In vertebrate embryos, the histone demethylase KDM4B affects otic placode proliferation, intercellular adhesion, and invagination by directly regulating Dlx3 expression.
Abstract
In vertebrates, the inner ear arises from the otic placode, a thickened swathe of ectoderm that invaginates to form the otic vesicle. We report that histone demethylase KDM4B is dynamically expressed during early stages of chick inner ear formation. A loss of KDM4B results in defective invagination and striking morphological changes in the otic epithelium, characterized by abnormal localization of adhesion and cytoskeletal molecules and reduced expression of several inner ear markers, including Dlx3. In vivo chromatin immunoprecipitation reveals direct and dynamic occupancy of KDM4B and its target, H3K9me3, at regulatory regions of the Dlx3 locus. Accordingly, coelectroporations of DLX3 or KDM4B encoding constructs, but not a catalytically dead mutant of KDM4B, rescue the ear invagination phenotype caused by KDM4B knockdown. Moreover, a loss of DLX3 phenocopies a loss of KDM4B. Collectively, our findings suggest that KDM4B play a critical role during inner ear invagination via modulating histone methylation of the direct target Dlx3.
Introduction
The vertebrate inner ear is a complex structure comprised of the auditory (cochlear) and balance (vestibular) systems, which are responsible for perception of sound and gravitational/angular acceleration (Driver and Kelley, 2009), respectively. Arising from a simple epithelium, the otic placode, the inner ear undergoes extensive morphogenesis and differentiation, ultimately giving rise to many different cell types that are arranged to form an organ of exquisite complexity (Christophorou et al., 2010; Chen and Streit, 2013). Comprised of cells as diverse as mechanosensory hair cells, sensory neurons, and support cells, defects in inner ear development lead to hearing loss and balance disorders, which are among the most common of live birth defects (Stevens et al., 2013; Van Kerschaver et al., 2013; Emmett and West, 2014).
The otic placode begins as a thickening of the ectoderm adjacent to the developing hindbrain. As development proceeds, the placode invaginates to form the otic pit that subsequently develops into a cup that pinches off the ectoderm and closes dorsally to form the otic vesicle (otocyst). Invagination of placode cells to form the otocyst requires extensive cell shape changes, cell movements, and rearrangements that require remodeling of cell–matrix and cell–cell interactions among otic cells, as well as with the adjacent hindbrain (Gerchman et al., 1995; Brown et al., 1998; Moro-Balbás et al., 2000; Visconti and Hilfer, 2002). As development proceeds, the otic vesicle becomes regionalized, undergoes extensive proliferation, and finally differentiates into the vestibular and cochlear components of the inner ear (Barald and Kelley, 2004; Neves et al., 2013). In the chick embryo, formation of a detectable otic placode is first apparent next to rhombomeres 5 and 6 of the hindbrain by Hamburger and Hamilton stage 10, by which time otic specification is complete. Invagination begins at stage 12, resulting in the formation of an otic cup that closes to form the internalized otic vesicle by stages 18–20 (Alvarez and Navascués, 1990; Streit, 2002; Barald and Kelley, 2004; Sai and Ladher, 2008; Sai et al., 2014).
Understanding the molecular mechanisms that regulate the spatiotemporal expression of genes involved in otic placode development is critical for elucidating both normal development and defects that may lead to hearing impairment. Numerous transcription factors, including Pax2, Dlx3, Dlx5, Nkx5.1, SOHo1, and Sox10, have been implicated in various aspects of ear development, primarily with respect to their roles in otic specification and subsequent differentiation (Deitcher et al., 1994; Baker and Bronner-Fraser, 2001; Brown et al., 2005; Barembaum and Bronner-Fraser, 2010; Christophorou et al., 2010). For example, Pax2 expression initiates within the early otic placode at stage 8 and plays an essential role in its specification (Christophorou et al., 2010; Groves and Fekete, 2012), whereas Sox10 is expressed later and is likely involved in differentiation events (Cheng et al., 2000; Dutton et al., 2009). In contrast to the role of transcriptional regulation during ear development, virtually nothing is known about the role of epigenetic regulators therein.
In this study, we report that lysine-specific demethylase 4B, KDM4B (also known as JmjD2B), is expressed in the developing otic placode, consistent with a possible role as an epigenetic modifier of inner ear development. The KDM4/JumonjiD2 family of histone lysine demethylases removes H3K9me3/me2 and H3K36me3/me2 methylation marks (Couture et al., 2007; Tan et al., 2008) and thus activates or represses expression of target genes, respectively. Despite their likely importance as epigenetic regulators, surprisingly little is known about the developmental roles of the KDM4 family or other epigenetic modifiers in vivo (Strobl-Mazzulla et al., 2010). Here, we show that KDM4B is necessary for proper invagination of the otic placode, with its loss resulting in malformed placodes that fail to form vesicles of the proper size and shape. In vivo chromatin immunoprecipitation (ChIP) further demonstrates that the Dlx3 gene, previously hypothesized to play a role in placode invagination (Brown et al., 2005), is a direct target of KDM4B. These results reveal for the first time an epigenetic contribution to the control of inner ear invagination in vertebrate embryos via modulation of histone methylation.
Results
KDM4B is expressed during chick otic placode specification/invagination, and its loss of function results in abnormal maintenance of H3K9me3 marks
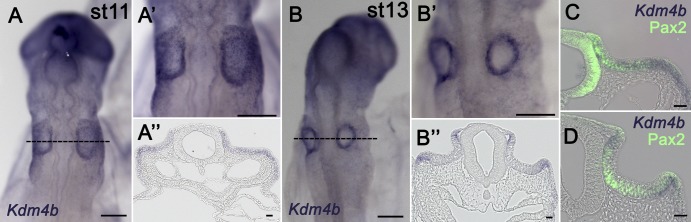
As we previously detected Kdm4b expression by quantitative PCR (qPCR) in the early embryos at the time of otic placode formation (Strobl-Mazzulla et al., 2010), we analyzed the spatial expression of Kdm4b by whole-mount in situ hybridization (ISH). We confirmed that Kdm4b is expressed in presumptive otic ectoderm by stage 9 (6 somite stage [ss]), persists through stage 9 (8ss), and by stage 12 (16ss) is confined to the border of the otocyst (Fig. 1 and Fig. S1). We compared the expression of Kdm4b transcripts by whole-mount ISH from stages 11 and 13 with the early inner ear marker PAX2. At stage 11 (Fig. 1 A), Kdm4b expression overlaps with PAX2 (Fig. 1 C). However, by stage 13, Kdm4b expression shifts to the edges of the invaginating otocyst (Fig. 1 B), only minimally overlapping with a few PAX2-positive cells (Fig. 1 D). These observations indicate that Kdm4b is dynamically expressed during the invagination phases of otic development, raising the possibility that histone demethylation via KDM4B may occur in a stage- and site-specific manner.
Figure 1.
Kdm4b is expressed during otic placode specification/invagination. (A and B) ISH of chick embryos at stages 11 (A) and 13 (B) with an antisense probe for Kdm4b transcripts. A’ and B’ are magnifications in the region of interest. A’’ and B’’ are transverse sections, as indicated by the dotted lines in A and B. (C and D) Immunostaining with the PAX2 antibody on transverse sections through Kdm4b in situ hybridized embryos reveal overlap of PAX2 and Kdm4b expression throughout the otic ectoderm at stage 11 (C) that diminishes by stage 13 (D), when kdm4b becomes restricted to the edges. Bars, 200 µm.
Next, we assessed the distribution of the KDM4B substrate H3K9me3 by immunohistochemistry at similar stages of otic development. At stage 11, H3K9me3-positive cells were detected throughout the otic placode (Fig. 2 A); however, by stage 13, the mark was largely confined to cells along the edges of the invaginating otic vesicle (Fig. 2 B). These results reveal in vivo dynamic spatial distribution of both the H3K9me3 epigenetic mark and Kdm4b transcripts in the otic and invaginating placode.
Figure 2.
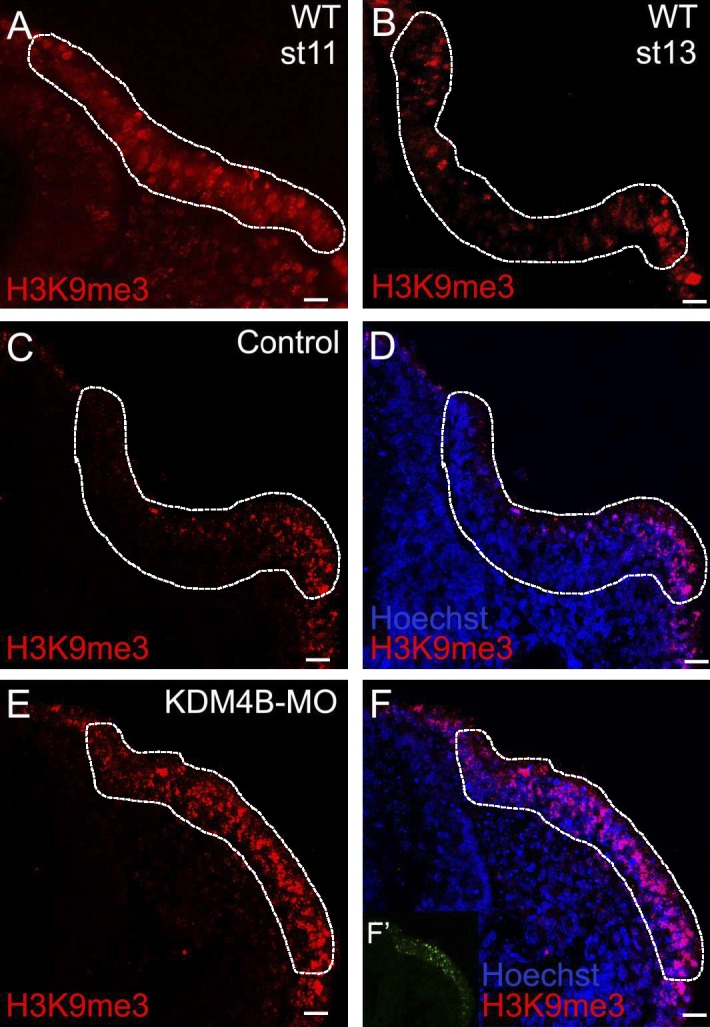
Kdm4b loss of function results in abnormal maintenance of H3K9me3 marks. (A and B) H3K9me3 immunostaining in wild-type (WT) embryos at stages 11 (A) and 13 (B), showing the distribution of the epigenetic mark within the otic placode. At stage 11, H3K9me3 is relatively uniform, whereas by stage 13, staining is restricted to the rim of the otic ectoderm. (C–F) Electroporation of KDM4B-MO (F’) followed by immunostaining for H3K9me3 at stage 13 (E and F) shows that the distribution of the epigenetic mark increases after KDM4B knockdown compared with the control side (C and D). Otic placode territories are indicated by dashed outlines. Bars, 20 µm.
The correlation of the spatial localization of Kdm4b at stages 11 and 13 (Fig. 1) with changes in H3K9me3 distribution raised the intriguing possibility that KDM4B may be responsible for removal of the triple methylation mark during otic placode invagination. To test this, we performed KDM4B loss-of-function experiments by electroporating a fluorescein-tagged antisense morpholino (MO) into the right otic ectoderm of stage 8 embryos, leaving the left side as an internal control. Immunostaining revealed the persistence of the H3K9m3 epigenetic mark at stage 13 throughout the entire placode region on the KDM4B-MO–treated side (Fig. 2, E and F) compared with the control side, where the mark was restricted to the edges of the otocyst (Fig. 2, C and D). These results show that KDM4B is necessary for removal of the repressive H3K9me3 methylation mark in the developing ear.
KDM4B loss of function results in ear deformities and reduction in marker gene expression
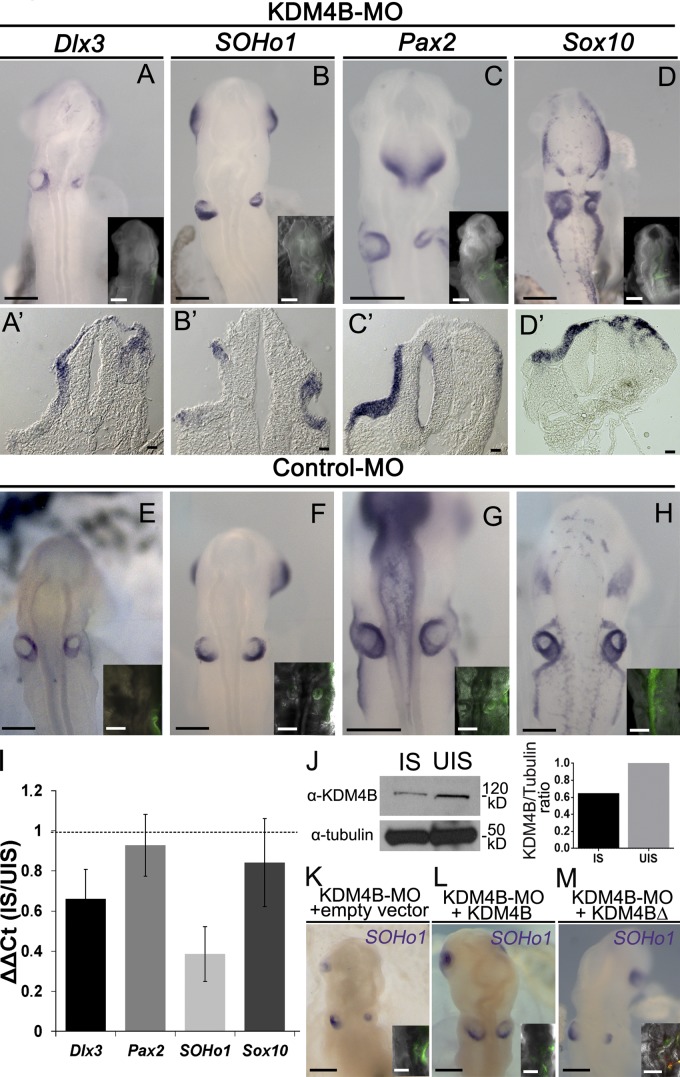
The dynamic expression of Kdm4b and its substrate is consistent with the possibility that KDM4B plays an important role in regulating otic vesicle development. To further test this, we performed loss-of-function experiments at stage 8 as described in the previous section and examined embryos at stage 13 for the expression of the ear markers Dlx3, SOHo1, Pax2, and Sox10 by ISH. Whole-mount and section analyses revealed that KDM4B-MO–electroporated otic vesicles were deformed in shape and exhibited a reduction in the expression domains of all inner ear markers examined when compared with the uninjected side of the same embryo (Fig. 3, A–D) or with control-MO–treated embryos (Fig. 3, E–H). Embryos were categorized according to the severity of the ear reduction phenotype as strong, mild, or none (Fig. S2, A–C). Some of the phenotypes observed were so severe that there was no sign of an otic vesicle, and SOHo1 expression was completely absent (Fig. S2 A). For all of the genes analyzed, we observed a significant increase in the severity of the phenotypes compared with control-MO–treated embryos (Fig. S2 D). To further analyze the reduction of the otic domain, we quantified the expression of Dlx3, Pax2, SOHo1, and Sox10 transcripts by qPCR from stage 13 otic vesicles that were electroporated with KDM4B-MO compared with uninjected vesicles. The results show a consistent reduction in the expression of SOHo1 and Dlx3 genes caused by the depletion of KDM4B, with less obvious changes for Sox10 and Pax2 (Fig. 3 I). It is interesting to note that on the dissected tissue used for this experiment, SOHo1 and Dlx3 are exclusively expressed in the otic placode cells, whereas Sox10 and Pax2 are also expressed in surrounding tissues such as neural crest and epibranchial placode cells, respectively. To demonstrate the specificity of the MO, we performed a Western blot analysis that confirmed a marked reduction of KDM4B protein after MO knockdown on the injected compared with uninjected sides of the same embryos (Fig. 3 J). In addition, we performed rescue experiments in which KDM4B-MO was coelectroporated with an expression vector containing a KDM4B coding sequence. The result showed a significant attenuation (P < 0.01) in the severity of the phenotypes compared with KDM4B-MO plus an empty vector (Fig. 3, K and L; and Fig. S2 E). However, when a vector containing a catalytically dead mutant version of KDM4B (KDM4BΔ) was coelectroporated with the KDM4B-MO, there was no rescue, demonstrating that catalytic activity of the enzyme plays a critical role (Fig. 3 M and Fig. S2 E).
Figure 3.
KDM4B loss of function results in ear deformities and reduction in marker gene expression. (A–H) Electroporation of KDM4B-MO into the right side of the embryo (green staining in inset) causes a dramatic reduction in the otic placode domain as indicated by the expression of the otic placode markers Soho1, Pax2, Sox10, and Dlx3 in embryos examined at stages 12/13 (A–D), compared with the left uninjected side and control-MO–treated embryos (E–H). Transverse sections (A’–D’) reveal disorganization of the otic tissue. (I) RT-qPCR analyses show a consistent reduction in Dlx3 and SOHo1 expression, but moderate for Sox10 and Pax2, on the KDM4B-MO–injected side (IS) compared with the uninjected side (UIS). Vertical bars represent the ΔΔCt ratio between the injected and uninjected side from the same pool of embryos. Vertical error bars on data points represent the standard error of mean obtained from three independent samples, including eight otic vesicles on each. Horizontal dotted line indicates the threshold ΔΔCt ratio (injected side/uninjected side = 1). (J) Western blot analysis, performed from 16 isolated otic vesicles, reveals a 36% reduction of KDM4B protein expression on the KDM4B-MO–injected side compared with the uninjected side. (K–M) Electroporation of KDM4B-MO (green fluorescence in inset), together with a vector containing the coding region of KDM4B, rescues the depletion of SOHo1 as assayed by ISH and the otic placode invagination (L). In contrast, coelectroporation of KDM4B-MO plus an empty vector (K) or catalytically dead mutant of Kdm4b (KDM4BΔ; M) failed to rescue the loss-of-function phenotype (see Fig. S2 for phenotypes quantitation). Bars: (A–H and K–M) 200 µm; (A’–D’) 20 µm.
Collectively, these results demonstrate that KDM4B demethylation activity is required for the correct expression of several ears markers, and its perturbation causes profound defects in inner ear formation, highlighting the importance of epigenetic regulation during this process.
Depletion of KDM4B protein transiently increases cell proliferation and cell death
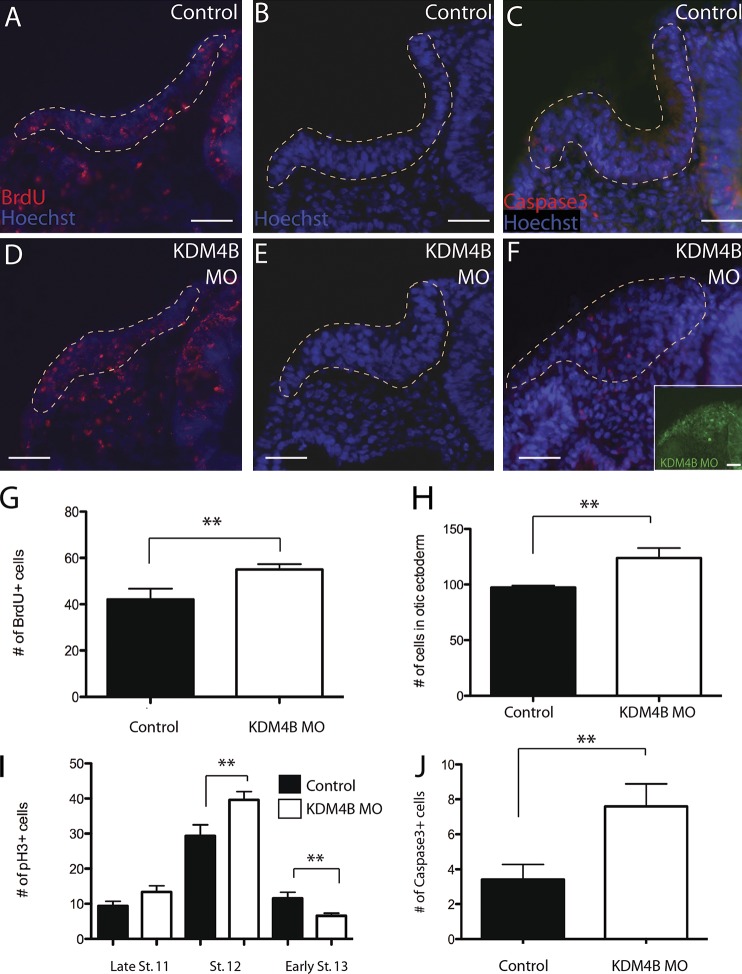
The striking change in otic vesicle morphology and ear marker gene expression after a reduction of KDM4B levels suggests that proper levels of this demethylase are required for the cellular mechanisms underlying inner ear growth and morphogenesis. To assess whether the otic phenotype of KDM4B-MO–treated embryos might result from a defect in cell proliferation, we performed BrdU incorporation assays to assess S-phase occupancy, as well as cell counts using Hoechst staining and phosphohistone H3 (pH3) immunostaining to analyze mitotic activity at different stages (Fig. 4). At stage 11, BrdU incorporation assays revealed a slight but significant increase in the number of S-phase cells (P = 0.0488), with control otic vesicle sections containing a mean of 42 ± 4.76 positive cells and KDM4B-MO–injected vesicles containing 55 ± 2.27 BrdU-positive cells (Fig. 4, A, D, and G). Total cell counts revealed a corresponding increase in the mean number of cells within the otic placode domain at stage 12, with control otic sections containing a mean of 97.4 ± 1.63 cells, whereas KDM4B-MO–injected vesicles had a mean of 123.8 ± 9.02 cells (P = 0.0205; Fig. 4, B, E, and H). In addition, mitotic activity was examined using pH3 immunohistochemistry at stages 11–13 (Fig. 4 I). There was no significant increase (P = 0.1059) in the number of pH3-positive cells on the MO-injected side at stage 11, by which time the otic placode is already specified. By stage 12, however, a significant increase in the number of pH3-positive cells was observed (P = 0.0322), with control otic sections containing a mean of 29.4 ± 3.14 dividing cells and KDM4B-MO otic sections containing a mean of 39.6 ± 2.38 dividing cells (Fig. 4 I). Interestingly, the total number of pH3-positive cells significantly declined at stage 13 (P = 0.0286), by which time the otic pit has formed, with KDM4B-MO vesicles exhibiting 6.6 ± 0.75, whereas control otic vesicles contained a mean of 11.6 ± 1.72. To determine if this reduction in cell proliferation is accompanied by cell death, we performed Caspase3 immunostaining in sections through the ear of KDM4B-MO–treated embryos (Fig. 4, C, F, and J). Accordingly, by stages 12 and 13, we observed a significant increase in cell death upon depletion of KDM4B (P = 0.0491).
Figure 4.
Depletion of KDM4B protein transiently increases cell proliferation and cell death. BrdU assays (A, D, and G), cell counts (B, E, and H), and pH3 immunostaining (I) performed on KDM4B-MO–treated embryos reveal a transient increase in cell proliferation at stage 12. At stages 12/13, there was an increase in cell death observed with Caspase3 immunostaining (C, F, and J) on the KDM4B-MO–treated side compared with the control side. Dashed lines outline the otic placodes in A–F. Error bars indicate ±SEM. **, P < 0.05 with Student’s t test. Bars, 40 µm.
Taken together with the morphological changes noted in Figs. 2 and 3, the results suggest that loss of KDM4B function causes a striking change in otic morphology accompanied by a transient increase in the number of placode cells in S phase and undergoing mitosis when the otic cup normally undergoes invagination, followed by cell death.
Knockdown of KDM4B affects the polarization of adhesion molecules on otic placode cells
During otic placode invagination, epithelial cells undergo shape changes mediated intracellularly by cytoskeletal changes and extracellularly by cell adhesion molecules. This leads to progressive invagination, ending with closure to form the otocyst (Gerchman et al., 1995; Moro-Balbás et al., 2000). Morphometric studies have shown that otic invagination is biphasic. Phase one involves a basal expansion to make the initial depression, and phase two comprises an apical constriction that guides the gradual hollowing of the otic vesicle (Alvarez and Navascués, 1990; Sai and Ladher, 2008; Sai et al., 2014). During apical constriction phases, epithelial cells express cadherin adhesion molecules, well known for their involvement in cell–cell adhesion events during embryonic development (Martin and Goldstein, 2014).
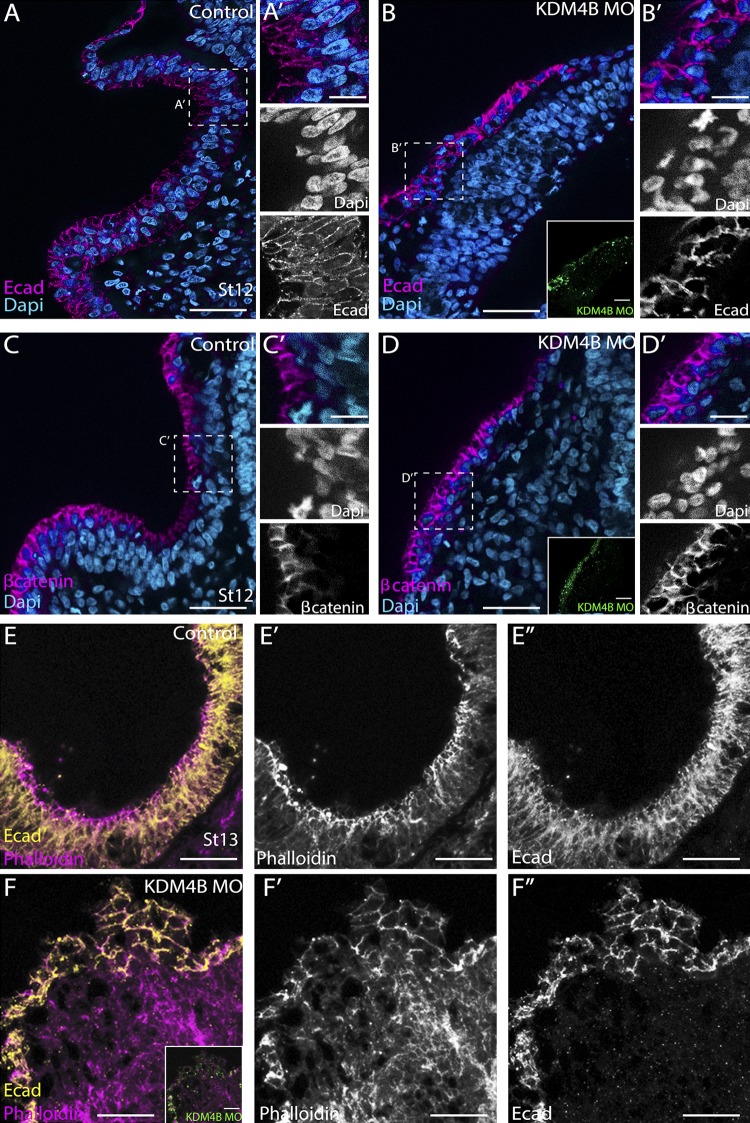
The profound changes in epithelial cell shape within the otic placode after KDM4B knockdown raised the intriguing possibility that this phenotype might be accompanied by changes in adhesive interactions between the cells. To examine this possibility, we analyzed the effects of KDM4B knockdown on the localization of E-cadherin (E-cad), the cadherin most prominently expressed throughout the embryonic ectoderm (Fig. 5). At stage 12, E-cad was enriched uniformly along cell membranes in control otic vesicles (Fig. 5 A), clearly revealing cell shapes that were oriented along the apical–basal axis. In stark contrast, KDM4B-MO–treated otic cells exhibited a nonuniform and disorganized distribution of E-cad along their membranes (Fig. 5 B), revealing perturbed cell shapes and suggesting that cadherin-mediated adhesion was disrupted. To further investigate this, we analyzed the distribution of β-catenin, a component of the cadherin-mediated adhesion complex. At stage 12, β-catenin was enriched along the apical surface of control otic vesicles, during which time the otic placode was invaginating (Fig. 5 C). In contrast, β-catenin localization in KDM4B-MO–treated otic tissue was not apically enriched, but instead, it was disorganized (Fig. 5 D), in agreement with observations of E-cad distribution.
Figure 5.
Depletion of KDM4B affects the polarization of adhesion molecules on otic placode cells. Subcellular localization of E-cad, β-catenin, and phalloidin staining by stages 12 (A–D) and 13 (E and F) at the onset of otic invagination on the control- (A, C, and E) and KDM4B-MO–treated sides (B, D, and F). The results show mislocalization of E-cad and β-catenin at stage 12 as well as disorganization of actin filaments, as revealed by phalloidin staining, on the KDM4B-MO–injected compared with the uninjected side in stage 13. Bars: (A–F) 50 µm; (A’–D’) 20 µm.
We next assessed E-cad distribution and apical constriction at stage 13. Although on the control side the distribution of E-cad was polarized, localized to the apical side of invaginating placode cells (Fig. 5 E), E-cad was not apically enriched on the KDM4B-MO–electroporated side. Instead, it appeared evenly distributed along the cell membrane of all placodal cells, which appeared to have lost polarity (Fig. 5 F). Indeed, the morphology of the otic placode was disorganized and invagination failed. During normal otic placode development, actin filaments exhibit an apical-to-basal polarity and upon invagination are rearranged and become enriched apically (Sai and Ladher, 2008). After KDM4B loss of function, however, phalloidin staining revealed a loss of actin filament polarity (Fig. 5 F) compared with the control side (Fig. 5 E). To quantitate the change of cell polarity during the invagination process, we measured the nuclear angles, toward a horizontal median, along the mediolateral axis in transverse sections stained with Hoechst. This analysis revealed a lack of a proper nuclear orientation and columnar morphology on the KDM4B-MO–treated side compared with the untreated side (Fig. S4).
Thus, actin filament polarity as well as E-cad and β-catenin distribution appear mislocalized after reduction of KDM4B, accompanied by misalignment of otic cells. Collectively, the results suggest that the failure of proper otic invagination after exposure to KDM4B-MO may reflect impairment of cell–cell interactions and cell polarization.
KDM4B binds to the Dlx3 regulatory region and is required for demethylation of H3K9me3 at the Dlx3 promoter
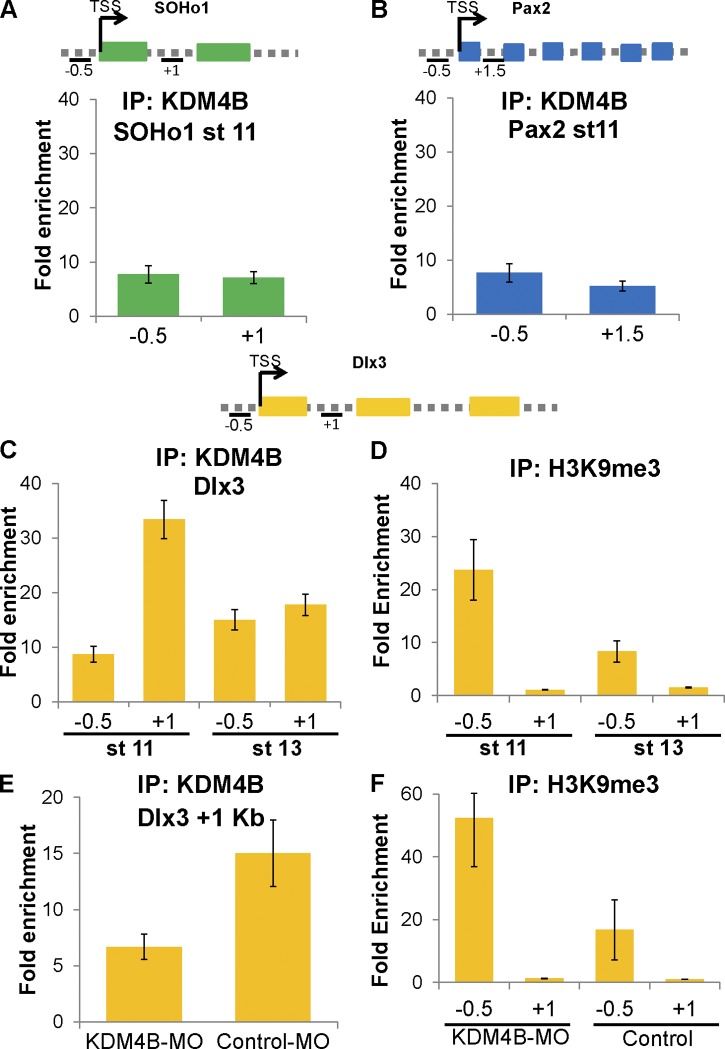
To test whether KDM4B may directly regulate otic marker gene expression by histone demethylation, which in turn impacts otic invagination, we examined whether the histone demethylase directly interacts with regulatory regions of key otic transcription factors. To this end, we performed in vivo ChIP-qPCR on ∼30–40 dissected otic placodes from embryos at stage 11 using an anti-KDM4B antibody. We tested the occupancy of the KDM4B protein on regulatory regions of SOHo1, Pax2, and Dlx3 genes (Fig. 6). Although no binding was observed to the known chromatic regions of SOHo1 and Pax2 (Fig. 6, A and B), we observed consistent binding to the first intron (1 kbp from the transcription start site [TSS]), but not to the promoter of the Dlx3 gene at stage 11 (Fig. 6 C). Interestingly, this strong binding did not persist at stage 13 (Fig. 6 C), suggesting dynamic and transient regulation mediated by KDM4B during inner ear development.
Figure 6.
Endogenous KDM4B binds to a regulatory region of Dlx3 locus during inner ear invagination. (A–C) In vivo ChIP-qPCR assays were used to assess KDM4B binding to genes implicated in ear development such as Soho1 (A), Pax2 (B), and Dlx3 (C). The vertical axes represent fold enrichment (specific ChIP/IgG enriched), and the numbers on the horizontal axes represent distances in kilobase pairs with respect to the TSS on the analyzed genes. Error bars show standard deviation. Schematic diagrams at the top show primer locations for each analyzed gene. One representative sample out of three independent experiments is depicted. The results show that KDM4B strongly associates with the Dlx3 gene at the first intron (C), but not to SOHo1 (A) or Pax2 (B) at stage 11. This association was greatly diminished at stage 13 (C). (D) The results show high occupancy of H3K9me3 at −0.5 kb from the TSS of Dlx3 at stage 11, though the repressive mark is clearly reduced by stage 13. (E) ChIP assays were performed on ∼20–30 stage 11 otic placodes from KDM4B-MO– and control-MO–treated embryos. The results show a lack of KDM4B binding to the Dlx3 locus on KDM4B-MO– compared with control-MO–treated embryos. (F) H3K9me3 occupancy was greatly increased when knocking down KDM4B on the promoter region of Dlx3.
We next assayed for H3K9me3 occupancy and observed pronounced enrichment of this epigenetic mark in the promoter region (−0.5 kbp from the TSS) of Dlx3 at stage 11, also greatly diminished by stage 13 (Fig. 6 D). To demonstrate specificity, we repeated the ChIP with both anti-KDM4B and anti-H3K9me3 after KDM4B-MO knockdown. As predicted, the KDM4B occupancy on the Dlx3 locus at stage 11 was greatly reduced after treatment with KDM4B-MO– compared with control-MO–treated embryos (Fig. 6 E). Reciprocally, H3K9me3 occupancy on the Dlx3 promoter clearly increased in KDM4B-MO–treated embryos, consistent with removal of demethylase activity. Collectively, these results demonstrate that KDM4B is recruited in proximity to the first intron of Dlx3, and its activity is responsible for demethylation of the H3K9me3 mark at the promoter of the Dlx3 gene.
DLX3 is sufficient to rescue invagination defects after KDM4B knockdown
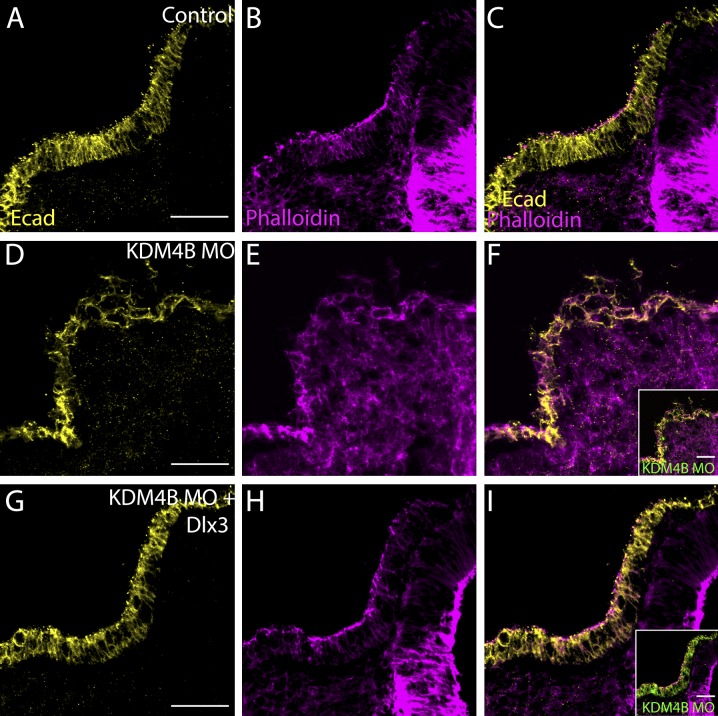
The ChIP data suggest that KDM4B may affect otic invagination via regulation of Dlx3 expression. To test this hypothesis, we next asked whether adding back DLX3 would rescue the effects of KDM4B loss. To this end, we coelectroporated KDM4B-MO together with an expression construct containing the Dlx3 coding sequence into otic ectoderm at stage 8. Embryos were sorted for severity of phenotypes and immunostained for E-cad and phalloidin at stage 12/13 to assess the invagination phenotype. Remarkably, Dlx3 expression was sufficient to rescue the severity of the phenotypes compared with KDM4B-MO plus empty vector (Fig. 7 and Fig. S5). Phenotype quantitation showed that 80% of the analyzed embryos exhibited no otic invagination defects and displayed rescued otic vesicle size (Fig. S5, B and D), whereas 36% and 47% of embryos electroporated with KDM4B-MO plus empty vector exhibited strong and mild invagination phenotypes, respectively (Fig. S5, A and D). Immunohistochemistry revealed that E-cad distribution, actin polarity, and invagination (Fig. 7, A–I), as well as cell death (Fig. S5, E–G and K), were strikingly rescued after coelectroporation of KDM4B-MO with the Dlx3 expression construct. In contrast, coelectroporation of KDM4B-MO plus an expression vector containing the Dlx5 gene, another Dlx factor that is expressed during otic placode development, was not sufficient to rescue invagination defects (Fig. S5, C, D, and H–J). Collectively, these data show that KDM4B histone demethylase epigenetically influences otic placode invagination via modulating Dlx3 gene expression.
Figure 7.
Coelectroporation of KDM4B-MO with pCIG-Dlx3 was sufficient to rescue otic invagination. Using immunohistochemistry, the control side (A–C) shows normal invagination as visualized by E-cad and phalloidin staining, but disrupted invagination on the KDM4B-MO–treated side (D–F). Coexpression of Dlx3 (G–I) with KDM4B-MO was sufficient to rescue otic invagination, leading to a proper arrangement of actin filaments and correct localization of E-cad. Bars, 50 µm.
Knockdown of Dlx3 causes similar phenotypes to KDM4B loss
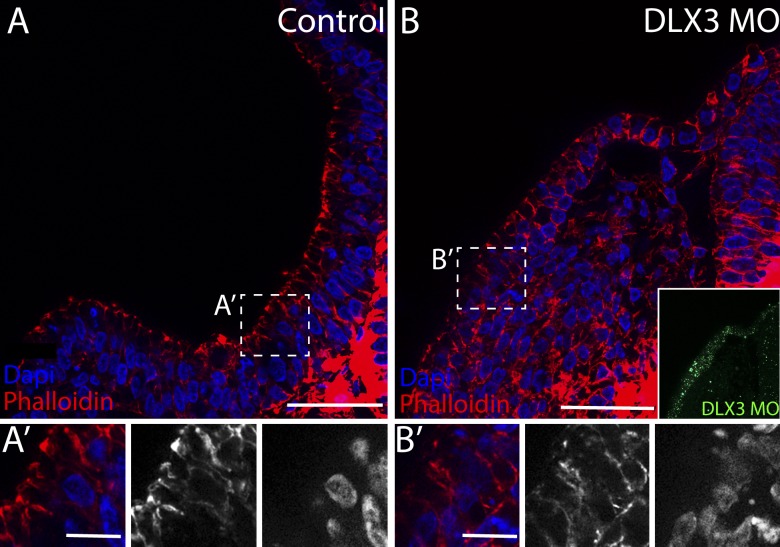
As Dlx3 expression was sufficient to rescue otic defects after knockdown of KDM4B, we postulated that DLX3 plays a key functional role during otic placode invagination. To test this hypothesis, we performed DLX3 loss-of-function experiments by electroporating a fluorescein-tagged antisense MO into the otic ectoderm at stage 8. At stage 12, DLX3 loss resulted in a failure in otic placode invagination (Fig. 8 B) when compared with the control side (Fig. 8 A). Apical accumulation of actin, as viewed by phalloidin, was also compromised compared with the control (Fig. 8, A′ and B′), indicating that the cellular rearrangements necessary for invagination failed to occur in a timely manner. These data show that Dlx3 is one of the key genes required for proper otic placode morphogenesis.
Figure 8.
Knockdown of Dlx3 results in failure of otic invagination. The control side (A) shows normal invagination as seen by apical accumulation of actin filaments viewed by phalloidin staining. The electroporation of DLX3-MO compromise otic invagination (B), indicated by irregular rearrangement of actin filaments. (A’ and B’) Inset views indicate the lack of apical accumulation of actin filaments on the injected side. Bars: (A and B) 60 µm; (A’ and B’) 20 µm.
Discussion
After its induction, the otic placode undergoes morphogenesis that transforms it from a flat ectodermal disk to a hollow internalized spherical vesicle. Experiments using otic placode explants have provided evidence that the invagination process occurs as a consequence of extrinsic signals that trigger a specific transcriptional program (Sai and Ladher, 2008). In addition, epigenetic modifications are gaining recognition as key regulators in the fine-tuning of gene expression and cell differentiation. In this study, we show that in addition to external signaling events, intrinsic epigenetic programs are necessary for proper otic morphogenesis. Our results demonstrate that the histone demethylase KDM4B is critical for otic invagination, with its loss causing abnormal ear morphogenesis. Moreover, we show that KDM4B functions by directly controlling expression of the Dlx3 gene, in a spatiotemporally regulated manner, through H3K9me3 removal. These data reveal for the first time an epigenetic contribution to the control of inner ear invagination in vertebrate embryos via modulation of histone methylation.
Our finding that Dlx3 is a direct target of KDM4B is consistent with the similar spatiotemporal expression patterns of Kdm4b (present study) and Dlx3 (Brown et al., 2005) transcripts during inner ear development. Like Kdm4b, Dlx3 is highly expressed throughout the entire chick otic placode at stage 11, and its expression becomes restricted to the rim of the otic pit as invagination proceeds (Brown et al., 2005). Our results show that KDM4B abundantly occupies the first intron of Dlx3, from which it exercises its demethylase activity on the H3K9me3 mark located at the promoter region of the gene. We speculate that the first intron of Dlx3 may act as a KDM4B-recruiting element, thus allowing H3K9me3 removal and gene activation. Related to this, some studies have suggested that first introns may play a vital role in transcriptional regulation of genes (Majewski and Ott, 2002; Li et al., 2012). As a general trend, first introns are longer, highly conserved, and enriched in CpG dinucleotides compared with other introns (Majewski and Ott, 2002; Kalari et al., 2006; Bradnam and Korf, 2008; Park et al., 2014), suggesting that they may contain regulatory elements. The methylation of CpG dinucleotide is also known to be involved in transcriptional regulation of genes by modifying chromatin structure and/or altering binding sites of transcription factors (Siegfried et al., 1999; Campanero et al., 2000). Dlx3 is no exception, as its first intron is embedded within a large CpG island, suggesting that this may play a critical role in recruitment of transcription factors necessary for its regulation. Interestingly, Park et al. (2014) have shown that first introns are enriched in active transcriptional chromatin marks such as H3K4me3/me1. Related to this, there is evidence that double Tudor domains, present in KDM4 proteins, may recognize the H3K4me3 marks necessary for their recruitment to target chromatin regions (Sims et al., 2007; Lee et al., 2008).
Interestingly, Dlx3 has been suggested to play a role in the regulation of extracellular matrix and adhesion molecules during the otic invagination process (Gerchman et al., 1995; Moro-Balbás et al., 2000; Visconti and Hilfer, 2002). Consistent with this, injection of function-blocking antibodies to perturb the extracellular matrix prevents association of the otic primordium with the hindbrain and inhibits subsequent invagination (Moro-Balbás et al., 2000; Visconti and Hilfer, 2002). Oriented actin filaments and adhesion molecules, like E-cad and adherent junction proteins, have been shown to play an essential role in collective migration processes, including otic placode invagination (Borges et al., 2011; Theveneau et al., 2013; Sai et al., 2014). Otic invagination occurs in a biphasic manner, initiating with basal expansion to generate the initial depression followed by apical constriction to drive the deepening of the otocyst (Sai et al., 2014). During the first phase, extrinsic FGF signaling coordinates the depletion of basal actin filaments producing basal expansion of otic placode cells (Sai and Ladher, 2008). The second phase, which is regulated in a RhoA–Rho-associated protein kinase–dependent manner, results in myosin II activation necessary to direct the contraction of the apical actin networks that results in apical constriction (Sai et al., 2014). These observed cytoskeletal changes necessary for proper otic placode invagination are similar to those perturbed by a KDM4B loss of function. In the present study, we noted a clear mislocalization of actin filaments and E-cad molecules, along with improper ear invagination.
The morphological changes observed after KDM4B loss correlated with a transient increase in cell proliferation at stages 11/12, which is the transitional stage during which the ectodermal invagination process initiates and gradually leads to the internalization of the developing ear. Then, at stage 13, we noted a cessation in cell proliferation and a significant increase in cell death. Collectively, we propose that after a loss of KDM4B, otic placode cells are unable to exit the cell cycle, and that this lack of early differentiation/invagination leads to apoptosis. Cell death normally occurs concomitant with invagination movements and proliferative activity in the otic epithelium (Represa et al., 1990). Based on this, we hypothesize that alteration in the proliferation/death balance may also contribute to defects in otic morphological development, mainly by affecting the cell–cell interactions necessary for correct tissue folding.
The present data support an important role for Dlx3 in otic invagination. Our loss-of-function experiments show that a DLX3 loss results in failure in otic invagination and striking morphological changes, including irregular accumulation of actin filaments. These observations are consistent with previous work in zebrafish showing that MO-mediated reduction of Dlx3b (Dlx3 homologous) delays specification of the otic placode and impairs complete maturation of the otic vesicle, resulting in formation of a smaller vesicle (Liu et al., 2003; Hans et al., 2004). Although the mechanistic link between Dlx genes and ear morphogenesis has yet to be elucidated, clues to its function may be gleaned from other systems. For example, during interneuron migration, Dlx genes are known to repress PAK3 (member of the p21-activated serine/threonine kinases family) that is required for cytoskeletal rearrangements (Cobos et al., 2007). PAK kinases are major downstream effectors of the Rho GTPases Rac1 and Cdc42 (Bokoch, 2003; Hofmann et al., 2004). In turn, Rho GTPases are thought to act as integrators of intrinsic and extrinsic signals to orchestrate cytoskeletal changes that influence cell motility (Luo, 2000; Govek et al., 2005) and apical constriction (Martin and Goldstein, 2014). Accordingly, in the developing otic placode, we speculate that intrinsic epigenetic regulation of the Dlx3 gene may affect the expression of analogous targets that in turn may control cytoskeletal rearrangements during ear development.
In summary, we provide strong evidence that early epigenetic influences are critical for regulating expression of transcription factors, most notably Dlx3, in the developing ear. Perturbation of this epigenetic fine-tuning leads to major downstream defects in ear formation. These results highlight the importance of and close connection between transcriptional and epigenetic regulatory programs during development in general and of the forming ear in particular.
Materials and methods
Embryos
Fertilized chicken eggs were obtained from local commercial sources and incubated at 38°C to the desired stages according to the criteria of Hamburger and Hamilton.
Electroporation, MOs, and rescue experiments
An antisense MO oligomer to KDM4B was designed against a sequence near the ATG codon (5′-GTTTTCAGACCCCATATTTCCAGGA-3′) and against DLX3 near the ATG codon (5′-TCTTGTCGAAGGAGCCGCTCATCGC-3′). As a control, we used the standard control-MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′; Gene Tools, LLC). Injections of fluorescein-tagged MO (1–2-mM plus 0.3 µg/µl of plasmid DNA) and vectors, on the right side of the embryos, were performed by air pressure using a glass micropipette targeted to the presumptive otic placode region at stage 8, located lateral to somites 1 and 2. Electroporation was performed with five pulses of 8 V in intervals (30 ms on and 100 ms off). For rescue experiments, the construct included the coding region of KDM4B, the catalytically dead mutant (KDM4BΔ), Dlx3, or Dlx5 in a pCI-IRES-H2BGFP vector. The pCIG-Dlx3 and pCIG-Dlx5 constructs were provided by M. Barembaum (Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, CA). For each embryo, 1-mM MO and 1 µg/µl DNA was used for KDM4B rescue and 1-mM MO and 1.5 µg/µl DNA for Dlx3 and Dlx5 rescue. KDM4BΔ was obtained by fusion PCR (Table S1) mutating specific amino acids (H189A, F186A, and E191Q), described to be necessary for the demethylation activity of KDM4B (Fodor et al., 2006; Beyer et al., 2008; Kawazu et al., 2011), and then cloned into pCIG-IRES-H2B-RFP vector. We have confirmed that both vectors, containing KDM4B and KDM4BΔ, exhibit strong protein expression, as they were immunodetected in the nucleus of electroporated cells by using an anti-KDM4B antibody (rabbit anti-KDM4B; Novus Biologicals). Moreover, we have also demonstrated that KDM4B, but not KDM4BΔ, overexpression was able to erase the H3K9me3 modification on electroporated cells (Fig. S3).
After electroporation, eggs were sealed with tape and incubated to reach the desired stages. Embryos were removed from eggs, placed in PBS, and viewed and photographed as whole mounts using a fluorescence stereomicroscope to assay electroporation efficiency within the otic domain.
ISH
Whole-mount ISH was performed as previously described (Acloque et al., 2008). Embryos were fixed overnight in 4% PFA at 4°C, washed in PBSw-DEPC (PBS-diethylpyrocarbonate containing 0.1% Tween), and dehydrated in MeOH/PBSw-DEPC series at RT before being stored at −20°C in 100% MeOH. After being rehydrated step-wise with decreasing concentrations of MeOH/ PBSw-DEPC Tween 0.1% and then washed three times with PBSw-DEPC, embryos were permeabilized using 10 µg/µl proteinase K at RT. Embryos were then hybridized at 70°C overnight with the desired digoxigenin (DIG)-labeled RNA probe. DIG-labeled RNA probes were synthesized from pCI plasmids containing the following cDNAs: Kdm4b, Dlx3, Pax2, SOHo1, and Sox10. Hybridized probes were detected using an alkaline phosphatase–conjugated anti-DIG antibody (1:2,000; Roche) in the presence of nitroblue tetrazolium/5-bromo-4-chloro-3-indolyphosphate substrates (Roche). Whole-mount pictures were taken at RT using AxioVision software (Carl Zeiss) with a microscope (Stemi SVII; Carl Zeiss), or with the software Image-Pro Plus 5.1 Biological (Media Cybernetics) with a camera (Evolution VF; Media Cybernetics) in a microscope (SMZ800; Nikon). After ISH, some embryos were fixed in 4% PFA in PBS, washed, embedded in gelatin, and cryostat sectioned at a thickness of 14–16 µm. They were photographed using the AxioVision software with a microscope (Axioskop 2 Plus; Carl Zeiss) and processed using Photoshop (CS3; Adobe).
Western blot
Otic vesicles were dissected from 16 unilaterally electroporated embryos with KDM4B-MO and snap frozen in liquid nitrogen and stored at −80°C. Afterward, the injected and uninjected pool of otic vesicles were lysed by homogenization using a plunger and 150 µl of protein extraction buffer (50-mM Tris-HCl, pH 7.5, 100-mM NaCl, and 5-mM EDTA) supplemented with protease inhibitor (Complete Protease Inhibitor; Roche) and 1% of NP-40. Subsequently, they were centrifuged for 5 min at 1,000 g at 4°C, and the supernatant containing the proteins was collected. Proteins were quantified by Bradford, and an equal amount was run into an SDS-PAGE and transferred to a membrane for blotting. Rabbit anti-KDM4B (Novus Biologicals) was diluted 1:1,000 to test the reduction in KDM4B protein, and a mouse antitubulin (Sigma-Aldrich) was diluted 1:2,000 as a loading control. Both primary antibodies were incubated overnight at 4°C. Finally, secondary conjugated goat anti–mouse HRP or anti–rabbit HRP was diluted 1:10,000 and incubated for 45 min at RT for posterior developing by using ECL plus reagents according to the manufacturer’s instructions (GE Healthcare).
Immunohistochemistry
Fixed embryos were washed in 1× TBST (500-mM Tris-HCl, pH 7.4, 1.5-M NaCl, 10-mM CaCl2, and 0.5% Triton X-100) and subsequently blocked in 10% donkey serum in 1× TBST for 3–5 h at RT. Embryos were then incubated in either mouse anti–E-cad (1:1,000; 610181; BD), mouse anti–β-catenin (1:1,000; 6301; Abcam), rabbit anti-H3K9me3 (1:500; 8898; Abcam), rabbit anti-FITC (1:500; 71–1900; Invitrogen), goat anti-FITC (1:500; NB600-493; Novus Biologicals), or rabbit anti-Caspase3 (1:500; AF835; R&D Systems) overnight at 4°C. The secondary antibodies used were goat or donkey anti–mouse, donkey anti–goat, and anti–rabbit Alexa Fluor 350, 488, 594, and 647 (1:1,000; Molecular Probes), which were incubated for 45 min to 3 h at RT. After washes, embryos were briefly postfixed for 10 min at RT in 4% PFA, washed in 1× PBS, incubated in 15% sucrose/1× PBS for 1 h, and then embedded in 7.5% gelatin and 15% sucrose for cryosectioning. Sections of embryos were then incubated in 1:100 phalloidin 568 and/or Hoechst 3358 (1:2,000). Sections were then imaged on one of the following microscopes: Axioskop 2 Plus with AxioVision software; Zen software (Carl Zeiss) and a photon confocal microscope (LSM 710 2; Carl Zeiss) using a plan-apochromat/.95 korr 40× oil objective; an Image.M2 Apoptome.2 microscope (Carl Zeiss) using a plan-apochromat/.95 korr 40× oil objective; or the NIS-Elements AR4.00 software (Nikon) on a microscope (Eclipse E600; Nikon) using plan 10× 0.25 and plan 40× 0.65 objectives.
qPCR
Three different batches of embryos were electroporated with KDM4B-MO unilaterally at stage 8 and incubated to stage 13. Otic placodes from the injected and the uninjected side were dissected separately. Total RNA was extracted using an RNAqueous-Micro isolation kit (Ambion) according to the manufacturer’s instruction. After DNaseI-amplification grade (Invitrogen) treatment, 1 µg of total RNA was used for cDNA synthesis using SuperScriptII (Invitrogen) and random hexamers (Roche). qPCR was performed using a 96-well plate in MX3005P equipment (Agilent Technologies). Each reaction was performed using Fast Start Syber green (Roche) in a 15-µl volume. All the used primers (Table S1) were tested to have amplification efficiency ranging from 95 to 100%. The subsequent quantification method was done using the ΔΔCt method (threshold cycle). For a reference gene, we used a qPCR primer designed for the GAPDH gene. Gene expression levels are represented relative to GAPDH expression.
Proliferation assays
BrdU powder (Sigma-Aldrich) was dissolved (10 mM) in Ringer’s solution and applied directly under the vitelline membrane via a glass needle adjacent to the KDM4B-MO–electroporated embryos at stage 11. Embryos were incubated at 38°C for 45 min and then fixed in 4% PFA for 25 min at RT for cryosectioning. Cryosections were briefly rehydrated with 1× PBS, incubated with 2-N HCl for 10 min at 37°C, and then immediately washed in PBS with Tween, blocked at RT, and incubated at 4°C overnight in mouse anti-BrdU (1:250; B8434; Sigma-Aldrich). For mitotic counts, sections of embryos treated with KDM4B-MO were incubated with an antibody against pH3 (1:500; EMD Millipore) and then detected using the secondary Alexa Fluor 568 goat anti–rabbit (1:1,000). Nuclei were detected by incubating sections in Hoechst 3358 with the secondary. All cell counts were performed within the otic ectoderm of KDM4B-MO–injected and –uninjected sides, and statistical significance was determined using Student’s t test (Prism; GraphPad Software).
ChIP
Approximately 20–40 otic placodes/otic pits were dissected from stage 11 and stage 13 embryos. The tissue was homogenized in nuclei extraction buffer (0.25% NP-40, 0.25% Triton X-100, 10-mM Tris-HCl, pH 7.5, 3-mM CaCl2, 0.25-M sucrose, 1-mM DTT, 0.2-mM PMSF, and EDTA-free protease inhibitor [Roche]). Cells were cross-linked with 1% formaldehyde for 10 min at RT, and the formaldehyde was then inactivated by the addition of 125-mM glycine for 5 min. Cross-linked cells were centrifuged, and the pellet was washed three times in cold PBS containing a protease inhibitor. The final pellet was snap frozen in liquid nitrogen and stored at –80°C until use. Cells were resuspended in nuclei extraction buffer and rehomogenized until the nuclei went to the solution. After the centrifugation, the pellet was resuspended in SDS-lysis buffer (1% SDS, 50-mM Tris-HCl, pH 8.0, 10-mM EDTA, and EDTA-free protease inhibitor), and 2 vol of ChIP dilution buffer (0.01% SDS, 1.2-mM EDTA, 16.7-mM Tris-HCl, pH 8.0, 167-mM NaCl, 1-mM DTT, 0.2-mM PMSF, and stock of Complete EDTA-free protease inhibitor) was added. Chromatin was sheared to 300–800-bp fragments using a sonicator (amp: 30%; cycle: 30 s on and 30 s off) for 15 min and centrifuged, and the supernatant was split in three tubes for input sample, mock control (rabbit anti-IgG; Abcam), and target antibodies of rabbit anti-KDM4B (Novus Biologicals) or rabbit anti-H3K9me3 (Abcam), all bound to protein A magnetic beads (Invitrogen). IgG and target antibodies (10 µg) were incubated overnight at 4°C, and after extensive washes with radioimmunoprecipitation assay buffer (50-mM Hepes-KOH, pH 8, 500-mM LiCl, 1-mM EDTA, 1% NP-40, 0.7% Na-deoxycholate, 1-mM DTT, 0.2-mM PMSF, and a stock of complete protease inhibitor), the complexes were resuspended in an elution buffer (50-mM Tris-HCl, pH 8.0, 10-mM EDTA, and 1% SDS). The magnetic beads were eluted by incubating at 65°C for 15 min and vortexing every 2 min and then were spun down at 16,000 g for 1 min at RT. The supernatant was reverse cross-linked by heating at 65°C overnight. Immunoprecipitated DNA was treated with 0.2 µg/ml RNaseA and 0.2 µg/ml proteinase K, both for 1 h. Phenol/chloroform/isoamyl alcohol extraction was performed followed by EtOH precipitation. Finally, the purified DNA was used as a template for qPCR analyses (see Table S1 for a list of primers). Each sample was loaded in triplicate, and the results were expressed as fold enrichment (specific ChIP/IgG enriched).
Online supplemental material
Fig. S1 shows the expression of Kdm4b at early stages of otic development by ISH. Fig. S2 shows examples and quantitates the otic phenotypes observed in KDM4B-MO–treated embryos and rescued with a full-length or catalytically dead mutant of KDM4B. Fig. S3 shows that electroporation with a vector containing the full length of KDM4B, but not the catalytically dead mutant KDM4BΔ, is capable of erasing the H3K9me3 mark. Fig. S4 shows that the depletion of KDM4B affects proper nuclear orientation in the invaginating otic placode. Fig. S5 shows that coelectroporation of KDM4B-MO with pCIG-Dlx3 was sufficient to rescue cell death and otic vesicle size, whereas coelectroporation of KDM4B-MO with pCIG-Dlx5 is not enough to rescue the phenotype. Table S1 lists all of the primers used for qPCR. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201503071/DC1.
Supplementary Material
Acknowledgments
We thank Dr. M. Barembaum for pCIG-Dlx3 and pCIG-Dlx5 constructs and for insight throughout this study.
This work was supported by grants from the Consejo Nacional de Investigaciones Científicas (CONICET-PIP D4574) and Agencia Nacional de Promocion Cientifica y Tecnologica (PICT 2011-0500) to P.H. Strobl-Mazzulla and by the Consejo Nacional de Investigaciones Científicas-National Science Foundation grant (CONICET-NSF D2445) to P.H. Strobl-Mazzulla, as well as by the following supplements from the National Institutes of Health: DC011577 to R.A. Uribe and DC011577 and DE16459 to M.E. Bronner.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ChIP
- chromatin immunoprecipitation
- DEPC
- diethylpyrocarbonate
- DIG
- digoxigenin
- ISH
- in situ hybridization
- MO
- morpholino
- qPCR
- quantitative PCR
- TSS
- transcription start site
References
- Acloque H., Wilkinson D.G., and Nieto M.A.. 2008. In situ hybridization analysis of chick embryos in whole-mount and tissue sections. Methods Cell Biol. 87:169–185. 10.1016/S0091-679X(08)00209-4 [DOI] [PubMed] [Google Scholar]
- Alvarez I.S., and Navascués J.. 1990. Shaping, invagination, and closure of the chick embryo otic vesicle: scanning electron microscopic and quantitative study. Anat. Rec. 228:315–326. 10.1002/ar.1092280311 [DOI] [PubMed] [Google Scholar]
- Baker C.V., and Bronner-Fraser M.. 2001. Vertebrate cranial placodes I. Embryonic induction. Dev. Biol. 232:1–61. 10.1006/dbio.2001.0156 [DOI] [PubMed] [Google Scholar]
- Barald K.F., and Kelley M.W.. 2004. From placode to polarization: new tunes in inner ear development. Development. 131:4119–4130. 10.1242/dev.01339 [DOI] [PubMed] [Google Scholar]
- Barembaum M., and Bronner-Fraser M.. 2010. Pax2 and Pea3 synergize to activate a novel regulatory enhancer for spalt4 in the developing ear. Dev. Biol. 340:222–231. 10.1016/j.ydbio.2009.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer S., Kristensen M.M., Jensen K.S., Johansen J.V., and Staller P.. 2008. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J. Biol. Chem. 283:36542–36552. 10.1074/jbc.M804578200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokoch G.M. 2003. Biology of the p21-activated kinases. Annu. Rev. Biochem. 72:743–781. 10.1146/annurev.biochem.72.121801.161742 [DOI] [PubMed] [Google Scholar]
- Borges R.M., Lamers M.L., Forti F.L., Santos M.F., and Yan C.Y.. 2011. Rho signaling pathway and apical constriction in the early lens placode. Genesis. 49:368–379. 10.1002/dvg.20723 [DOI] [PubMed] [Google Scholar]
- Bradnam K.R., and Korf I.. 2008. Longer first introns are a general property of eukaryotic gene structure. PLoS ONE. 3:e3093 10.1371/journal.pone.0003093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.W., Beck-Jefferson E., and Hilfer S.R.. 1998. A role for neural cell adhesion molecule in the formation of the avian inner ear. Dev. Dyn. 213:359–369. [DOI] [PubMed] [Google Scholar]
- Brown S.T., Wang J., and Groves A.K.. 2005. Dlx gene expression during chick inner ear development. J. Comp. Neurol. 483:48–65. 10.1002/cne.20418 [DOI] [PubMed] [Google Scholar]
- Campanero M.R., Armstrong M.I., and Flemington E.K.. 2000. CpG methylation as a mechanism for the regulation of E2F activity. Proc. Natl. Acad. Sci. USA. 97:6481–6486. 10.1073/pnas.100340697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., and Streit A.. 2013. Induction of the inner ear: stepwise specification of otic fate from multipotent progenitors. Hear. Res. 297:3–12. 10.1016/j.heares.2012.11.018 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Cheung M., Abu-Elmagd M.M., Orme A., and Scotting P.J.. 2000. Chick sox10, a transcription factor expressed in both early neural crest cells and central nervous system. Brain Res. Dev. Brain Res. 121:233–241. 10.1016/S0165-3806(00)00049-3 [DOI] [PubMed] [Google Scholar]
- Christophorou N.A., Mende M., Lleras-Forero L., Grocott T., and Streit A.. 2010. Pax2 coordinates epithelial morphogenesis and cell fate in the inner ear. Dev. Biol. 345:180–190. 10.1016/j.ydbio.2010.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I., Borello U., and Rubenstein J.L.. 2007. Dlx transcription factors promote migration through repression of axon and dendrite growth. Neuron. 54:873–888. 10.1016/j.neuron.2007.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture J.-F., Collazo E., Ortiz-Tello P.A., Brunzelle J.S., and Trievel R.C.. 2007. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat. Struct. Mol. Biol. 14:689–695. 10.1038/nsmb1273 [DOI] [PubMed] [Google Scholar]
- Deitcher D.L., Fekete D.M., and Cepko C.L.. 1994. Asymmetric expression of a novel homeobox gene in vertebrate sensory organs. J. Neurosci. 14:486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver E.C., and Kelley M.W.. 2009. Specification of cell fate in the mammalian cochlea. Birth Defects Res. C Embryo Today. 87:212–221. 10.1002/bdrc.20154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton K., Abbas L., Spencer J., Brannon C., Mowbray C., Nikaido M., Kelsh R.N., and Whitfield T.T.. 2009. A zebrafish model for Waardenburg syndrome type IV reveals diverse roles for Sox10 in the otic vesicle. Dis. Model. Mech. 2:68–83. 10.1242/dmm.001164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmett S.D., and West K.P. Jr. 2014. Gestational vitamin A deficiency: a novel cause of sensorineural hearing loss in the developing world? Med. Hypotheses. 82:6–10. 10.1016/j.mehy.2013.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor B.D., Kubicek S., Yonezawa M., O’Sullivan R.J., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., and Jenuwein T.. 2006. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 20:1557–1562. 10.1101/gad.388206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerchman E., Hilfer S.R., and Brown J.W.. 1995. Involvement of extracellular matrix in the formation of the inner ear. Dev. Dyn. 202:421–432. 10.1002/aja.1002020411 [DOI] [PubMed] [Google Scholar]
- Govek E.E., Newey S.E., and Van Aelst L.. 2005. The role of the Rho GTPases in neuronal development. Genes Dev. 19:1–49. 10.1101/gad.1256405 [DOI] [PubMed] [Google Scholar]
- Groves A.K., and Fekete D.M.. 2012. Shaping sound in space: the regulation of inner ear patterning. Development. 139:245–257. 10.1242/dev.067074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans S., Liu D., and Westerfield M.. 2004. Pax8 and Pax2a function synergistically in otic specification, downstream of the Foxi1 and Dlx3b transcription factors. Development. 131:5091–5102. 10.1242/dev.01346 [DOI] [PubMed] [Google Scholar]
- Hofmann C., Shepelev M., and Chernoff J.. 2004. The genetics of Pak. J. Cell Sci. 117:4343–4354. 10.1242/jcs.01392 [DOI] [PubMed] [Google Scholar]
- Kalari K.R., Casavant M., Bair T.B., Keen H.L., Comeron J.M., Casavant T.L., and Scheetz T.E.. 2006. First exons and introns—a survey of GC content and gene structure in the human genome. In Silico Biol. (Gedrukt). 6:237–242. [PubMed] [Google Scholar]
- Kawazu M., Saso K., Tong K.I., McQuire T., Goto K., Son D.O., Wakeham A., Miyagishi M., Mak T.W., and Okada H.. 2011. Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS ONE. 6:e17830 10.1371/journal.pone.0017830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Thompson J.R., Botuyan M.V., and Mer G.. 2008. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat. Struct. Mol. Biol. 15:109–111. 10.1038/nsmb1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Chen D., and Zhang J.. 2012. Analysis of intron sequence features associated with transcriptional regulation in human genes. PLoS ONE. 7:e46784 10.1371/journal.pone.0046784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Chu H., Maves L., Yan Y.L., Morcos P.A., Postlethwait J.H., and Westerfield M.. 2003. Fgf3 and Fgf8 dependent and independent transcription factors are required for otic placode specification. Development. 130:2213–2224. 10.1242/dev.00445 [DOI] [PubMed] [Google Scholar]
- Luo L. 2000. Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 1:173–180. 10.1038/35044547 [DOI] [PubMed] [Google Scholar]
- Majewski J., and Ott J.. 2002. Distribution and characterization of regulatory elements in the human genome. Genome Res. 12:1827–1836. 10.1101/gr.606402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A.C., and Goldstein B.. 2014. Apical constriction: themes and variations on a cellular mechanism driving morphogenesis. Development. 141:1987–1998. 10.1242/dev.102228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro-Balbás J.A., Gato A., Alonso M.I., Martín P., and de la Mano A.. 2000. Basal lamina heparan sulphate proteoglycan is involved in otic placode invagination in chick embryos. Anat. Embryol. (Berl.). 202:333–343. 10.1007/s004290000119 [DOI] [PubMed] [Google Scholar]
- Neves J., Abelló G., Petrovic J., and Giraldez F.. 2013. Patterning and cell fate in the inner ear: a case for Notch in the chicken embryo. Dev. Growth Differ. 55:96–112. 10.1111/dgd.12016 [DOI] [PubMed] [Google Scholar]
- Park S.G., Hannenhalli S., and Choi S.S.. 2014. Conservation in first introns is positively associated with the number of exons within genes and the presence of regulatory epigenetic signals. BMC Genomics. 15:526 10.1186/1471-2164-15-526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Represa J.J., Moro J.A., Gato A., Pastor F., and Barbosa E.. 1990. Patterns of epithelial cell death during early development of the human inner ear. Ann. Otol. Rhinol. Laryngol. 99:482–488. 10.1177/000348949009900613 [DOI] [PubMed] [Google Scholar]
- Sai X., and Ladher R.K.. 2008. FGF signaling regulates cytoskeletal remodeling during epithelial morphogenesis. Curr. Biol. 18:976–981. 10.1016/j.cub.2008.05.049 [DOI] [PubMed] [Google Scholar]
- Sai X., Yonemura S., and Ladher R.K.. 2014. Junctionally restricted RhoA activity is necessary for apical constriction during phase 2 inner ear placode invagination. Dev. Biol. 394:206–216. 10.1016/j.ydbio.2014.08.022 [DOI] [PubMed] [Google Scholar]
- Siegfried Z., Eden S., Mendelsohn M., Feng X., Tsuberi B.Z., and Cedar H.. 1999. DNA methylation represses transcription in vivo. Nat. Genet. 22:203–206. 10.1038/9727 [DOI] [PubMed] [Google Scholar]
- Sims R.J. III, Millhouse S., Chen C.F., Lewis B.A., Erdjument-Bromage H., Tempst P., Manley J.L., and Reinberg D.. 2007. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell. 28:665–676. 10.1016/j.molcel.2007.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens G., Flaxman S., Brunskill E., Mascarenhas M., Mathers C.D., and Finucane M.. Global Burden of Disease Hearing Loss Expert Group . 2013. Global and regional hearing impairment prevalence: an analysis of 42 studies in 29 countries. Eur. J. Public Health. 23:146–152. 10.1093/eurpub/ckr176 [DOI] [PubMed] [Google Scholar]
- Streit A. 2002. Extensive cell movements accompany formation of the otic placode. Dev. Biol. 249:237–254. 10.1006/dbio.2002.0739 [DOI] [PubMed] [Google Scholar]
- Strobl-Mazzulla P.H., Sauka-Spengler T., and Bronner-Fraser M.. 2010. Histone demethylase JmjD2A regulates neural crest specification. Dev. Cell. 19:460–468. 10.1016/j.devcel.2010.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H., Wu S., Wang J., and Zhao Z.K.. 2008. The JMJD2 members of histone demethylase revisited. Mol. Biol. Rep. 35:551–556. 10.1007/s11033-007-9121-3 [DOI] [PubMed] [Google Scholar]
- Theveneau E., Steventon B., Scarpa E., Garcia S., Trepat X., Streit A., and Mayor R.. 2013. Chase-and-run between adjacent cell populations promotes directional collective migration. Nat. Cell Biol. 15:763–772. 10.1038/ncb2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kerschaver E., Boudewyns A.N., Declau F., Van de Heyning P.H., and Wuyts F.L.. 2013. Socio-demographic determinants of hearing impairment studied in 103,835 term babies. Eur. J. Public Health. 23:55–60. 10.1093/eurpub/cks010 [DOI] [PubMed] [Google Scholar]
- Visconti R.P., and Hilfer S.R.. 2002. Perturbation of extracellular matrix prevents association of the otic primordium with the posterior rhombencephalon and inhibits subsequent invagination. Dev. Dyn. 223:48–58. 10.1002/dvdy.1237 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.