ/δ mKO: black discontinuous line, PGC-1α mTg: grey continuous line, PPARβ/δ mKO + PGC-1α mTg: grey discontinuous line. Values are mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. In d and e *p < 0.05, **p < 0.01, ***p < 0.001 CON vs. PGC-1α mTg; ¶p < 0.05, ¶¶p < 0.01 CON vs. PPARβ/δ mKO+ PGC-1α mTg; §p < 0.05, §§§p < 0.001 PPARβ/δ mKO vs. PGC-1α mTg; ‡‡p < 0.01 PPARβ/δ mKO vs. PPARβ/δ mKO+ PGC-1α mTg.
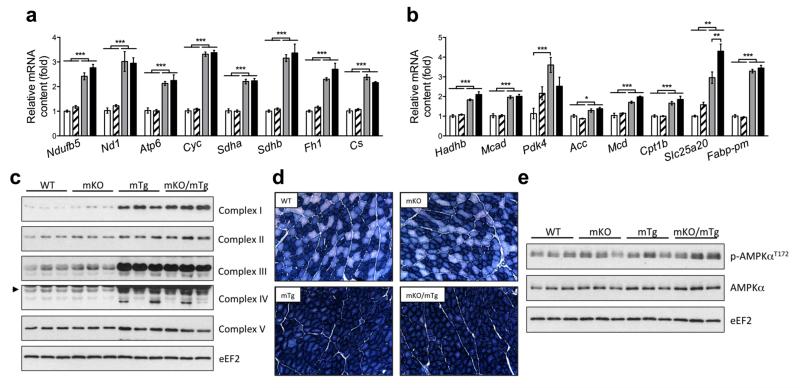
Next, we determined the relevance of PGC-1α and PPARβ/δ interaction in the regulation of skeletal muscle metabolism. We therefore determined the mRNA levels of genes regulating skeletal muscle oxidative metabolism, several of which have been suggested to be both PGC-1α and PPARβ/δ targets. Interestingly, we observed that PPARβ/δ deletion in skeletal muscle did not change the transcript abundance of genes involved in the TCA cycle, β-oxidation and electron transport chain (ETC) (Fig. 4a, b). In contrast, most of these genes were strongly up-regulated by PGC-1α overexpression in a PPARβ/δ independent manner (Fig. 4a, b). The assessment of protein content of different components of mitochondrial complexes supports mRNA data, though the overall effects were milder (Fig. 4c, ESM Fig. 2a). We then assessed the metabolic muscle phenotype by determining the proportion of oxidative fiber using NADH staining, which revealed a higher oxidative activity and proportion of oxidative fibers in PGC-1α mTg and PPARβ/δ mKO+PGC-1α mTg mice independent of a functional PPARβ/δ gene (Fig. 4d). Total protein content and phosphorylation levels of the key metabolic regulator AMP-activated protein kinase (AMPK) were not different in PPARβ/δ mKO, PGC-1α mTg or PPARβ/δ mKO+PGC-1α mTg mice, suggesting no altered energy status in any of these models (Fig. 4e, ESM Fig. 2b, c). We finally explored the relevance of the PGC-1α-PPARβ/δ axis in the context of the PPARβ/δ agonist GW0742-induced gene expression. As shown in the ESM Fig. 2d, GW0742 enhanced the expression of the PPARβ/δ targets Angptl4 and Ucp3, whereas it did not affect the mRNA levels of the key regulators of oxidative metabolism. Moreover, the effects of PGC-1α overexpression on gene expression were not affected by GW0742 (ESM Fig. 2d). Finally, as expected, PPARβ/δ gene ablation likewise abrogated any effect of the synthetic ligand (ESM Fig. 2d, Fig. 1c and 1d).
Fig. 4.
Oxidative metabolism of gastrocnemius is enhanced by PGC-1α even in the absence of PPARβ/δ. (a and b) mRNA levels of genes regulating oxidative and fatty acid metabolism (n = 6 per group). (c) Western blot analysis of key proteins regulating the ETC (n = 6 per group). (d) Assessment of oxidative muscle fibers (dark blue) via NADH staining (n = 3 per group). (e) Western blot analysis of AMPK phosphorylation status (n = 6 per group). In a and b CON: white, PPARβ/δ mKO: white with stripes, PGC-1α mTg: grey, PPARβ/δ mKO + PGC-1α mTg: black. Values are mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
The oxidative phenotype of skeletal muscle is strongly linked to physical activity levels and it has been associated with health beneficial effects in metabolic diseases and other pathologies. Even though the molecular mechanisms controlling exercise-induced adaptation in skeletal muscle have not been fully elucidated, PGC-1α is thought to promote mitochondrial function, myofibrillar gene expression, vascularization and other gene programs that are characteristic of oxidative muscle fibers [1]. Interestingly, PPARβ/δ is able to recapitulate several of these effects [7], though the functional interaction between PGC-1α and PPARβ/δ has not been elucidated in this tissue so far. We now provide strong evidence indicating almost complete PPARβ/δ independence of PGC-1α overexpression on the metabolic phenotype of skeletal muscle.
Importantly, supporting our hypothesis, contrary to the effects observed in PGC-1α muscle-specific transgenic mice, the enhancement of skeletal muscle oxidative metabolism is weaker in a bona fide muscle-specific PPARβ/δ gain-of-function mouse model [14]. Moreover, ligand-based activation of PPARβ/δ only increases exercise performance in trained mice, but not in sedentary animals [10]. Interestingly, oxidative metabolism and exercise performance can be boosted by fusing the PPARβ/δ protein to the heterologous VP16 activation domain that strongly increases its transcriptional activity in the absence of ligand or coactivator recruitment [13]. These data demonstrate that the reported functions of PPARβ/δ upstream and downstream of PGC-1α thereby are dispensable for PGC-1α function in an overexpression context. These observations are consistent with cell culture-based experiments showing that PGC-1α strongly increased oxidative metabolism in the absence of PPARβ/δ in skeletal muscle cells [21]. It appears hence that PGC-1α regulates skeletal muscle oxidative metabolism by increasing the transcriptional activity of alternative transcription factors, some of which might even compensate for the loss of PPARβ/δ. In fact, Pparα, Errα and Errγ were significantly up-regulated in both PGC-1α mTg and PPARβ/δ mKO+ PGC-1α mTg skeletal muscle, suggesting that these transcription factors might play a more relevant function in this context. Importantly, our results indicate that PPARβ/δ deletion by itself does not result in a compensatory activation of such related transcription factors. In fact, PPARβ/δ mKO animals do not exhibit an up-regulation of PPARs, ERRs and TFAM in skeletal muscle. In addition, several target genes of these transcription factors were unaltered in PPARβ/δ mKO mice.
The contribution of PPARβ/δ to the regulation of skeletal muscle metabolism seems to be more relevant in the context of ligand-induced activation. Accordingly, PPARβ/δ activation with synthetic ligands is an efficient treatment for metabolic disorders [13, 17, 18, 22], though it remains unclear whether this effect is mediated by skeletal muscle PPARβ/δ. Conversely, overexpression of PGC-1α in skeletal muscle is insufficient to evoke similar therapeutic benefits in young mice and even accelerates the development of insulin resistance when such animals are fed a high-fat diet [23], unless the mice are concomitantly exercised [24]. In old animals however, overexpression of PGC-1α in muscle prevents age-induced insulin resistance [25]. These findings indicate that in some pathological settings, PPARβ/δ activation might be more relevant than PGC-1α, in particular in the absence of physical activity.
Surprisingly, in our study, PPARβ/δ mKO animals had a similar phenotype compared to CON mice, with minimal or no changes in body composition, blood parameters and gene expression. In contrast, Shuler et al. have reported higher body weight and fat, in addition to increased serum levels of glucose, insulin and TG in the same mouse model [11]. Intriguingly, similar discrepancies have been reported in global PPARβ/δ KO mouse models in regard to whole body metabolism assessed under basal conditions [18, 26-29]. These differences in the phenotype of PPARβ/δ KO mouse models in chow fed sedentary condition might stem from different environmental factors (e.g. diet and temperature) that could lead to a partial PPARβ/δ activation in CON animals and thus lead to more pronounced phenotypic differences in metabolic parameters when compared to KO mice. Importantly, most of the effects of skeletal muscle PPARβ/δ deletion reported by Schuler et al. on energy metabolism are observed following high-fat diet feeding and/or in old mice [11]. Moreover, in the same study, the phenotype of adult PPARβ/δ mKO mice fed chow diet is rather mild and not substantially different to our results, which is reflected by the magnitude and variability of the data [11].
During exercise, skeletal muscle exerts a bigger impact on whole body metabolism. Accordingly, PGC-1α mTg mice exhibit a higher VO2 and lower RER during treadmill running, reflecting an enhanced oxidative capacity and increased fatty acid oxidation [4]. Interestingly, while the PGC-1α-mediated improvement in VO2 during exercise is maintained in the absence of a functional PPARβ/δ gene, knockout of PPARβ/δ attenuated the decrease in the RER in PPARβ/δ mKO+ PGC-1α mTg mice. In line with our observations, it has been shown that PPARβ/δ overexpression in skeletal muscle does not affect VO2 and RER during treadmill running [20]. Moreover, PPARβ/δ has been proposed to specifically regulate fatty acid metabolism and only to a smaller extent other oxidative metabolic genes in cultured muscle cells [21]. Surprisingly, the effect of PPARβ/δ knockout on RER during maximal exercise appears to be unrelated to mRNA level of genes controlling fatty acid transport and oxidation. Interestingly, PPARβ/δ deletion attenuated the up-regulation of Pdk4 induced by PGC-1α overexpression. Importantly, skeletal muscle PDK4 has been extensively shown to be a key regulator of fatty acid oxidation during exercise [30], suggesting a possible mechanism by which PPARβ/δ modulates RER and thus energy substrate utilization during maximal exercise. It should be noted that PPARβ/δ knockout induced the up-regulation of Pdk4, an effect that supports the idea that this nuclear receptor can actively repress target genes in the absence of ligand [16, 31]. Together, these data suggest that the effects of skeletal muscle PGC-1α on VO2 are not dependent of PPARβ/δ, even though this nuclear receptor appears to be partially involved in the PGC-1α-mediated increase in β-oxidation during exercise. In addition, our findings support previous data suggesting that PGC-1α-controlled lactate metabolism is predominantly regulated by ERRα and not PPARβ/δ [19].
In summary, our results reveal important insights into the regulatory networks that control skeletal muscle plasticity. Here, we show that in normal/physiological conditions, PPARβ/δ is dispensable for the effect of PGC-1α on skeletal muscle remodeling. Importantly, the different therapeutic effects of PPARβ/δ and PGC-1α in the context of metabolic diseases during sedentary vs. exercise/aging state, strongly suggest that the relative importance of these molecules in controlling the metabolic phenotype of skeletal muscle varies significantly depending of the physiological and pathological context. Therefore, we hope that these findings will allow a more targeted dissection and modulation of skeletal muscle plasticity in health and disease in the future.
Supplementary Material
Acknowledgments
Funding This project was funded by by the Swiss National Science Foundation, the Muscular Dystrophy Association USA (MDA), the SwissLife ‘Jubiläumsstiftung für Volksgesundheit und medizinische Forschung’, the Swiss Society for Research on Muscle Diseases (SSEM), the Swiss Diabetes Association, the Roche Research Foundation, the United Mitochondrial Disease Foundation (UMDF), the Association Française contre les Myopathies (AFM), the Neuromuscular Research Association Basel (NeRAB), the Gebert-Rüf Foundation “Rare Diseases” Program, the University of Basel and the Biozentrum.
Abbreviations
- AMPK
AMP-activated protein kinase
- ANGPTL4
angiopoietin-like 4
- CHOL
Cholesterol
- CON
control mice
- ERR
estrogen related receptors
- ETC
electron transport chain
- GLUT4
glucose transporter 4
- GTT
glucose tolerance tests
- HK2
hexokinase 2
- ITT
Insulin tolerance tests
- LDHA
lactate dehydrogenase A
- LDHB
lactate dehydrogenase B
- MCT1
monocarboxylic acid transporters 1
- MCT4
monocarboxylic acid transporters 4
- mKO
PPARβ/δ muscle-specific knockout
- mTg
PGC-1α muscle-specific transgenic
- PFK
phosphofructokinase
- PGC-1α
PPARγ coactivator-1α
- PGC-1β
PPARγ coactivator-1β
- PPARα
peroxisome proliferator-activated receptor α
- PPARβ/δ
peroxisome proliferator-activated receptor β/δ
- PPARγ
peroxisome proliferator-activated receptor γ
- PRC
PGC-1-related coactivator
- RER
respiratory exchange ratio
- TBC1D1
TBC1 domain family member 1
- TBC1D4
TBC1 domain family member 4
- TBP
TATA binding protein
- TFAM
mitochondrial transcription factor A
- TG
triglycerides
- UCP3
uncoupling protein 3
- VO2peak
peak oxygen consumption
Footnotes
Duality of interest The authors declare that there is no duality of interest associated with this manuscript.
References
- [1].Handschin C. Regulation of skeletal muscle cell plasticity by the peroxisome proliferator-activated receptor gamma coactivator 1alpha. J Recept Signal Transduct Res. 2010;30:376–384. doi: 10.3109/10799891003641074. [DOI] [PubMed] [Google Scholar]
- [2].Summermatter S, Baum O, Santos G, Hoppeler H, Handschin C. Peroxisome proliferator-activated receptor {gamma} coactivator 1{alpha} (PGC-1{alpha}) promotes skeletal muscle lipid refueling in vivo by activating de novo lipogenesis and the pentose phosphate pathway. J Biol Chem. 2010;285:32793–32800. doi: 10.1074/jbc.M110.145995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Pérez-Schindler J, Handschin C. New insights in the regulation of skeletal muscle PGC-1α by exercise and metabolic diseases. Drug Discovery Today: Disease Models. 2013;10:e79–e85. [Google Scholar]
- [4].Calvo JA, Daniels TG, Wang X, et al. Muscle-specific expression of PPARgamma coactivator-1alpha improves exercise performance and increases peak oxygen uptake. J Appl Physiol. 2008;104:1304–1312. doi: 10.1152/japplphysiol.01231.2007. [DOI] [PubMed] [Google Scholar]
- [5].Lin J, Wu H, Tarr PT, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- [6].Summermatter S, Thurnheer R, Santos G, et al. Remodeling of calcium handling in skeletal muscle through PGC-1alpha: impact on force, fatigability, and fiber type. Am J Physiol Cell Physiol. 2012;302:C88–99. doi: 10.1152/ajpcell.00190.2011. [DOI] [PubMed] [Google Scholar]
- [7].Ehrenborg E, Krook A. Regulation of skeletal muscle physiology and metabolism by peroxisome proliferator-activated receptor delta. Pharmacol Rev. 2009;61:373–393. doi: 10.1124/pr.109.001560. [DOI] [PubMed] [Google Scholar]
- [8].Dressel U, Allen TL, Pippal JB, Rohde PR, Lau P, Muscat GE. The peroxisome proliferator-activated receptor beta/delta agonist, GW501516, regulates the expression of genes involved in lipid catabolism and energy uncoupling in skeletal muscle cells. Mol Endocrinol. 2003;17:2477–2493. doi: 10.1210/me.2003-0151. [DOI] [PubMed] [Google Scholar]
- [9].Perez-Schindler J, Summermatter S, Salatino S, et al. The corepressor NCoR1 antagonizes PGC-1alpha and estrogen-related receptor alpha in the regulation of skeletal muscle function and oxidative metabolism. Mol Cell Biol. 2012;32:4913–4924. doi: 10.1128/MCB.00877-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Narkar VA, Downes M, Yu RT, et al. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134:405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Schuler M, Ali F, Chambon C, et al. PGC1alpha expression is controlled in skeletal muscles by PPARbeta, whose ablation results in fiber-type switching, obesity, and type 2 diabetes. Cell Metab. 2006;4:407–414. doi: 10.1016/j.cmet.2006.10.003. [DOI] [PubMed] [Google Scholar]
- [12].Hondares E, Pineda-Torra I, Iglesias R, Staels B, Villarroya F, Giralt M. PPARdelta, but not PPARalpha, activates PGC-1alpha gene transcription in muscle. Biochem Biophys Res Commun. 2007;354:1021–1027. doi: 10.1016/j.bbrc.2007.01.092. [DOI] [PubMed] [Google Scholar]
- [13].Wang YX, Zhang CL, Yu RT, et al. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2004;2:e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Luquet S, Lopez-Soriano J, Holst D, et al. Peroxisome proliferator-activated receptor delta controls muscle development and oxidative capability. Faseb J. 2003;17:2299–2301. doi: 10.1096/fj.03-0269fje. [DOI] [PubMed] [Google Scholar]
- [15].Gaudel C, Schwartz C, Giordano C, Abumrad NA, Grimaldi PA. Pharmacological activation of PPARbeta promotes rapid and calcineurin-dependent fiber remodeling and angiogenesis in mouse skeletal muscle. Am J Physiol Endocrinol Metab. 2008;295:E297–304. doi: 10.1152/ajpendo.00581.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Adhikary T, Kaddatz K, Finkernagel F, et al. Genomewide analyses define different modes of transcriptional regulation by peroxisome proliferator-activated receptor-beta/delta (PPARbeta/delta) PLoS One. 2011;6:e16344. doi: 10.1371/journal.pone.0016344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tanaka T, Yamamoto J, Iwasaki S, et al. Activation of peroxisome proliferator-activated receptor delta induces fatty acid beta-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc Natl Acad Sci U S A. 2003;100:15924–15929. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lee CH, Olson P, Hevener A, et al. PPARdelta regulates glucose metabolism and insulin sensitivity. Proc Natl Acad Sci U S A. 2006;103:3444–3449. doi: 10.1073/pnas.0511253103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Summermatter S, Santos G, Perez-Schindler J, Handschin C. Skeletal muscle PGC-1alpha controls whole-body lactate homeostasis through estrogen-related receptor alpha-dependent activation of LDH B and repression of LDH A. Proc Natl Acad Sci U S A. 2013;110:8738–8743. doi: 10.1073/pnas.1212976110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gan Z, Burkart-Hartman EM, Han DH, et al. The nuclear receptor PPARbeta/delta programs muscle glucose metabolism in cooperation with AMPK and MEF2. Genes Dev. 2011;25:2619–2630. doi: 10.1101/gad.178434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kleiner S, Nguyen-Tran V, Bare O, Huang X, Spiegelman B, Wu Z. PPAR{delta} agonism activates fatty acid oxidation via PGC-1{alpha} but does not increase mitochondrial gene expression and function. J Biol Chem. 2009;284:18624–18633. doi: 10.1074/jbc.M109.008797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Salvado L, Serrano-Marco L, Barroso E, Palomer X, Vazquez-Carrera M. Targeting PPARbeta/delta for the treatment of type 2 diabetes mellitus. Expert Opin Ther Targets. 2012;16:209–223. doi: 10.1517/14728222.2012.658370. [DOI] [PubMed] [Google Scholar]
- [23].Choi CS, Befroy DE, Codella R, et al. Paradoxical effects of increased expression of PGC-1alpha on muscle mitochondrial function and insulin-stimulated muscle glucose metabolism. Proc Natl Acad Sci U S A. 2008;105:19926–19931. doi: 10.1073/pnas.0810339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Summermatter S, Shui G, Maag D, Santos G, Wenk MR, Handschin C. PGC-1alpha improves glucose homeostasis in skeletal muscle in an activity-dependent manner. Diabetes. 2013;62:85–95. doi: 10.2337/db12-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wenz T, Rossi SG, Rotundo RL, Spiegelman BM, Moraes CT. Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proc Natl Acad Sci U S A. 2009;106:20405–20410. doi: 10.1073/pnas.0911570106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [26].Feng X, Luo Z, Ma L, et al. Angiotensin II receptor blocker telmisartan enhances running endurance of skeletal muscle through activation of the PPAR-delta/AMPK pathway. J Cell Mol Med. 2011;15:1572–1581. doi: 10.1111/j.1582-4934.2010.01085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].He H, Yang D, Ma L, et al. Telmisartan prevents weight gain and obesity through activation of peroxisome proliferator-activated receptor-delta-dependent pathways. Hypertension. 2010;55:869–879. doi: 10.1161/HYPERTENSIONAHA.109.143958. [DOI] [PubMed] [Google Scholar]
- [28].Akiyama TE, Lambert G, Nicol CJ, et al. Peroxisome proliferator-activated receptor beta/delta regulates very low density lipoprotein production and catabolism in mice on a Western diet. J Biol Chem. 2004;279:20874–20881. doi: 10.1074/jbc.M312802200. [DOI] [PubMed] [Google Scholar]
- [29].Peters JM, Lee SS, Li W, et al. Growth, adipose, brain, and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor beta(delta) Mol Cell Biol. 2000;20:5119–5128. doi: 10.1128/mcb.20.14.5119-5128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Peters SJ. Regulation of PDH activity and isoform expression: diet and exercise. Biochem Soc Trans. 2003;31:1274–1280. doi: 10.1042/bst0311274. [DOI] [PubMed] [Google Scholar]
- [31].Lee CH, Kang K, Mehl IR, et al. Peroxisome proliferator-activated receptor delta promotes very low-density lipoprotein-derived fatty acid catabolism in the macrophage. Proc Natl Acad Sci U S A. 2006;103:2434–2439. doi: 10.1073/pnas.0510815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.