Synaptonemal complex (SC) assembly requires polySUMOylation of Ecm11, which promotes polymerization of Zip1, the transverse filament, whereas the N terminus of Zip1 activates Ecm11 polySUMOylation, suggesting that this positive feedback loop underpins SC assembly.
Abstract
During meiotic prophase I, proteinaceous structures called synaptonemal complexes (SCs) connect homologous chromosomes along their lengths via polymeric arrays of transverse filaments (TFs). Thus, control of TF polymerization is central to SC formation. Using budding yeast, we show that efficiency of TF polymerization closely correlates with the extent of SUMO conjugation to Ecm11, a component of SCs. HyperSUMOylation of Ecm11 leads to highly aggregative TFs, causing frequent assembly of extrachromosomal structures. In contrast, hypoSUMOylation leads to discontinuous, fragmented SCs, indicative of defective TF polymerization. We further show that the N terminus of the yeast TF, Zip1, serves as an activator for Ecm11 SUMOylation. Coexpression of the Zip1 N terminus and Gmc2, a binding partner of Ecm11, is sufficient to induce robust polySUMOylation of Ecm11 in nonmeiotic cells. Because TF assembly is mediated through N-terminal head-to-head associations, our results suggest that mutual activation between TF assembly and Ecm11 polySUMOylation acts as a positive feedback loop that underpins SC assembly.
Introduction
During meiotic prophase I, prominent meiosis-specific chromosomal structures called synaptonemal complexes (SCs) play a critical role in successful chromosome segregation at meiosis I (Zickler and Kleckner, 1999; Lake and Hawley, 2012). SCs keep pairs of homologous chromosomes tightly aligned along their lengths. The SC is highly conserved among most eukaryotes and consists of very similar substructures. Chromatin loops of each chromosome are bound to rigid chromosomal axes called the axial elements, which later form the lateral elements of the SCs. The aligned axes are closely juxtaposed through the central region of the SC, where oligomeric arrays of transverse filaments (TFs) lie perpendicular to the lateral elements, serving as a proteinaceous connection between homologs. In the middle of this central region runs an electron-dense linear substructure called the central element.
Budding yeast has one TF protein called Zip1 (Roeder, 1997). Zip1 possesses a long α-helical coiled-coil region flanked by N- and C-terminal regions (Sym et al., 1993; Lake and Hawley, 2012). The N-terminal domain of Zip1 lies in the middle of the central region of the SC, whereas the C-terminal domain localizes with the lateral elements, leading to the proposal that the N-terminal domains of Zip1 are bound head-to-head at the center of the SC and the C-terminal domains are anchored to the lateral elements (Dong and Roeder, 2000; Voelkel-Meiman et al., 2013). Thus, controlling chromosomal recruitment and polymerization of Zip1 is crucial for controlling SC assembly. Chromosomal assembly of Zip1 is initiated by the synapsis initiation complex (Zip2, 3, 4, and Spo16; Chua and Roeder, 1998; Agarwal and Roeder, 2000; Perry et al., 2005; Tsubouchi et al., 2006; Shinohara et al., 2008). SUMO ligase Zip3 and the prolyl-isomerase Fpr3 are known to render SC assembly dependent on DSBs (Macqueen and Roeder, 2009).
Small ubiquitin-like modifier (SUMO) has emerged as an important regulator of SC formation (Cheng et al., 2006; de Carvalho and Colaiácovo, 2006; Hooker and Roeder, 2006). Many lateral element proteins, including Red1, Pdr5, and Top2, are SUMOylated (Stead et al., 2003; Cheng et al., 2006; Takahashi et al., 2006; Eichinger and Jentsch, 2010). Zip3 has been shown to possess SUMO E3 ligase activity, whereas both Zip1 and Red1 have SUMO chain–binding activities (Cheng et al., 2006; Lin et al., 2010). SUMOylation of the SUMO E2 ligase (Ubc9) is also proposed to regulate SC assembly by controlling the formation of oligomeric SUMO chains (Klug et al., 2013).
In budding yeast, Ecm11 and Gmc2 are emerging as key regulators for TF assembly (Brar et al., 2012; Humphryes et al., 2013). Ecm11 undergoes prominent SUMOylation that is Gmc2 dependent and is essential for efficient SC assembly. SUMOylated Ecm11 is localized to the central element of the SC (Voelkel-Meiman et al., 2013). These observations raise the possibility that TF assembly is controlled through SUMOylation of Ecm11.
In this study, we found that the extent of Ecm11 SUMOylation closely correlates with the efficiency of TF oligomerization. We further show that Ecm11 SUMOylation and Zip1 assembly reciprocally activate each other, leading to the proposal that mutual activation of Ecm11 SUMOylation and TF oligomerization establishes a positive feedback mechanism that promotes SC assembly.
Results and discussion
Ecm11–SUMO conjugates are the prominent SUMOylated species during early meiosis
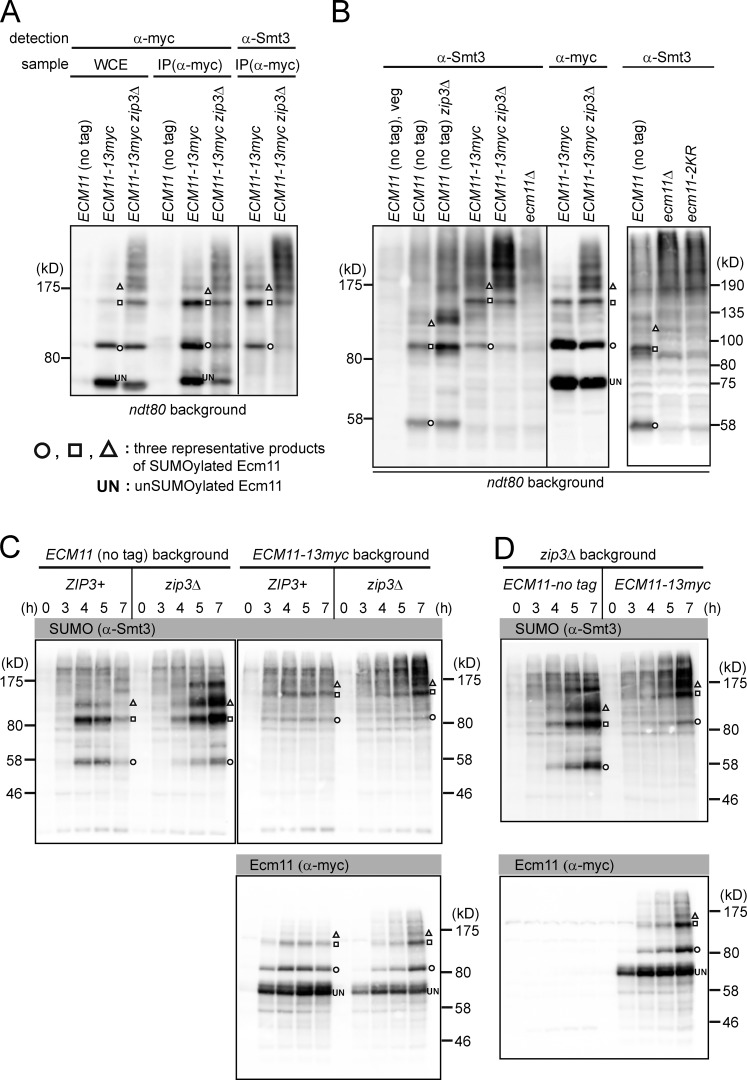
Previous work implied that oligomeric SUMO chains accumulate during prophase I (Cheng et al., 2006). We considered the possibility that these polySUMO species could in fact be polySUMOylated Ecm11 and not free SUMO chains. To test this possibility, we used two kinds of ECM11 alleles: wild type and ECM11-13myc, which has an extra 21 kD attached to Ecm11. Also used were the zip3 and ndt80 mutations, which cause hyperSUMOylation of Ecm11 and late prophase I arrest, respectively (Xu et al., 1995; Agarwal and Roeder, 2000; Humphryes et al., 2013). Ecm11-13myc showed a ladder-like migration pattern caused by the attachment of multiple SUMO molecules, which was more prominent by the zip3 mutation as shown previously (Fig. 1 A, left; Humphryes et al., 2013). The anti-myc immunoprecipitation products reacted with anti-Smt3 antibodies (Smt3 is the only SUMO in budding yeast) and showed a migration pattern identical to the ladder-like bands of Ecm11-13myc (Fig. 1 A, right).
Figure 1.
Polymeric SUMO chains formed during early meiosis derive from Ecm11. (A) Whole-cell extract (WCE) from BR1919 ndt80 diploids 22 h into meiosis was subjected to immunoprecipitation with anti-myc antibodies. Whole-cell extract and immunoprecipitates were subjected to Western blotting with indicated antibodies. IP, immunoprecipitate. (B) Whole-cell extract was subjected to Western blotting with indicated antibodies, and the patterns of signal were compared side by side. BR1919 ndt80 diploids at the 20-h time point were used. (C) SK1 strains with indicated genotypes were subjected to Western blotting as in B. (D) Wild-type and ECM11-13myc strains were compared side by side. Theoretical molecular masses: Ecm11, 34 kD; Ecm11-13myc, 55 kD.
We then tested if the prominent ladder-like SUMO signals are related to Ecm11. We reasoned that, if such polySUMO chains are conjugated with Ecm11, the ladder-like bands should change their migration speed as a group when the molecular size of Ecm11 is increased, which was achieved by attaching myc epitopes. Consistent with Cheng et al. (2006), SUMO antibodies identified a meiosis-specific ladder of bands in wild type, which was enhanced by the zip3 mutation (Fig. 1 B). Upon addition of 13myc to Ecm11, the ladder collectively increased in molecular mass, yielding a pattern that matches that of the SUMO-conjugated Ecm11 ladder. The majority, if not all, of the prominent SUMO ladder disappeared in the ecm11-null mutant, or in a mutant in which the Ecm11 SUMO targeting sites are mutated (ecm11-2KR; i.e., K5R K101R ; Fig. 1 B, right).
Similar observations were obtained in a second strain background (Fig. 1, C and D). Furthermore, when Ecm11 was attached to different tags that add smaller masses (3, 7, and 12 kD), the SUMO ladder shifted upward accordingly (Fig. S1 A).
Collectively, our results indicate that the entities previously inferred to be free polySUMO chains are actually polySUMOylated Ecm11 species. The observation that Ecm11 SUMOylation becomes further pronounced in the absence of Zip3 rules out the possibility that Zip3 functions as the SUMO E3 ligase responsible for Ecm11 SUMOylation.
E3 SUMO ligases Siz1 and Siz2 facilitate polySUMOylation of Ecm11 and SC assembly
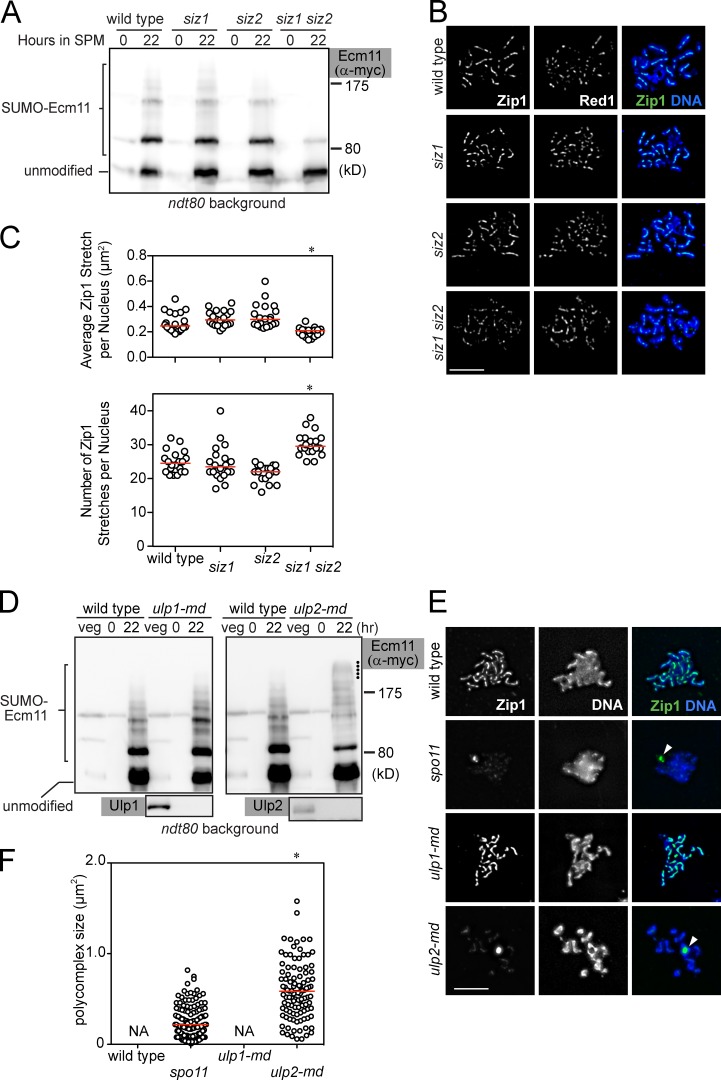
We next examined the involvement of two SUMO E3 ligases, Siz1 and Siz2, in Ecm11 SUMOylation (Johnson and Gupta, 2001; Takahashi et al., 2001). SUMOylation of Ecm11 was substantially reduced only when both SIZ1 and SIZ2 were deleted (Fig. 2 A and Fig. S1 B). We also examined chromosomal assembly of Zip1 in the absence of Siz1, Siz2, or both (Fig. S1 C and Materials and methods). In brief, we measured the size and number of Zip1 stretches; a Zip1 stretch is defined as one continuous immunostaining Zip1 structure, which is typically a long line in wild type. If SC assembly is defective, Zip1 staining tends to become discontinuous, which is reflected as a decrease in the size of individual Zip1 stretches and an increase in the number of stretches per nucleus. The siz1 siz2 double mutant exhibited a substantial reduction in Zip1 linearity (Fig. 2, B and C; and Fig. S1 D).
Figure 2.
SUMO E3 ligases Siz1 and Siz2 and the SUMO-protease Ulp2 play opposing roles in regulating Zip1 assembly and SUMOylation of Ecm11. (A) SUMOylation of Ecm11 at the 22-h time point was analyzed as in Fig. 1. SPM, sporulation media. (B) Meiotic chromosomes were spread and visualized for indicated proteins and DNA (DAPI). Cells at the 20-h time point are shown. Bar, 5 µm. (C) Zip1 localization pattern was quantitatively analyzed for cells 20 h into meiosis. See Fig. S2 C and Materials and methods for details. Red bars, median values. *, P < 0.001 (Mann-Whitney test), in comparison to wild type. (D) SUMOylation of Ecm11 was analyzed as in A. Dots indicate the location of Ecm11 attached with polySUMO chains longer than those in wild type. (E) Meiotic chromosomes were analyzed as in B. White arrowhead, PC. Bar, 5 µm. (F) PC size was quantitatively analyzed (Materials and methods). In wild type and ulp1-mn, PC formation was barely seen. Red bars, median values. NA, not applicable. *, P < 0.001 (Mann-Whitney test) in comparison to the spo11 mutant.
To further examine the effect of the siz1 siz2 mutations on extrachromosomal assemblies of Zip1 (i.e., polycomplexes [PCs]), we used two mutant backgrounds in which PCs are frequently formed: spo11 and zip3. In both mutants, the siz1 siz2 mutations reduced both the size of the PCs and the fraction of nuclei carrying a PC (Fig. S1 E).
From these results, we conclude that Siz1 and Siz2 are important for promoting efficient Ecm11 SUMOylation, as well as chromosomal and extrachromosomal assemblies of Zip1.
SUMO protease Ulp2 prevents hyperSUMOylation of Ecm11 and extrachromosomal assembly of TF
Ulp1 and Ulp2 are SUMO-specific proteases involved in deSUMOylation (Li and Hochstrasser, 1999, 2000; Schwienhorst et al., 2000). We found that meiosis-specific depletion (md; see Yeast strains) of Ulp2 caused hyperSUMOylated Ecm11 to accumulate (Fig. 2 D, right, marked with dots), whereas Ulp1 depletion had little impact (Fig. 2 D, left). Furthermore, Ulp2 depletion, but not Ulp1 depletion, caused frequent PC formation: 1.8% (2/110) in wild type; 63% (80/127) in spo11; 1.0% (1/105) in ulp1-md; and 88% (88/100) in ulp2-md. The size of the PCs was substantially bigger in Ulp2-depleted cells than in the spo11 mutant (Fig. 2, E and F), in which double-strand breaks (DSBs) are not induced and chromosomal assembly of Zip1 is defective. Thus, Ulp2 is important for controlling the extent of Ecm11 SUMOylation; hyperSUMOylated Ecm11 and extrachromosomal Zip1 assembly are concurrent in the absence of Ulp2.
Extent of Ecm11 SUMOylation correlates with efficiency of Zip1 assembly
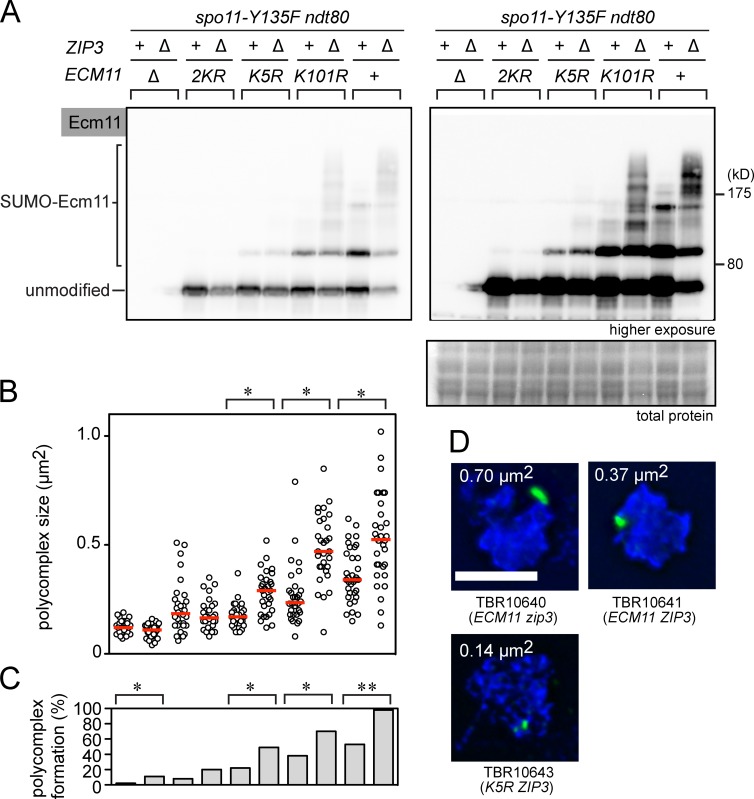
To further investigate the relationship between the levels of Ecm11 SUMOylation and the efficiency of Zip1 assembly, we constructed a series of mutant strains showing various levels of Ecm11 SUMOylation. Mutating one of two canonical SUMO targeting sites, K5 and K101, leads to a partial reduction in Ecm11 SUMOylation, with the K5R mutation causing a bigger reduction than K101R; mutating both sites (2KR; i.e., K5R K101R) leads to a near-complete loss of SUMOylation (Zavec et al., 2008; Humphryes et al., 2013). These mutations and the null mutation were combined with the zip3 mutation, which enhances Ecm11 SUMOylation.
Initially, we used a condition in which meiotic DSB formation is normal (SPO11+; Fig. S2). Overall, the size of PCs closely correlated with the level of Ecm11 SUMOylation (Fig. S2, A and B), although this interpretation was complicated by two considerations. First, in some strains such as wild type and K101R, the majority of Zip1 assembles between chromosomes as normal SCs, whereas in zip3, Zip1 assembles as both PCs and SCs (Fig. S2, C and D). Second, the absence of Zip3 compromises chromosomal localization of Zip1, likely because Zip3 is also involved in synapsis initiation (Fig. S2 D). Thus, we introduced a mutation that prevents meiotic DSB formation (spo11-Y135F) so that Zip1 assembly now occurs exclusively as PCs (Bergerat et al., 1997; Chua and Roeder, 1998; Diaz et al., 2002). This analysis uses PC formation as a surrogate for Zip1 oligomerization, thus allowing us to uniformly assess Zip1 assembly in all the strains used. We note that the configuration of Zip1 polymers in a PC, although formed in a pathological condition, is similar to that found in SCs (Dong and Roeder, 2000).
Ecm11 was conjugated with various levels of polySUMO chains depending on the combination of mutations used (Fig. 3 A). The frequency and size of PCs showed a strong correlation with the degree of Ecm11 SUMOylation. For example, in the ECM11 zip3 strain, nearly all the spread nuclei contained a large PC (e.g., Fig. 3 D, 0.70 µm2); in the ECM11 ZIP3 strain, ∼60% of nuclei contained a medium-sized PC (e.g., Fig. 3 D, 0.37 µm2); and in K5R ZIP3, only ∼20% of spread nuclei had a PC, which was also much smaller (e.g., Fig. 3 D, 0.14 µm2).
Figure 3.
Levels of Ecm11 SUMOylation correlate with efficiency of extrachromosomal Zip1 assembly. (A) SUMOylation of Ecm11 at the 22-h time point was analyzed as in Fig. 1 in the spo11-Y135F background. Total protein, Ponceau S stain. (B) The size of PCs was quantitatively analyzed (Materials and methods). Red bars, median values. *, P < 0.001 (Mann-Whitney test). (C) The ratio of chromosome spreads carrying a PC was measured with the sample used in B. 50 or more spreads were examined per strain. *, P < 0.05; **, P < 0.01 (χ2 test). (D) Three representative images showing examples of PCs. Bar, 5 µm.
These observations, along with the phenotypes of the siz1 siz2 double mutant (Fig. 2, A–C) and the ulp2-mn mutant (Fig. 2, D–F), suggest that efficient Zip1 assembly necessitates Ecm11 and is controlled by the efficiency of Ecm11 SUMOylation and perhaps the length of polySUMO chains. Thus, we propose that Ecm11 SUMOylation serves as a molecular throttle that controls the efficiency of SC assembly. The degree of Ecm11 polySUMOylation is likely to be maintained by the balance between SUMO ligases and SUMO proteases. These two classes of enzymes might communicate with the chromosomal environment to control SUMOylation of Ecm11. In this context, Zip3 is likely involved, directly or indirectly, in shortening the SUMO chain. It is currently unclear whether overall efficiency of Ecm11 SUMOylation or the length of SUMO chain is more relevant for promoting Zip1 assembly.
PolySUMOylation of Ecm11 requires the N terminus of Zip1
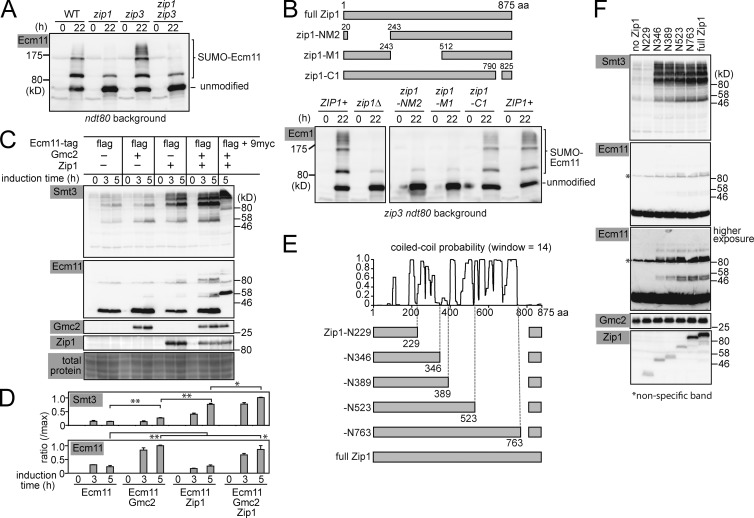
In the zip3 mutant, oversized PCs are frequently formed. To test if hyperSUMOylation of Ecm11 requires Zip1, zip3 was combined with the zip1-null mutation and Ecm11 SUMOylation was examined (Fig. 4 A). The combination of zip1 and zip3 led to a substantial reduction in SUMOylation, the level of which was comparable to that of the zip1 single mutant. Thus, hyperSUMOylation of Ecm11 requires Zip1.
Figure 4.
Zip1 N terminus and Gmc2 Activate Ecm11 SUMOylation in vegetative cells. (A) SUMOylation of Ecm11 was analyzed as in Fig 1. (B) Schematic of Zip1 truncation constructs. The effect of indicated zip1 mutations on Ecm11 SUMOylation was assessed by Western blotting. (C) Induction of indicated proteins and their impact on Ecm11 SUMOylation were examined by Western blotting. (D) Quantitative analysis depicting the ratio of SUMOylated Ecm11 as a ratio of maximum SUMOylated Ecm11 (see Materials and methods for quantification). The results are presented as the mean of two independent experiments. Error bar, SEM. *, P < 0.05; **, P < 0.01 (t test). (E) Schematic of Zip1 truncation constructs and their relationship to the high-probability locations forming coiled-coil structures, calculated by COILS (Lupas et al., 1991). (F) The impact of truncating Zip1 on Ecm11 SUMOylation was evaluated by Western blotting using cells harvested 4 h after induction. The antibody that recognizes the C terminus of Zip1 was used, which rendered the constructs carrying a larger fragment of the C terminus more recognizable (Fig. S3 A).
The N terminus head-to-head association of Zip1 molecules is central to the adhesion of homologous chromosomes. Thus, a collection of zip1 mutant alleles used in previous studies were tested for their effect on Ecm11 SUMOylation (Fig. 4 B; Tung and Roeder, 1998). Both Zip1-NM2 and Zip1-M1 are defective in closing the zipper structure of the SC. In contrast, Zip1-C1 does not interact with axial elements, resulting in failure to associate with chromosomes and frequent PC formation. Ecm11 SUMOylation was barely affected in the zip1-C1 mutant, whereas the level dropped to that of the zip1-null mutant in zip1-NM2 and -M1 cells (Fig. 4 B). Thus, the N terminus of Zip1 is necessary for promoting Ecm11 SUMOylation.
Zip1 N terminus and Gmc2 are sufficient for polySUMOylation of Ecm11
To further understand the mechanisms of SC assembly, we set out to identify the minimal conditions that support efficient Ecm11 SUMOylation and Zip1 assembly in vegetative cells in which other meiosis-specific proteins do not exist. The Ecm11, Gmc2, and Zip1 proteins were conditionally produced by using the galactose-inducible promoter. When Ecm11 was produced along with Gmc2, more Ecm11 was detected, possibly suggesting a role for Gmc2 in stabilizing Ecm11 (Fig. 4, C and D). When Zip1 was produced along with Ecm11, a substantial increase in Ecm11 SUMOylation was detected. Ecm11 SUMOylation reached the maximum level when Ecm11 was produced with both Gmc2 and Zip1. The robust SUMO signal derives from Ecm11, because these signals collectively shifted upward when Ecm11 was fused to a tag (9myc; Fig. 4 C, rightmost lane).
We further set out to determine the minimum region of Zip1 that can support the activation of Ecm11 SUMOylation. A truncation series of ZIP1 alleles was created so that each construct encodes various lengths of N-terminal Zip1 (Fig. 4 E). The C-terminal end of each truncation construct was fixed and corresponded to an interruption in the predicted coiled-coil regions of Zip1 (Lupas et al., 1991). A small C terminus region, consisting of 825–875 aa, was found to be essential (most likely for nuclear transport; see Materials and methods) and thus was attached to every construct. Each truncated Zip1 was produced in vegetative cells, along with Ecm11 and Gmc2 (Fig. 4 F and Fig. S3). Although robust Ecm11 SUMOylation was seen with full-length Zip1, a comparable level of SUMOylation was detected with the truncation carrying 1–346 aa (Zip1-N346) but not in the 1–229 aa construct (Zip1-N229). Thus, the N-terminal 346 aa is sufficient for activating Ecm11 SUMOylation.
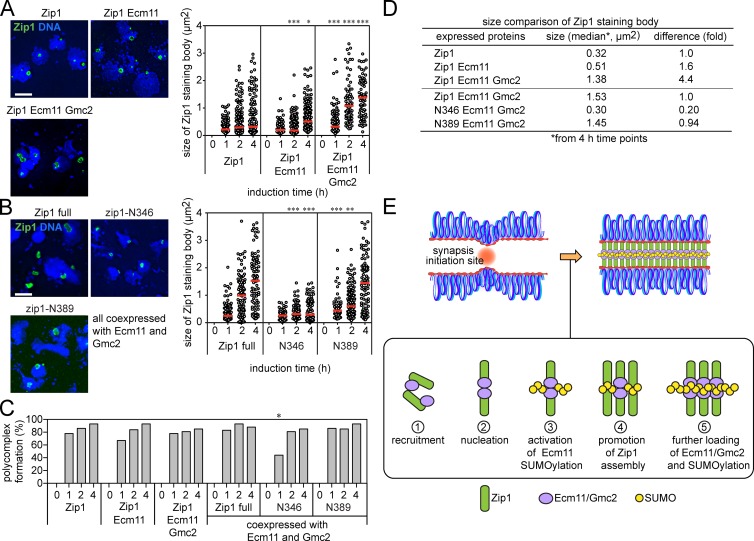
We then set out to examine how Ecm11 SUMOylation affects Zip1 assembly in vegetative cells. Nuclei were spread and Zip1 localization was examined cytologically in vegetative cells producing Zip1 alone, Zip1 and Ecm11, or Zip1, Ecm11, and Gmc2 (Fig. 5, A and C). 1 h after the start of protein induction, a nuclear body similar to a PC was seen in the majority of cells. The overall size of the PCs became progressively larger when Zip1 was coproduced with Ecm11 (1.6-fold at 4 h; Fig. 5 D), although they remained similar or slightly smaller than PCs in the cells producing only Zip1 at earlier time points. However, when both Ecm11 and Gmc2 were produced with Zip1, the size of PCs became even larger (4.4-fold at 4 h; Fig. 5 D), indicating that both Ecm11 SUMOylation and Gmc2 contribute to efficient development of Zip1 structures. We confirmed that the mitotic induction of these meiosis-specific proteins barely affected the cell cycle (Fig. S3, B and C), implying that the results presented in Fig. 5 A are not associated with cell-cycle abnormality.
Figure 5.
Ecm11 and Gmc2 promote Zip1 assembly in vegetative cells. (A and B) Representative images of Zip1 assembly bodies as visualized on chromosome spreads by immunostaining. Bar, 5 µm. Red bars, median values. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Mann-Whitney test), in comparison to Zip1 (top) or Zip1 full (bottom) at corresponding time points. (C) The ratio of chromosome spreads carrying a Zip1 assembly body was measured with the samples used in A and B. 100 spreads were examined for each time point. *, P < 0.05 (χ2 test). (D) Summary of Zip1 assembly body size. (E) Positive feedback loop between polySUMOylation of Ecm11 and transverse filament assembly.
We then applied this minimal assembly system to examine two truncated Zip1 proteins, Zip1-N346 and -N389 (Fig. 5, B and C). Zip1-N389 developed Zip1 PCs of similar sizes to those of full-length Zip1 (0.94-fold at 4 h; Fig. 5 D), although the kinetics of assembly was slightly delayed. In Zip1-N346, the size of the Zip1 bodies was substantially smaller than those of both full-length Zip1 and Zip1-N389 (0.20-fold at 4 h; Fig. 5, B and D). These observations suggest two things. First, the N-terminal 389 aa of Zip1 is sufficient for activating Ecm11 SUMOylation and promoting self-assembly. Second, Zip1-N346 is sufficient for activating Ecm11-SUMOylation but incapable of promoting self-assembly, thus suggesting that the domain necessary for activating Ecm11 SUMOylation is separable from that for promoting Zip1 self-assembly.
The discovery that the N terminus of Zip1 serves as an activator of Ecm11 SUMOylation provides insights into how TF assembly progresses (Fig. 5 E). At the beginning, an initiating mechanism recruits Zip1 and central element proteins (Ecm11 and Gmc2) to synapsis initiation sites (Fig. 5 E, step 1). It is interesting to note that Ecm11/Gmc2 can be recruited to synapsis initiation sites independently of Zip1 (Humphryes et al., 2013). Then, initial Zip1 nucleation occurs in association with Ecm11/Gmc2 (Fig. 5 E, step 2), followed by activation of Ecm11 SUMOylation by the Zip1 N terminus (Fig. 5 E, step 3). The ensuing formation of SUMO chains facilitates the recruitment and assembly of more Zip1 molecules (Fig. 5 E, step 4). This causes further recruitment of Ecm11/Gmc2 and SUMOylation of Ecm11 (Fig. 5 E, step 5), establishing a positive feedback loop. Both Zip1 N-termini and Ecm11/Gmc2 are located at the central element. Thus, we propose that the central element acts as the control center for SC assembly by regulating the extent of Ecm11 SUMOylation.
Materials and methods
Yeast strains
Strains used are listed in Table S1. Strains used in each figure are as follows. Fig. 1 A: TBR5896, 5296, and 6088; Fig. 1, B and C: TBR6621, 9451, 9446, 9450, and 11045; Fig. 2 A: TBR5296, 7613, 7611, and 7706; Fig. 2, B and C: TBR2065, 7569, 7567, and 7609; Fig. 2 D: TBR5296, 7829, and 7789; Fig. 2, E and F: TBR2065, 309, 7783, and 7785; Fig. 3: TBR10639, 10638, 10647, 10646, 10643, 10642, 10645, 10644, 10641, and 10640; Fig. 4 A: TBR5296, 6090, 6088, and 8904; Fig. 4 B: TBR6088, 8904, 9467, 9468, and 9469; Fig. 4, C and D: TBR9290, 6458, 9286, 9293, and 9455; Fig. 4 F: TBR9892, 9885, 9917, 9887, 9889, 9891, and 9916; Fig. 5 A: TBR7496, 9286, and 9293; Fig. 5 B: TBR9916, 9917, and 9887; Fig. S1 A: TBR7556, 11099, 11101, and 11103; Fig. S1 B: TBR5296 and 7706; Fig. S1 D: TBR2065 and 7609; Fig. S1 E: TBR11114, 11113, 6088, and 7885; Fig. S2: TBR5921, 7507, 5636, 7350, 5641, 7348, 5649, 7349, 5296, and 6088; Fig. S3 A: TBR9884, 9885, 9917, 9887, 9888, 9889, 9890, and 9891; Fig. S3, B and C: TBR7496, 9286, and 9293. Gene deletions, promoter swapping, and N and C terminus epitope tagging were performed using PCR-mediated techniques as described previously (Longtine et al., 1998). For construction of md alleles, the CLB2 promoter was inserted immediately upstream of the start codon of the gene of interest by PCR-mediated genetic manipulation (Lee and Amon, 2003). For construction of strains producing Ecm11 with various tags of different sizes (Fig. S1 A), the DNA fragment encoding C-terminal Ecm11 fused to 13myc, along with the KAN marker, was first cloned by PCR using the genomic DNA of TBR9446 as the template. The 13myc part was then replaced with a synthetic DNA sequence encoding 1xmyc, 4xmyc, or 7xmyc with the remaining elements (3′ end of the ECM11 gene, 3′UTR, and the KAN marker) retained. For construction of Zip1 truncation series, a DNA fragment encoding 825–875 aa of Zip1 and the Kluyveromyces lactis TRP1 gene were cloned next to each other, and the plasmid was used as a template for standard PCR-mediated genetic manipulation as above. The C terminus fragment of Zip1 carries a stretch of highly basic amino acids (KKRRRK, 870–875 aa), likely serving as NLS.
Meiotic induction
For BR1919 strains, a single colony grown on YPADU (1% yeast extract, 2% peptone, 0.3 mM adenine, 2% glucose, and 0.2 mM uracil) was resuspended in 2 ml liquid YPADU and grown overnight. This was supplemented with 5 ml fresh YPADU and grown for a further 8 h before centrifuging, washing with water, and resuspending in 50 ml of 2% potassium acetate. Cells were harvested as indicated in each experiment.
In SK1 strains, a single colony was resuspended in 10 ml liquid YPADU and grown to saturation. These cells were resuspended into 100 ml BYTA (1% yeast extract, 2% tryptone, 1% potassium acetate, and 50 mM potassium phthalate; Blitzblau and Hochwagen, 2011) to an OD600 of 0.5. After incubation for 12 h, cells were washed once and resuspended in 100 ml of 2% potassium acetate to an OD600 of 1.9. Cells were harvested as indicated in each experiment.
Protein induction in mitotic cells
Single colonies were inoculated in 2 ml complete medium with 2% glucose for 12 h and resuspended in 5 ml fresh complete medium. After 3 h of incubation, cells were collected from 5 ml of the culture, washed once, and then resuspended in 10 ml complete medium with 2% raffinose. The culture was further incubated for 3 h. After washing once, cells were resuspended in complete medium carrying 2% galactose for induction of appropriate proteins.
Statistics
Statistical analysis was performed using Prism and InStat software (GraphPad).
Cytology
Images were captured on the Deltavision IX70 system (Applied Precision; Olympus) with a 100× objective lens (NA 1.40) and a camera (CoolSNAP HQ2; Photometrics) using softWoRx software at RT. 7 consecutive z-slices with an interval of 0.2 µm were acquired, then processed by deconvolution using the constrained iterative deconvolution algorithm within softWoRx. The deconvolved z-slices were projected using a maximum-intensity algorithm to form the final processed image. Meiotic chromosomes were surface spread as described previously (Dresser and Giroux, 1988), except that glass slides were not precoated with plastic. In brief, meiotic cells from appropriate time points were digested with zymolyase in the presence of 1 M sorbitol to form spheroplasts. Cells were gently washed once, and chromosome spreads were prepared by resuspending spheroplasts in buffered solution without sorbitol (22 mM MES) and 3% PFA. Cell suspension was spotted onto clean glass slides and left for 20 min for spread chromosomes to settle. The slides with chromosome spreads were washed with PBS once and subjected to standard immunostaining protocols. Primary antibodies used were Zip1 and Red1 (rabbit/mouse, 400-fold dilution, S. Roeder (Yale University, New Haven, CT); Sym et al., 1993; Smith and Roeder, 1997). Secondary antibodies were Alexa Fluor 488 or 594 (Invitrogen).
Quantitative analysis of chromosomal Zip1 localization
Zip1 distribution was quantified using various tools within softWoRx. Projected spread images of pachytene nuclei or equivalent (determined by the presence of condensed chromatin via DAPI staining) were used to obtain signal intensity values for the spread-containing region of the image in the green channel. The 90th percentile value for signal intensity was calculated and used as a threshold value for the Polygon Finder tool in softWoRx, which identified continuous regions of Zip1 localization (stretches). The region incorporating the whole spread area (and a threshold perimeter of 0.78 µm for polygons) was specified, and polygons were calculated. Polygons identified outside the DAPI-stained area were manually deselected and not included in the data. Polygon number and area were recorded and used to evaluate Zip1 distribution. ≥20 spread projections were analyzed per strain.
Quantitative analyses of Zip1 PC area
Zip1 PC size was quantified using various tools within softWoRx. Projected spread images of pachytene nuclei or equivalent (determined by the presence of condensed chromatin via DAPI staining) were used to obtain signal intensity values for the spread-containing region of the image in the green channel. The Polygon Finder tool in softWoRx, which identified Zip1 stretches, was used to apply threshold values and identify regions of high Zip1 signal intensity as PCs. The selected area of the PC was calculated. 20 or more spread projections were analyzed per strain.
Immunoprecipitation
Native whole-cell extracts were prepared using 50 ml sporulating culture. Pelleted cells were resuspended in 400 µl lysis buffer (1 mM DTT, 0.05% Igepal CA-630, 200 mM NaCl, 10 mM EDTA, 10% glycerol, and 50 mM Tris-HCl, pH 8.0) containing protease inhibitors (1 mM PMSF and 1× protease inhibitor, EDTA-free [Roche], 50 mM N-ethylmaleimide, and 100 µM MG132). Cells were lysed by beating six times for 20 s in the presence of zirconia/silica beads at 4°C. Antiflag or antimyc antibody was incubated with whole-cell extract at 1:125 dilution. Bound proteins were retrieved using Protein G–coated Dynabeads (Invitrogen). Beads were washed, and bound proteins were eluted by boiling with sample loading buffer. The immunoprecipitates were subjected to Western blotting.
Quantification of Western blot
Western blot signals (Fig. 4 D) were quantitatively acquired by ImageQuant LAS4000 (GE) and normalized according to the total protein signal for each lane (Ponceau S stain). For Smt3 signal, the total amount of signal >46 kD of each lane was measured and normalized, and the amount of the signal at time zero of each strain background was subtracted. For both Smt3 and Ecm11, the y-axis represents the ratio to the maximum value of each experiment; in Smt3, the amount of signal at 5 h of the strain producing Ecm11, Gmc2, and Zip1 was set to 1.0, whereas in Ecm11, it is the 5-h time point of the strain producing Ecm11 and Gmc2.
Protein detection
Western blotting was performed as previously described (Hooker and Roeder, 2006). Antibodies used were Zip1 (rabbit, 5,000-fold dilution; S. Roeder; Sym and Roeder, 1994; Dong and Roeder, 2000), Smt3 (rabbit, 5,000-fold dilution; S. Roeder; Hooker and Roeder, 2006), flag (mouse, 5,000-fold dilution; Sigma-Aldrich), HA (mouse, 3,000-fold dilution; Covance), and myc (mouse, 3,000-fold dilution; Covance).
Online supplemental material
Fig. S1 shows the relationship between Ecm11 and polySUMO ladder migration and the effect of the siz1 siz2 mutations on Ecm11 SUMOylation. Fig. S2 shows the relationship between the level of Ecm11 SUMOylation and Zip1 assembly in the presence of Spo11. Fig. S3 shows the induction levels of various truncated Zip1 proteins in vegetative cells and the effect of the induction of Zip1, Ecm11, and Gmc2 on the cell cycle. Table S1 shows a list of the yeast strains used in this work, along with their respective genotypes. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201506103/DC1.
Supplementary Material
Acknowledgments
We thank Shirleen Roeder and Angelika Amon for strains and antibodies.
This work was supported by the Biotechnology and Biological Sciences Research Council (BB/I009159/1) and the Medical Research Council (doctoral studentship to B. Argunhan).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- DSB
- double-strand break
- md
- meiosis-specific depletion
- PC
- polycomplex
- SC
- synaptonemal complex
- SUMO
- small ubiquitin-like modifier
- TF
- transverse filament
References
- Agarwal S., and Roeder G.S.. 2000. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 102:245–255. 10.1016/S0092-8674(00)00029-5 [DOI] [PubMed] [Google Scholar]
- Bergerat A., de Massy B., Gadelle D., Varoutas P.C., Nicolas A., and Forterre P.. 1997. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 386:414–417. 10.1038/386414a0 [DOI] [PubMed] [Google Scholar]
- Blitzblau H.G., and Hochwagen A.. 2011. Genome-wide detection of meiotic DNA double-strand break hotspots using single-stranded DNA. Methods Mol. Biol. 745:47–63. 10.1007/978-1-61779-129-1_4 [DOI] [PubMed] [Google Scholar]
- Brar G.A., Yassour M., Friedman N., Regev A., Ingolia N.T., and Weissman J.S.. 2012. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 335:552–557. 10.1126/science.1215110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.H., Lo Y.H., Liang S.S., Ti S.C., Lin F.M., Yeh C.H., Huang H.Y., and Wang T.F.. 2006. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 20:2067–2081. 10.1101/gad.1430406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua P.R., and Roeder G.S.. 1998. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell. 93:349–359. 10.1016/S0092-8674(00)81164-2 [DOI] [PubMed] [Google Scholar]
- de Carvalho C.E., and Colaiácovo M.P.. 2006. SUMO-mediated regulation of synaptonemal complex formation during meiosis. Genes Dev. 20:1986–1992. 10.1101/gad.1457806 [DOI] [PubMed] [Google Scholar]
- Diaz R.L., Alcid A.D., Berger J.M., and Keeney S.. 2002. Identification of residues in yeast Spo11p critical for meiotic DNA double-strand break formation. Mol. Cell. Biol. 22:1106–1115. 10.1128/MCB.22.4.1106-1115.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., and Roeder G.S.. 2000. Organization of the yeast Zip1 protein within the central region of the synaptonemal complex. J. Cell Biol. 148:417–426. 10.1083/jcb.148.3.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresser M.E., and Giroux C.N.. 1988. Meiotic chromosome behavior in spread preparations of yeast. J. Cell Biol. 106:567–573. 10.1083/jcb.106.3.567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger C.S., and Jentsch S.. 2010. Synaptonemal complex formation and meiotic checkpoint signaling are linked to the lateral element protein Red1. Proc. Natl. Acad. Sci. USA. 107:11370–11375. 10.1073/pnas.1004248107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker G.W., and Roeder G.S.. 2006. A role for SUMO in meiotic chromosome synapsis. Curr. Biol. 16:1238–1243. 10.1016/j.cub.2006.04.045 [DOI] [PubMed] [Google Scholar]
- Humphryes N., Leung W.K., Argunhan B., Terentyev Y., Dvorackova M., and Tsubouchi H.. 2013. The Ecm11-Gmc2 complex promotes synaptonemal complex formation through assembly of transverse filaments in budding yeast. PLoS Genet. 9:e1003194 10.1371/journal.pgen.1003194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.S., and Gupta A.A.. 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 106:735–744. 10.1016/S0092-8674(01)00491-3 [DOI] [PubMed] [Google Scholar]
- Klug H., Xaver M., Chaugule V.K., Koidl S., Mittler G., Klein F., and Pichler A.. 2013. Ubc9 sumoylation controls SUMO chain formation and meiotic synapsis in Saccharomyces cerevisiae. Mol. Cell. 50:625–636. 10.1016/j.molcel.2013.03.027 [DOI] [PubMed] [Google Scholar]
- Lake C.M., and Hawley R.S.. 2012. The molecular control of meiotic chromosomal behavior: events in early meiotic prophase in Drosophila oocytes. Annu. Rev. Physiol. 74:425–451. 10.1146/annurev-physiol-020911-153342 [DOI] [PubMed] [Google Scholar]
- Lee B.H., and Amon A.. 2003. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science. 300:482–486. 10.1126/science.1081846 [DOI] [PubMed] [Google Scholar]
- Li S.J., and Hochstrasser M.. 1999. A new protease required for cell-cycle progression in yeast. Nature. 398:246–251. 10.1038/18457 [DOI] [PubMed] [Google Scholar]
- Li S.J., and Hochstrasser M.. 2000. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20:2367–2377. 10.1128/MCB.20.7.2367-2377.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F.M., Lai Y.J., Shen H.J., Cheng Y.H., and Wang T.F.. 2010. Yeast axial-element protein, Red1, binds SUMO chains to promote meiotic interhomologue recombination and chromosome synapsis. EMBO J. 29:586–596. 10.1038/emboj.2009.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., and Stock J.. 1991. Predicting coiled coils from protein sequences. Science. 252:1162–1164. 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- Macqueen A.J., and Roeder G.S.. 2009. Fpr3 and Zip3 ensure that initiation of meiotic recombination precedes chromosome synapsis in budding yeast. Curr. Biol. 19:1519–1526. 10.1016/j.cub.2009.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J., Kleckner N., and Börner G.V.. 2005. Bioinformatic analyses implicate the collaborating meiotic crossover/chiasma proteins Zip2, Zip3, and Spo22/Zip4 in ubiquitin labeling. Proc. Natl. Acad. Sci. USA. 102:17594–17599. 10.1073/pnas.0508581102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder G.S. 1997. Meiotic chromosomes: it takes two to tango. Genes Dev. 11:2600–2621. 10.1101/gad.11.20.2600 [DOI] [PubMed] [Google Scholar]
- Schwienhorst I., Johnson E.S., and Dohmen R.J.. 2000. SUMO conjugation and deconjugation. Mol. Gen. Genet. 263:771–786. 10.1007/s004380000254 [DOI] [PubMed] [Google Scholar]
- Shinohara M., Oh S.D., Hunter N., and Shinohara A.. 2008. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat. Genet. 40:299–309. 10.1038/ng.83 [DOI] [PubMed] [Google Scholar]
- Smith A.V., and Roeder G.S.. 1997. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell Biol. 136:957–967. 10.1083/jcb.136.5.957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead K., Aguilar C., Hartman T., Drexel M., Meluh P., and Guacci V.. 2003. Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J. Cell Biol. 163:729–741. 10.1083/jcb.200305080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M., and Roeder G.S.. 1994. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 79:283–292. 10.1016/0092-8674(94)90197-X [DOI] [PubMed] [Google Scholar]
- Sym M., Engebrecht J.A., and Roeder G.S.. 1993. Zip1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 72:365–378. 10.1016/0092-8674(93)90114-6 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Kahyo T., Toh-E A., Yasuda H., and Kikuchi Y.. 2001. Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J. Biol. Chem. 276:48973–48977. 10.1074/jbc.M109295200 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Yong-Gonzalez V., Kikuchi Y., and Strunnikov A.. 2006. SIZ1/SIZ2 control of chromosome transmission fidelity is mediated by the sumoylation of topoisomerase II. Genetics. 172:783–794. 10.1534/genetics.105.047167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi T., Zhao H., and Roeder G.S.. 2006. The meiosis-specific Zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with Zip2. Dev. Cell. 10:809–819. 10.1016/j.devcel.2006.04.003 [DOI] [PubMed] [Google Scholar]
- Tung K.S., and Roeder G.S.. 1998. Meiotic chromosome morphology and behavior in zip1 mutants of Saccharomyces cerevisiae. Genetics. 149:817–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K., Taylor L.F., Mukherjee P., Humphryes N., Tsubouchi H., and MacQueen A.J.. 2013. SUMO localizes to the central element of synaptonemal complex and is required for the full synapsis of meiotic chromosomes in budding yeast. PLoS Genet. 9:e1003837 10.1371/journal.pgen.1003837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Ajimura M., Padmore R., Klein C., and Kleckner N.. 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:6572–6581. 10.1128/MCB.15.12.6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavec A.B., Comino A., Lenassi M., and Komel R.. 2008. Ecm11 protein of yeast Saccharomyces cerevisiae is regulated by sumoylation during meiosis. FEMS Yeast Res. 8:64–70. 10.1111/j.1567-1364.2007.00307.x [DOI] [PubMed] [Google Scholar]
- Zickler D., and Kleckner N.. 1999. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33:603–754. 10.1146/annurev.genet.33.1.603 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.