Protein delivery based on bacterial type III secretion allows controllable injection of bacterial, viral, and human proteins into target cells, functional interaction studies, targeting of proteins to subcellular locations, and systems-level signaling analysis of a given translocated protein.
Abstract
Methods enabling the delivery of proteins into eukaryotic cells are essential to address protein functions. Here we propose broad applications to cell biology for a protein delivery tool based on bacterial type III secretion (T3S). We show that bacterial, viral, and human proteins, fused to the N-terminal fragment of the Yersinia enterocolitica T3S substrate YopE, are effectively delivered into target cells in a fast and controllable manner via the injectisome of extracellular bacteria. This method enables functional interaction studies by the simultaneous injection of multiple proteins and allows the targeting of proteins to different subcellular locations by use of nanobody-fusion proteins. After delivery, proteins can be freed from the YopE fragment by a T3S-translocated viral protease or fusion to ubiquitin and cleavage by endogenous ubiquitin proteases. Finally, we show that this delivery tool is suitable to inject proteins in living animals and combine it with phosphoproteomics to characterize the systems-level impact of proapoptotic human truncated BID on the cellular network.
Introduction
In cell biology, protein function is addressed by various methods including cDNA transfection, microinjection, and proteofection of purified proteins. Although these methods are informative, they often result in massive overrepresentation of the protein of interest and/or highly heterogeneous cell populations, making functional dynamics studies and –omics approaches difficult to interpret. They can also be costly when used on a large scale.
Bacteria have developed sophisticated nanomachines enabling the delivery of virulence proteins into eukaryotic cells (translocation). The type III secretion (T3S) system of certain gram-negative bacteria functions like a nanosyringe that injects substrate proteins into target cells (Cornelis and Wolf-Watz, 1997; Fig. 1 a). Delivered proteins harbor a short N-terminal secretion signal (Michiels et al., 1990). In bacteria, they bind to chaperones that stabilize them, prevent premature interactions, and favor secretion (Wattiau and Cornelis, 1993; Gauthier and Finlay, 2003). An ATPase associated with the base of the T3S apparatus participates in directing substrates to be secreted into a thin needle-like structure. These proteins travel unfolded or only partially folded (Feldman et al., 2002) and subsequently refold in the host cell, where they exert their virulence activity toward various host proteins and cellular machineries. Over 100 different effector proteins are known (Mota and Cornelis, 2005), displaying a large repertoire of biochemical activities that modulate the functions of host regulatory molecules.
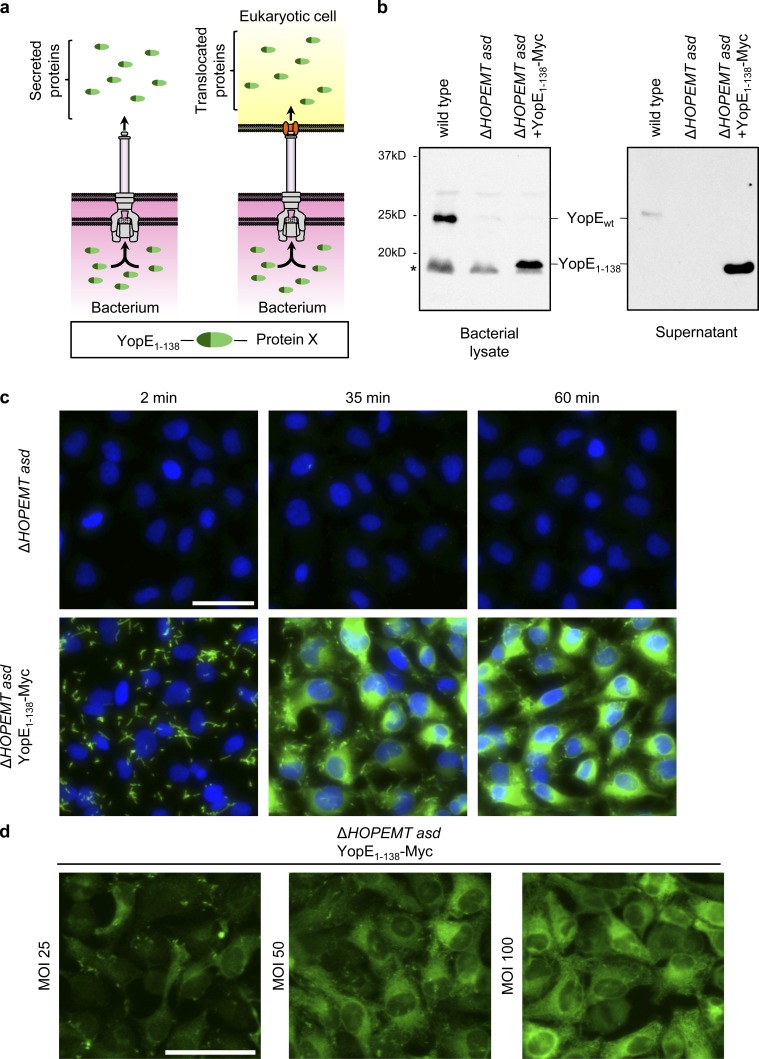
Figure 1.
Characterization of T3S-based protein delivery. (a) Schematic representation of T3S-dependent protein secretion into the supernatant (in vitro secretion) or eukaryotic cells (protein translocation). (b) Bacterial lysate or in vitro secretion (supernatant) of indicated strains revealed by Western blot using an anti-YopE antibody. Asterisk indicates a nonspecific band. (c) Anti-Myc immunofluorescence staining of HeLa cells infected with the indicated strains at an MOI of 100. Anti-Myc staining is shown in green and nuclei in blue. (d) Anti-Myc staining of HeLa cells infected for 45 min with the indicated strain at different MOIs. Anti-Myc staining is shown in green. Bars, 50 µm.
On a few occasions, immunologists and infection biologists have exploited T3S to deliver hybrid peptides and proteins into target cells. Viral and bacterial epitopes (Sory et al., 1992; Van Damme et al., 1992; Rüssmann et al., 1998, 2003; Chen et al., 2006) as well as peptides from human tumors (Chaux et al., 1999) have been delivered by T3S with the aim of vaccination. Bordetella pertussis adenylate cyclase (Sory and Cornelis, 1994), murine DHFR (Feldman et al., 2002), or a phosphorylatable tag (Garcia et al., 2006) were used as reporters of translocation to identify the secretion signal requirements for T3S. More recently, an elegant Yersinia enterocolitica–based delivery toolbox was developed to investigate Rho-GTPase affecting T3S effectors from Shigella flexneri and Escherichia coli (Wölke et al., 2011). Functional nanobodies (Blanco-Toribio et al., 2010) or nuclear proteins as cre-recombinase and MyoD (Bichsel et al., 2011, 2013) were also delivered inside target cells in vitro, whereas an S. flexneri–based T3S was used to release IL-10 and IL-1 receptor antagonists in vivo (Chamekh et al., 2008). Although these studies were successful, T3S-based protein delivery has never been shown to be applicable to a broad range of eukaryotic cargos, and its utilization in cell biology therefore remained limited.
Here we optimize the delivery of heterologous proteins into target cells via T3S and propose protocols manageable by noninfection biologists. We show that the delivery of bacterial effectors proteins as well as viral and eukaryotic proteins fused to the N-terminal fragment of the Y. enterocolitica T3S substrate YopE is rapid, homogeneous in all cells, and can be tuned by the MOI. We demonstrate that translocated proteins can be targeted to the nucleus by a nuclear localization signal (NLS) or to a specific subcellular localization after fusion to specific nanobodies. Furthermore, we show that they can be cleaved from the YopE fragment by T3S-translocated tobacco etch virus (TEV) protease or by an ubiquitin-dependent mechanism. Finally, we show that this delivery system is suitable to inject functional eukaryotic proteins in living animals, and that it can be combined with phosphoproteomics to gain new biological insights into the mechanism of apoptosis.
Results
A protein delivery method based on T3S of YopE fusion proteins
We took advantage of Y. enterocolitica-Ysc-T3S to develop an infection protocol that enables effective secretion and delivery of proteins into eukaryotic cells (Fig. 1 a). This method is based on the delivery of proteins fused to the N-terminal fragment of YopE, a Y. enterocolitica effector with Rho GTPase activating protein (GAP) activity (Von Pawel-Rammingen et al., 2000). First, the translocation of endogenous effectors was abolished by using a strain deleted for all known effectors named YopH, O, P, E, M, and T (ΔHOPEMT; Iriarte and Cornelis, 1998; Fig. S1 a). Furthermore, this strain was deleted for the aspartate-β-semialdehyde dehydrogenase gene (Δasd) to make it unable to grow in absence of exogenous meso-2,6-diaminopimelic acid (Hoang et al., 1997; Kudryashev et al., 2013) and thereby compatible with biosafety level 1 (Fig. S1 a). The two cloning plasmids, pBad_Si_1 and pBad_Si_2, were optimized for high protein secretion and to facilitate cloning (Fig. S1 b). The N-terminal 138-amino-acid fragment of YopE (YopE1–138), which contains a T3S signal (Feldman et al., 2002) and a chaperone-binding site for the chaperone SycE (Boyd et al., 2000b), is optimal for heterologous T3S substrate translocation (Wölke et al., 2011). Therefore, it was systematically fused to proteins of interest, and SycE was coexpressed. Notably, the Rho-GAP domain of YopE is not in the YopE1–138 fragment. The SycE-YopE1–138 DNA sequence containing the endogenous promoters of YopE and SycE was cloned from the purified pYV virulence plasmid from Y. enterocolitica (pYV40; Sory et al., 1995; Fig. S1 b). The production of SycE and all YopE1–138 fusion proteins was then induced by a rapid temperature shift from growth at room temperature to 37°C (Fig. S2 a). Cloning was further facilitated by adding a multiple cloning site at the 3′ end of YopE1–138, followed by a Myc tag, a 6xHis tag and a stop codon (Fig. S1 c).
Protein secretion was first addressed by monitoring the secretion of full-length YopE and YopE1–138-Myc in an in vitro secretion assay (Fig. 1 a and Fig. S2 a). Secretion, artificially induced by a temperature shift to 37°C and deprivation of calcium, was monitored by precipitating proteins from the supernatant and performing a Western blot analysis with an anti-YopE antibody (Fig. 1 b). As expected, YopE was secreted by wild-type bacteria, but not by the ΔHOPEMT asd strain. A band corresponding to YopE1–138-Myc-His was found in the supernatant of the ΔHOPEMT asd strain encoding YopE1–138-Myc-His (hereafter named YopE1–138-Myc; Fig. 1 b), showing that it was successfully secreted. The fact that YopE1–138-Myc was more effectively secreted than YopE was likely caused by the absence of other T3S substrates in the ΔHOPEMT asd strain and to a higher copy number of this plasmid.
Protein translocation into eukaryotic cells was monitored by immunofluorescence staining of Myc after infection of HeLa cells by the strain encoding YopE1–138-Myc. At 2 min, Myc was only detected in bacteria (Fig. 1 c). In contrast, almost all cells were Myc positive at 35 or 60 min. Intensity was homogeneous within the cell population and increasing over time, showing that YopE1–138-Myc accumulated in cells (Fig. 1 c and Fig. S2 b). Interestingly, low MOIs led to weaker levels of protein translocation, but the number of recipient cells was still nearly maximal (Fig. 1 d and Fig. S2 c), indicating that protein levels in target cells was controllable via the MOI. As reported for YopE (Isaksson et al., 2009), YopE1–138-Myc was excluded from the nuclei (Fig. 1 d). Furthermore, in agreement with data showing that Y. enterocolitica can infect different cell types (Boyd et al., 2000a), YopE1–138-Myc was delivered into murine fibroblasts, Jurkat T-lymphocytes, and primary human umbilical vein endothelial cells (Fig. S3 a). Altogether, these data indicated that YopE1–138-Myc is effectively secreted and delivered into cells and that Y. enterocolitica–based delivery is fast, homogenous, and finely controllable.
Translocation of type III and type IV bacterial effectors
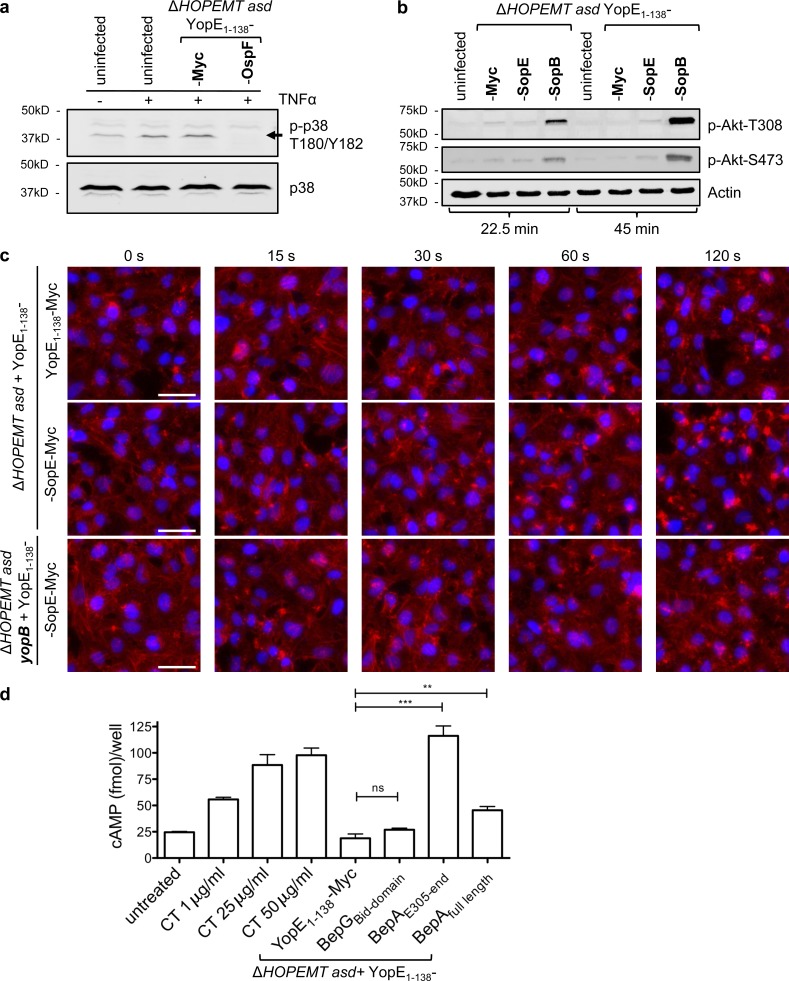
We tested whether translocated heterologous T3S effectors fused to YopE1–138 were functionally active in cells. We first monitored the activity of OspF, an S. flexneri effector harboring a phosphothreonine lyase activity that dephosphorylates MAP kinases p38 and ERK (Li et al., 2007). TNF-induced p38 phosphorylation present in uninfected cells or cells infected with YopE1–138-Myc-expressing bacteria was abolished in cells infected by YopE1–138-OspF–expressing bacteria, showing that delivered OspF was fully functional (Fig. 2 a). We also confirmed the effect of translocated YopE1–138-SopB on Akt activation. SopB is a Salmonella enterica effector that protects epithelial cells from apoptosis by inducing a sustained activation of Akt (Norris et al., 1998). Our results show that Akt was activated by YopE1–138-SopB, whereas the translocation of YopE1–138-Myc or YopE1–138-SopE had no effect (Fig. 2 b).
Figure 2.
Delivery of type III and type IV effectors into eukaryotic cells. (a) TNF-induced phosphorylation of p38 is abolished by translocated OspF. Phospho-p38 (indicated by the arrow) and total p38 revealed by Western blot analysis of HeLa cells infected for 75 min with the indicated strains at an MOI of 100. Cells were stimulated with TNF for the last 30 min of infection. (b) Translocated SopB induces Akt phosphorylation at T308 and S473. Phospho-Akt and actin Western blot analysis of HeLa cells infected for 22.5 or 45 min with the indicated strains at an MOI of 100. (c) Translocated SopE induces a dramatic remodelling of F-actin. HeLa cells were infected with the indicated strains at an MOI of 100 for the indicated time periods. After fixation, cells were stained for nuclei (blue) and F-actin (red). Bars, 50 µm. (d) T3S-based protein delivery of BepA type IV effector. Measurements of cAMP levels in HeLa cells infected for 2.5 h with the indicated strains at an MOI of 100. As positive control, cholera toxin (CT) was added for 1 h at indicated concentrations. Data correspond to the mean of n = 3 independent experiments, and error bars are standard errors of the mean. Statistical analysis was performed using a Mann-Whitney test (**, P < 0.01; ***, P < 0.001; ns, not significant).
Multiple T3S effectors also target the actin cytoskeleton. The S. enterica SopE effector is a guanine nucleotide exchange factor that interacts with Cdc42, promoting actin cytoskeletal remodeling (Hardt et al., 1998). Although the translocation of YopE1–138-Myc into HeLa cells had no impact, infection with the YopE1–138-SopE–expressing strain induced dramatic changes in the F-actin network (Figs. 2 c and S2 d). These changes were strictly dependent on T3S, as infection by a YopB deletion mutant, unable to form translocation pores in the eukaryotic cell membrane (Håkansson et al., 1996; Fig. S4), had no effect on actin (Figs. 2 c, S2 d, and S3 b). YopE1–138-SopE–induced F-actin foci were detected around 60 s postinfection (p.i.) in most cells (Figs. 2 c and S3 b), whereas at later time points, a characteristic SopE-induced F-actin meshwork (Hardt et al., 1998) was found (Figs. 3 b and S2 d).
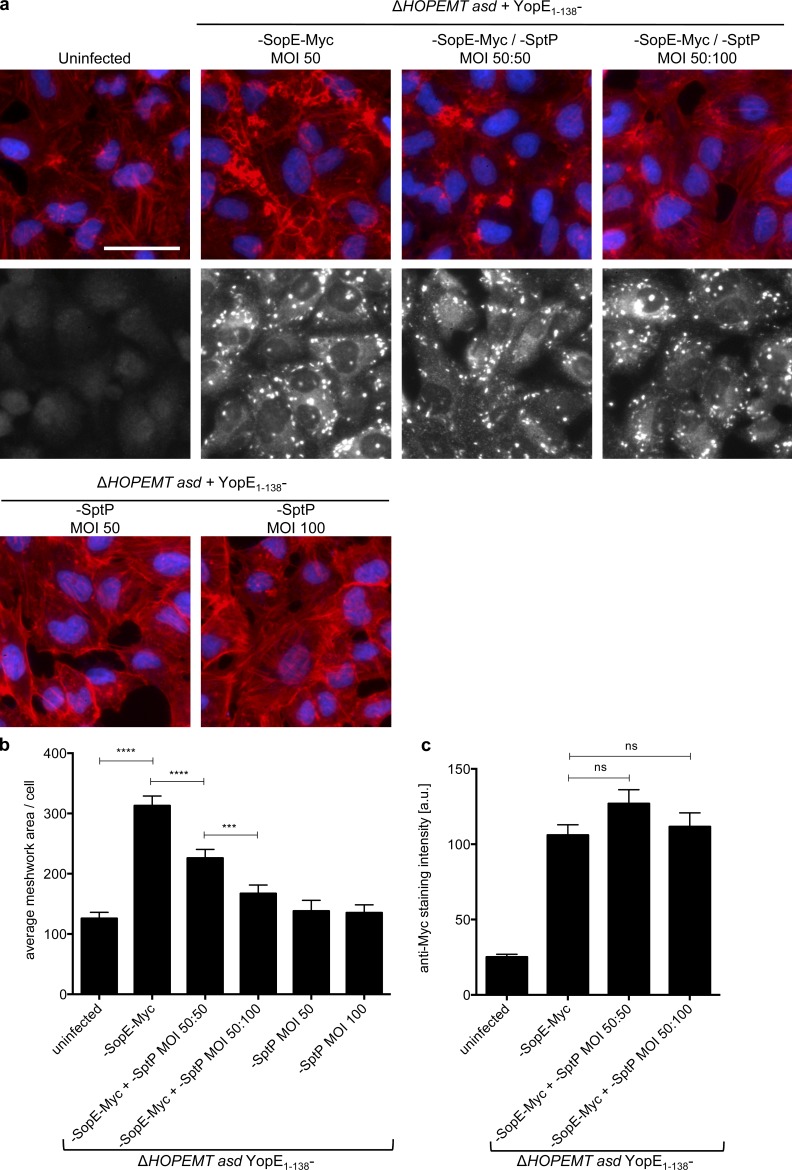
Figure 3.
Use of T3S-based protein delivery for functional interaction studies. (a) When coinjected, SptP abolishes the activity of SopE. HeLa cells were infected with indicated combinations of strains at indicated MOI for 1 h. After fixation, cells were stained for nuclei (blue) and F-actin (red). Translocation of YopE1–138-SopE-Myc was monitored by an anti-Myc staining (gray images), which also stains bacteria (bright dots). Bar, 50 µm. (b) Quantification of the F-actin meshwork area per cell of images shown in panel a. (c) Quantification of the Myc staining intensity of images shown in panel a (gray images). Image analysis was performed on n = 60 cells per condition. Error bars indicate standard errors of the mean, and statistical analysis was performed using a Mann-Whitney test (***, P < 0.001; ****, P < 0.0001; ns, not significant).
Several bacteria use type IV secretion to inject proteins into cells. As recently proposed for a Legionella effector (Rothmeier et al., 2013), we tested whether type IV effectors could also be delivered by our Y. enterocolitica–based protein delivery. The Bartonella henselae full-length BepA, BepAE305-end, containing the C-terminal Bid domain, and the Bid domain of BepG, another type IV effector used as control, were cloned into pBad_Si_2. As BepA induces the production of cAMP (Pulliainen et al., 2012), the level of cAMP was measured after infection. Although the translocation of the Bid domain of BepG failed to induce cAMP, full-length BepA and BepAE305-end triggered cAMP production in expected amounts (Pulliainen et al., 2012; Fig. 2 d), showing that type IV effectors fused to YopE1–138 were effectively translocated and active. Altogether, our results demonstrate that Y. enterocolitica–based protein delivery is suitable for type III and IV effectors.
To explore additional applications in cell biology, we tested whether our tool of T3S-based protein delivery was suitable to address functional interactions of multiple T3S-delivered proteins in cells. We took advantage of the antagonistic activities of the effectors SopE and SptP on the actin cytoskeleton (Stebbins and Galán, 2001). During S. enterica infection, SopE translocation is followed by the translocation of SptP, which then functions as a GAP for Cdc42. Whereas YopE1–138-SopE-Myc alone triggered massive F-actin rearrangements, coinfection with YopE1-138-SptP–expressing bacteria abolished this effect in a dose-dependent manner (Fig. 3, a and b). An anti-Myc staining showed that this inhibition was not caused by a reduced level of translocated YopE1–138-SopE-Myc (Fig. 3, a and c). This result shows that coinfecting cells with at least two Y. enterocolitica strains can be used to address functional interactions between translocated proteins.
Translocation of eukaryotic proteins into epithelial cells
To extend the use of Y. enterocolitica–based protein delivery to eukaryotic cargos, we monitored the secretion of proteins including cell-cycle proteins (Mad2, CDK1, CDKN2A/INK4A, CDKN2B/INK4B, and CDKN2C/INK4C), apoptosis-related proteins (Bad, FADD, caspase-3 [CASP] p17 and p12, zebrafish BID, and zebrafish t-BID), signaling proteins (TRAF6, TIFA, and the GPCR Gα subunit GNA12), an anti-GFP nanobody (VHHGFP4), and a VHHGFP4 fusion construct for targeted protein degradation (Caussinus et al., 2011; Slmb-VHHGFP4; Fig. S5). Although Western blot analysis revealed differences in efficiency, all proteins were secreted by correspondingly engineered bacteria, suggesting that this method allows considerable variability in the size and structure of secreted proteins.
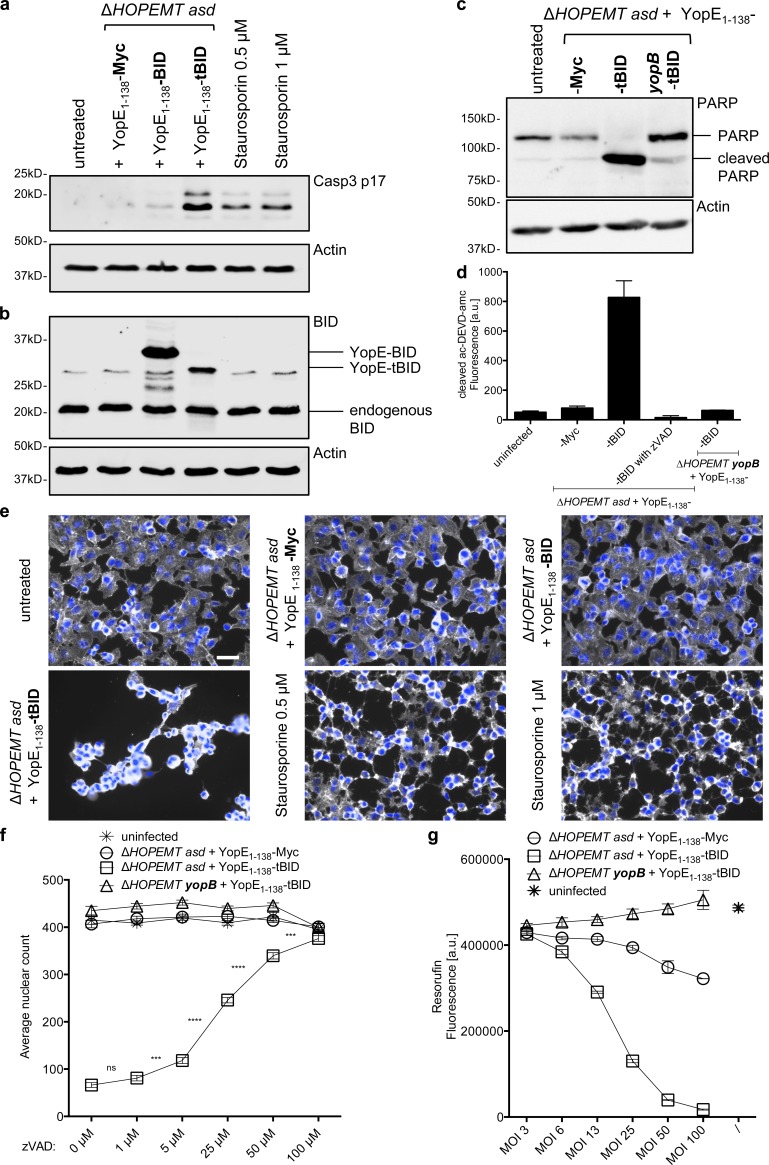
Next, we examined the functional consequences of the translocation of human BH3 interacting-domain death agonist (BID) into HeLa cells. BID is a proapoptotic member of the Bcl-2 protein family and a mediator of mitochondrial damage induced by caspase-8. Caspase-8 cleaves BID, and truncated BID (tBID) translocates to mitochondria, where it triggers cytochrome c release (Li et al., 1998). The latter leads to CASP3 activation by cleavage into the 17- and 12-kD subunits. Data showed that translocated YopE1–138-tBID triggered CASP3 (Fig. 4 a) and PARP (Fig. 4 c) cleavage, CASP3 activation (Fig. 4 d) and massive cell death (Fig. 4, e–g). Effects were even more pronounced than after treatment with the apoptosis inducer staurosporine (Fig.4, a and e), strictly dependent on YopB (Fig. 4, c, d, f, and g) and blocked by the pan-caspase inhibitor z-VAD (Fig. 4, d and f). To compare the amounts of translocated proteins to endogenous BID, HeLa cells were lysed with digitonin, which does not lyse bacteria (Lee et al., 1998; Fig. S4, a and c), and analyzed using an anti-BID antibody (Fig. 4 b). Translocated YopE1–138-tBID reached the level of endogenous BID, whereas delivered YopE1–138-BID was present in even higher amount (2.5-fold). Altogether, these results show that Y. enterocolitica–based T3S can deliver functional eukaryotic proteins into cells, opening entirely new perspectives for this method in cell biology.
Figure 4.
Delivery of human tBID into cells induces massive apoptosis. (a) Translocated tBID induces CASP3 cleavage. Cleavage of CASP3 into the p17 subunit was monitored by Western blot analysis of HeLa cells infected for 60 min with the indicated strains at an MOI of 100 or treated with staurosporine. (b) Translocated YopE1–138-BID fusion proteins reach endogenous levels. Digitonin-lysed HeLa cells, infected for 1 h with the indicated strains as shown in panel a at an MOI of 100, were analyzed by Western blot analysis with an anti-BID antibody to compare endogenous BID, translocated YopE1–138-BID and YopE1-138-tBID levels. (c) Translocated tBID induces PARP cleavage. Cleavage of PARP was monitored by Western blot analysis of HeLa cells infected for 60 min with the indicated strains at an MOI of 100. (d) Delivery of t-BID activates CASP3 as monitored by measuring the fluorogenic substrate ac-DEVD-amc after incubation for 1 h with cell lysates of HeLa cells infected for 60 min with the indicated strains at an MOI of 100. Pan-caspase inhibitor z-VAD was added at 50 µM when indicated. Error bars indicate standard errors of the mean. (e) Translocated tBID induces massive cell loss. HeLa cells were infected with the indicated strains at an MOI of 100 for 1 h or treated with staurosporine. After fixation, cells were stained for nuclei (blue) and F-actin (gray). Bar, 50 µm. (f) Delivery of t-BID leads to a strong reduction in cell number, assessed by counting nuclei. HeLa cells were infected for 60 min with the indicated strains at an MOI of 100. Cells were preincubated 1 h before infection with the indicated concentrations of z-VAD. z-VAD blocks the effect of translocated t-BID in a dose-dependent manner. Automated image analysis was performed on n = 18 images per condition. Error bars indicate standard errors of the mean. Statistical analysis was performed using a Mann-Whitney test (***, P < 0.001; ****, P < 0.0001; ns, not significant). (g) Metabolic activity of HeLa cells 24 h after infection with the indicated strains at indicated MOIs assessed by resazurin assay. 1 h p.i. penicillin and streptomycin were added. Error bars indicate standard errors of the mean.
Delivery of fluorescent proteins, nuclear relocalization, and nanobody-dependent targeting of fusion proteins
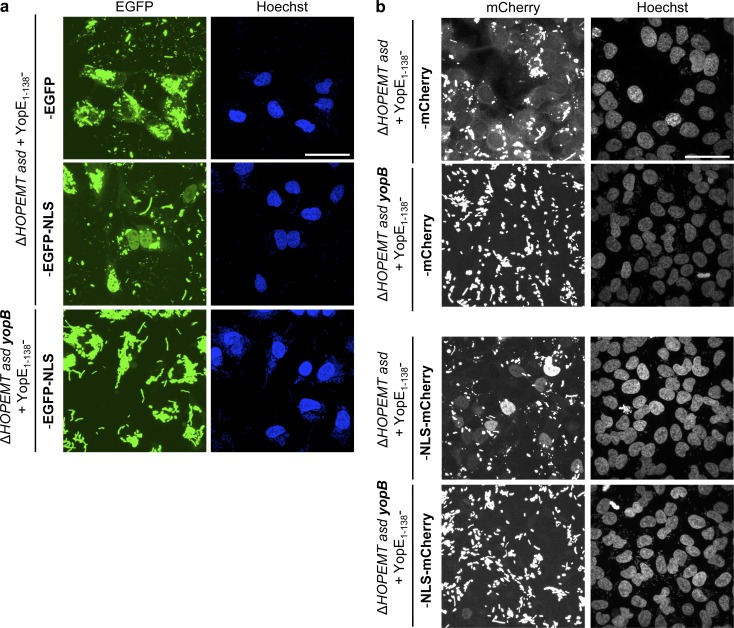
The delivery of fluorescent proteins can be used to monitor T3S-mediated protein transport or to determine the subcellular localization of translocated fusion proteins. Unfortunately, GFP is a notoriously poor T3S substrate (Jacobi et al., 1998), and use of split GFP was proposed instead (Van Engelenburg and Palmer, 2010). Recently, mCherry fused to an NLS was delivered successfully by a plant pathogenic bacterium to plant cells (Sharma et al., 2013). First, we addressed the ability of the YopE1–138-based tool to deliver GFP into cells. Infection of HeLa cells with YopE1–138-GFP–expressing bacteria revealed a weak cytosolic staining, indicating that GFP proteins were secreted into target cells (Fig. 5 a). To extend these results, we concentrated fluorescent signals into nuclei by adding an NLS (SV40 large T-antigen) to the fusion protein. As predicted, a strong nuclear relocalization of GFP was observed, confirming that YopE-based T3S enables some level of GFP delivery into cells and that an NLS sequence is efficient to redirect proteins to the nucleus (Figs. 5 a and S3 c). As expected, a ΔyopB mutant failed to inject GFP into cells. For mCherry, protein translocation was also observed, and addition of an NLS enhanced nuclear localization of the fusion protein (Figs. 5 b and S3 d). Altogether, these results show a weak but reproducible translocation of GFP, document successful delivery of mCherry via T3S by a human pathogen in mammalian cells, and confirm the ability of an NLS to relocate proteins to the nucleus.
Figure 5.
Delivery of EGFP and mCherry fusion proteins and nuclear relocalization. (a) T3S-based delivery of EGFP fusion proteins into cells. EGFP localization was monitored by confocal microscopy of HeLa cells infected with the indicated strains at an MOI of 100 for 4 h. Nuclei were stained with Hoechst. The YopE1–138 fragment does not prevent the nuclear localization of translocated EGFP-NLS. Bar, 50 µm. (b) T3S-based delivery of mCherry fusion proteins into cells. mCherry localization was monitored by confocal microscopy of HeLa cells infected with the indicated strains at an MOI of 50 for 4 h. Nuclei were stained with Hoechst. The YopE1–138 fragment does not prevent the nuclear localization of translocated NLS-mCherry. Bar, 50 µm. Fluorescent images correspond to maximum intensity z projections.
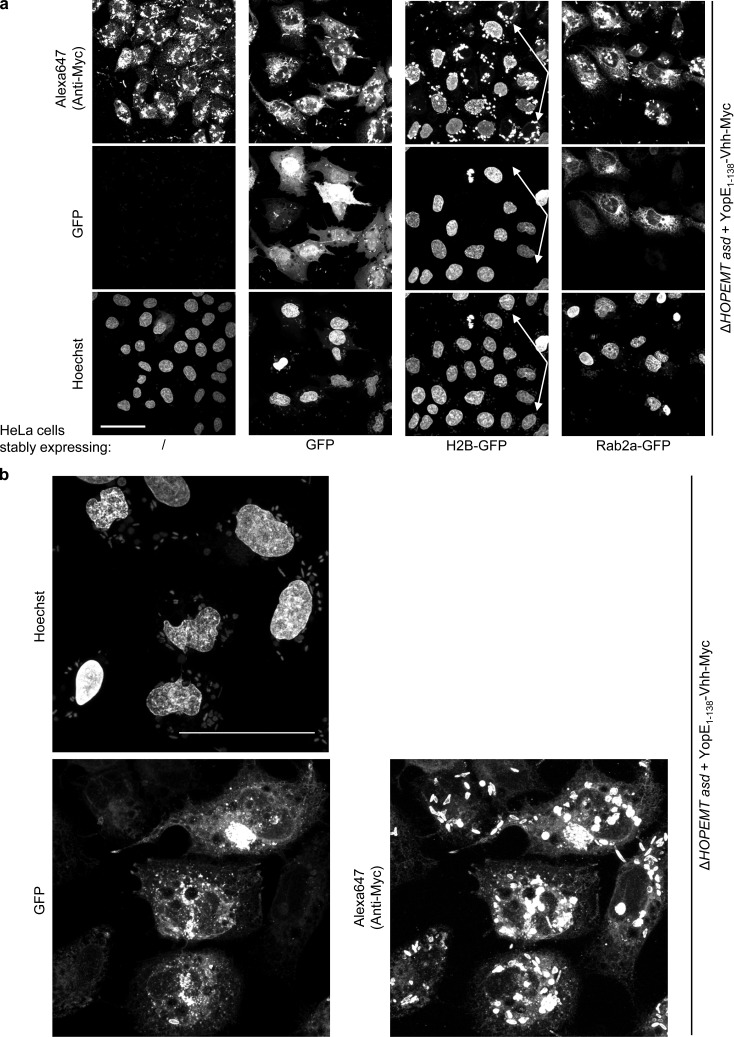
Besides conventional antibodies, camelids produce antibodies lacking light chains (Hamers-Casterman et al., 1993). The N-terminal antigen-binding domain of these heavy-chain antibodies is known as a nanobody or VHH. Nanobodies, which have successfully been delivered by E. coli via T3S (Blanco-Toribio et al., 2010), constitute a valuable tool to target specific intracellular proteins. We took advantage of a nanobody recognizing EGFP, VHHGFP4 (Saerens et al., 2005), to direct translocated YopE1–138-VHHGFP4-Myc to the intracellular location of EGFP, histone 2B-GFP (H2B-GFP; Silljé et al., 2006), and EGFP-Rab2a, stably expressed in different HeLa cell lines. In absence of EGFP, translocated YopE1–138-VHHGFP4-Myc (Fig. 6 a) was detected in the cytoplasm, reflecting the distribution of YopE1–138-Myc (Fig. 1, c and d). In EGFP-expressing cells, the nanobody fusion protein was mostly localized to the cytoplasm, suggesting that the presence of YopE may slightly prevent nuclear localization in some cases. In contrast, in cells expressing H2B-GFP, translocated YopE1–138-VHHGFP4-Myc was exclusively found in nuclei. Interestingly, VHHGFP4-Myc failed to localize to the nuclei of HeLa cells that did not express H2B-GFP (marked with arrows in Fig. 6 a), attesting the specificity of the interactions. In these cells, a weaker cytosolic staining was observed instead. These results show consistent colocalization between the nanobody fusion protein and the EGFP constructs and illustrate that intracellular targeting of proteins can be achieved by translocated nanobodies. Finally, in an EGFP-Rab2a–expressing cell line, translocated YopE1–138-VHHGFP4-Myc colocalized with EGFP-Rab2a on perinuclear structures (Fig. 6, a and b). Altogether, these data indicate that nanobody fusion proteins can be functionally delivered into eukaryotic cells by T3S and that a fusion partner can be targeted to different subcellular localizations by a specific nanobody.
Figure 6.
Nanobody-dependent targeting of fusion proteins. (a) Nanobody-dependent sub-cellular localization of a translocated Myc-tagged fusion protein. Control HeLa and stable HeLa cell lines expressing EGFP, H2B-GFP, or EGFP-Rab2a were infected at MOI 50 with YopE1–138-VHHGFP4-Myc-encoding bacteria for 4 h (2 h p.i., gentamicin was added) and analyzed by confocal microscopy for EGFP, Myc (stained by Alexa Fluor 647), and Hoechst for the nuclei. Arrows mark HeLa cells lacking H2B-GFP expression. Bar, 50 µm. Fluorescent images correspond to maximum intensity z projections. (b) Stable HeLa cell line expressing EGFP-Rab2a were infected as in panel a and observed at 60× by confocal microscopy for EGFP, Myc (stained by Alexa Fluor 647), and Hoechst for the nuclei. Bar, 50 µm.
Intracellular cleavage of the YopE1–138 fragment from T3S-delivered proteins
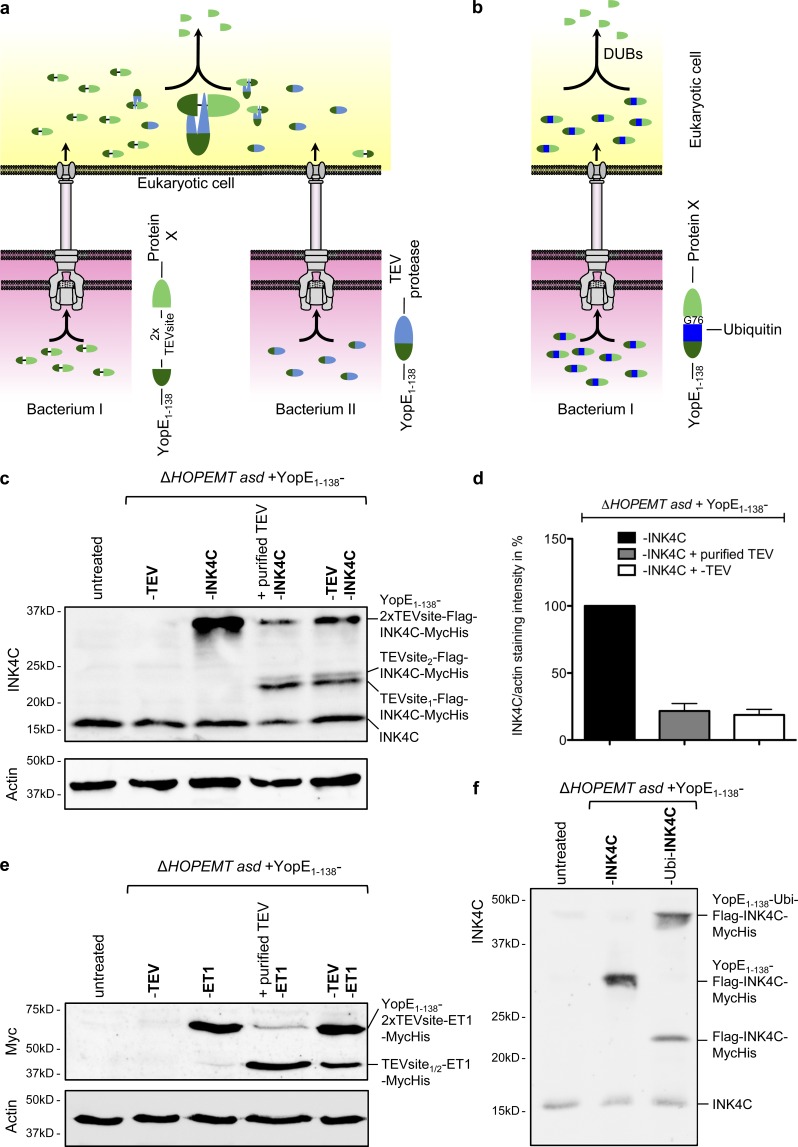
As the YopE1-138 N-terminal fusion part might prevent some fusion proteins from being fully functional, we implemented the option of cleaving off the YopE1–138 fragment into our Y. enterocolitica–based tool. First, we took advantage of the TEV protease activity, which cleaves proteins at the consensus site ENLYFQ\S (Carrington and Dougherty, 1988). Two TEV cleavage sites were introduced between YopE1–138 and two proteins of interest (Fig. 7 a), namely, INK4C and the transcription regulator ET1 (Weber et al., 2002). In addition, the S219V variant of the TEV protease (Kapust et al., 2001), which is resistant to autoinactivation, was fused to YopE1–138, and cells were coinfected with the YopE1–138-proteinX and YopE1–138-TEV–expressing strains. To exclusively lyse human cells and thus restrict analysis to translocated proteins, infected HeLa cells were lysed with digitonin (Fig. S4 c). Western blot analysis with anti-INK4C or anti-Myc antibodies revealed that the YopE1–138-2xTEV-cleavage-sites-Flag-INK4C-MycHis and YopE1–138-2xTEV-cleavage-sites-ET1-Myc were both effectively translocated into cells (Fig. 7, c and e). Remarkably, when cells were coinfected with YopE1–138-TEV–expressing bacteria, the intensity of the band corresponding to the full-length fusion protein was decreased, whereas a second band consistent with the protein of interest plus the N-terminal remnants of the TEV cleavage site (likely one serine) was visible (Fig. 7, c and e). In the case of the YopE1–138-2xTEV-cleavage-sites-Flag-INK4C-MycHis fusion protein, the presence of an additional weaker band was observed (Fig. 7 c). The size of the corresponding protein was compatible with cleavage at the site being closer to the YopE fragment, resulting in S-ENLYFQS-Flag-INK4C-MycHis. A similar pattern was also found when cell lysates were directly incubated overnight with purified TEV protease, indicative of TEV-mediated cleavage in injected cells. Although cleavage was not complete, it occurred already 2 h p.i. Furthermore, overnight digestion with purified TEV protease failed to provide a better yield (Fig. 7 c–e). Altogether, these results demonstrate that coinfections with YopE1–138-TEV–expressing bacteria enable the intracellular cleavage of YopE1–138, and thereby the release of nearly native proteins after T3S.
Figure 7.
Cleavage of the YopE1–138 fragment from the translocated fusion protein. (a) Schematic representation of the strategy developed to cleave off the YopE1–138-fragment after T3S-dependent protein delivery into HeLa cells. HeLa cells are coinfected with two different Y. enterocolitica strains. One strain delivers the TEV protease fused to YopE1–138, whereas the other strain delivers a protein of interest fused to YopE1–138 via a linker containing a double TEV protease cleavage site. After protein delivery into the eukaryotic cell, TEV cleaves off the YopE1–138 fragment from the protein of interest X. (b) Schematic representation of the strategy developed to cleave off the YopE1–138 fragment after T3S-dependent protein delivery into HeLa cells by fusion to ubiquitin. Ubiquitin is processed at its C terminus (after G76) by a group of endogenous ubiquitin-specific C-terminal proteases (DUBs), leading to the liberation of the protein C-terminally fused to ubiquitin. (c) TEV-mediated cleavage of the YopE1–138 fragment from the INK4C fusion protein. Digitonin-lysed HeLa cells infected for 2 h with the indicated strains each at an MOI of 100 were analyzed by Western blot with an anti-INK4C antibody for the presence of YopE1–138-2xTEVsite–Flag–INK4C–MycHis or its cleaved form Flag–INK4C–MycHis. In lane 4, cell lysate was incubated overnight with purified TEV protease. (d) Quantification of TEV-mediated cleavage. Cleavage was measured by quantifying the band corresponding to YopE1–138–2x TEV site–Flag–INK4C from panel c normalized to the actin staining intensity. Intensity of the lane 3 band was set to 100%. Data correspond to the mean of n = 2 independent experiments, and error bars indicate the standard error of the mean. (e) TEV-mediated cleavage of the YopE1–138 fragment from the ET1-Myc fusion protein. Digitonin-lysed HeLa cells infected for 1 h with the indicated strains at an MOI of 100 were analyzed by Western blotting with an anti-Myc antibody for the presence of YopE1–138–2x TEV site–ET1–Myc or its cleaved form ET1-Myc. In lane 4, cell lysate was incubated overnight with purified TEV protease. (f) Cleavage of YopE1–138–ubiquitin–Flag–INK4C–MycHis after translocation into HeLa cells. Digitonin-lysed HeLa cells infected for 1 h with the indicated strains each at an MOI of 100 were analyzed by Western blot with an anti-INK4C antibody for the presence of YopE1–138–ubiquitin–Flag–INK4C–MycHis or its cleaved form Flag–INK4C–MycHis.
An alternative approach consisted in incorporating ubiquitin into the fusion protein of interest. Indeed, ubiquitin is processed at its C terminus by a group of endogenous ubiquitin-specific C-terminal proteases (deubiquitinating enzymes, DUBs) and was previously used to monitor cytosolic uptake of proteins fused to transduction domains (Loison et al., 2005). It was shown that C-terminal ubiquitin fusion proteins were freed from ubiquitin upon microinjection into eukaryotic cells (Inomata et al., 2009). As the cleavage is supposed to happen at the very C terminus of ubiquitin (after G76), the protein of interest should be free of additional amino acid sequence (Inomata et al., 2009). This method was tested on the YopE1–138-ubiquitin-Flag-INK4C-MycHis fusion protein. In control cells infected by YopE1–138-Flag-INK4C-MycHis–expressing bacteria, a band corresponding to YopE1–138-Flag-INK4C-MycHis was found, indicative of efficient translocation of the protein (Fig. 7 f). When cells were infected for 1 h with YopE1–138-ubiquitin-Flag-INK4C-MycHis–expressing bacteria, an additional band corresponding to the size of Flag-INK4C-MycHis was visible, indicating that part of the fusion protein was cleaved. This result shows that the introduction of ubiquitin into the fusion protein enables to cleave off the YopE1–138 fragment without a need for an exogenous protease.
In vivo translocation of z-BIM in zebrafish embryos induces apoptosis
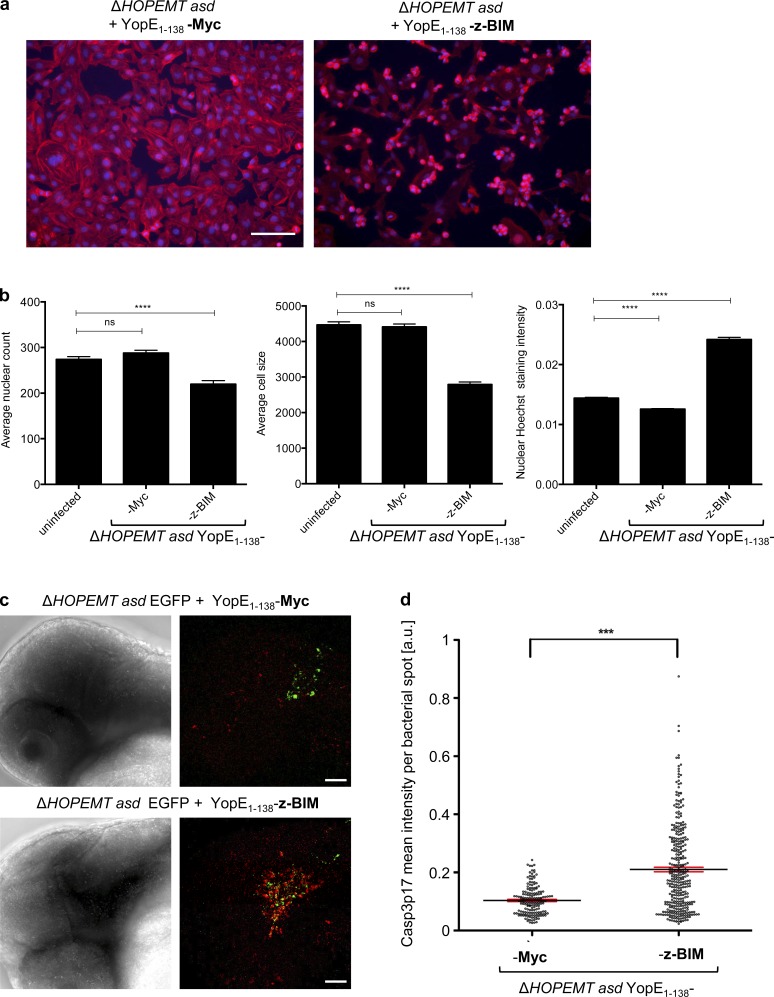
An interesting feature of T3S-based protein delivery is its potential use in living animals. We directly tested this opportunity by infecting zebrafish embryos. As apoptosis is an important cellular process that can be easily visualized in vivo, we investigated the impact of the delivery of z-BIM, the best-characterized apoptosis inducer in zebrafish (Jette et al., 2008). Although z-BIMhas low sequence similarity tohuman BIM, we found that HeLa cells infected with YopE1–138-z-BIM–expressing bacteria showed a clear reduction of cell number and cell size as well as nuclear condensation (Fig. 8, a and b). In vivo infections of zebrafish embryos were then performed by microinjecting EGFP and YopE1–138-Myc or EGFP and YopE1–138-z-BIM–expressing bacteria into the hindbrain (Benard et al., 2012). Embryos were fixed 5.5 h p.i., permeabilized, and stained for CASP3 p17. Upon infection with the control strain, bacteria were visible in the hindbrain region (Fig. 8 c), but no induction of apoptosis was detected. In contrast, YopE1–138-z-BIM–delivering bacteria induced a strong increase in cleaved CASP3. This observation was confirmed by automated image analysis (Fig. 8 d). This result indicates that translocated YopE1–138-z-BIM is functional in developing zebrafish and validates the use of our T3S tool to deliver eukaryotic proteins into living animals.
Figure 8.
T3S-dependent delivery of zebrafish BIM induces apoptosis. (a) Translocated z-BIM induces apoptosis in HeLa cells. HeLa cells were infected with the indicated strains at an MOI of 100 for 1 h. After fixation, cells were stained for nuclei (blue) and F-actin (red). Bar, 50 µm. (b) Automated quantification of mean nuclear count, cell size and nuclear staining intensity as marker for nuclear condensation of n = 84 images as in panel a. Error bars indicate standard errors of the mean. Statistical analysis was performed using a Mann-Whitney test (***, P < 0.001; ****, P < 0.0001; ns, not significant). (c) Translocated z-BIM induces apoptosis in zebrafish embryos. Zebrafish embryos were infected with the EGFP-expressing Y. enterocolitica control strain or YopE1–138-z-BIM delivering strain by injecting 400 bacteria into the hindbrain region. After 5.5 h, embryos were fixed, stained for CASP3 p17 subunit (red), and analyzed by fluorescent microscopy for the presence of bacteria (green). Fluorescent images correspond to maximum intensity z projections. Bars, 50 µm. (d) Automated image analysis of maximum intensity z projections of recorded z-stack images as shown in panel c. In brief, bacteria were detected via the GFP channel. Around each bacterial spot, a circle with a radius of 10 pixels was created and CASP3 p17 staining intensity was measured. Statistical analysis was performed using a Mann-Whitney test (***, P < 0.001). Data represent the mean of n = 14 infected embryos for YopE1–138-Myc and n = 19 for YopE1–138-z-BIM, and error bars (red) indicate standard errors of the mean.
Phosphoproteomics reveals the global impact of tBID on protein phosphorylation
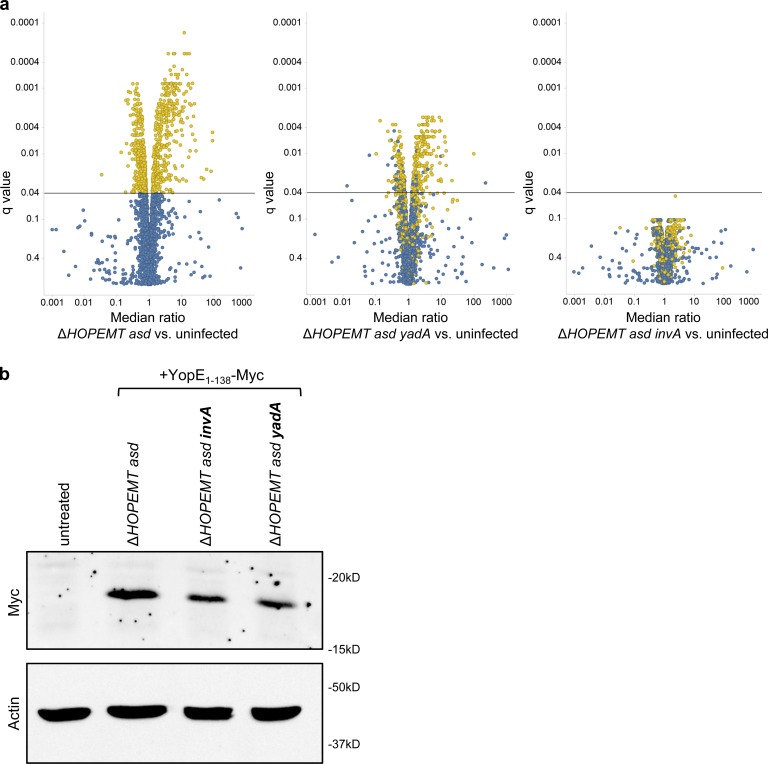
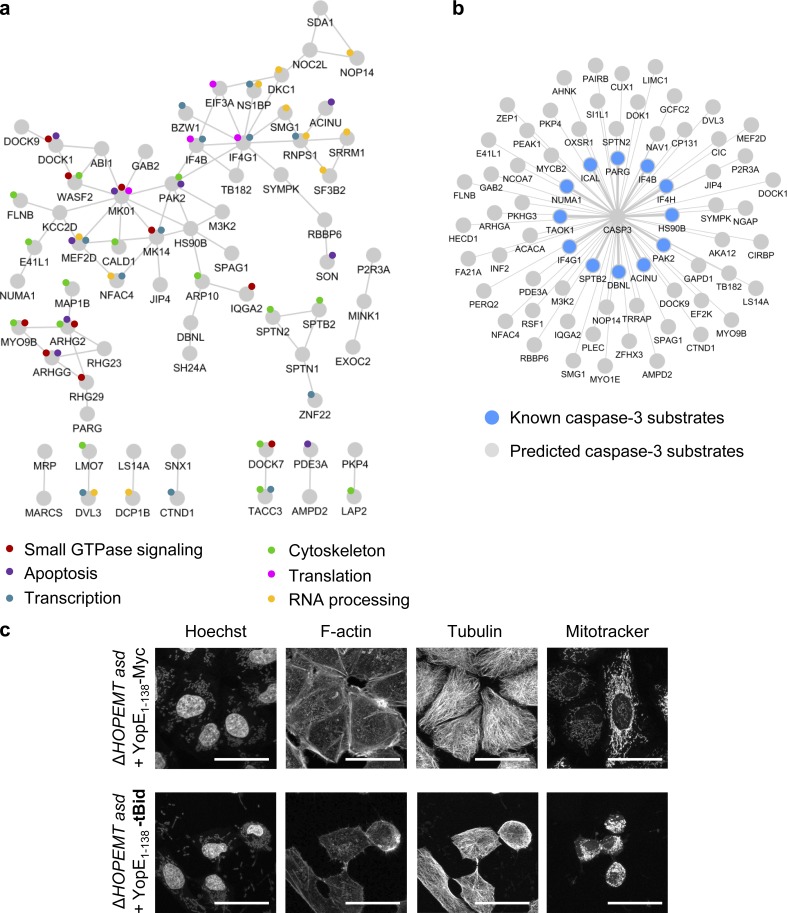
T3S-based protein delivery was used to gain new biological insights into the mechanism of apoptosis triggered by tBID. Although tBID is a central player of apoptosis, the systems-level impact of this factor on cellular signaling remains poorly characterized. As phosphorylation is a widespread posttranslational modification, we used a label-free approach of liquid chromatography tandem mass spectrometry (LC-MS/MS)–based phosphoproteomics to determine the impact of translocated tBID on cellular signaling in HeLa cells. The process of Y. enterocolitica infection itself may lead to protein phosphorylation (Schulte et al., 2000; Uliczka et al., 2009). Therefore, we first determined the background phosphorylation changes triggered by YopE1–138-Myc–encoding bacteria compared with uninfected cells. Phosphopeptides were enriched, quantified, and identified as previously described in Schmutz et al. (2013) and in Materials and methods. A volcano plot representing q-values and phosphorylation ratios between infected and uninfected cells showed that control infection had a significant impact on phosphorylation (Fig. 9 a, left). As this background phosphorylation may interfere with tBID-induced phosphorylation changes, we minimized it by making use of two mutants deleted for the adhesion molecules invasin A (InvA) or YadA, which are known to trigger host signaling (Uliczka et al., 2009). As expected, infection by the ΔHOPEMT asd yadA–deleted strain (encoding Yop1–138-Myc) triggered fewer phosphorylation changes than ΔHOPEMT asd Yop1–138-Myc–delivering bacteria (Fig. 9 a, middle). Interestingly, even fewer changes were observed upon infection with the invA-deleted strain (no more changes with q < 0.04; Fig. 9 a, right), showing that the latter was optimal for system-wide signaling studies. Although translocation efficiency was reduced in absence of InvA (Fig. 9 b), tBID delivery was still able to induce massive cell death (Fig. S3 e). Phosphoproteomics was then performed to measure the impact of tBID delivered by the invA-deleted strain on HeLa cells. Data revealed 240 tBID-dependent phosphorylation changes (q-values < 0.04); 218 phosphopeptides underwent an increase in phosphorylation whereas 22 were less phosphorylated after tBID delivery (Table S4). These phosphopeptides corresponded to 187 different phosphoproteins that we defined as the tBID phosphoproteome. The STRING database was combined with Gene Ontology analysis (Huang da et al., 2008) to create a functional protein-protein interaction network of the tBID phosphoproteome (Fig. 10 a and Table S5). Data showed that phosphorylation of proteins involved in diverse cellular processes are affected by tBID. These include the cytoskeleton (i.e., SPTN2, SPTB2, and FLNB), small GTPase signaling (i.e., DOCK1, DOCK9, and ARHG2), RNA processing (i.e., SF3B2 and SRRM1), and protein synthesis (i.e., IF4B, IF3A, and IF4G1). Interestingly, 12 known CASP3 substrates and 58 putative ones predicted by an in silico tool based on a position-specific scoring matrix approach (Ayyash et al., 2012) were identified in the phosphoproteome of tBID (Fig. 10 b and Table S1). Among known substrates, spectrin (SPTB2) is a cytoskeletal protein that lines the intracellular side of the plasma membrane. Its cleavage by CASP3 leads to membrane collapse and apoptosis (Wang et al., 1998). In the same line, CASP3-mediated cleavage of plectin disrupts the cytoskeleton and induces morphological changes and cell detachment (Liu et al., 2011). Finally, cleavage of ACINU and NUMA1 triggers chromatin condensation and nuclear matrix reorganization. Interestingly, phosphorylation changes of these proteins occurred concomitantly with cell rounding up and nuclei condensation observed after tBID delivery (Fig. 10 c). Altogether, our results show that tBID-induced apoptosis triggers hundreds of phosphorylation events involved in multiple cellular functions and confirms the central role of CASP3 in this process. Besides providing a valuable dataset for apoptosis, these results illustrate how T3S-based protein delivery combined with –omics studies can reveal the systems-level impact of a protein on a cellular network.
Figure 9.
Characterization of the cellular impact of T3S-based delivery by phosphoproteomics. (a) Volcano plot representation of the impact of T3S-based delivery on phosphorylation in the presence or in absence of YadA or InvA compared with uninfected cells. HeLa cells were infected for 30 min with the indicated strains at a MOI of 100. Yellow dots represent in all three panels the phosphopeptides associated with a q-value < 0.04 in control infection compared with uninfected cells. Phosphoproteomic analysis was performed in independent triplicates. (b) Digitonin-lysed HeLa cells, infected for 30 min with the indicated strains at an MOI of 100, were analyzed by Western blot analysis with an anti-Myc antibody to detect delivery of YopE1–138-Myc by corresponding strains.
Figure 10.
Analysis of t-BID-dependent phosphoproteome. (a) Representation of the functional protein interaction network of the tBID phosphoproteome. Proteins, containing at least one phosphopeptide undergoing a change in phosphorylation after tBID delivery (light gray; q-value < 0.04) are represented in a STRING network (high-confidence, score 0.7). Only proteins with at least one connection in STRING are shown. Colored circles depict the biological annotation of proteins as obtained from DAVID (Table S5). (b) Graphical representation of CASP3 known (color) and predicted substrates (gray) as shown in Table S1. (c) Confocal images of HeLa cells infected with either ΔHOPEMT asd + YopE1–138 or ΔHOPEMT asd + YopE1–138-tBID reveal the induction of an apoptotic phenotype after tBID delivery. Cells were stained for the nuclei with Hoechst, for F-actin with phalloidin, for tubulin with an anti-tubulin antibody, and for mitochondria with Mitotracker. Bars, 40 µm.
Discussion
Here we provide optimized protocols for making use of Y. enterocolitica T3S to inject heterologous type III and IV bacterial effectors as well as viral and mammalian proteins in cells and in developing animals. We show that first effects of translocated proteins can already be observed within seconds after infection, indicating that this method is particularly well suited to study short-lived cellular events that are triggered or controlled by a given protein. As protein translocation via T3S can be temporally controlled, it allows investigating the temporal dynamics of cellular and signaling changes. In contrast to cDNA transfection, T3S-based protein delivery is homogenous within the cell population and can be gradually tuned with the MOI. Unlike proteofection, protein electroporation (Freund et al., 2013), or cell-penetrating peptides, T3S-dependent protein delivery does not require protein purification. We show that the amounts of translocated tBID and BID reach and exceed (2.5-fold) endogenous levels of BID, respectively. According to a deep proteome dataset estimating that HeLa cells contain 4.4 × 105 copies of BID per cell (Nagaraj et al., 2011), our results suggest that T3S can inject 105–106 proteins per cell. We further show that T3S-delivered INK4C can exceed the level of endogenous INK4C by a factor of 5 (Fig. 7, c and f). As deep proteome data indicate that HeLa cells contain 5 times 105 copies of INK4C per cell (Nagaraj et al., 2011), this suggests that T3S can inject more than 106 proteins per cell. These numbers are compatible with the translocation yield of the S. typhimurium T3S substrate SipA (6 ± 3 × 103 molecules per bacterium; Schlumberger et al., 2005) or nanobodies via E. coli T3S (105–106 molecules per infected cell; Blanco-Toribio et al., 2010). Interestingly, the described method is not limited to single protein delivery. Coinfection protocols can be performed to inject multiple proteins in cells and study functional interactions. We also show that T3S-based protein delivery is suitable to inject functionally active eukaryotic proteins into living animals.
So far, the use of T3S-based protein delivery was hampered by the difficulties of proteins to pass through the needle of the injectisome. For example, constraints on YopE length and strength of protein folding were previously described for the translocation of DHFR (Feldman et al., 2002) and GFP (Van Engelenburg and Palmer, 2010). We found that the YopE1–138/SycE-dependent strategy described here allows the translocation of various bacterial, viral and eukaryotic proteins ranging from a few kilodaltons to more than 60 kD, including proteins characterized by strong folds such as β-barrels. This extends by far the list of previously described heterologous cargo for T3S and thus opens up the possibility for broad applications of this technology in cell biology. Although most fusion proteins tested in our study were secreted successfully, the proteins TIAM1 and clathrin heavy chain (65 and 191 kD, respectively) were not secreted. This indicates that the method, although valid for a large variety of substrates, is not yet applicable to all proteins. However, the suitability of substrates for translocation can easily be assessed for individual proteins by performing the described in vitro-secretion assay before translocation into cells.
Protein localization strongly contributes to protein function. We found that YopE1–138 does not prevent the nuclear localization of mCherry and EGFP tagged with an NLS, indicating that injected proteins containing an NLS will be effectively targeted to the nucleus. Noticeably, the strategy presented here allows the delivery of the fluorescent protein mCherry into eukaryotic cells while conferring weak but reproducible levels of EGFP translocation (Jacobi et al., 1998). Besides nuclear targeting via an NLS, we also show that a T3S-delivered protein can be directed to different subcellular localizations, including the cytoplasm, the nucleus, and a perinuclear compartment, through fusion to a nanobody recognizing GFP (provided that the recipient cells express GFP-tagged proteins in the corresponding compartments).
We describe a TEV-protease–dependent strategy to cleave off the YopE1–138 fragment after translocation into cells. TEV is a very specific protease, which is not toxic to cells (Henrichs et al., 2005). In an alternative way, simple fusion to YopE1–138-ubiquitin leads to its cleavage after delivery to the eukaryotic cells without the need for additional proteins. Ubiquitin was reported to be processed by endogenous proteases, so-called DUBs, at the very C terminus, thereby releasing the fused protein in its native amino acid sequence. These approaches thus offer the option of releasing native heterologous proteins after T3S-based delivery.
Finally, we used T3S-based protein delivery to gain new insights into the mechanism of apoptosis. Our tool was combined with phosphoproteomics to reveal the systems-level impact of tBID on protein phosphorylation. We found that tBID-induced apoptosis is associated with numerous phosphorylation changes in proteins involved in several cellular processes affected in apoptotic cells such as the cytoskeleton and protein translation (Clemens et al., 2000; Elmore, 2007). Among proteins of the phosphoproteome, 70 known or predicted CASP3 substrates were identified. This number corresponds to 37% of our dataset whereas only 14% of the human proteins have been predicted to be CASP3 substrates (Ayyash et al., 2012), strongly suggesting that CASP3 substrates are overrepresented in the tBID phosphoproteome. This is in line with recent publications reporting a complex interplay between phosphorylation and CASP3 cleavage of proteins in apoptosis (Dix et al., 2012; Turowec et al., 2014). In conclusion, our results reveal new possible players involved in early apoptosis and emphasize the value of T3S-based protein delivery combined with phosphoproteomics to gain new molecular insights into complex biological processes.
Materials and methods
Bacterial strains and growth conditions
The bacterial strains used in this study are listed in Table S3. All E. coli strains were grown on LB agar plates and in LB broth. E. coli Top10 was used for plasmid purification and cloning and E. coli Sm10 λ pir was used for conjugation. Ampicillin was used at 200 µg/ml (Y. enterocolitica) or 100 µg/ml (E. coli) to select for expression vectors. Streptomycin was used at 100 µg/ml to select for suicide vectors. Y. enterocolitica MRS40, a non-ampicillin–resistant E40 derivate (Sory and Cornelis, 1994), and all derived strains were routinely grown on brain heart infusion (BHI; Difco). Nalidixic acid (35 µg/ml) was added to all Y. enterocolitica strains. All Y. enterocolitica asd strains were additionally supplemented with 100 µg/ml meso-2,6-diaminopimelic acid (mDAP; Sigma-Aldrich).
Construction of plasmids
The pBad_Si2 and pBad_Si_1 plasmids (Fig. S1, a and b) were used to fuse the proteins of interest to the N-terminal 138 amino acids of YopE. pBad_Si2 was constructed by cloning the SycE-YopE1–138 fragment containing the endogenous promoters of YopE and SycE from purified pYV40 into the KpnI–HindIII site of pBad-MycHisA (Invitrogen). Additional modifications include the elimination of the NcoI–BglII fragment of pBad-MycHisA by digestion, Klenow fragment treatment, religation, and the addition the XbaI–XhoI–BstBI–(HindIII) cleavage sites at the 3′ end of the YopE1–138 fragment (Fig. S1 c). pBad_Si1 is identical to pBad_Si2 except for the insertion of an EGFP marker, amplified from pEGFP-C1 vector (Clontech), into the NcoI–BglII site, under the arabinose inducible promoter.
Full-length genes (Table S3) were amplified with the specific primers listed in Table S2 and cloned into pBad_Si2 or, in case of z-BIM, into pBad_Si1. For genes of bacterial species, purified genomic DNA was used as template (S. flexneri M90T, S. enterica subsp. enterica serovar Typhimurium SL1344, Bartonella henselae ATCC 49882). For human genes, a universal cDNA library (Clontech) was used if not otherwise stated (Table S3). Zebrafish genes were amplified from a zebrafish cDNA library. Ligated plasmids were cloned in E. coli Top10. Sequenced plasmids were electroporated into the desired Y. enterocolitica strain by using the settings of standard E. coli electroporation.
In vitro secretion
Induction of the yop regulon was performed by shifting the bacterial culture to 37°C in BHI supplemented with 4 mg/ml glucose as carbon source and 20 mM sodium oxalate (secretion-permissive conditions; Cornelis et al., 1987). Pellet and supernatant were separated by centrifugation at 20,800 g for 10 min at 4°C. The pellet was taken as total bacteria fraction. Proteins in the supernatant were precipitated with trichloroacetic acid 10% (wt/vol) final for 1 h at 4°C. After centrifugation (20,800 g for 15 min) and elimination of the supernatant, the resulting pellet was washed in ice-cold acetone overnight. The samples were centrifuged again, the supernatant was discarded, and the pellet was air-dried and resuspended in 1× SDS loading buffer. Secreted proteins were analyzed by SDS–PAGE. Typicaly, each lane was loaded with proteins secreted by 3 × 108 bacteria. Detection of specific secreted proteins by immunoblotting was performed using 12.5% SDS-PAGE gels. For detection of proteins in total bacterial extracts, the equivalent of 2 × 108 bacteria was loaded per lane, if not stated otherwise. Proteins were also separated on 12.5% SDS-PAGE gels before detection by immunoblotting. Immunoblotting was performed using rat monoclonal antibodies against YopE (13A9; raised against full-length YopE; 1:1,000; Grosdent et al., 2002). In brief, this monoclonal antibody was generated by intraperitoneal injection of 109 Y. enterocolitica W22703, grown under secretion inducing condition and mixed with Freud’s adjuvant, into LOU rats (Bodeus et al., 1990). After 3 mo, the same procedure was repeated. Three days later, the spleen was removed and splenic cells were fused with IR983F azaguanine-resistant cells (Bodeus et al., 1990). Hybrid clones were grown and supernatants were analyzed for the identification of a YopE-detecting antibody. The antibody was preabsorbed twice overnight against Y. enterocolitica ΔHOPEMT asd to reduce background staining. Detection was performed with secondary antibodies directed against rat antibodies and conjugated to horseradish peroxidase (1:5,000; 3010-05; Southern Biotech), before development with ECL chemiluminescent substrate (LumiGlo, KPL).
Cell culture and infections
HeLa CCL-2, HEK 293T, and Swiss 3T3 fibroblasts were cultured in DMEM supplemented with 10% FCS and 2 mM l-glutamine. Human umbilical vein cells were isolated and cultivated as described previously (Dehio et al., 1997). Jurkat cells were cultured in RPMI 1640 supplemented with 10% FCS and 2mM l-glutamine. Y. enterocolitica were grown in BHI plus additives overnight at RT, diluted in fresh BHI to an OD600 of 0.2 and grown for 2h at RT before a temperature shift to a 37°C waterbath shaker for further 30 min or for 1 h in case of EGFP or mCherry translocation. Finally, bacteria were collected by centrifugation (6,000 rcf, 30 s) and washed once with DMEM supplemented with 10 mM Hepes and 2 mM l-glutamine. Cells, seeded in 96-well (for immunofluorescence) or 6-well (for Western blotting) plates, were infected at indicated MOIs in DMEM supplemented with 10 mM Hepes and 2 mM l-glutamine. After adding bacteria, plates were centrifuged for 1 min at 1,750 rpm and placed at 37°C for indicated time periods. Extracellular bacteria were killed by gentamicin (100 mg/ml) if indicated. In case of immunofluorescence analysis, infection assays were stopped by PFA fixation. For Western blot analysis of cellular proteins, cells were washed and lysed as described in the next paragraph. Z-VAD(OMe)-FMK (zVAD; Santa Cruz) was dissolved at 10 mM in DMSO and added at the concentration indicated 1h before the infection.
Western blot analysis of cellular proteins after infection
Cells were infected as described in the previous paragraph, washed twice with ice-cold PBS, and Phospho-safe lysis buffer (Novagen) was added to lyse the cells. After incubation on ice, the cells were centrifuged (16,000 rcf, 25 min, 4°C). Supernatants were collected and analyzed for total protein content by BCA assay (Pierce), samples were standardized on total protein content before SDS-PAGE and Western blot analysis using anti-Phospho-Akt (Ser473 and T308, both rabbit, 4058 and 2965; Cell Signaling), anti-actin (mouse, MAB1501; Millipore), anti-Myc (9E11, mouse, sc-47694; Santa Cruz), anti-PARP (rabbit, 9542; Cell Signaling), and anti-CASP3 p17 (rabbit, 9664; Cell Signaling) antibodies. Detection was performed with secondary antibodies directed against murine or rabbit antibodies and conjugated to horseradish peroxidase (1:5,000; NA931V and NA934V; GE Healthcare), before development with ECL substrate (LumiGlo or LumiGlo reserve; KPL).
TNF stimulation and Western blot analysis of phospho-p38
HeLa cells, seeded in 6-well plates, were infected with an MOI of 100. Gentamicin was added 30 min p.i and TNF 45 min p.i. (10 ng/ml). After 75 min, cells were washed twice with ice-cold PBS, and Phospho-safe lysis buffer (Novagen) was added to lyse the cells. After incubation on ice, the cells were centrifuged (16,000 rcf, 25 min, 4°C). Supernatants were collected and analyzed for total protein content by BCA assay (Pierce) before SDS-PAGE and Western blot analysis using anti-p38 (rabbit, 9212; Cell Signaling) and anti-phospho-p-38 (Thr180/Tyr182; rabbit, 4631; Cell Signaling) antibodies. Detection was performed with secondary antibodies directed against rabbit antibodies conjugated to horseradish peroxidase (1:5,000, NA934V; GE Healthcare), before development with ECL chemiluminescent substrate (LumiGlo or LumiGlo reserve; KPL).
Western blot analysis of T3S-translocated proteins into infected cells
HeLa cells in 6-well plates were infected at an MOI of 100 as described in the Cell culture and infections paragraph. In coinfection experiments, the two bacterial suspensions were mixed in a tube at a ratio of 1:1 (if not otherwise indicated) and added to the cells at the MOI of 100 for each strain (if not otherwise stated). At the end of the infection assay, cells were washed twice with ice-cold PBS and collected by scraping in ice-cold PBS. After centrifugation (16,000 rcf, 5 min, 4°C), pellets were dissolved in 0.002% digitonin supplemented with a protease inhibitor cocktail (Roche complete; Roche). Dissolved pellets were incubated for 5 min on ice and then centrifuged (16,000 rcf, 25 min, 4°C). Supernatants were collected and analyzed for total protein content by BCA assay (Pierce) before SDS-PAGE and Western blot analysis using an anti-Myc (9E11, mouse, sc-47694; Santa Cruz), anti-BID (rabbit, 2002; Cell Signaling), or anti-INK4C (mouse, 2896; Cell Signaling) antibody. Control lysis was performed with a lysis buffer containing 0.2% Triton (in 20 mM Tris, pH 8, 10 mM EDTA, and 100 mM NaCl) and repeated sonication at 4°C (Hielscher).
Immunofluorescence
Cells, seeded in 96-well plates (Corning), were infected as described in the Cell culture and infections paragraph. After fixation with 4% PFA, they were washed three times with PBS and incubated with 5% goat serum in PBS 0.3% Triton X-100 for 1h at RT. The primary antibody (anti-Myc, 1:100; Santa Cruz) was diluted in PBS with 1% BSA and 0.3% Triton X-100 and cells were incubated overnight at 4°C. Cells were washed four times with PBS before the secondary antibody (AF 488 goat anti–mouse, A11029, 1:250; Invitrogen), diluted in PBS 1% BSA and 0.3% Triton X-100, was added. If needed, Hoechst DNA staining (1:2500; 62249; Thermo Fisher Scientific) and/or F-actin staining (Dy647-Phalloidin; DyeOmics) were included. In some cases, staining for DNA and/or F-actin was applied directly after PFA fixation. Cells were incubated for 1h at RT, washed three times with PBS and analyzed by automated image analysis as described in the next paragraph. In case of confocal microscopy, 25,000 HeLa CCL-2 cells were seeded per well of an 8-well µ-Slide (80826; Ibidi) and grown to confluency for 24 h before infection. After fixation in 4% PFA for 10 min, cells were permeabilized in blocking buffer (0.3% Triton X-100/5% goat serum) for 1 h. Samples were incubated overnight with an anti–mouse tubulin-β antibody (mouse, MA5-11732, 1:100; Thermo Fisher Scientific) in antibody dilution buffer (1× PBS/1% BSA/0.3% Triton X-100) followed by secondary antibody application using an Alexa Fluor 647–conjugated goat anti-mouse antibody (A21236; Invitrogen) in antibody dilution buffer. In parallel, F-actin and DNA were stained for 1 h at room temperature using FITC-phalloidin (Invitrogen) and Hoechst (62249; Thermo Fisher Scientific), respectively. Mitochondria were stained using Mitotracker (Life Technologies) according to the manufacturer’s instructions. Images were acquired by a Point Scanning Confocal LSM700 Inverted microscope (Carl Zeiss) using a Plan Apo 40× magnification/1.3 NA oil immersion objective and Zen 2010 software. Images were acquired at room temperature in PBS. Filter cubes used were blue (excitation: G 365, dichroic: FT 395, emission: BP 445/50; filter 49 DAPI), green (exc.: BP 470/40, dichroic: FT 495, em.: BP 525/50; filter 38H eGFP), and red (exc.: BP 546/12, dichroic: FT 560, em.: BP 575-640; filter 20 Rhodamine). The confocal stacks were analyzed (maximum intensity projection) using ImageJ software.
Automated microscopy and image analysis
Images were automatically acquired with an ImageXpress Micro automated cellular imaging microscope (Molecular Devices), equipped with a fast laser autofocus with a precision motorized Z-stage and a Lumencor Spectra X light engine and CFI Super Fluor 10× air objective 0.5 NA (Nikon) or CFI Plan Fluor ELWD 20× C 0.45 NA air objective (Nikon). Images were acquired at room temperature and in PBS. Filter cubes (Semrock) were BrightLine, DAPI-5060B (Hoechst); BrightLine, GFP- 3035B (EGFP, Alexa Fluor 488, FITC); BrightLine, TXRED-4040B (RFP, mCherry); and BrightLine, CY5-4040A (Alexa Fluor 647, Dy47). Camera used was a CoolSNAP HQ monochrome charge coupled device camera (Photometrics). MetaXpress software controlled all components of the system.
Quantification of anti-Myc staining, EGFP, and mCherry intensities was performed using MetaXpress (Molecular Devices). Circular regions were manually chosen in the cytoplasm or nucleus excluding cell areas containing bacteria, and used for quantification. Cell number, cell size, nuclear Hoechst intensity staining, and F-actin meshwork were quantified using CellProfiler (Carpenter et al., 2006). In brief, the Hoechst and the actin stainings were used to automatically identify cell nuclei and cell area, respectively. F-actin foci or cables as seen within the actin meshwork triggered by SopE were quantified by specific analysis modules of CellProfiler (see source code files in the Online supplemental material).
cAMP level determination
HeLa cells, seeded in 96-well plates, were infected as described in the cell culture and infections paragraph. Before infection (30 min), cells were incubated with fresh DMEM supplemented with 10 mM Hepes, 2 mM l-glutamine, and 100 µM 3-Isobutyl-1-methylxanthin (IBMX; Sigma-Aldrich). Gentamicin was added 60 min p.i., and cells were further incubated at 37°C for another 90 min. cAMP concentrations were determined by a competitive ELISA assay performed accordingly to the manufacturer’s instructions (cAMP Biotrak, RPN225; GE Healthcare). As positive control, cells were incubated with cholera toxin (C8052; Sigma-Aldrich) for 1 h.
Ac-DEVD-amc and resazurin assays
HeLa cells infected at an MOI of 100 for 1 h were lysed with a buffer containing 0.2% Triton (in 20 mM Tris, pH 8, 10 mM EDTA, and 100 mM NaCl) on ice. Cell lysates were mixed 1:1 with 0.2 mM ac-DEVD-amc (Alexis, dissolved at 10 mM in DMSO) in a caspase assay buffer (10 mM Pipes, pH 7.2, 10 mM NaCl, 10% sucrose, 0.1% CHAPS, 1 mM EDTA, and 10 mM DTT) and incubated for 1 h at 37°C before fluorescence was measured at an excitation of 380 nm and an emission at 460 nm.
For the resazurin assay, cells were infected at an MOI of 100. 1 h after the infection, penicillin and streptomycin were added. Cells were further incubated for 23 h. Resazurin was dissolved in PBS to a concentration of 0.1 mg/ml, filter-sterilized and added 1:6 into the assay. After 2 h incubation at 37°C, the fluorescence was measured at an excitation of 528 nm and an emission at 600 nm.
Zebrafish embryo infections, imaging, and automated image quantification
All animal experiments were performed according to approved guidelines. Zebrafish were maintained in standard conditions. Embryos were staged by hours postfertilization at 28.5°C. The wild-type AB/EK and EK/TL zebrafish lines were used in this study. Infection protocol followed guidelines given in Benard et al. (2012). In brief, 12 h after fertilization, embryos were maintained in E3 medium containing 0.2 mM N-phenylthiourea to prevent pigment formation. Then, 2 d postfertilization embryos were anesthetized by 0.2 mg/ml tricaine and aligned on 1% agar plates in E3 using a hair loop tool (Benard et al., 2012). Y. enterocolitica bacteria were grown in BHI supplemented with 0.4% arabinose, antibiotics, and mDAP overnight at RT, diluted in fresh BHI with 0.5% arabinose and other additives to an OD600 of 0.2, and grown for 2 h at RT before a temperature shift to 37°C in a water bath shaker for 45 min. Finally, bacteria were collected by centrifugation (6,000 rcf, 30 s) and washed once with PBS. The OD600 was set to 2 in PBS containing mDAP. 1–2 nL of this suspension was injected into the hindbrain of aligned zebrafish embryos using an Femtojet Microinjector device (Eppendorf) and Femtotips II (Eppendorf). Before injection, the tip of the needle was broken off with fine tweezers. The injection time was set to 0.2 s, the compensation pressure to 15 hPa, and the injection pressure was adjusted between 600 and 800 hPa. Drop size, and thus the inoculum, was checked by microscopy and by control plating. After microinjection, the fish were collected in E3 containing tricaine and N-phenylthiourea and incubated for 30 min at 37°C and then for 5 h at 28°C. EGFP bacteria were observed with fluorescence binocular (Leica) 1 h p.i. in zebrafish hindbrains, and embryos that were not properly injected were discarded. At the end of infection, fish were fixed with 2% PFA for 1 h on ice and then incubated with fresh 2% PFA overnight at 4°C. Antibody staining was performed as described previously (Herwig et al., 2011). In brief, embryos were washed four times with PBS 0.1% Tween (PBS-T) for 5 min and permeabilized with PBS-T + 0.5% Triton X-100 for 30 min at RT. Embryos were incubated in blocking solution (PBS-T + TritonX-100, 5% goat serum, and 1% BSA) at 4°C overnight. Cleaved CASP3 antibody (Asp175, 9664; Cell Signaling) was diluted 1:100 in blocking solution and incubated under shaking at 4°C in the dark. Fish were washed seven times with PBS-T for 30 min before the secondary antibody (goat anti–rabbit AF647, 1:500; A21244; Invitrogen), diluted in blocking solution, was added and incubated at 4°C overnight. Larvae were washed with PBS-T four times for 30 min at 4°C, incubated overnight, and further washed three or four times. Embryos were mounted in 0.7% low melting agarose (Sigma-Aldrich) in 35-mm glass-bottom dishes (MatTek). Confocal images were recorded at RT on a Leica TCS SP5 II point scanning confocal microscope (stand Leica DMI 6000B with adaptive focus, equipped with Leica HyD hybrid detectors and AOBS as a tunable dichroic for all lasers) using a 40× water immersion objective (HCX PLAN APO CS with a 1.10 NA). Confocal stacks were acquired with the LAS AF version 2.6.0.7266 software using a z-step size of 1.2 µm. EGFP and AF647 channels were recorded sequentially. The confocal stacks were analyzed (maximum intensity projection) using ImageJ software. Automated image quantification was performed via CellProfiler (Carpenter et al., 2006) on maximum intensity z projections of recorded z-stack images. In brief, bacteria were detected via the GFP channel. Around bacterial spot, a 10-pixel-radius circle was created and the CASP3 p17 staining intensity measured. Overlapping regions were separated equally among connecting members.
Generation of stable cell lines
To produce the pMDK124 lentiviral vector (a gift of O. Pertz, University of Basel, Basel, Switzerland) encoding EGFP-Rab2A (pSL052), pMDK124 was digested with EcoRI and BamHI overnight at 37°C and gel purified. PCR was performed with gene-specific primers that contain 15-bp extensions complementary to the vector-digested ends. EGFP-Rab2A was amplified from templates provided by F.A. Barr (Short et al., 2001). prSL045 (5′-CGACTCTAGAGGATCCGCCACCATGGTGAGCAAGGGCGAGGA-3′) and prSL046 (5′-GATTGTCGACGAATTCTCAACAGCAGCCGCCCCCAG-3′) were used to PCR-amplify EGFP-Rab2A. Using the gel-purified PCR products and digested pMDK124, In-Fusion recombination was performed with 200 ng vector and PCR product using In-Fusion enzyme mixture from In-Fusion HD cloning kit (Clontech), incubated 15 min at 50°C, and then transferred to ice. 5 µl of the reaction mixture was transformed into 100 µl E. coli DH5α and plated on ampicillin containing LA plates. Helper plasmids pVSV, pMDL, and pRev are gifts from Oliver Pertz’s group.
3 × 106 HEK 293T cells were grown in a 10-cm dish and incubated at 37°C, 5% CO2 for at least 6–8 h. 2.2 µg lentiviral vector pMDK124 encoding EGFP-Rab2A, 0.75 µg pVSV, 1.5 µg pMDL, and 0.5 µg pREV in 600 µl DMEM without FCS were then mixed with 25 µl Fugene HD (Promega) in 600 µl DMEM without FCS and incubated for 15 min at RT. The DNA-fugene complex was then added to cells in 5 ml freshly replaced medium. The medium was again replaced the following day. 2 d later, the supernatant of HEK293T containing viruses was collected and filtered through a 0.45-µm membrane filter. Viral containing supernatant (1-ml or 2-ml volumes) were added to HeLa cells in presence of fresh DMEM with 10% FCS to a total volume of 3 ml in a 6-well plate, and polybrene (H9268; Sigma-Aldrich) was added at a final concentration of 5 µg/ml. The medium was exchanged with fresh DMEM with 10% FCS the next day and used for further experiments.
Sample preparation for phosphoproteomics
For each biological condition, two 6-well plates of HeLa CCL-2 cells were grown to confluency. Per condition, three independent biological replicates were performed. Cells were infected for 30 or 45 min as described above. The plates were put on ice and washed twice with ice-cold PBS. Samples were then collected in 8 M urea (AppliChem), 0.1 M ammoniumbicarbonate (Sigma-Aldrich), 0.1% RapiGest (Waters), and PhosSTOP (Roche). The samples were briefly vortexed, sonicated at 4°C (Hielscher), shaken for 5 min on a thermomixer, and centrifuged for 20 min at 4°C and 169,000 g. Supernatants were collected and stored at −80°C for further processing. BCA protein assay (Pierce) was used to measure protein concentration.
Phosphopeptide enrichment
Disulfide bonds were reduced with tris(2-carboxyethyl)phosphine at a final concentration of 10 mM at 37°C for 1 h. Free thiols were alkylated with 20 mM iodoacetamide (Sigma-Aldrich) at RT for 30 min in the dark. The excess of iodoacetamide was quenched with N-acetyl cysteine at a final concentration of 25 mM for 10 min at room temperature. Lys-C endopeptidase (Wako) was added to a final enzyme/protein ratio of 1:200 (wt/wt) and incubated for 4h at 37°C. The solution was subsequently diluted with 0.1 M ammoniumbicarbonate (Sigma-Aldrich) to a final concentration below 2 M urea and digested overnight at 37°C with sequencing-grade modified trypsin (Promega) at a protein/enzyme ratio of 50:1. Peptides were desalted on a C18 Sep-Pak cartridge (Waters) and dried under vacuum. Phosphopeptides were isolated from 2 mg of total peptide mass with TiO2 as described previously (Bensimon et al., 2010). In brief, dried peptides were dissolved in an 80% acetonitrile and 2.5% trifluoroacetic acid (TFA) solution saturated with phthalic acid. Peptides were added to the same amount of equilibrated TiO2 (5-µm bead size; GL Sciences) in a blocked Mobicol spin column (MoBiTec) that was incubated for 30 min with end-over-end rotation. The column was washed twice with the saturated phthalic acid solution, twice with 80% acetonitrile and 0.1% TFA, and finally twice with 0.1% TFA. The peptides were eluted with a 0.3 M NH4OH solution. The pH of the eluates was adjusted to be below 2.5 with 5% TFA solution and 2 M HCl. Phosphopeptides were again desalted with microspin C18 cartridges (Harvard Apparatus).
LC-MS/MS analysis
Chromatographic separation of peptides was performed using an EASY nano-LC system (Thermo Fisher Scientific), equipped with a heated RP-HPLC column (75 µm × 45 cm) packed in-house with 1.9 µm C18 resin (Reprosil-AQ Pur; Dr. Maisch). Aliquots of 1 µg total phosphopeptide samples were analyzed per LC-MS/MS run using a linear gradient ranging from 98% solvent A (0.15% formic acid) and 2% solvent B (98% acetonitrile, 2% water, and 0.15% formic acid) to 30% solvent B over 120 min at a flow rate of 200 nl/min. Mass spectrometry analysis was performed on a dual pressure LTQ-Orbitrap mass spectrometer equipped with a nanoelectrospray ion source (both Thermo Fisher Scientific). Each MS1 scan (acquired with the Orbitrap) was followed by collision-induced dissociation (acquired in the LTQ) of the 20 most abundant precursor ions with dynamic exclusion for 30 s. For phosphopeptide analysis, the 10 most abundant precursor ions were subjected to collision-induced dissociation with enabled multistage activation. Total cycle time was ∼2 s. For MS1, 106 ions were accumulated in the Orbitrap cell over a maximum time of 300 ms and scanned at a resolution of 60,000 FWHM (at 400 m/z). MS2 scans were acquired using the normal scan mode, a target setting of 104 ions, and accumulation time of 25 ms. Singly charged ions and ions with unassigned charge state were excluded from triggering MS2 events. The normalized collision energy was set to 32%, and one microscan was acquired for each spectrum.
Label-free quantification and database searching
The acquired raw files were imported into the Progenesis software tool (Nonlinear Dynamics, Version 4.0) for label-free quantification using the default parameters. MS2 spectra were exported directly from Progenesis in mgf format and searched using the MASCOT algorithm (version 2.4; Matrix Science) against a decoy database (Perkins et al., 1999) containing normal and reverse sequences of the predicted SwissProt entries of Homo sapiens and commonly observed contaminants (in total 41,250 sequences) generated using the SequenceReverser tool from the MaxQuant software (version 1.0.13.13). To identify proteins originating from Y. enterocolitica, nonphosphopeptide-enriched samples were searched against the same database above including predicted SwissProt entries of Y. enterocolitica. The precursor ion tolerance was set to 10 ppm and fragment ion tolerance was set to 0.6 D. The search criteria were set as follows: full tryptic specificity was required (cleavage after lysine or arginine residues unless followed by proline), two missed cleavages were allowed, and carbamidomethylation (C) was set as fixed modification and phosphorylation (S,T,Y) or oxidation (M) as a variable modification for TiO2 enriched or not enriched samples, respectively. Finally, the database search results were exported as an xml-file and imported back to the Progenesis software for MS1 feature assignment. For phosphopeptide quantification, a csv file containing the MS1 peak abundances of all detected features was exported and for not enriched samples, a csv file containing all protein measurements based on the summed feature intensities of all identified peptides per protein was created. Importantly, the Progenesis software was set in a way that proteins identified by similar sets of peptides are grouped together and that only nonconflicting peptides with specific sequences for single proteins in the database were used for protein quantification. Both files were further processed using the in-house developed SafeQuant v1.0 R script (see online supplemental material files; also available at https://github.com/eahrne/SafeQuant/). In brief, the software sets the identification level false discovery rate to 1% (based on the number of decoy protein sequence database hits) and normalizes the identified MS1 peak abundances (extracted ion chromatogram [XIC]) across all samples, i.e., the summed XIC of all confidently identified peptide features is scaled to be equal for all LC-MS runs. Next, all quantified phosphopeptides/proteins are assigned an abundance ratio for each time point, based on the median XIC per time point. The statistical significance of each ratio is given by its q-value (false discovery rate adjusted p-values), obtained by calculating modified t-statistic p-values (Smyth, 2004) and adjusting for multiple testing (Ting et al., 2009). The location of the phosphorylated residues was automatically assigned by MASCOT (score >10). All data are shown in Table S4.
Online supplemental material
Fig. S1 describes the toolbox of T3S-based protein delivery and provides vector maps. Fig. S2 illustrates the protocols of protein delivery and in vitro secretion and provides the quantification of kinetics and dose–response of the T3S-based protein delivery. Fig. S3 shows examples of target cells and quantifications of T3S-based protein delivery. Fig. S4 shows the T3S-dependent delivery of SopE into eukaryotic cells and a control experiment for digitonin-based cell lysis. Fig. S5 shows various eukaryotic proteins secreted by T3S. Table S1 lists the known or predicted CASP3 substrates in the tBID phosphoproteome. Table S2 contains a list of primers used in the study and Table S3 contains a list of strains used in this study. Table S4 describes the changes in protein phosphorylation revealed by phosphoproteomics, and Table S5 shows the ontology analysis of the tBID phosphoproteome. All source codes for computational methods are available online, including CellProfiler pipelines and the SafeQuant algorithm. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201502074/DC1.
Supplementary Material
Acknowledgments
We thank Prof. C. Dehio for human umbilical vein cells. We thank Prof. O. Pertz for the pVSV, pMDL, pRev, and pMDK124 plasmids. We thank Prof. F.A. Barr for providing the EGFP-Rab2A template. We thank the Biozentrum Imaging Core facility for technical assistance.
This work was supported by the Swiss National Science Foundation (grant 310030_138377 to C. Arrieumerlou) and InfectX. S.J. Ittig and C.A. Kasper were partly founded by the commission for technology and innovation (grant 16084.1). S.J. Ittig was further supported by the Dr. Hans Altschüler foundation and the Dr. Arnold U. and Susanne Huggenberger-Bischoff foundation.
S.J. Ittig and C. Arrieumerlou submitted a patent application on the method described here.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- asd
- aspartate-β-semialdehyde dehydrogenase
- BHI
- brain heart infusion
- BID
- BH3 interacting-domain death agonist
- DUBs
- deubiquitinating enzymes
- GAP
- GTPase activating protein
- H2B-GFP
- histone 2B-GFP
- LC-MS/MS
- liquid chromatography tandem mass spectrometry
- mDAP
- meso-2,6-diaminopimelic acid
- NLS
- nuclear localization signal
- p.i.
- postinfection
- T3S
- type 3 secretion
- tBID
- truncated BID
- TEV
- tobacco etch virus
- TFA
- trifluoroacetic acid
- XIC
- extracted ion chromatogram
References
- Ayyash M., Tamimi H., and Ashhab Y.. 2012. Developing a powerful in silico tool for the discovery of novel caspase-3 substrates: a preliminary screening of the human proteome. BMC Bioinformatics. 13:14 10.1186/1471-2105-13-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard E.L., van der Sar A.M., Ellett F., Lieschke G.J., Spaink H.P., and Meijer A.H.. 2012. Infection of zebrafish embryos with intracellular bacterial pathogens. J. Vis. Exp. 61:3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensimon A., Schmidt A., Ziv Y., Elkon R., Wang S.Y., Chen D.J., Aebersold R., and Shiloh Y.. 2010. ATM-dependent and -independent dynamics of the nuclear phosphoproteome after DNA damage. Sci. Signal. 3:rs3 10.1126/scisignal.2001034 [DOI] [PubMed] [Google Scholar]
- Bichsel C., Neeld D.K., Hamazaki T., Wu D., Chang L.J., Yang L., Terada N., and Jin S.. 2011. Bacterial delivery of nuclear proteins into pluripotent and differentiated cells. PLoS ONE. 6:e16465 10.1371/journal.pone.0016465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichsel C., Neeld D., Hamazaki T., Chang L.J., Yang L.J., Terada N., and Jin S.. 2013. Direct reprogramming of fibroblasts to myocytes via bacterial injection of MyoD protein. Cell Reprogram. 15:117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Toribio A., Muyldermans S., Frankel G., and Fernández L.A.. 2010. Direct injection of functional single-domain antibodies from E. coli into human cells. PLoS ONE. 5:e15227 10.1371/journal.pone.0015227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodeus M., Sory M.-P., Fang J.C., Janssens M., Delferriere N., Cornelis G.R., Wauters G., and Burtonboy G.. 1990. Production of rat hybridomas directed against Yersinia enterocolitica. In Rat Hybridoma and Rat Monoclonal Antibodies. Bazin H., editor. CRC Press Inc., Boca Raton, FL: 335–338. [Google Scholar]
- Boyd A.P., Grosdent N., Tötemeyer S., Geuijen C., Bleves S., Iriarte M., Lambermont I., Octave J.N., and Cornelis G.R.. 2000a Yersinia enterocolitica can deliver Yop proteins into a wide range of cell types: development of a delivery system for heterologous proteins. Eur. J. Cell Biol. 79:659–671. 10.1078/0171-9335-00098 [DOI] [PubMed] [Google Scholar]
- Boyd A.P., Lambermont I., and Cornelis G.R.. 2000b Competition between the Yops of Yersinia enterocolitica for delivery into eukaryotic cells: role of the SycE chaperone binding domain of YopE. J. Bacteriol. 182:4811–4821. 10.1128/JB.182.17.4811-4821.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., et al. . 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington J.C., and Dougherty W.G.. 1988. A viral cleavage site cassette: identification of amino acid sequences required for tobacco etch virus polyprotein processing. Proc. Natl. Acad. Sci. USA. 85:3391–3395. 10.1073/pnas.85.10.3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussinus E., Kanca O., and Affolter M.. 2011. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol. 19:117–121. 10.1038/nsmb.2180 [DOI] [PubMed] [Google Scholar]
- Chamekh M., Phalipon A., Quertainmont R., Salmon I., Sansonetti P., and Allaoui A.. 2008. Delivery of biologically active anti-inflammatory cytokines IL-10 and IL-1ra in vivo by the Shigella type III secretion apparatus. J. Immunol. 180:4292–4298. 10.4049/jimmunol.180.6.4292 [DOI] [PubMed] [Google Scholar]
- Chaux P., Luiten R., Demotte N., Vantomme V., Stroobant V., Traversari C., Russo V., Schultz E., Cornelis G.R., Boon T., and van der Bruggen P.. 1999. Identification of five MAGE-A1 epitopes recognized by cytolytic T lymphocytes obtained by in vitro stimulation with dendritic cells transduced with MAGE-A1. J. Immunol. 163:2928–2936. [PubMed] [Google Scholar]
- Chen L.M., Briones G., Donis R.O., and Galán J.E.. 2006. Optimization of the delivery of heterologous proteins by the Salmonella enterica serovar Typhimurium type III secretion system for vaccine development. Infect. Immun. 74:5826–5833. 10.1128/IAI.00375-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens M.J., Bushell M., Jeffrey I.W., Pain V.M., and Morley S.J.. 2000. Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells. Cell Death Differ. 7:603–615. 10.1038/sj.cdd.4400695 [DOI] [PubMed] [Google Scholar]
- Cornelis G.R., and Wolf-Watz H.. 1997. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol. Microbiol. 23:861–867. 10.1046/j.1365-2958.1997.2731623.x [DOI] [PubMed] [Google Scholar]
- Cornelis G., Vanootegem J.C., and Sluiters C.. 1987. Transcription of the yop regulon from Y. enterocolitica requires trans acting pYV and chromosomal genes. Microb. Pathog. 2:367–379. 10.1016/0882-4010(87)90078-7 [DOI] [PubMed] [Google Scholar]
- Dehio C., Meyer M., Berger J., Schwarz H., and Lanz C.. 1997. Interaction of Bartonella henselae with endothelial cells results in bacterial aggregation on the cell surface and the subsequent engulfment and internalisation of the bacterial aggregate by a unique structure, the invasome. J. Cell Sci. 110:2141–2154. [DOI] [PubMed] [Google Scholar]
- Dix M.M., Simon G.M., Wang C., Okerberg E., Patricelli M.P., and Cravatt B.F.. 2012. Functional interplay between caspase cleavage and phosphorylation sculpts the apoptotic proteome. Cell. 150:426–440. 10.1016/j.cell.2012.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore S. 2007. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 35:495–516. 10.1080/01926230701320337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M.F., Müller S., Wüest E., and Cornelis G.R.. 2002. SycE allows secretion of YopE-DHFR hybrids by the Yersinia enterocolitica type III Ysc system. Mol. Microbiol. 46:1183–1197. 10.1046/j.1365-2958.2002.03241.x [DOI] [PubMed] [Google Scholar]
- Freund G., Sibler A.P., Desplancq D., Oulad-Abdelghani M., Vigneron M., Gannon J., Van Regenmortel M.H., and Weiss E.. 2013. Targeting endogenous nuclear antigens by electrotransfer of monoclonal antibodies in living cells. MAbs. 5:518–522. 10.4161/mabs.25084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J.T., Ferracci F., Jackson M.W., Joseph S.S., Pattis I., Plano L.R., Fischer W., and Plano G.V.. 2006. Measurement of effector protein injection by type III and type IV secretion systems by using a 13-residue phosphorylatable glycogen synthase kinase tag. Infect. Immun. 74:5645–5657. 10.1128/IAI.00690-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier A., and Finlay B.B.. 2003. Translocated intimin receptor and its chaperone interact with ATPase of the type III secretion apparatus of enteropathogenic Escherichia coli. J. Bacteriol. 185:6747–6755. 10.1128/JB.185.23.6747-6755.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosdent N., Maridonneau-Parini I., Sory M.P., and Cornelis G.R.. 2002. Role of Yops and adhesins in resistance of Yersinia enterocolitica to phagocytosis. Infect. Immun. 70:4165–4176. 10.1128/IAI.70.8.4165-4176.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håkansson S., Schesser K., Persson C., Galyov E.E., Rosqvist R., Homblé F., and Wolf-Watz H.. 1996. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 15:5812–5823. [PMC free article] [PubMed] [Google Scholar]
- Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E.B., Bendahman N., and Hamers R.. 1993. Naturally occurring antibodies devoid of light chains. Nature. 363:446–448. 10.1038/363446a0 [DOI] [PubMed] [Google Scholar]
- Hardt W.D., Chen L.M., Schuebel K.E., Bustelo X.R., and Galán J.E.. 1998. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 93:815–826. 10.1016/S0092-8674(00)81442-7 [DOI] [PubMed] [Google Scholar]
- Henrichs T., Mikhaleva N., Conz C., Deuerling E., Boyd D., Zelazny A., Bibi E., Ban N., and Ehrmann M.. 2005. Target-directed proteolysis at the ribosome. Proc. Natl. Acad. Sci. USA. 102:4246–4251. 10.1073/pnas.0408520102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herwig L., Blum Y., Krudewig A., Ellertsdottir E., Lenard A., Belting H.G., and Affolter M.. 2011. Distinct cellular mechanisms of blood vessel fusion in the zebrafish embryo. Curr. Biol. 21:1942–1948. 10.1016/j.cub.2011.10.016 [DOI] [PubMed] [Google Scholar]
- Hoang T.T., Williams S., Schweizer H.P., and Lam J.S.. 1997. Molecular genetic analysis of the region containing the essential Pseudomonas aeruginosa asd gene encoding aspartate-beta-semialdehyde dehydrogenase. Microbiology. 143:899–907. 10.1099/00221287-143-3-899 [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Stephens R., Baseler M.W., Lane H.C., and Lempicki R.A.. 2008. DAVID gene ID conversion tool. Bioinformation. 2:428–430. 10.6026/97320630002428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomata K., Ohno A., Tochio H., Isogai S., Tenno T., Nakase I., Takeuchi T., Futaki S., Ito Y., Hiroaki H., and Shirakawa M.. 2009. High-resolution multi-dimensional NMR spectroscopy of proteins in human cells. Nature. 458:106–109. 10.1038/nature07839 [DOI] [PubMed] [Google Scholar]
- Iriarte M., and Cornelis G.R.. 1998. YopT, a new Yersinia Yop effector protein, affects the cytoskeleton of host cells. Mol. Microbiol. 29:915–929. 10.1046/j.1365-2958.1998.00992.x [DOI] [PubMed] [Google Scholar]
- Isaksson E.L., Aili M., Fahlgren A., Carlsson S.E., Rosqvist R., and Wolf-Watz H.. 2009. The membrane localization domain is required for intracellular localization and autoregulation of YopE in Yersinia pseudotuberculosis. Infect. Immun. 77:4740–4749. 10.1128/IAI.00333-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobi C.A., Roggenkamp A., Rakin A., Zumbihl R., Leitritz L., and Heesemann J.. 1998. In vitro and in vivo expression studies of yopE from Yersinia enterocolitica using the gfp reporter gene. Mol. Microbiol. 30:865–882. 10.1046/j.1365-2958.1998.01128.x [DOI] [PubMed] [Google Scholar]
- Jette C.A., Flanagan A.M., Ryan J., Pyati U.J., Carbonneau S., Stewart R.A., Langenau D.M., Look A.T., and Letai A.. 2008. BIM and other BCL-2 family proteins exhibit cross-species conservation of function between zebrafish and mammals. Cell Death Differ. 15:1063–1072. 10.1038/cdd.2008.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapust R.B., Tözsér J., Fox J.D., Anderson D.E., Cherry S., Copeland T.D., and Waugh D.S.. 2001. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14:993–1000. 10.1093/protein/14.12.993 [DOI] [PubMed] [Google Scholar]
- Kudryashev M., Stenta M., Schmelz S., Amstutz M., Wiesand U., Castaño-Díez D., Degiacomi M.T., Münnich S., Bleck C.K., Kowal J., et al. . 2013. In situ structural analysis of the Yersinia enterocolitica injectisome. eLife. 2:e00792 10.7554/eLife.00792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee V.T., Anderson D.M., and Schneewind O.. 1998. Targeting of Yersinia Yop proteins into the cytosol of HeLa cells: one-step translocation of YopE across bacterial and eukaryotic membranes is dependent on SycE chaperone. Mol. Microbiol. 28:593–601. 10.1046/j.1365-2958.1998.00822.x [DOI] [PubMed] [Google Scholar]
- Li H., Zhu H., Xu C.J., and Yuan J.. 1998. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 94:491–501. 10.1016/S0092-8674(00)81590-1 [DOI] [PubMed] [Google Scholar]
- Li H., Xu H., Zhou Y., Zhang J., Long C., Li S., Chen S., Zhou J.M., and Shao F.. 2007. The phosphothreonine lyase activity of a bacterial type III effector family. Science. 315:1000–1003. 10.1126/science.1138960 [DOI] [PubMed] [Google Scholar]
- Liu Y.H., Ho C.C., Cheng C.C., Chao W.T., Pei R.J., Hsu Y.H., and Lai Y.S.. 2011. Cytokeratin 18-mediated disorganization of intermediate filaments is induced by degradation of plectin in human liver cells. Biochem. Biophys. Res. Commun. 407:575–580. 10.1016/j.bbrc.2011.03.066 [DOI] [PubMed] [Google Scholar]
- Loison F., Nizard P., Sourisseau T., Le Goff P., Debure L., Le Drean Y., and Michel D.. 2005. A ubiquitin-based assay for the cytosolic uptake of protein transduction domains. Mol. Ther. 11:205–214. 10.1016/j.ymthe.2004.10.010 [DOI] [PubMed] [Google Scholar]
- Michiels T., Wattiau P., Brasseur R., Ruysschaert J.M., and Cornelis G.. 1990. Secretion of Yop proteins by Yersiniae. Infect. Immun. 58:2840–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota L.J., and Cornelis G.R.. 2005. The bacterial injection kit: type III secretion systems. Ann. Med. 37:234–249. 10.1080/07853890510037329 [DOI] [PubMed] [Google Scholar]
- Nagaraj N., Wisniewski J.R., Geiger T., Cox J., Kircher M., Kelso J., Pääbo S., and Mann M.. 2011. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7:548 10.1038/msb.2011.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris F.A., Wilson M.P., Wallis T.S., Galyov E.E., and Majerus P.W.. 1998. SopB, a protein required for virulence of Salmonella dublin, is an inositol phosphate phosphatase. Proc. Natl. Acad. Sci. USA. 95:14057–14059. 10.1073/pnas.95.24.14057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D.N., Pappin D.J., Creasy D.M., and Cottrell J.S.. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 20:3551–3567. [DOI] [PubMed] [Google Scholar]
- Pulliainen A.T., Pieles K., Brand C.S., Hauert B., Böhm A., Quebatte M., Wepf A., Gstaiger M., Aebersold R., Dessauer C.W., and Dehio C.. 2012. Bacterial effector binds host cell adenylyl cyclase to potentiate Gαs-dependent cAMP production. Proc. Natl. Acad. Sci. USA. 109:9581–9586. 10.1073/pnas.1117651109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothmeier E., Pfaffinger G., Hoffmann C., Harrison C.F., Grabmayr H., Repnik U., Hannemann M., Wölke S., Bausch A., Griffiths G., et al. . 2013. Activation of Ran GTPase by a Legionella effector promotes microtubule polymerization, pathogen vacuole motility and infection. PLoS Pathog. 9:e1003598 10.1371/journal.ppat.1003598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüssmann H., Shams H., Poblete F., Fu Y., Galán J.E., and Donis R.O.. 1998. Delivery of epitopes by the Salmonella type III secretion system for vaccine development. Science. 281:565–568. 10.1126/science.281.5376.565 [DOI] [PubMed] [Google Scholar]
- Rüssmann H., Gerdemann U., Igwe E.I., Panthel K., Heesemann J., Garbom S., Wolf-Watz H., and Geginat G.. 2003. Attenuated Yersinia pseudotuberculosis carrier vaccine for simultaneous antigen-specific CD4 and CD8 T-cell induction. Infect. Immun. 71:3463–3472. 10.1128/IAI.71.6.3463-3472.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saerens D., Pellis M., Loris R., Pardon E., Dumoulin M., Matagne A., Wyns L., Muyldermans S., and Conrath K.. 2005. Identification of a universal VHH framework to graft non-canonical antigen-binding loops of camel single-domain antibodies. J. Mol. Biol. 352:597–607. 10.1016/j.jmb.2005.07.038 [DOI] [PubMed] [Google Scholar]
- Schlumberger M.C., Müller A.J., Ehrbar K., Winnen B., Duss I., Stecher B., and Hardt W.D.. 2005. Real-time imaging of type III secretion: Salmonella SipA injection into host cells. Proc. Natl. Acad. Sci. USA. 102:12548–12553. 10.1073/pnas.0503407102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz C., Ahrné E., Kasper C.A., Tschon T., Sorg I., Dreier R.F., Schmidt A., and Arrieumerlou C.. 2013. Systems-level overview of host protein phosphorylation during Shigella flexneri infection revealed by phosphoproteomics. Mol. Cell. Proteomics. 12:2952–2968. 10.1074/mcp.M113.029918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte R., Grassl G.A., Preger S., Fessele S., Jacobi C.A., Schaller M., Nelson P.J., and Autenrieth I.B.. 2000. Yersinia enterocolitica invasin protein triggers IL-8 production in epithelial cells via activation of Rel p65-p65 homodimers. FASEB J. 14:1471–1484. 10.1096/fj.14.11.1471 [DOI] [PubMed] [Google Scholar]
- Sharma S., Sharma S., Hirabuchi A., Yoshida K., Fujisaki K., Ito A., Uemura A., Terauchi R., Kamoun S., Sohn K.H., et al. . 2013. Deployment of the Burkholderia glumae type III secretion system as an efficient tool for translocating pathogen effectors to monocot cells. Plant J. 74:701–712. 10.1111/tpj.12148 [DOI] [PubMed] [Google Scholar]
- Short B., Preisinger C., Körner R., Kopajtich R., Byron O., and Barr F.A.. 2001. A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J. Cell Biol. 155:877–883. 10.1083/jcb.200108079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silljé H.H., Nagel S., Körner R., and Nigg E.A.. 2006. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 16:731–742. 10.1016/j.cub.2006.02.070 [DOI] [PubMed] [Google Scholar]
- Smyth G.K. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:e3. [DOI] [PubMed] [Google Scholar]
- Sory M.P., and Cornelis G.R.. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14:583–594. 10.1111/j.1365-2958.1994.tb02191.x [DOI] [PubMed] [Google Scholar]
- Sory M.P., Kaniga K., Goldenberg S., and Cornelis G.R.. 1992. Expression of the eukaryotic Trypanosoma cruzi CRA gene in Yersinia enterocolitica and induction of an immune response against CRA in mice. Infect. Immun. 60:3830–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sory M.P., Boland A., Lambermont I., and Cornelis G.R.. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. USA. 92:11998–12002. 10.1073/pnas.92.26.11998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins C.E., and Galán J.E.. 2001. Structural mimicry in bacterial virulence. Nature. 412:701–705. 10.1038/35089000 [DOI] [PubMed] [Google Scholar]
- Ting L., Cowley M.J., Hoon S.L., Guilhaus M., Raftery M.J., and Cavicchioli R.. 2009. Normalization and statistical analysis of quantitative proteomics data generated by metabolic labeling. Mol. Cell. Proteomics. 8:2227–2242. 10.1074/mcp.M800462-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turowec J.P., Zukowski S.A., Knight J.D., Smalley D.M., Graves L.M., Johnson G.L., Li S.S., Lajoie G.A., and Litchfield D.W.. 2014. An unbiased proteomic screen reveals caspase cleavage is positively and negatively regulated by substrate phosphorylation. Mol. Cell. Proteomics. 13:1184–1197. 10.1074/mcp.M113.037374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uliczka F., Kornprobst T., Eitel J., Schneider D., and Dersch P.. 2009. Cell invasion of Yersinia pseudotuberculosis by invasin and YadA requires protein kinase C, phospholipase C-gamma1 and Akt kinase. Cell. Microbiol. 11:1782–1801. 10.1111/j.1462-5822.2009.01371.x [DOI] [PubMed] [Google Scholar]
- Van Damme M., Sory M.P., Biot T., Vaerman J.P., and Cornelis G.R.. 1992. Oral immunization against cholera toxin with a live Yersinia enterocolitica carrier in mice. Gastroenterology. 103:520–531. [DOI] [PubMed] [Google Scholar]
- Van Engelenburg S.B., and Palmer A.E.. 2010. Imaging type-III secretion reveals dynamics and spatial segregation of Salmonella effectors. Nat. Methods. 7:325–330. 10.1038/nmeth.1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Pawel-Rammingen U., Telepnev M.V., Schmidt G., Aktories K., Wolf-Watz H., and Rosqvist R.. 2000. GAP activity of the Yersinia YopE cytotoxin specifically targets the Rho pathway: a mechanism for disruption of actin microfilament structure. Mol. Microbiol. 36:737–748. 10.1046/j.1365-2958.2000.01898.x [DOI] [PubMed] [Google Scholar]
- Wang K.K., Posmantur R., Nath R., McGinnis K., Whitton M., Talanian R.V., Glantz S.B., and Morrow J.S.. 1998. Simultaneous degradation of alphaII- and betaII-spectrin by caspase 3 (CPP32) in apoptotic cells. J. Biol. Chem. 273:22490–22497. 10.1074/jbc.273.35.22490 [DOI] [PubMed] [Google Scholar]
- Wattiau P., and Cornelis G.R.. 1993. SycE, a chaperone-like protein of Yersinia enterocolitica involved in Ohe secretion of YopE. Mol. Microbiol. 8:123–131. 10.1111/j.1365-2958.1993.tb01209.x [DOI] [PubMed] [Google Scholar]
- Weber W., Fux C., Daoud-el Baba M., Keller B., Weber C.C., Kramer B.P., Heinzen C., Aubel D., Bailey J.E., and Fussenegger M.. 2002. Macrolide-based transgene control in mammalian cells and mice. Nat. Biotechnol. 20:901–907. 10.1038/nbt731 [DOI] [PubMed] [Google Scholar]
- Wölke S., Ackermann N., and Heesemann J.. 2011. The Yersinia enterocolitica type 3 secretion system (T3SS) as toolbox for studying the cell biological effects of bacterial Rho GTPase modulating T3SS effector proteins. Cell. Microbiol. 13:1339–1357. 10.1111/j.1462-5822.2011.01623.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.