Mutations in the seipin complex components Fld1 and Ldb16 result in the loss of lipid droplet identity and phospholipid packing defects, revealing a role of this complex in the stabilization of ER–lipid droplet contact sites.
Abstract
Lipid droplets (LDs) are storage organelles consisting of a neutral lipid core surrounded by a phospholipid monolayer and a set of LD-specific proteins. Most LD components are synthesized in the endoplasmic reticulum (ER), an organelle that is often physically connected with LDs. How LD identity is established while maintaining biochemical and physical connections with the ER is not known. Here, we show that the yeast seipin Fld1, in complex with the ER membrane protein Ldb16, prevents equilibration of ER and LD surface components by stabilizing the contact sites between the two organelles. In the absence of the Fld1/Ldb16 complex, assembly of LDs results in phospholipid packing defects leading to aberrant distribution of lipid-binding proteins and abnormal LDs. We propose that the Fld1/Ldb16 complex facilitates the establishment of LD identity by acting as a diffusion barrier at the ER–LD contact sites.
Introduction
In virtually all eukaryotic cells, lipid droplets (LDs) play central roles in lipid and energy metabolism and their deregulation is associated with metabolic disorders such as obesity, diabetes, and lipodystrophy (Krahmer et al., 2013). At structural level, LDs are rather unique: a hydrophobic core composed of neutral lipids, mainly triglycerides and sterol esters, surrounded by a monolayer of phospholipids acting as surfactants and a specific set of proteins (Fujimoto and Parton, 2011; Thiam et al., 2013b; Pol et al., 2014). This structural organization of LDs favors the binding of proteins with hydrophobic α-helical hairpins or amphipathic helices (AHs), whereas it precludes the association of integral membrane proteins with luminal domains. Proteins with AHs are thought to be recruited to LDs directly from the cytosol, whereas the ones with hydrophobic hairpins are first targeted to the ER before concentrating at the LD monolayer (Thiam et al., 2013b; Pol et al., 2014). In both cases, the targeting appears to be highly regulated. This set of LD-specific proteins, mostly consisting of lipid-modifying enzymes and regulatory proteins, to a large extent determines many of the LD properties. The ER is also involved in the synthesis of most of the lipids both at the surface and in the hydrophobic core of LDs. Moreover, a large fraction of (in mammals) or all (in yeast) LDs are continuous with the ER (Jacquier et al., 2011; Wilfling et al., 2013). Therefore, how these biochemically and physically connected organelles achieve and maintain their identity is a major question in cell biology.
Depending on the cell type or metabolic state, LDs vary widely in number, size, and composition (Yang et al., 2012). The molecular mechanisms controlling these aspects of LD biology are largely unknown, but changes in phospholipid biosynthesis were shown to influence LD morphology (Guo et al., 2008; Krahmer et al., 2011; Fei et al., 2011b). Phospholipid imbalances, particularly defects leading to a decrease in the levels of phosphatidylcholine (PC), induce the formation of abnormally large LDs (i.e., “supersized”). For example, mutations in CHO2 or OPI3, components of the phosphatidylethanolamine N-methyltransferase pathway for PC biosynthesis, lead to supersized LDs in the yeast Saccharomyces cerevisiae (Fei et al., 2011b). In this case, the supersized LDs appear to form by coalescence of smaller ones as a consequence of both a decrease in levels of PC, which acts as a surfactant to prevent LDs coalescence (Krahmer et al., 2011), together with increased amounts of phosphatidic acid (PA), which is thought to have fusogenic properties (Chernomordik and Kozlov, 2005; Zeniou-Meyer et al., 2007). Consistent with these data, activation of an alternative pathway for PC biosynthesis, the Kennedy pathway, restores the PA/PC ratio and LD morphology. Similarly, depletion of the Kennedy pathway rate-limiting enzyme CCT1 (CTP:phosphocholine cytidylyltransferase) in Drosophila melanogaster cells also induces the coalescence of small LDs into supersized LDs (Guo et al., 2008; Krahmer et al., 2011). Whether a general imbalance in phospholipid composition is the only or the major mechanism leading to the formation of supersized LDs is unclear.
Seipin is an evolutionarily conserved ER membrane protein that has been implicated in regulating LD morphology, but its function is not well understood. It was originally identified as being mutated in patients with Berardinelli-Seip congenital lipodystrophy (Magré et al., 2001). These patients display almost complete absence of adipose tissue, ectopic fat accumulation, and altered glucose metabolism, a phenotype recapitulated in mice and flies upon targeted ablation of seipin (Cui et al., 2011; Tian et al., 2011; Chen et al., 2012; Prieur et al., 2013).
At the cellular level, the major defect caused by seipin mutations is observed in LDs, which appear smaller and aggregated (Szymanski et al., 2007; Boutet et al., 2009; Fei et al., 2011a). In S. cerevisiae lacking the seipin homolog Fld1, a fraction of cells displays similar LD defects (Szymanski et al., 2007; Fei et al., 2008, 2011b; Wang et al., 2014). In the rest of fld1Δ cells, LDs are still abnormal, but instead of small and aggregated, they appear in reduced number and supersized. Although resembling those observed in the N-methyltransferase pathway mutants opi3Δ or cho2Δ, supersized LDs in fld1Δ cells are not rescued by stimulation of the Kennedy pathway, indicating that they are caused by a different defect (Fei et al., 2011b; Wang et al., 2014). Interestingly, the distinct morphologies of abnormal LDs in fld1Δ cells can be manipulated by inositol, a phospholipid precursor with a central role in glycerolipid metabolism (Henry et al., 2012). At low inositol concentrations supersized LDs are seen in a large fraction of fld1Δ cells; in contrast, if inositol concentration is high the frequency of supersized LDs decreases with a concomitant increase in LD aggregates (Fei et al., 2011b; Wang et al., 2014). This observation suggests that the LD defects in FLD1 mutants might derive from abnormal phospholipid homeostasis. However, robust and consistent changes in global phospholipid composition have not been detected in seipin mutants, in yeast, or in any other cell type (Szymanski et al., 2007; Fei et al., 2008, 2011a,b; Wang et al., 2014).
In yeast, Fld1 binds to and is necessary for the stability of Ldb16, another ER membrane protein (Wang et al., 2014). Fluorescence microscopy experiments suggest that the two proteins localize to the ER–LD contact sites (Szymanski et al., 2007; Fei et al., 2008; Wang et al., 2014). Moreover, the LD phenotypes of fld1Δ and ldb16Δ are remarkably similar, further indicating they have a common unknown function in LD formation (Wang et al., 2014).
Here we show that the Fld1/Ldb16 complex is required for the identity of LDs in S. cerevisiae. In the absence of this complex, incorporation of phospholipids into the monolayer of nascent LDs is entirely dependent on the ER phospholipid pools, leading to the generation of membrane defects and abnormal localization of LD and other lipid-binding proteins. In fld1Δ and ldb16Δ mutants, these defects are a consequence of abnormal ER–LD contact sites, as demonstrated by electron tomography. We propose that stabilization of ER–LD contact sites by the Fld1/Ldb16 complex establishes a diffusion barrier necessary for normal LD morphology and identity.
Results
Fld1/Ldb16 complex mutants are defective in uncoupling LD biogenesis from phospholipid synthesis
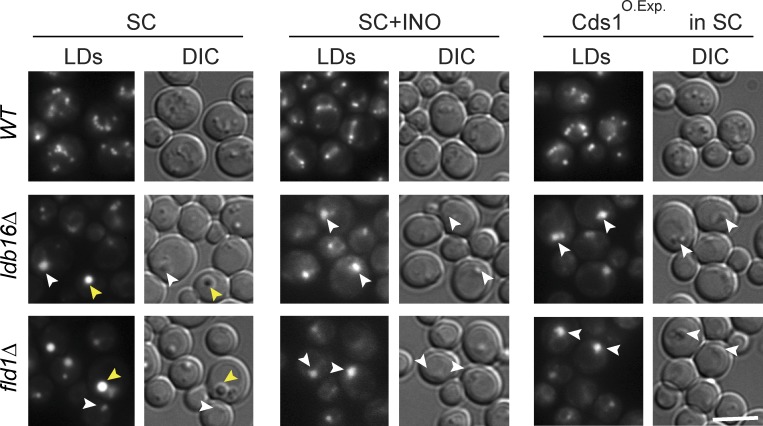
The changes in LD morphology induced by inositol supplementation are unique to fld1Δ and ldb16Δ mutants, and its presence does not induce LD aggregates in wt cells or in other mutants with LD defects (Fei et al., 2011b; Wang et al., 2014). Inositol is a precursor of phosphatidylinositol, and, if supplemented to the yeast growth media, it strongly stimulates phosphatidylinositol synthesis (Henry et al., 2012). This raises the possibility that LD morphology in fld1Δ and ldb16Δ cells is sensitive to changes in phospholipid synthesis (Cartwright et al., 2015). To directly test this hypothesis, we stimulated phospholipid biosynthesis in an inositol-independent manner by overexpressing the CDP-diacylglycerol synthase CDS1, which converts PA into CDP-DAG, a key step in the synthesis of most phospholipids in yeast (Shen et al., 1996). Overexpression of CDS1 did not affect LD morphology in wt cells (Fig. 1). In contrast, increasing Cds1 levels in fld1Δ and ldb16Δ cells resulted in complete disappearance of supersized LDs and formation of LD aggregates. These were indistinguishable from the ones induced by inositol supplementation (Fig. 1), suggesting that the effect is caused by the stimulation of phospholipid synthesis. Thus, together with previous experiments (Fei et al., 2011b; Wang et al., 2014; Cartwright et al., 2015), these data indicate that the Fld1/Ldb16 complex uncouples LD growth from ER phospholipid pools.
Figure 1.
Fld1/Ldb16 complex mutants are defective in uncoupling LD morphology from phospholipid synthesis. Analysis of LD morphology in cells with the indicated genotype. Cells were grown in SC or SC supplemented with 75 µM inositol to early stationary phase. Yellow arrowheads indicate supersized LDs; white arrowheads indicate LD aggregates. LDs were stained with MDH. Bar, 5 µm.
Abnormal LD composition in Fld1/Ldb16 complex mutants
As a first step in dissecting the mechanism by which Fld1/Ldb16 complex uncouples LD biogenesis from phospholipid biosynthesis, we purified LDs from cells grown in presence or absence of inositol and analyzed their composition. To assess the purity of the isolated organelles, the presence of several marker proteins in the different fractions was analyzed by Western blotting (Fig. S1 A). A specific enrichment of LD proteins with negligible amounts of ER proteins was detected in the LD fraction. Negative stain electron microscopy of isolated LDs also indicated minimal, if any, microsomal contamination (Fig. S1 B). These data confirmed the high quality of the LD preparations and also indicated that the buoyant properties of LDs from wt, fld1Δ, and ldb16Δ are not drastically different. Interestingly, the morphology of isolated LDs recapitulated well the in vivo situation. Whereas LDs isolated from wt cells were regular in size and highly circular in cross section, the ones from ldb16Δ cells were extremely irregular, both in size and circularity.
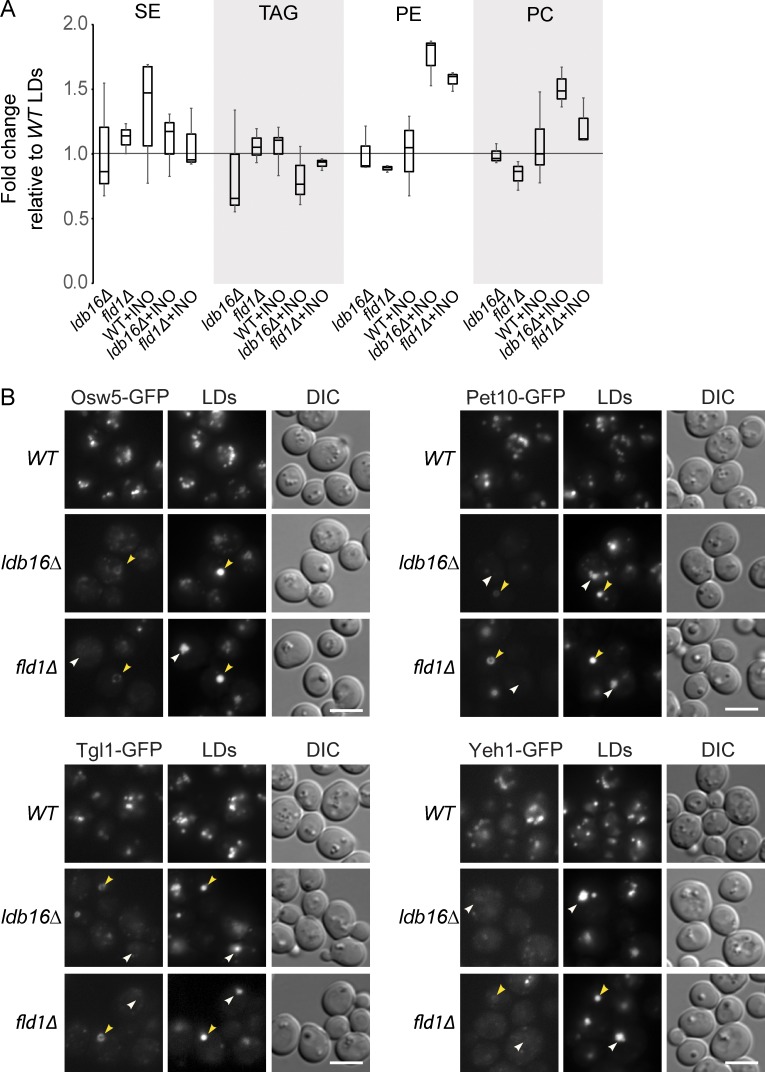
To characterize the LDs of ldb16Δ or fld1Δ cells, we first determined their lipid composition. The LDs isolated from wt and mutant cells grown in regular minimal medium (no inositol supplementation) showed a similar composition, both at the level of phospho- and neutral lipids (Fig. 2 A). Similarly, the composition LDs isolated from wt cells grown with or without inositol supplementation did not change. In contrast, LDs isolated from ldb16Δ or fld1Δ cells grown in inositol-supplemented medium, although maintaining similar neutral lipid content, were highly enriched in PC and phosphatidylethanolamine (PE), the major ER phospholipids (Fig. 2 A). The changes were specific to the LD fraction as the whole-cell lipid composition was indistinguishable between mutant and wt cells (Fig. S1 C). Given the high purity of the isolated LDs, this increase in phospholipids cannot be attributed to ER contamination of the LD fraction. Instead, these data are consistent with the increased surface to volume ratio of LDs in the aggregates observed in fld1Δ and ldb16Δ cells grown under conditions stimulating phospholipid synthesis.
Figure 2.
Abnormal composition of LDs in Fld1/Ldb16 complex mutants. (A) Lipid composition of LDs isolated from cells with the indicated genotype and grown in SC or SC supplemented with 75 µM of inositol (+INO). LDs from wt cells grown in SC media were used as reference. The result of at least three independent experiments is shown in the graph; whiskers represent the maximum and minimum values. (B) Localization of the LD-specific proteins Osw5, Pet10, Tgl1, and Yeh1 in wt, ldb16Δ, and fld1Δ cells grown in YPD until early stationary phase. All proteins were expressed from their endogenous locus as C-terminal GFP fusions. Yellow arrowheads indicate supersized LDs and white arrowheads indicate LD aggregates. LDs are stained with the neutral lipid dye MDH. Bar, 5 µm.
Next we compared the LD proteome of wt and mutant cells using label-free quantitative mass spectrometry. Protein composition of LDs isolated from fld1Δ and ldb16Δ mutants were similar between them but very distinct from LDs of wt cells. Among the 30–40 high-confidence LD proteins (Grillitsch et al., 2011; Currie et al., 2014), 27 were strongly reduced or completely absent from ldb16Δ and fld1Δ LDs (Table S1) suggesting a global defect in protein targeting in these mutants. To validate the proteomic data, many of these proteins were expressed from their endogenous locus as C-terminal GFP fusions and their localization evaluated by fluorescence microscopy. In agreement with the mass spectrometric data, all tested LD proteins were strongly reduced or completely absent from the LD surface in ldb16Δ or fld1Δ mutants (Fig. 2 B and not depicted). The changes did not appear to be caused by an overall decrease in protein levels, which for the tested proteins were comparable in wt and mutant cells (Fig. S1 A and not depicted). Importantly, the magnitude of the defect varied depending on the LD morphology. Although LD proteins were decreased but still visible in supersized LDs, they were undetectable in the aggregates (Fig. 2 B), suggesting that these morphologically distinct LDs have monolayers with different properties. Altogether, these data show that Ldb16 and Fld1 are required for the localization of LD-specific proteins.
Lipid-driven relocalization of Opi1 in Fld1/Ldb16 complex mutants
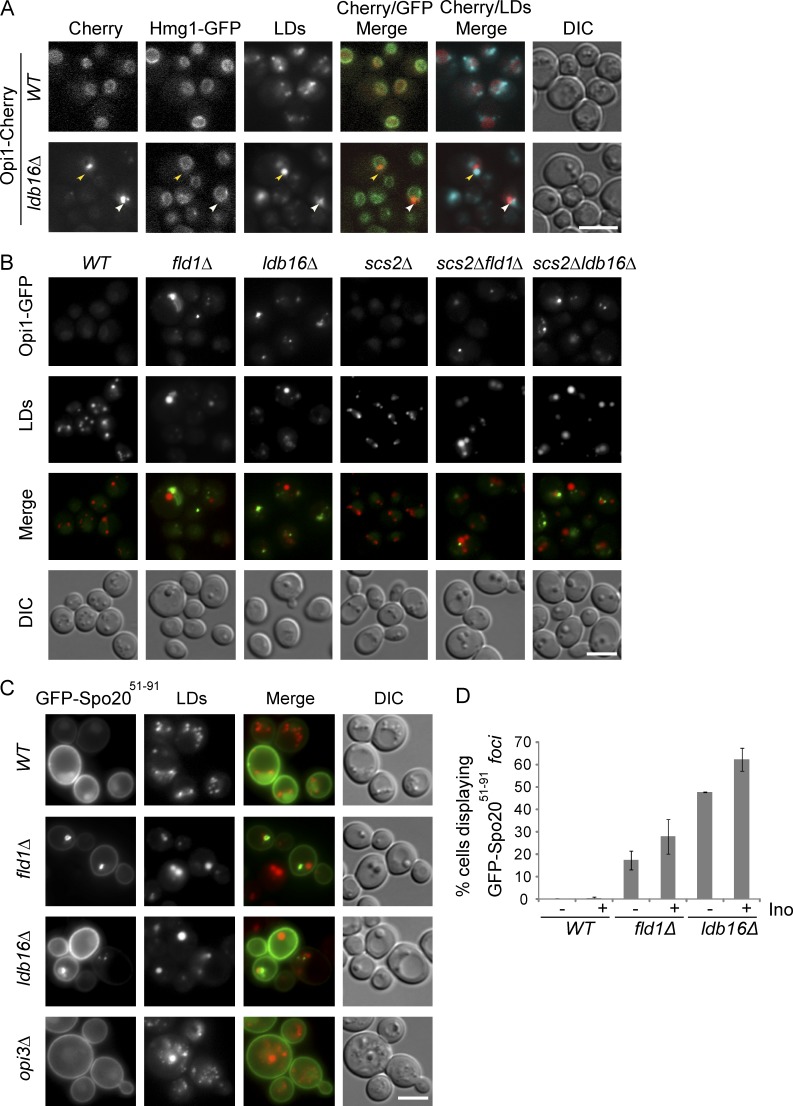
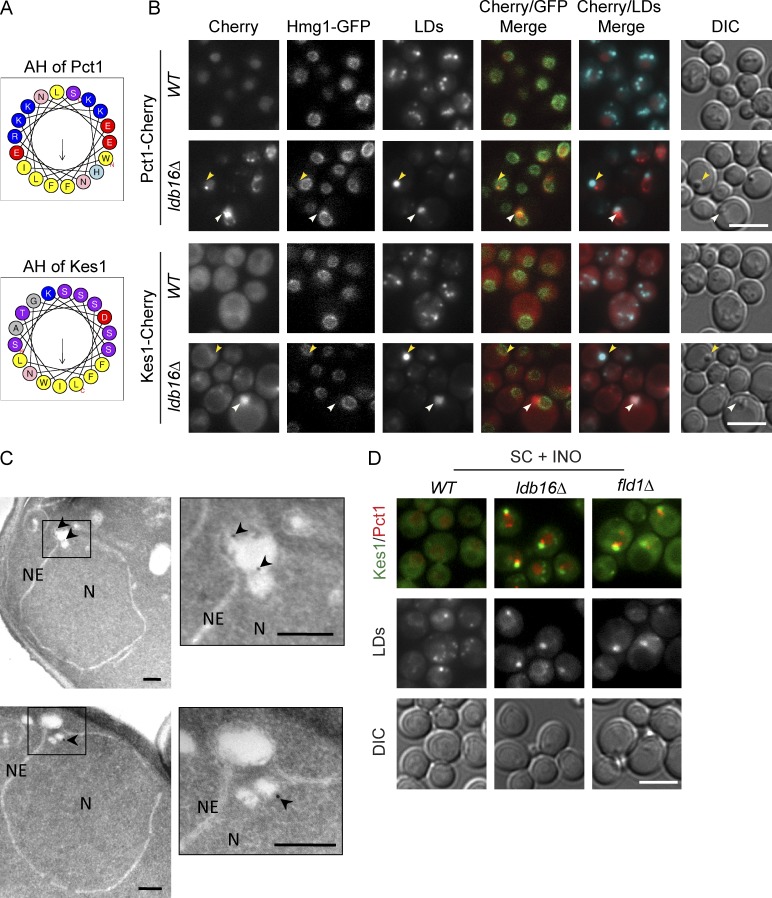
The proteomic analysis of LDs revealed a second major distinction between organelles isolated from wt and ldb16Δ or fld1Δ mutant cells. A group of peripheral membrane proteins that in wt cells do not associate with LDs was highly enriched in organelles isolated from ldb16Δ and fld1Δ mutants (Table S2). The most abundant of these proteins was Opi1, a transcriptional repressor whose activity is controlled by association with the membrane of the ER (Loewen et al., 2004). In the nucleus, Opi1 represses the activation of many genes, including phospholipid biosynthetic genes (Henry et al., 2012). Under conditions of active phospholipid synthesis, Opi1 is bound to the ER membrane in an inactive state. The membrane association of Opi1 involves a bipartite signal: through its FFAT motif, Opi1 binds to the ER membrane protein Scs2; through a short lysine-rich segment (Q2), Opi1 is thought to bind to PA, a precursor for most cellular phospholipids as well as triglycerides (Loewen et al., 2003, 2004). To validate the mass spec data, we analyzed the localization of endogenous Opi1 C-terminally tagged with mCherry (Opi1-Cherry). In wt cells, Opi1-Cherry was uniformly distributed at the nuclear ER as expected (Fig. 3 A; Loewen et al., 2003). In cells lacking Ldb16 or Fld1, Opi1-Cherry still overlapped with the nuclear ER marker Hmg1-GFP; however, its distribution was uneven forming 1–3 foci/cell often in close proximity to abnormal aggregated or supersized LDs (Fig. 3 A and not depicted).
Figure 3.
Lipid-driven relocalization of Opi1 in Fld1/Ldb16 complex mutants. (A) Localization of Opi1 in wt and ldb16Δ cells grown in SC media to early stationary phase. Opi1 was expressed from the endogenous locus as C-terminal mCherry fusion (Opi1-Cherry). Nuclear envelope was labeled by endogenously expressed Hmg1-GFP. LDs were stained with the neutral lipid dye MDH. Yellow arrowheads indicate supersized LDs; white arrowhead indicates LD aggregates. Bar, 5 µm. (B) Localization of endogenously expressed Opi1-GFP in cells with the indicated genotype grown to early stationary phase in SC media. LDs were stained with the neutral lipid dye MDH. Bar, 5 µm. (C) Localization of GFP-Spo2051–91 expressed from a 2 µm plasmid in cells with the indicated genotype. LDs were stained with the neutral lipid dye MDH. Bar, 5 µm. (D) Percentage of cells with the indicated genotype displaying abnormal foci of GFP-Spo2051–91. Late logarithmic cells grown in SC media (−) or SC supplemented with 75 µM inositol (+) are shown. The mean of two independent experiments is graphed; error bars represent SD. For each genotype and condition, at least 100 cells/experiment were analyzed.
Next, we investigated the determinants leading to Opi1 relocalization in ldb16Δ and fld1Δ cells. First, we tested the involvement of its membrane-bound partner Scs2. In scs2Δ cells, Opi1-GFP is mostly nuclear (Fig. 3 B), as previously shown (Loewen et al., 2003). In contrast, the localization of Opi1-GFP in ldb16Δ scs2Δ and fld1Δ scs2Δ double mutants is similar to that in single ldb16Δ and fld1Δ mutants (Fig. 3 B), suggesting that Scs2 is not involved in Opi1 foci formation. Moreover, although enriched in proximity of LD aggregates in ldb16Δ and fld1Δ mutants, Scs2 is normally distributed throughout the ER (Fig. S2 A). A similar distribution is observed for a GFP-fusion of the Opi1 FFAT domain (GFP-Opi1FFAT; Fig. S2 B). Thus, the aberrant distribution of Opi1 in ldb16Δ and fld1Δ mutants is independent of Scs2.
Next we tested whether the abnormal Opi1 distribution in the mutants relied on its lipid-binding activity, putatively to PA. A short Opi1 fragment rich in basic amino acids, Opi1Q2, was shown to bind to PA both in vivo and in vitro (Loewen et al., 2004). In wt cells, Opi1Q2 fused to GFP (GFP-Opi1Q2) localized mostly to the nucleus, as previously shown (Loewen et al., 2004). In ldb16Δ cells, besides the nuclear localization, GFP-Opi1Q2 formed foci resembling the Opi1-GFP although at a much lower frequency (Fig. S2 C). These results suggested that abnormal PA distribution could be the cause of Opi1 relocalization in seipin complex mutants. Therefore, we analyzed the distribution of GFP-Spo2051–91, a 40–amino acid fragment containing the AH of the SNARE Spo20 widely used as a PA biosensor (Nakanishi et al., 2004). In wt cells, GFP-Spo2051–91 localizes at the cell periphery (Fig. 3 C), as expected (Nakanishi et al., 2004). Besides the peripheral staining, fld1Δ and ldb16Δ mutant cells frequently displayed additional GFP-Spo2051–91 foci. These were often apposed to LDs and were reminiscent of those observed for Opi1-GFP. The GFP-Spo2051–91 spots were specific to fld1Δ and ldb16Δ cells and were not present in other mutants displaying supersized LDs, such as opi3Δ (Fig. 3 C). Curiously, in inositol-supplemented media, GFP-Spo2051–91 foci were detected at higher frequency (Fig. 3 D). Altogether, these data indicate that aberrant Opi1 distribution is driven by its lipid-binding domain. However, it is unlikely that abnormal PA accumulation is the cause of the defect as the relocalization of both Opi1-GFP and GFP-Spo2051–91 increases in presence of inositol, a condition that favors PA consumption (Fig. S2 D; Loewen et al., 2004).
Relocalization of amphipathic helix-containing proteins in seipin complex mutants
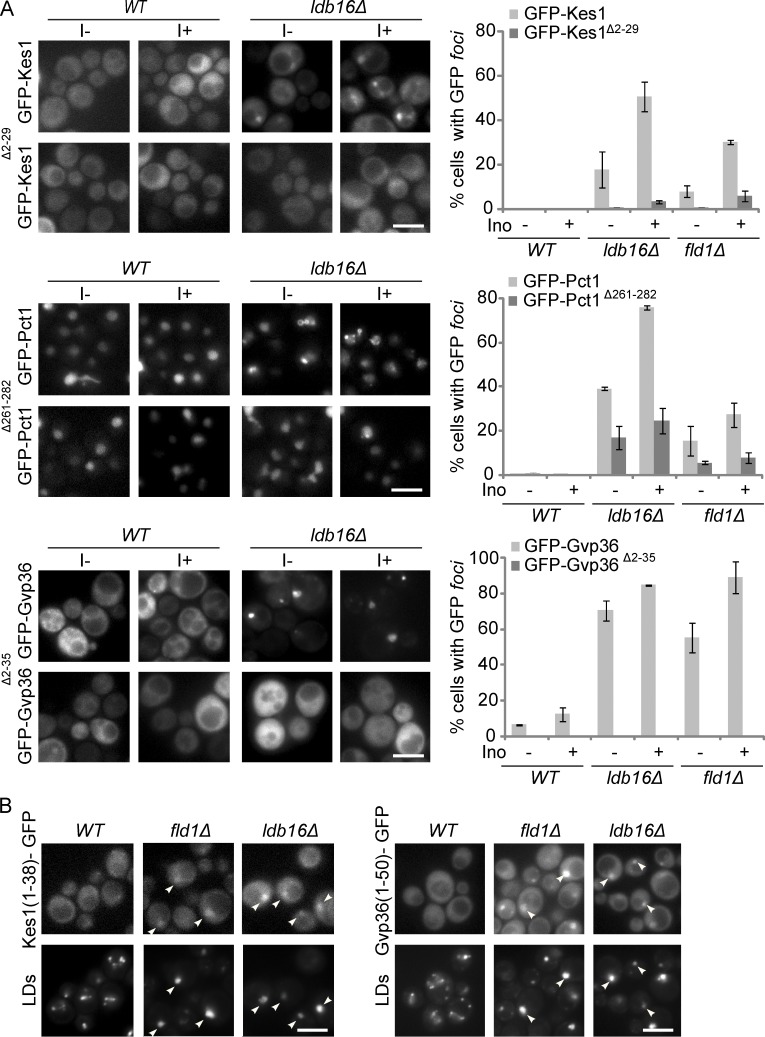
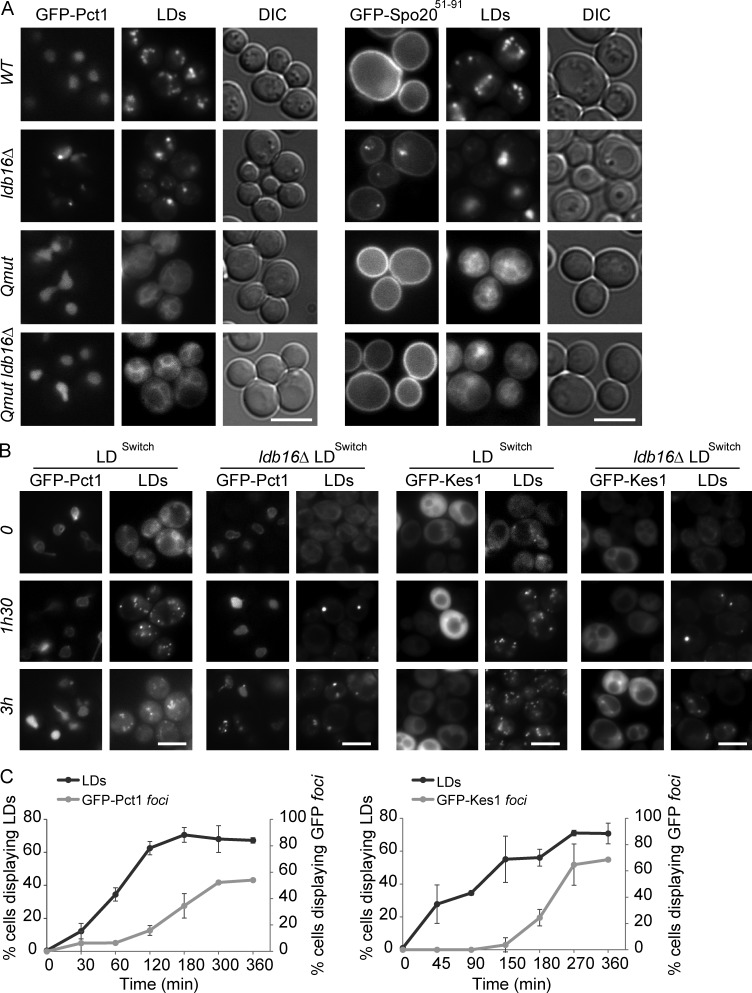
The other peripheral membrane proteins copurifying ectopically with ldb16Δ and fld1Δ LDs were very diverse in function and normal subcellular localization. However, they all contain or are predicted to contain an AH, a common motif involved in membrane association (Table 1; Gautier et al., 2008). These AH-containing proteins were expressed from their endogenous loci as C-terminal GFP fusions, and their distribution was analyzed by fluorescence microscopy (Fig. S3 A). When compared with wt cells, in ldb16Δ and fld1Δ mutants all the tested proteins were dramatically relocalized appearing as one to three foci/cell overlapping or proximal to LDs (Fig. S3 A and not depicted). These results confirmed the proteomic analysis of isolated LDs. Next, we asked whether the change in localization of those proteins required their AHs. In all tested cases, deletion of the AH abolished or dramatically decreased the relocalization of the proteins in ldb16Δ and fld1Δ mutants (Fig. 4 A). The drop in ectopic localization was observed irrespective of the inositol concentration (Fig. 4 A), which influences the morphology of LDs in the mutants, and was not caused by changes in the steady-state levels of proteins (Fig. S3 B). Finally, endogenous expression of the sole AH motif fused to GFP was sufficient for LD relocalization in ldb16Δ and fld1Δ cells (Fig. 4 B). Altogether, these results indicate that the relocalization of several proteins in ldb16Δ and fld1Δ cells is mediated by their AHs.
Table 1. AH-containing proteins ectopically localized to LDs in ldb16Δ and fld1Δ cells.
| Protein | Localization | Function | Type of AH | Reference |
|---|---|---|---|---|
| Pct1 | Nuclear envelope/nucleus | Cholinephosphate cytidylyltransferase | “Canonical” | Cornell and Taneva, 2006 |
| Kes1 | Cytoplasm/Golgi | Oxysterol-binding protein | ALPS | Drin et al., 2007 |
| Vps13 | Cytoplasm/endosomes (?) | Unknown | ALPS | Drin et al., 2007 |
| Gvp36 | Cytoplasm/Golgi vesicles | Unknown (BAR domain–containing protein) | AH (ALPS type) in N-BAR domain | Gautier et al., 2008 |
Figure 4.
Relocalization of amphipathic helix-containing proteins in seipin complex mutants. (A) Localization of GFP-tagged Kes1, Pct1, Gvp36, and the corresponding counterparts lacking the AH Kes1Δ2–29, Pct1Δ261–282, and Gvp36Δ2–35 in cells with the indicated genotype. The GFP fusion proteins were expressed from a centromeric plasmid. Cells in early stationary phase grown in SC media (I−) or SC supplemented with 75 µM inositol (I+) were imaged. LDs were stained with the neutral lipid dye MDH. Bar, 5 µm. On the right the percentage of cells with the indicated genotype displaying abnormal foci of GFP-tagged proteins. The mean of three independent experiments is displayed; error bars represent SD. At least 100 cells/experiment were analyzed per genotype. (B) Localization of Kes1(1–38)-GFP and Gvp36(1–50)-GFP in cells with the indicated genotype grown in SC supplemented with inositol. Kes1(1–38)-GFP and Gvp36(1–50)-GFP were expressed from the endogenous loci and encode for AH of Kes1 and Gvp36, respectively. LDs were stained with the neutral lipid dye MDH. Bar, 5 µm.
Seipin complex mutants display phospholipid-packing defects
AHs do not act as simple membrane anchors but can play an active role in deforming lipid membranes or sensing membrane packing defects, thereby controlling membrane-related processes (Drin and Antonny, 2010; Campelo and Kozlov, 2014). Moreover, the chemical properties of AHs can vary significantly and determine the binding preferences to target membranes. Intriguingly, Kes1 and Pct1, two of the ectopically localized proteins in the mutant cells, contain AHs with very distinct chemical properties. The AH of Pct1, the yeast CTP:phosphocholine cytidylyltransferase, has a highly charged polar face (Fig. 5 A). In contrast, the AH of the lipid transfer protein Kes1 has a polar face enriched in serine/threonines and poor in charged residues (Fig. 5 A). This latter type of AH is also known as ALPS (amphipathic lipid packing sensor) domain and was shown to recognize lipid packing defects arising either from membrane curvature or accumulation of conical lipids in flat membranes (Drin et al., 2007; Vamparys et al., 2013; Vanni et al., 2013; Campelo and Kozlov, 2014).
Figure 5.
Phospholipid packing defects in Fld1/Ldb16 complex mutants. (A) Helical-wheel representation of the AH of Pct1 and Kes1 as predicted and drawn by Heliquest (Gautier et al., 2008). Both AHs display a well-defined face enriched in hydrophobic residues (yellow). In contrast, the polar faces are very distinct: in Pct1, it is enriched in charged residues (blue and red), whereas in Kes1, the abundance of noncharged serine and threonine residues (purple) dominates. (B) Localization of Pct1 and Kes1 in wt and ldb16Δ cells grown in SC media to early stationary phase. Proteins were expressed from their endogenous locus as C-terminal mCherry fusions. Nuclear envelope is labeled by endogenously expressed Hmg1-GFP. LDs are stained with the neutral lipid dye MDH. Yellow arrowheads indicate supersized LDs; white arrowheads indicate LD aggregates. Bar, 5 µm. (C) Electron micrographs showing immunolocalization of endogenously expressed Pct1-GFP in ldb16Δ cells. Logarithmic cultures in YPD were processed for immuno-EM as described in the Materials and methods section. Arrowheads point to 12 nm immunogold particles specifically labeling Pct1-GFP in association with nuclear LDs. NE, nuclear envelope; N, nucleus. Magnified insets are shown on the right of each micrograph. Bars, 200 nm. (D) Localization of endogenously expressed Kes1-GFP and Pct1-mCherry in wt, ldb16Δ, and fld1Δ cells. Late logarithmic cultures in SC media or SC supplemented with 75 µM inositol (SC + INO) were imaged. LDs are stained with the neutral lipid dye MDH. Bar, 5 µm.
Given the distinct properties of their AHs, the distribution of Kes1 and Pct1 was analyzed in detail. In wt cells, both Pct1-Cherry and Kes1-Cherry had a diffuse distribution as expected (MacKinnon et al., 2009; Fig. 5 B). In ldb16Δ and fld1Δ mutants, Pct1-Cherry formed foci proximal, but not completely overlapping with abnormal LDs, both supersized and aggregated (Fig. 5 B). Using immuno-EM, we confirmed that in mutant cells, Pct1 concentrated at the surface of a subset of LDs (Fig. 5 C and Table S3, top). Interestingly, Pct1 was detected primarily in LDs present in the nucleus, which have been previously seen in fld1Δ mutants (Cartwright et al., 2015). Consistent with the different chemistry of its AH, the foci formed by Kes1-Cherry were distinct from the ones of Pct1. Kes1-Cherry foci perfectly overlapped with the LD aggregates (Fig. 5 B) The association of Kes1 with LD aggregates was confirmed by immuno-EM (Fig. S3 C and Table S3). Remarkably, by fluorescence microscopy, Kes1 was never seen at supersized LDs, indicating that the monolayers of aggregated and supersized LDs have different properties.
In cells coexpressing Pct1 and Kes1 as Cherry and GFP fusions, respectively, we confirmed that, in ldb16Δ and fld1Δ mutants, these proteins relocalize to proximal but distinct regions, likely as a consequence of the different chemical properties of their AHs (Fig. 5 D). Accordingly, Vps13 and Gvp36, that like Kes1 contain AHs of the ALPS type also localize to LD aggregates (Fig. S3 D). In sum, these data indicate that in ldb16Δ and fld1Δ mutants, LDs display phospholipid packing defects that are recognized by AHs with different specificities.
Membrane defects in seipin complex mutants are caused by LD assembly
Mutations in FLD1 and LDB16 might have a general effect on membrane properties causing the observed defects in LD morphology and protein localization. Alternatively, the membrane defects might derive specifically from abnormal LD assembly in these mutants. To discriminate between these possibilities, we analyzed the distribution of Pct1, GFP-Spo2051–91, and Kes1 in cells lacking LDs, as is the case of the quadruple mutant are1Δ are2Δ dga1Δ lro1Δ (Qmut) unable to synthesize neutral lipids (Sandager et al., 2002; Sorger et al., 2004). The distribution of these proteins was indistinguishable between wt and Qmut cells (Fig. 6 A and Fig. S4, A and B). Importantly, the abnormal localization of Pct1, GFP-Spo2051–91 and Kes1 induced by ldb16Δ and fld1Δ mutations was completely reverted in the absence of LDs, such as the quintuple mutants are1Δ are2Δ dga1Δ lro1Δ ldb16Δ and are1Δ are2Δ dga1Δ lro1Δ fld1Δ (Fig. 6 A and Fig. S4, A and B).
Figure 6.
LD assembly causes phospholipid-packing defects in seipin complex mutants. (A) Localization of GFP-Pct1 and GFP-Spo2051–91 in cells with (wt and ldb16Δ) and without LDs (are1Δ are2Δ dga1Δ lro1Δ and are1Δ are2Δ dga1Δ lro1Δ ldb16Δ). Cells were grown in SC media up to early stationary phase. LDs were stained with the neutral lipid dye MDH. Bar, 5 µm. (B) Localization of GFP-Pct1 and GFP-Kes1 at the indicated time points upon induction of LD formation. LDs were induced by expression of the triglyceride-synthesizing enzyme Dga1 from a regulatable promoter in are1Δ are2Δ lro1Δ (LDSwitch) or are1Δ are2Δ lro1Δ ldb16Δ (ldb16Δ LDSwitch) cells. LDs were stained with the neutral lipid dye MDH. Bar, 5 µm. (C) Kinetics of LD formation and appearance of GFP-Pct1 (left) or GFP-Kes1 (right) foci in ldb16Δ LDSwitch cells. Cells were grown in minimal media supplemented with 75 µM inositol until early stationary phase. The percentages of ldb16Δ LDSwitch cells displaying LDs (black line) or foci of the indicated GFP-tagged protein (gray line). Expression of Dga1 was induced at time zero. The mean of two independent experiments is displayed; error bars represent SD. At least 100 cells/time point were analyzed in each experiment.
Next we followed LD assembly in cells lacking are1Δ are2Δ lro1Δ in which DGA1 is under the control of an inducible promoter (LDSwitch) either in presence or absence of LDB16 and FLD1. Time-course experiments showed that relocalization of Pct1 and Kes1 follows the appearance of LDs (Fig. 6, B and C). Moreover, the abnormal LD morphology in seipin complex mutants was not reverted by additional deletion of Pct1 or any of the other proteins localizing ectopically to LDs (Fig. S4 C and not depicted). These data show that the membrane defects are a consequence of abnormal LD formation in ldb16Δ and fld1Δ mutants. Moreover, they suggest that the seipin complex organizes ER membrane domains required specifically during LD assembly.
Destabilized ER–LD contact sites in seipin complex mutants
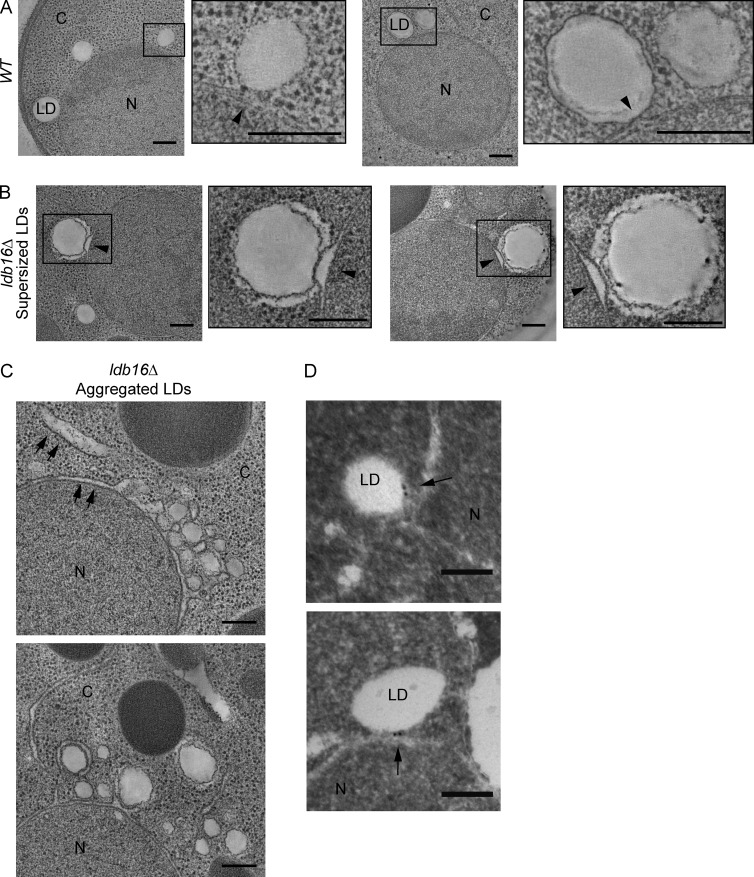
To gain further insight into the membrane defects induced during LD biogenesis in ldb16Δ and fld1Δ mutants, we performed dual-axis electron tomography of high-pressure frozen and freeze-substituted cells. This approach allowed us to resolve at high definition LDs, adjacent ER regions, and ER–LD contact sites. In wt cells, LDs had a smooth contour, were invariably in contact with the ER, and regions previously described as ER–LD bridges were seen (Fig. 7 A and Videos 1 and 2). The structure of the ER at the contact sites was highly regular, with the luminal space showing a uniform width throughout. This was in striking contrast with the LDs observed in ldb16Δ mutants, irrespective of whether they were supersized or aggregated. In both cases, the contour of LDs appeared frequently wrinkled and irregular and ER–LD contact sites were noticeably abnormal. In the case of supersized LDs, the ER membrane contacting the LD protruded in a nipple-like structure toward the LD resulting in an abnormal expansion of ER luminal width (Fig. 7 B and Videos 3 and 4). The luminal expansion was circumscribed to the contact site with supersized LDs, and other ER regions appeared largely normal. In the case of aggregated LDs, observed in cells with active phospholipid synthesis, contact sites had a peculiar appearance. Overall, the contact site was enlarged with prominent ER protrusions molded around the LDs in the aggregate (Fig. 7 C). If in most cases ER and LDs were apposed but clearly identified as separate structures, regions with appearance of ER–LD bridges were also detected (Fig. 7 C and Videos 5, 6, and 7). Expansion of ER luminal width was frequently observed and in most cases was not restricted to the ER contact sites with the aggregated LDs, and bloated ER cisternae could be seen away from the aggregates (Fig. 7 C and Video 7). The expanded ER lumen was electrotranslucent, with an appearance similar to the neutral lipid core of LDs. Abnormal ER–LD contact sites in this mutant suggest that Ldb16 functions at interface of the two organelles, where it appears to localize with Fld1 based on fluorescent microscopy experiments (Szymanski et al., 2007; Fei et al., 2008; Wang et al., 2014). Indeed, using immuno-EM, we detected that functional Ldb16-GFP expressed from its endogenous locus localizes specifically at the ER–LD contact sites (Fig. 7 D and Table S4). Altogether, these data indicate that Fld1/Ldb16 complex is required for the stability of ER–LD contact sites.
Figure 7.
Destabilized ER–LD contact sites in Seipin complex mutants. (A) 2D tomograms derived from 250-nm-thick sections of wt cells. Magnified insets on the right of each tomogram show ER–LD contact sites (arrowhead). Bars, 200 nm. (B) 2D tomograms derived from a 250-nm-thick sections of ldb16Δ cells. Magnified insets on the right of each tomogram show the contact sites between ER and supersized LDs (arrowhead). Bars, 200 nm. (C) 2D tomograms derived from a 250-nm-thick sections of ldb16Δ cells. Arrows indicate ER regions with enlarged luminal width. Bars, 200 nm. (D) Electron micrographs showing immunolocalization of endogenously expressed Ldb16-GFP in wt cells. Arrows point to 12-nm immunogold particles labeling specifically Ldb16-GFP. Bars, 100 nm. C, cytoplasm; N, nucleus.
Discussion
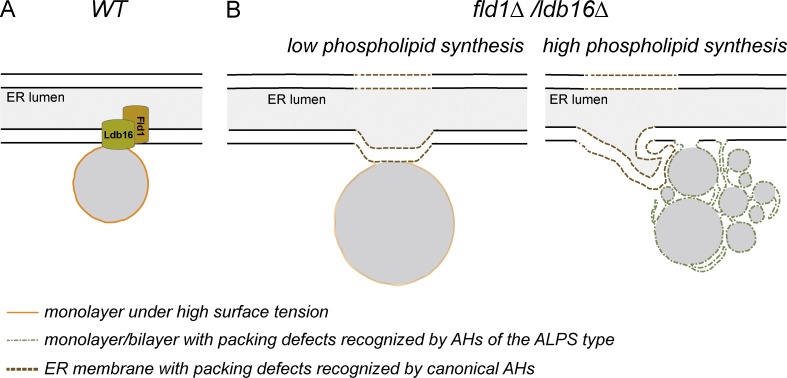
Here we characterized the function of the yeast seipin complex in LD biogenesis. LD assembly in the absence of Fld1/Ldb16 led to membrane defects in LDs and proximal ER regions and to abnormal LDs, which were either supersized or small and aggregated. Moreover, we showed that the determinant between these contrasting LD morphologies was the availability of ER phospholipid pools, indicating that Fld1/Ldb16 is important during LD biogenesis to couple the amounts of monolayer and core lipids. Structural analysis indicated that the defects were the result of abnormal ER/LD contact sites in ldb16Δ and fld1Δ mutants. We propose that stabilization of ER–LD contact sites by the seipin complex establishes a diffusion barrier controlling LD assembly and membrane identity (Fig. 8).
Figure 8.
Fld1/Ldb16 complex stabilizes and establishes a diffusion barrier at the ER–LD contact sites. (A) The scheme illustrates a contact site between the ER and a LD. Although connected with the ER membrane, the LD monolayer has different properties such as a higher surface tension (in orange). The presence of the Fld1/Ldb16 complex at the contact sites prevents the equilibration of the two membrane systems. (B) In the absence of this complex, phospholipids freely diffuse between the two organelles. Under low-synthesis conditions, phospholipids can become limiting and LDs coalesce into a supersized one. The mild surface tension (faint orange) in these LDs still allows the targeting of LD proteins, albeit at lower efficiency (left). Under high-phospholipid-synthesis conditions, ER membrane and LDs equilibrate. The low surface tension of these LDs prevents their coalescence and the recruitment of LD proteins. Instead, these aggregates display phospholipid packing defects (green), which recruit ALPS-motif–containing proteins (right). Both low and high phospholipid synthesis leads to phospholipid defects in membranes adjacent to LDs, which are recognized by proteins containing canonical AHs (brown).
Changes in phospholipid composition, in particular defects in the PC synthesis, lead to abnormal LD morphology (Guo et al., 2008; Krahmer et al., 2011; Fei et al., 2011b). This is unlikely the reason of LD abnormalities in fld1Δ and ldb16Δ mutants, as we and others could not detect significant changes in their lipidome (Fei et al., 2008, 2011b; Wang et al., 2014). In contrast, we showed that the morphology of LDs in these mutants is determined by phospholipid synthesis, as previously suggested (Cartwright et al., 2015). In agreement with our results, unperturbed fld1Δ and ldb16Δ cells display mainly LD aggregates during logarithmic growth, a condition favoring phospholipid synthesis, whereas the supersized LD phenotype dominates as cells approach stationary phase, when phospholipid synthesis is reduced (Wang et al., 2014; unpublished data).
The effect of phospholipid availability on LD morphology correlates well with the content of phospholipids in LDs. Under conditions that stimulate phospholipid synthesis, LDs isolated from mutant cells had a dramatic increase in phospholipids. This was not because of ER contamination of the isolated LDs and was consistent with the increased surface-to-volume ratio of LDs in aggregates. However, according to our EM data, the highly wrinkled contour of LDs in the mutant cells also contributes to the increased amounts of phospholipids. Moreover, in the aggregates, structures resembling ER–LD bridges likely facilitate the free diffusion of phospholipids between ER and LDs. The combined effect of the changes resulted in a higher content of the major ER phospholipids in LD aggregates. Under low-synthesis conditions, phospholipids become limiting and supersized LDs, with lower surface-volume ratio, formed. These still displayed an irregular contour. Thus, this dependence of LD morphology on ER phospholipid pools, not seen for any other mutant with abnormal LDs, results from destabilized ER–LD contact sites in fld1Δ and ldb16Δ mutants.
AHs adsorb to membranes exposing phospholipid packing defects (Drin and Antonny, 2010; Campelo and Kozlov, 2014). The recruitment of AH-containing proteins to LDs and ER proximal regions indicates the presence of membrane packing defects in fld1Δ and ldb16Δ mutants. Importantly, these are a direct consequence of LD assembly, as in time-course experiments, their appearance follows LD biogenesis. Interestingly, proteins containing AHs of different chemistry are recruited to distinct subpopulations of LDs. The AH of Kes1, a prototypical ALPS motif, binds to membranes with phospholipid packing defects generated by high curvature or accumulation of conical phospholipids (such as PE) in a flat membrane (Drin et al., 2007; Vamparys et al., 2013; Vanni et al., 2013). In vitro studies demonstrated that Kes1 ALPS binds to small liposomes (in the range of 30–40 nm in diameter), but not to bigger ones (Drin et al., 2007). Crude measurements suggest that the diameter of individual LDs in the aggregates are bigger (>80 nm). Therefore, it is possible that the ectopic recruitment of ALPS-containing proteins to LD aggregates in fld1Δ and ldb16Δ LDs is caused by the combined effect of curvature and increased levels of PE, a conic-shaped lipid, present in these structures. On the other hand, the AHs of Pct1 and Spo20 appear canonical and likely associate with membranes by a combination of both hydrophobic and electrostatic interactions (Drin and Antonny, 2010). Although the Pct1 AH has not been extensively characterized biochemically, the AH of Spo20 (Spo2051–91) was shown to bind to PA-rich membranes in vivo (Nakanishi et al., 2004; Horchani et al., 2014); however, recent in vitro studies suggest a general preference for anionic phospholipids and not necessarily for PA (Horchani et al., 2014). A preference for PA, preferably with shorter acyl chains, was also observed for the Q2 region of Opi1 (Loewen et al., 2004; Hofbauer et al., 2014). We detected only a small increase in whole-cell levels of PA and no enrichment in LD lipid fractions; therefore, Spo2051–91 and Opi1Q2 likely recognize some other feature at the nuclear envelope of fld1Δ and ldb16Δ mutants. The fact that conditions stimulating PA consumption, such as inositol supplementation or Cds1 overexpression, augment the relocalization of Spo2051–91 and Opi1 to foci at the nuclear envelope further argue against a PA-driven process, as previously proposed (Fei et al., 2011b; Wolinski et al., 2015). In sum, we favor that unstable ER–LD contact sites in fld1Δ and ldb16Δ mutants, allowing uncontrolled phospholipid flow between the ER bilayer and LD monolayer, are prone to generate the packing defects recognized by AH-containing proteins. The spatial segregation of the different AHs possibly reflects differences in membrane curvature or surface tension.
The ectopic recruitment of AH-containing proteins to LDs observed in fld1Δ and ldb16Δ mutants can have several consequences. On one hand, it can deplete proteins from their site of function, potentially leading to a “loss of function” phenotype. This appears to be the case of Opi1, a negative regulator of Ino2/Ino4 required for transcription of most phospholipid biosynthetic genes (Henry et al., 2012). In fact, several Ino2/Ino4 targets, such as INO1 or OPI3, are upregulated in fld1Δ and ldb16Δ mutants (Hancock et al., 2006; Fei et al., 2011b; Wang et al., 2014; unpublished data). However, upregulation of Ino2/Ino4 target genes is not the main cause of fld1Δ and ldb16Δ phenotype, as LDs are mostly normal in opi1Δ cells. On the other hand, ectopic concentration of AH-containing proteins at LDs can lead to local “gain of function” of these proteins. The phenotype of fld1Δ and ldb16Δ mutations was not modified by deleting individually Pct1, Kes1, Gvp36, or Vps13, suggesting that the ectopic recruitment of each one of these proteins per se is not responsible for the LD defects. This was further supported by time-course experiments showing that protein recruitment followed the formation of abnormal LDs in fld1Δ and ldb16Δ cells.
A major consequence of the loss of Fld1/Ldb16 is the global reduction of LD-specific proteins at the monolayer of these organelles. Many LD proteins are originally targeted to the ER before concentrating in the LD monolayer (Yang et al., 2012; Thiam et al., 2013b). It is possible that the crowding of the LD monolayer because of ectopic localization of AH-containing proteins precludes proper targeting of LD proteins. However, under conditions of overexpression, the LD-specific protein Dga1 was shown to localize normally in fld1Δ cells (Jacquier et al., 2011), suggesting that the targeting of this protein is not defective. An alternative appealing possibility is that LD-specific proteins are able to target but are not retained/concentrated at the LD monolayer as a consequence of changes in surface tension owing to the free diffusion of phospholipids from the ER into LDs in FLD1 and LDB16 mutants. The wrinkled contour of LDs in these mutants indeed suggests changes in properties of the monolayer. Moreover, it is also consistent with the stronger protein targeting defect to LD aggregates, which have a higher content of phospholipids. In LDs that are not connected to the ER, the regulation of monolayer surface tension is essential in recruiting soluble proteins from the cytoplasm (Krahmer et al., 2011; Thiam et al., 2013a), in facilitating ER–LD reattachments (Wilfling et al., 2014), and consequently in generating LD identity. It is possible that regulation of LD monolayer surface tension also plays a central role in concentrating proteins in LDs that remain connected to the ER. By stabilizing ER–LD contact sites, the Fld1/Ldb16 complex is well positioned to serve that function.
In contrast with most organelle contact sites, in which the membranes of two organelles are closely apposed (Prinz, 2014), at ER–LD contact sites the membranes of the two organelles merge, as the LD monolayer is continuous with the outer leaflet of the ER bilayer. Although the phospholipid classes in the two organelles are similar, the biophysical properties and protein composition of ER bilayer and LD monolayer are very distinct. We postulate that stabilization of these unique contact sites by Fld1/Ldb16 establishes a diffusion barrier necessary to regulate LD surface tension and identity. Potentially, such a barrier can also facilitate the concentration of neutral lipids and their packaging into nascent LDs. Future studies should test these hypotheses directly. Both Fld1 and Ldb16 form oligomers, which in the case of Fld1 appear to have a toroid shape (Binns et al., 2010; Wang et al., 2014). Whether and how these oligomerization events are controlled at ER–LD contact sites should also be addressed in follow-up work.
Materials and methods
Reagents
The LD dyes Bodipy493/503 (Invitrogen) and monodansyl pentane (MDH; Abgent) were used at 1 µg/ml and 0.1 mM, respectively. Anti-HA (rat 3F10 monoclonal) antibody was purchased from Roche, anti-PGK1 (mouse) from Life Technologies, anti-GFP (rabbit) from Santa Cruz Biotechnology, Inc., and anti-RFP (mouse) from Abcam. Polyclonal anti-Usa1 antibody was raised against a C-terminal peptide of Usa1 and was previously described (Carvalho et al., 2006). Recombinant protein fragments were used to raise polyclonal antibodies anti-Dga1 (amino acids 123–272) and anti-Pet10 (amino acids 148–283). All the antibodies were raised in rabbits, and sera anti-Dga1 and anti-Pet10 were affinity purified.
Yeast strains
Protein tagging, promoter replacements, and individual gene deletions were performed by standard PCR-based homologous recombination (Longtine et al., 1998; Janke et al., 2004). Strains with multiple gene deletions were made either by PCR-based homologous recombination or by crossing haploid cells of opposite mating types, followed by sporulation and tetrad dissection using standard protocols (Guthrie and Fink, 1991). The strains used are isogenic either to BY4741 (Mata ura3Δ0 his3Δ1 leu2Δ0 met15Δ0) or to BY4742 (Matα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) unless otherwise specified and are listed in Table S5.
Plasmids and primers
The KES1 ORF (including 3′ UTR) was amplified by PCR from genomic DNA using primers 1688 and 1689. The obtained PCR fragment was digested using XhoI and BamHI and ligated to FLAG-GFP from pPC874 plasmid originating pPC1131. pPC1224, an equivalent plasmid encoding for Flag-GFP-Kes1Δ2–29 lacking Kes1 amphipathic helix was obtained by PCR mutagenesis using primer 1731. The PCT1 ORF (including 3′UTR) was amplified by PCR from genomic DNA using primers 1690 and 1691. The obtained PCR fragment was digested using XhoI and BamHI and ligated to FLAG-GFP from pPC874 plasmid originating pPC1130. pPC1226, an equivalent plasmid encoding for Flag-GFP-Pct1Δ261–282 lacking Pct1 amphipathic helix, was obtained by PCR mutagenesis using primer 1733. The GVP36 ORF (including 3′UTR) was amplified by PCR from genomic DNA using primers 1686 and 1687. The obtained PCR fragment was digested using XhoI and BamHI and ligated to FLAG-GFP from pPC874 plasmid originating pPC1129. To generate pPC1184, encoding Gvp36-GFP, a PCR fragment encoding genomic Gvp36-GFP was amplified using genomic DNA from yPC5554 as template and primers 1817 and 185, digested with XhoI–XbaI cloned in pRS416. The predicted amphipatic helix (amino acids 2–35) was deleted by mutagenesis on this plasmid using primer 1818 to obtain the plasmid pPC1183. The DNA fragment encoding for ADH1p-GAL4-ER-VP16 was isolated by EcoRI (blunted using Klenow) and NotI digestion from the plasmid described in Louvion et al. (1993), blunted using Klenow and ligated into SmaI–NotI-digested pRS415 giving rise to pPC924. The Q2 fragment from Opi1 containing residues 103–189 from Opi1 was amplified from genomic DNA using primers 1339 and 1340, digested with XhoI–HindIII, and cloned into the backbone of pPC670 in frame with FLAG-GFP originating pPC974. Plasmids and primers used in this study are listed in Tables S6 and S7, respectively.
Growth conditions
Strains were grown at 30°C in synthetic complete (SC; 0.17% yeast nitrogen base, 5 g/l ammonium sulfate, 2% glucose, and amino acids) or YPD liquid media (1% yeast extract, 2% peptone, and 2% glucose). For analysis of protein localization and LD morphology both late logarithmic phase (OD600 2–3) and early stationary phase (OD600 4–6) cultures were used. For phospholipids precursor treatment (I+), SC medium was supplemented with 75 µM inositol (Gaspar et al., 2006).
LD induction system
For LD induction time courses, strains GAL1-DGA1 are1Δ are2Δ lro1Δ and GAL1-DGA1 are1Δ are2Δ lro1Δ ldb16Δ, bearing a plasmid encoding for the chimeric Gal4-VP16 transcription factor fused to the hormone-binding domain of the human estrogen receptor, were used (Louvion et al., 1993). This chimeric GAL4-ERE-VP16 protein provides galactose-independent activation of transcription of genes driven by GAL promoters (DGA1 in this study) in response to β-estradiol. Cells were precultured in SC media lacking the appropriate amino acids until early stationary phase, diluted to OD 0.15 in 10 ml fresh media in 25-ml flasks at 30°C, and stimulated by addition of 100 nM β-estradiol. Time point 0 was imaged before induction with β-estradiol, and samples were acquired at the indicated time points, stained with MDH, and immediately imaged by live-cell fluorescence microscopy.
Fluorescence microscopy
Fluorescence microcopy was performed at RT in a wide-field AF6000LX microscope (Leica) with an Andor iXon EMCCD camera and controlled by LAS AF software (Leica) or in a Cell Observer HS (Carl Zeiss) with a Hamamatsu CMOS camera ORCA-Flash4.0 controlled by 3i Slidebook6.0 software. A 100× 1.40 oil immersion objective was used. GFP and Bodipy493/503, mCHERRY, and MDH signals were detected using GFP filter, RFP filter cube, and DAPI filters, respectively, with standard settings.
LD isolation
LD purification was performed as previously described (Leber et al., 1994; Connerth et al., 2009) with minor modifications. In brief, cells were grown in 500 ml YPD until stationary phase. Approximately 3,000 ODs of cells were centrifuged at 3,000g for 5 min (J J26-XP centrifuge, JLA8100 rotor), washed in milliQ water, preincubated in 0.1 M Tris-HCl, pH 9.5, and 10 mM DTT for 10 min at 30°C, washed, and resuspended in 50 ml spheroplasting buffer (1.2 M sorbitol and 50 mM Tris, pH 7.4). For spheroplast preparation, Zymolyase 20T (Seikagaku Biobusiness) was added (10 µg/OD600 unit cells) followed by incubation in a water bath at 30°C until a 10-fold drop in OD600 was observed (45–60 min). Spheroplasts were recovered by centrifugation (1,000 g, 4°C), washed with spheroplasting buffer, and resuspended in breaking buffer (BB; 10 mM MES, Tris, pH 6.9, 12% [wt/wt] Ficoll400, and 0.2 mM EDTA) at a final concentration of 0.3 g cells (wet weight)/ml. PMSF (1 mM) and Complete (Roche) were added before homogenization (loose-fitting pestle, 40 strokes) in a Dounce homogenizer on ice. The homogenate was centrifuged (5,000 g, 5 min) in J26-XP using rotor JS13.1. The resulting supernatant was transferred into 38ml Ultra-Clear centrifuge tubes (Beckman Coulter) and adjusted to 19 ml (cell lysate fraction), overlaid with an equal volume of BB and centrifuged (45 min, 30,000 rpm) in an optima L-100K centrifuge (Beckman Coulter) with an SW-32 swinging bucket rotor. The floating layer was collected from the top of the gradient and the LDs were further purified. The pellet was collected, resuspended in PBS, pH 7.4, with the help of a Dounce homogenizer, and fully dissolved with SDS–sample buffer (membranes fraction). For further purification of the LD fraction, the floating layer was gently resuspended in BB (five strokes in a Dounce homogenizer with loose-fitting pestle), adjusted to 19 ml with BB, transferred to a 38-ml Ultra-Clear tube, and overlaid with 19 ml of 10-mM MES-Tris, pH 6.9, 8% (wt/wt) Ficoll400, and 0.2 mM EDTA. Centrifugation was repeated as before (45 min, 30,000 rpm). The floating layer was collected and resuspended in 19 ml of 10-mM MES-Tris, pH 6.9, 0.6 M sorbitol, 8% (wt/wt) Ficoll400, and 0.2 mM EDTA, transferred to 38-ml Ultra-Clear tubes, overlaid with 19 ml of 10-mM MES-Tris, pH 6.9, 0.25 M sorbitol, and 0.2 mM EDTA, and centrifuged once more (30 min, 30,000 rpm). The recovered high-purity top LDs fraction was snap frozen, stored at −80°C, and used subsequently for proteomics and lipid analysis.
Immuno-EM
Yeast cells grown in YPD to OD600 ∼1 were fixed with 0.2% glutaraldehyde and 2% formaldehyde in 0.1 PHEM buffer (20 mM Pipes, 50 mM Hepes, pH 6.9, 20 mM EGTA, and 4 mM MgCl2) for 20 min at RT and overnight in the cold room with rolling. Cells were then processed for cryosectioning following the Tokuyasu method as described previously (Griffith et al., 2008). In brief, fixed cells were treated with 1% periodic acid in 0.1M PHEM for 60 min at room temperature, washed with 0.1 M PHEM, and embedded in 12% gelatin in the same buffer for 10 min at 37°C. Gelatin was allowed to solidify on ice for at least 60 min, and then cells were infiltrated in 2.3 M sucrose overnight at 4°C before processing into 1-mm3 cubes and freezing into liquid nitrogen. 50-nm cryosections were picked up in a 1:1 2% methyl cellulose and 2.3 M sucrose solution and stored at 4°C. For immunolabeling, ultrathin cryosections were blocked in blocking buffer containing 1% BSA mixed with 0.1% Aurion BSA-C, 0.1% glycine, 0.1% gelatin, and 1% Tween-20 in 0.1 M PHEM. As a primary antibody, rabbit anti-GFP antibody (Ab6556; Abcam) was used at 1:100 dilution in blocking buffer for 1 h and a 12-nm gold-conjugated anti–rabbit IgG antibody (Jackson ImmunoResearch Laboratories) as a secondary antibody at 1:30 dilution for 30 min. Cryosections were fixed with 1% glutaraldehyde, contrasted for 5 min in a drop of uranyl oxalate solution and for 5 min in a drop of uranyl acetate-methyl cellulose pH 4.0 solution on ice (Tokuyasu, 1986). Ultrathin sections were examined using a Tecnai Spirit transmission electron microscope (FEI Company) at 120 kV accelerating voltage. Micrographs were acquired using a CCD camera (MegaView III; Olympus) and the image acquisition analySIS software (Olympus). In all cases, the specificity of immunolabeling was accessed by comparing the number of immunogold particles in the strains expressing GFP-tagged Pct1, Kes1, or Ldb16 with a isogenic untagged strain.
EM tomography
Cells were cryoimmobilized by high-pressure freezing using a EM HPM100 (Leica). Freeze substitution of frozen samples was performed in an Automatic Freeze substitution System EM AFS-2 (Leica), using acetone containing 0.1% of uranyl acetate and 1% water, for 3 d at −90°C. On the fourth day, the temperature was slowly increased, by 5°C/h, to −45°C. At this temperature, samples were rinsed in acetone and then infiltrated and embedded in Lowicryl HM20 for 3 d. For electron tomography, serial sections were collected on Formvar-coated, palladium-copper slot grids. The grids with 250-nm-thick sections were placed in a high-tilt holder (Model 2020; Fischione Instruments), and the cells were recorded on a Tecnai F30 EM operating at 300 kV using the SerialEM software package (Mastronarde, 2005). Images were taken every degree over a ±60° range on an FEI Eagle 4K × 4K CCD camera at a magnification of 20,000× and a binning of 2 (pixel size 1.179 nm). The LDs were recorded as dual-axis tilt series. The tilted images were aligned by using the positions of the fiducial gold particles. The tomograms were generated using the R-weighted back-projection algorithm. For supersized LDs, serial sections were acquired. These were aligned and joined using the eTomo graphical user interphase (Ladinsky et al., 1994; Marsh et al., 2001; O’Toole et al., 2003; Höög et al., 2007). Tomograms were displayed as slices one-voxel thick, modeled, and analyzed with the IMOD software package (Kremer et al., 1996).
Protein mass spectrometry
Trichloroacetic acid precipitated proteins were resuspended in 6 M urea and 200 mM ammonium bicarbonate before reduction (10 mM dithiothreitol) and alkylation (20 mM iodoacetamide). Samples were diluted to 2 M urea and digested with trypsin (1:10 wt/wt) overnight at 37°C. Tryptic peptide mixtures were desalted using a C18 UltraMicroSpin column using three washes with 0.1% formic acid in water, followed by an elution step with 0.1% formic acid in a 1:1 mix of water and acetonitrile (Rappsilber et al., 2007).
Samples were analyzed in a LTQ-Orbitrap Velos Pro mass spectrometer (Thermo Fisher Scientific) coupled to nano-LC (Proxeon) equipped with a reversed-phase chromatography 12-cm column with an inner diameter of 75 µm, packed with 5-µm C18 particles (Nikkyo Technos). Chromatographic gradients were set from 93% buffer A, 7% buffer B to 65% buffer A, 35% buffer B in 60 min with a flow rate of 300 nl/min (buffer A: 0.1% formic acid in water; buffer B: 0.1% formic acid in acetonitrile). The instrument was operated in DDA mode and full mass spectrometry scans with 1 micro scans at resolution of 60,000 were used over a mass range of m/z 250–2,000 with detection in the Orbitrap. After each survey scan, the top 20 most intense ions with multiple charged ions above a threshold ion count of 5,000 were selected for fragmentation at normalized collision energy of 35%. Fragment ion spectra produced via collision-induced dissociation were acquired in the linear ion trap. All data were acquired with Xcalibur software v2.2.
Acquired data were analyzed using the Proteome Discoverer software suite (v1.3.0.339; Thermo Fisher Scientific), and the Mascot search engine (v2.3; Matrix Science) was used for peptide identification. Data were searched against database containing all yeast proteins according to the Saccharomyces Genome Database plus the most common contaminants (Bunkenborg et al., 2010). A precursor ion mass tolerance of 7 ppm at the MS1 level was used, and up to three miscleavages for trypsin were allowed. The fragment ion mass tolerance was set to 0.5 D. Oxidation of methionine and protein acetylation at the N terminus were defined as variable modifications. Carbamidomethylation on cysteines was set as a fix modification. The identified peptides were filtered using a false discovery rate <5%.
Protein areas were normalized intra- and intersamples by median of the log area. A linear modeling approach implemented in lmFit function and the empirical Bayes statistics implemented in eBayes and topTable functions of the Bioconductor limma package (Gentleman et al., 2004; Smyth, 2004) were used to perform a differential protein abundance analysis. The normalized protein areas of different yeast mutants were compared with wt samples. Protein p-values were calculated with limma and were adjusted with Benjamini–Hochberg method (Benjamini and Hochberg, 1995). A value of 0.05 was used as a cutoff.
Lipid analysis
Lipids from whole cells or from isolated LDs were extracted following the single-step modification of Folch et al. (1957) described by Atkinson et al. (1980): one volume of cells or LDs suspension was mixed with three volumes of chloroform/methanol 2:1 vol/vol and vortexed at high speed (with glass beads in case of whole cells) for 3 min followed by further shaking for 1 h. 1 vol of 0.9% NaCl was added, and brief vortexing was applied. Samples were centrifuged at 200 g, and the lower (chloroform) phase was recovered. The extracts were dried, dissolved in chloroform/methanol 2:1 vol/vol, and analyzed by TLC on silica gel 60 plates (Merk). Phospholipids were resolved with chloroform/ethyl acetate:acetone/isopropanol/ethanol/methanol/water/acetic acid (30:6:6:6:16:28:6:2 vol/vol). Neutral lipids were resolved with hexane/diethyl ether/acetic acid (80:20:1 vol/vol) to three fifths of the plate, dried, and followed by hexane/chloroform (9:1 vol/vol) to four fifths of the plate. DAG and free sterol were resolved with toluene/ethyl acetate/ethyl ether/ammonia-25% (80:10:10:0.2 vol/vol). Bands were stained in a chamber saturated with iodine vapor, scanned, and quantified by densitometry with Quantity One (Bio-Rad). Known standards were included on all plates for identification.
Steady-state lipid labeling for PA quantification
Cultures in SC were diluted to OD600 0.1 in SC or SC supplemented with 75 µM of inositol and grown for 24 h at 30°C in the presence of 1 µCi/ml [1-14C]acetate (45–60 mCi/mmol; Perkin Elmer). Lipids were extracted as described in the previous section with of 5 mM HCl, resolved by TLC with chloroform/methanol/acetic acid (65:25:8 vol/vol), scanned on a Typhoon Trio phosphorimager (Amersham Biosciences), and quantified with Quantity One (Bio-Rad).
Online supplemental material
Fig. S1 shows purity and lipid composition of LDs isolated from wt, fld1Δ, and ldb16Δ. Fig. S2 shows the distribution of Scs2, Scs2- and PA-binding domains of Opi1 and PA in fld1Δ and ldb16Δ mutants. Fig. S3 shows relocalization and levels of amphipathic-helix containing proteins in seipin complex mutants. Fig. S4 shows that LD assembly causes phospholipid-packing defects in seipin complex mutants. Videos 1 and 2 show the 3D reconstruction and segmentation of a 250-nm-thick section of a wt cell, respectively. Videos 3 and 4 show the 3D reconstruction and segmentation of serial sections of a ldb16Δ cell containing supersized LDs, respectively. Videos 5 and 6 show 3D reconstructions of serial sections of a ldb16Δ cell containing aggregated LDs. Video 7 shows the segmentation of a ldb16Δ cell in Video 6. Table S1 shows the LD-specific proteins reduced in LDs isolated from fld1Δ and ldb16Δ cells. Table S2 shows the peripheral membrane proteins increased in LDs isolated from fld1Δ and ldb16Δ cells. Tables S3 and S4 show the quantification of immuno-EM analysis. Tables S5, S6, and S7 list the yeast strains, plasmids, and primers used in this study, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201502070/DC1.
Supplementary Material
Acknowledgments
Preparation of EM tomography samples and immuno-EM was performed at the Centres Científics i Tecnològics University of Barcelona. EM tomography was performed at Electron Microscopy Core Facility, European Molecular Biology Laboratory (Heidelberg). Mass spectrometric measurements and data analysis were performed in the Centre for Genomic Regulation (CRG)/Universitat Pompeu Fabra Proteomics Unit, part of the “Plataforma de Recursos Biomoleculares y Bioinformàticos (ProteoRed-Instituto de Salud Carlos III, PT13/0001).” We thank S. Abreu and F. Reggiori for preliminary EM studies. We thank A. Curwin, T. Levine, F. Posas, and W. Prinz for reagents. We thank F. Campelo, O. Foresti, R. Klemm, and W. Prinz for discussions and critical reading of the manuscript.
P. Carvalho is supported by a European Research Council starting grant, the CRG, an International Early Career Award from the Howard Hughes Medical Institute, and the European Molecular Biology Organization Young Investigator Program. We acknowledge support of the Spanish Ministry of Economy and Competitiveness, "Centro de Excelencia Severo Ochoa 2013-2017," SEV-2012-0208.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AH
- amphipathic helix
- BB
- breaking buffer
- LD
- lipid droplet
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- SC
- synthetic complete
References
- Atkinson K.D., Jensen B., Kolat A.I., Storm E.M., Henry S.A., and Fogel S.. 1980. Yeast mutants auxotrophic for choline or ethanolamine. J. Bacteriol. 141:558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., and Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc., B. 57:289–300. [Google Scholar]
- Binns D., Lee S., Hilton C.L., Jiang Q.X., and Goodman J.M.. 2010. Seipin is a discrete homooligomer. Biochemistry. 49:10747–10755. 10.1021/bi1013003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet E., El Mourabit H., Prot M., Nemani M., Khallouf E., Colard O., Maurice M., Durand-Schneider A.M., Chrétien Y., Grès S., et al. 2009. Seipin deficiency alters fatty acid Delta9 desaturation and lipid droplet formation in Berardinelli-Seip congenital lipodystrophy. Biochimie. 91:796–803. 10.1016/j.biochi.2009.01.011 [DOI] [PubMed] [Google Scholar]
- Bunkenborg J., García G.E., Paz M.I., Andersen J.S., and Molina H.. 2010. The minotaur proteome: avoiding cross-species identifications deriving from bovine serum in cell culture models. Proteomics. 10:3040–3044. 10.1002/pmic.201000103 [DOI] [PubMed] [Google Scholar]
- Campelo F., and Kozlov M.M.. 2014. Sensing membrane stresses by protein insertions. PLOS Comput. Biol. 10:e1003556 10.1371/journal.pcbi.1003556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright B.R., Binns D.D., Hilton C.L., Han S., Gao Q., and Goodman J.M.. 2015. Seipin performs dissectible functions in promoting lipid droplet biogenesis and regulating droplet morphology. Mol. Biol. Cell. 26:726–739. 10.1091/mbc.E14-08-1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Goder V., and Rapoport T.A.. 2006. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 126:361–373. 10.1016/j.cell.2006.05.043 [DOI] [PubMed] [Google Scholar]
- Chen W., Chang B., Saha P., Hartig S.M., Li L., Reddy V.T., Yang Y., Yechoor V., Mancini M.A., and Chan L.. 2012. Berardinelli-seip congenital lipodystrophy 2/seipin is a cell-autonomous regulator of lipolysis essential for adipocyte differentiation. Mol. Cell. Biol. 32:1099–1111. 10.1128/MCB.06465-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik L.V., and Kozlov M.M.. 2005. Membrane hemifusion: crossing a chasm in two leaps. Cell. 123:375–382. 10.1016/j.cell.2005.10.015 [DOI] [PubMed] [Google Scholar]
- Connerth M., Grillitsch K., Köfeler H., and Daum G.. 2009. Analysis of lipid particles from yeast. Methods Mol. Biol. 579:359–374. 10.1007/978-1-60761-322-0_18 [DOI] [PubMed] [Google Scholar]
- Cornell R.B., and Taneva S.G.. 2006. Amphipathic helices as mediators of the membrane interaction of amphitropic proteins, and as modulators of bilayer physical properties. Curr. Protein Pept. Sci. 7:539–552. 10.2174/138920306779025675 [DOI] [PubMed] [Google Scholar]
- Cui X., Wang Y., Tang Y., Liu Y., Zhao L., Deng J., Xu G., Peng X., Ju S., Liu G., and Yang H.. 2011. Seipin ablation in mice results in severe generalized lipodystrophy. Hum. Mol. Genet. 20:3022–3030. 10.1093/hmg/ddr205 [DOI] [PubMed] [Google Scholar]
- Currie E., Guo X., Christiano R., Chitraju C., Kory N., Harrison K., Haas J., Walther T.C., and Farese R.V. Jr. 2014. High confidence proteomic analysis of yeast LDs identifies additional droplet proteins and reveals connections to dolichol synthesis and sterol acetylation. J. Lipid Res. 55:1465–1477. 10.1194/jlr.M050229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G., and Antonny B.. 2010. Amphipathic helices and membrane curvature. FEBS Lett. 584:1840–1847. 10.1016/j.febslet.2009.10.022 [DOI] [PubMed] [Google Scholar]
- Drin G., Casella J.F., Gautier R., Boehmer T., Schwartz T.U., and Antonny B.. 2007. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat. Struct. Mol. Biol. 14:138–146. 10.1038/nsmb1194 [DOI] [PubMed] [Google Scholar]
- Fei W., Shui G., Gaeta B., Du X., Kuerschner L., Li P., Brown A.J., Wenk M.R., Parton R.G., and Yang H.. 2008. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180:473–482. 10.1083/jcb.200711136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W., Li H., Shui G., Kapterian T.S., Bielby C., Du X., Brown A.J., Li P., Wenk M.R., Liu P., and Yang H.. 2011a Molecular characterization of seipin and its mutants: implications for seipin in triacylglycerol synthesis. J. Lipid Res. 52:2136–2147. 10.1194/jlr.M017566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W., Shui G., Zhang Y., Krahmer N., Ferguson C., Kapterian T.S., Lin R.C., Dawes I.W., Brown A.J., Li P., et al. 2011b A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 7:e1002201 10.1371/journal.pgen.1002201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J., Lees M., and Sloane Stanley G.H.. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226:497–509. [PubMed] [Google Scholar]
- Fujimoto T., and Parton R.G.. 2011. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb. Perspect. Biol. 3:3 10.1101/cshperspect.a004838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar M.L., Aregullin M.A., Jesch S.A., and Henry S.A.. 2006. Inositol induces a profound alteration in the pattern and rate of synthesis and turnover of membrane lipids in Saccharomyces cerevisiae. J. Biol. Chem. 281:22773–22785. 10.1074/jbc.M603548200 [DOI] [PubMed] [Google Scholar]
- Gautier R., Douguet D., Antonny B., and Drin G.. 2008. HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics. 24:2101–2102. 10.1093/bioinformatics/btn392 [DOI] [PubMed] [Google Scholar]
- Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J., Mari M., De Mazière A., and Reggiori F.. 2008. A cryosectioning procedure for the ultrastructural analysis and the immunogold labelling of yeast Saccharomyces cerevisiae. Traffic. 9:1060–1072. 10.1111/j.1600-0854.2008.00753.x [DOI] [PubMed] [Google Scholar]
- Grillitsch K., Connerth M., Köfeler H., Arrey T.N., Rietschel B., Wagner B., Karas M., and Daum G.. 2011. Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: lipidome meets proteome. Biochim. Biophys. Acta. 1811:1165–1176. 10.1016/j.bbalip.2011.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Walther T.C., Rao M., Stuurman N., Goshima G., Terayama K., Wong J.S., Vale R.D., Walter P., and Farese R.V.. 2008. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 453:657–661. 10.1038/nature06928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., and Fink G.. 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego, CA: 933 pp. [Google Scholar]
- Hancock L.C., Behta R.P., and Lopes J.M.. 2006. Genomic analysis of the Opi- phenotype. Genetics. 173:621–634. 10.1534/genetics.106.057489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry S.A., Kohlwein S.D., and Carman G.M.. 2012. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 190:317–349. 10.1534/genetics.111.130286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofbauer H.F., Schopf F.H., Schleifer H., Knittelfelder O.L., Pieber B., Rechberger G.N., Wolinski H., Gaspar M.L., Kappe C.O., Stadlmann J., et al. 2014. Regulation of gene expression through a transcriptional repressor that senses acyl-chain length in membrane phospholipids. Dev. Cell. 29:729–739. 10.1016/j.devcel.2014.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höög J.L., Schwartz C., Noon A.T., O’Toole E.T., Mastronarde D.N., McIntosh J.R., and Antony C.. 2007. Organization of interphase microtubules in fission yeast analyzed by electron tomography. Dev. Cell. 12:349–361. 10.1016/j.devcel.2007.01.020 [DOI] [PubMed] [Google Scholar]
- Horchani H., de Saint-Jean M., Barelli H., and Antonny B.. 2014. Interaction of the Spo20 membrane-sensor motif with phosphatidic acid and other anionic lipids, and influence of the membrane environment. PLoS One. 9:e113484 10.1371/journal.pone.0113484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier N., Choudhary V., Mari M., Toulmay A., Reggiori F., and Schneiter R.. 2011. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 124:2424–2437. 10.1242/jcs.076836 [DOI] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M.. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962. 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Krahmer N., Guo Y., Wilfling F., Hilger M., Lingrell S., Heger K., Newman H.W., Schmidt-Supprian M., Vance D.E., Mann M., et al. 2011. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 14:504–515. 10.1016/j.cmet.2011.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N., Farese R.V. Jr., and Walther T.C.. 2013. Balancing the fat: lipid droplets and human disease. EMBO Mol. Med. 5:905–915. 10.1002/emmm.201100671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Ladinsky M.S., Kremer J.R., Furcinitti P.S., McIntosh J.R., and Howell K.E.. 1994. HVEM tomography of the trans-Golgi network: structural insights and identification of a lace-like vesicle coat. J. Cell Biol. 127:29–38. 10.1083/jcb.127.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber R., Zinser E., Zellnig G., Paltauf F., and Daum G.. 1994. Characterization of lipid particles of the yeast, Saccharomyces cerevisiae. Yeast. 10:1421–1428. 10.1002/yea.320101105 [DOI] [PubMed] [Google Scholar]
- Loewen C.J., Roy A., and Levine T.P.. 2003. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22:2025–2035. 10.1093/emboj/cdg201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen C.J., Gaspar M.L., Jesch S.A., Delon C., Ktistakis N.T., Henry S.A., and Levine T.P.. 2004. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 304:1644–1647. 10.1126/science.1096083 [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Louvion J.F., Havaux-Copf B., and Picard D.. 1993. Fusion of GAL4-VP16 to a steroid-binding domain provides a tool for gratuitous induction of galactose-responsive genes in yeast. Gene. 131:129–134. 10.1016/0378-1119(93)90681-R [DOI] [PubMed] [Google Scholar]
- MacKinnon M.A., Curwin A.J., Gaspard G.J., Suraci A.B., Fernández-Murray J.P., and McMaster C.R.. 2009. The Kap60-Kap95 karyopherin complex directly regulates phosphatidylcholine synthesis. J. Biol. Chem. 284:7376–7384. 10.1074/jbc.M809117200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magré J., Delépine M., Khallouf E., Gedde-Dahl T. Jr., Van Maldergem L., Sobel E., Papp J., Meier M., Mégarbané A., Bachy A., et al. BSCL Working Group . 2001. Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat. Genet. 28:365–370. 10.1038/ng585 [DOI] [PubMed] [Google Scholar]
- Marsh B.J., Mastronarde D.N., Buttle K.F., Howell K.E., and McIntosh J.R.. 2001. Organellar relationships in the Golgi region of the pancreatic beta cell line, HIT-T15, visualized by high resolution electron tomography. Proc. Natl. Acad. Sci. USA. 98:2399–2406. 10.1073/pnas.051631998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51. 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Nakanishi H., de los Santos P., and Neiman A.M.. 2004. Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol. Biol. Cell. 15:1802–1815. 10.1091/mbc.E03-11-0798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole E.T., McDonald K.L., Mäntler J., McIntosh J.R., Hyman A.A., and Müller-Reichert T.. 2003. Morphologically distinct microtubule ends in the mitotic centrosome of Caenorhabditis elegans. J. Cell Biol. 163:451–456. 10.1083/jcb.200304035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol A., Gross S.P., and Parton R.G.. 2014. Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J. Cell Biol. 204:635–646. 10.1083/jcb.201311051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieur X., Dollet L., Takahashi M., Nemani M., Pillot B., Le May C., Mounier C., Takigawa-Imamura H., Zelenika D., Matsuda F., et al. 2013. Thiazolidinediones partially reverse the metabolic disturbances observed in Bscl2/seipin-deficient mice. Diabetologia. 56:1813–1825. 10.1007/s00125-013-2926-9 [DOI] [PubMed] [Google Scholar]
- Prinz W.A. 2014. Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics. J. Cell Biol. 205:759–769. 10.1083/jcb.201401126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., and Ishihama Y.. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2:1896–1906. 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- Sandager L., Gustavsson M.H., Ståhl U., Dahlqvist A., Wiberg E., Banas A., Lenman M., Ronne H., and Stymne S.. 2002. Storage lipid synthesis is non-essential in yeast. J. Biol. Chem. 277:6478–6482. 10.1074/jbc.M109109200 [DOI] [PubMed] [Google Scholar]
- Shen H., Heacock P.N., Clancey C.J., and Dowhan W.. 1996. The CDS1 gene encoding CDP-diacylglycerol synthase in Saccharomyces cerevisiae is essential for cell growth. J. Biol. Chem. 271:789–795. 10.1074/jbc.271.2.789 [DOI] [PubMed] [Google Scholar]
- Smyth G.K. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:e3. [DOI] [PubMed] [Google Scholar]
- Sorger D., Athenstaedt K., Hrastnik C., and Daum G.. 2004. A yeast strain lacking lipid particles bears a defect in ergosterol formation. J. Biol. Chem. 279:31190–31196. 10.1074/jbc.M403251200 [DOI] [PubMed] [Google Scholar]
- Szymanski K.M., Binns D., Bartz R., Grishin N.V., Li W.P., Agarwal A.K., Garg A., Anderson R.G., and Goodman J.M.. 2007. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. USA. 104:20890–20895. 10.1073/pnas.0704154104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam A.R., Antonny B., Wang J., Delacotte J., Wilfling F., Walther T.C., Beck R., Rothman J.E., and Pincet F.. 2013a COPI buds 60-nm lipid droplets from reconstituted water-phospholipid-triacylglyceride interfaces, suggesting a tension clamp function. Proc. Natl. Acad. Sci. USA. 110:13244–13249. 10.1073/pnas.1307685110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam A.R., Farese R.V. Jr., and Walther T.C.. 2013b The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 14:775–786. 10.1038/nrm3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Bi J., Shui G., Liu Z., Xiang Y., Liu Y., Wenk M.R., Yang H., and Huang X.. 2011. Tissue-autonomous function of Drosophila seipin in preventing ectopic lipid droplet formation. PLoS Genet. 7:e1001364 10.1371/journal.pgen.1001364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu K.T. 1986. Application of cryoultramicrotomy to immunocytochemistry. J. Microsc. 143:139–149. 10.1111/j.1365-2818.1986.tb02772.x [DOI] [PubMed] [Google Scholar]
- Vamparys L., Gautier R., Vanni S., Bennett W.F., Tieleman D.P., Antonny B., Etchebest C., and Fuchs P.F.. 2013. Conical lipids in flat bilayers induce packing defects similar to that induced by positive curvature. Biophys. J. 104:585–593. 10.1016/j.bpj.2012.11.3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanni S., Vamparys L., Gautier R., Drin G., Etchebest C., Fuchs P.F., and Antonny B.. 2013. Amphipathic lipid packing sensor motifs: probing bilayer defects with hydrophobic residues. Biophys. J. 104:575–584. 10.1016/j.bpj.2012.11.3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.W., Miao Y.H., and Chang Y.S.. 2014. Control of lipid droplet size in budding yeast requires the collaboration between Fld1 and Ldb16. J. Cell Sci. 127:1214–1228. 10.1242/jcs.137737 [DOI] [PubMed] [Google Scholar]
- Wilfling F., Wang H., Haas J.T., Krahmer N., Gould T.J., Uchida A., Cheng J.X., Graham M., Christiano R., Fröhlich F., et al. 2013. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell. 24:384–399. 10.1016/j.devcel.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F., Thiam A.R., Olarte M.J., Wang J., Beck R., Gould T.J., Allgeyer E.S., Pincet F., Bewersdorf J., Farese R.V. Jr., and Walther T.C.. 2014. Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. eLife. 3:e01607 10.7554/eLife.01607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolinski H., Hofbauer H.F., Hellauer K., Cristobal-Sarramian A., Kolb D., Radulovic M., Knittelfelder O.L., Rechberger G.N., and Kohlwein S.D.. 2015. Seipin is involved in the regulation of phosphatidic acid metabolism at a subdomain of the nuclear envelope in yeast. Biochim. Biophys. Acta. 1851:1450–1464. 10.1016/j.bbalip.2015.08.003 [DOI] [PubMed] [Google Scholar]
- Yang H., Galea A., Sytnyk V., and Crossley M.. 2012. Controlling the size of lipid droplets: lipid and protein factors. Curr. Opin. Cell Biol. 24:509–516. 10.1016/j.ceb.2012.05.012 [DOI] [PubMed] [Google Scholar]
- Zeniou-Meyer M., Zabari N., Ashery U., Chasserot-Golaz S., Haeberlé A.M., Demais V., Bailly Y., Gottfried I., Nakanishi H., Neiman A.M., et al. 2007. Phospholipase D1 production of phosphatidic acid at the plasma membrane promotes exocytosis of large dense-core granules at a late stage. J. Biol. Chem. 282:21746–21757. 10.1074/jbc.M702968200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.