Mice deficient for mitochondrial fission factor (Mff) die from severe cardiomyopathy, but cardiac function, tissue integrity, and mitochondrial respiration are robustly rescued by rebalancing mitochondrial dynamics through deletion of Mitofusin 1 (Mfn1).
Abstract
Defects in mitochondrial fusion or fission are associated with many pathologies, raising the hope that pharmacological manipulation of mitochondrial dynamics may have therapeutic benefit. This approach assumes that organ physiology can be restored by rebalancing mitochondrial dynamics, but this concept remains to be validated. We addressed this issue by analyzing mice deficient in Mff, a protein important for mitochondrial fission. Mff mutant mice die at 13 wk as a result of severe dilated cardiomyopathy leading to heart failure. Mutant tissue showed reduced mitochondrial density and respiratory chain activity along with increased mitophagy. Remarkably, concomitant deletion of the mitochondrial fusion gene Mfn1 completely rescued heart dysfunction, life span, and respiratory chain function. Our results show for the first time that retuning the balance of mitochondrial fusion and fission can restore tissue integrity and mitochondrial physiology at the whole-organ level. Examination of liver, testis, and cerebellum suggest, however, that the precise balance point of fusion and fission is cell type specific.
Introduction
Several debilitating diseases are caused by mutations in mitochondrial dynamics genes. Mutations in the mitochondrial fusion genes Mfn2 and Opa1 cause Charcot-Marie-Tooth neuropathy type 2A and dominant optic atrophy, respectively (Alexander et al., 2000; Delettre et al., 2000; Züchner et al., 2004). For mitochondrial fission, a Drp1 mutation has been shown to cause widespread neurological dysfunction and neonatal lethality (Waterham et al., 2007), and an Mff mutation has been found in two brothers with neuromuscular defects (Shamseldin et al., 2012).
In cultured cells, the balance between mitochondrial fusion and fission regulates mitochondrial morphology (Friedman and Nunnari, 2014; Mishra and Chan, 2014). This observation suggests that pharmacological agents that up- or down-regulate mitochondrial dynamics may have therapeutic value. Antagonists of mitochondrial fission have been developed (Cassidy-Stone et al., 2008; Qi et al., 2013) and may, in principle, benefit conditions associated with impaired mitochondrial fusion. To evaluate the feasibility of this approach, it is important to determine whether whole-organ physiology can be restored by resetting an optimal balance between mitochondrial fusion and fission.
In this study, we show that mice lacking the mitochondrial fission gene Mff die at ∼13 wk as a result of dilated cardiomyopathy. Mff is a mitochondrial and peroxisomal membrane protein that is a major receptor for Drp1, the effector of fission (Gandre-Babbe and van der Bliek, 2008; Otera et al., 2010; Losón et al., 2013). Heart-specific Drp1-null mice have a severe cardiac phenotype and die at 1–1.5 wk (Kageyama et al., 2014; Ishihara et al., 2015). Acute removal of Drp1 from adult heart similarly results in rapid heart failure (Ikeda et al., 2015; Song et al., 2015). Interestingly, removal of the mitochondrial fusion genes Mfn1 and Mfn2 also causes cardiac defects (Chen et al., 2011; Papanicolaou et al., 2012; Kasahara et al., 2013; Dorn et al., 2015; Song et al., 2015). Given the demonstrated importance of both mitochondrial fusion and fission in cardiomyocytes, we reasoned that the heart would be an ideal organ system to study how retuning of mitochondrial dynamics in vivo might regulate tissue physiology.
Results and discussion
Mff-deficient mice exhibit pleiotropic phenotypes
To examine the physiological role of Mff in mammals, we generated mice with a gene trap disruption of Mff (Fig. S1 A). Western blot analysis confirmed the lack or vast reduction of all Mff isoforms in homozygous gene trap (Mffgt) cells and tissues (Fig. S1, B and C). Loss of Mff resulted in a notable decrease in Drp1 and moderate reductions in other mitochondrial dynamics proteins (Fig. S1 D). The latter may result from a generalized reduction of mitochondria (see Fig. 4).
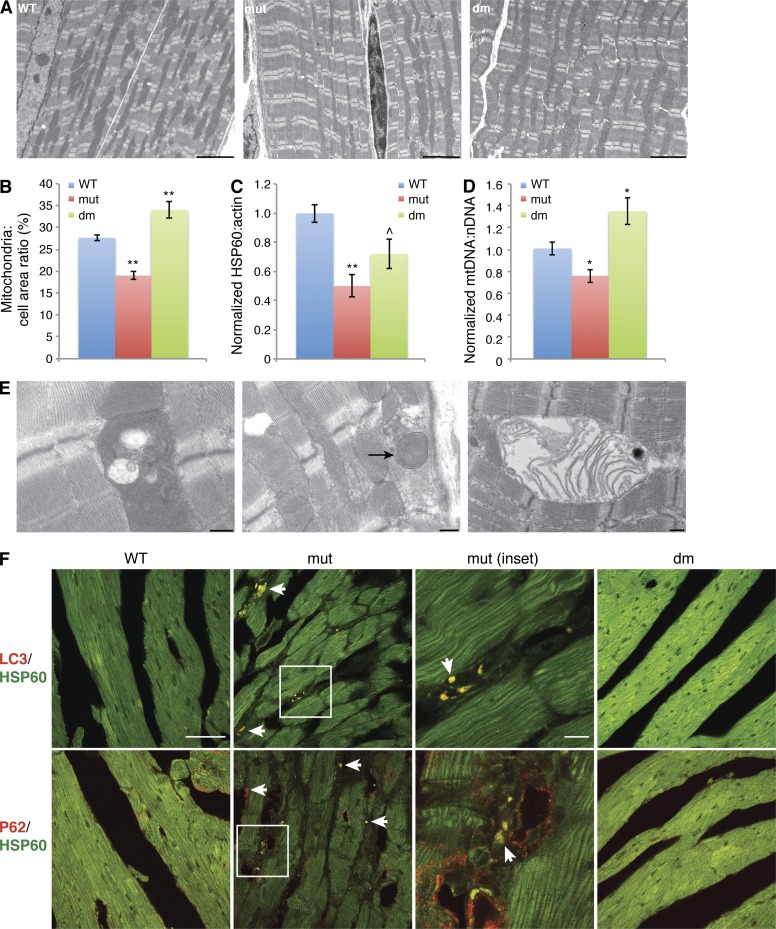
Figure 4.
Loss of mitochondria in Mffgt myocardium. (A) Low-magnification EM showing differences in mitochondrial mass. Bars, 5 µm. (B) Quantification of mitochondrial area in EM images. (C) HSP60/actin protein ratio, normalized to wild type (n ≥ 7). (D) mtDNA per cell, normalized to wild-type content (n ≥ 7). (E) EM of mutant myocardium, showing examples of defective mitochondria. Arrow indicates mitophagic structure. Bars, 0.5 µm. (F) Heart sections immunostained with LC3 or p62 (red) and HSP60 (green). Arrows indicate positive puncta. Mutant inset is enlarged. Bars: 10 µm; (mutant inset) 50 µm. Error bars = SEM. P-values as compared with wild type: ^, P ≤ 0.05; *, P ≤ 0.01; **, P ≤ 0.001. nDNA, nuclear DNA; WT, wild type.
Mffgt mice die prematurely with a mean life span of 13 wk (Fig. 1 A). The reduced life span results from dilated cardiomyopathy, as detailed in the following section. Mutants are notably smaller than their wild-type littermates (Fig. S1, E and F). In contrast to mitochondrial fusion–deficient mice that have metabolic signs of increased glycolysis (Chen et al., 2010), the Mff mutants have normal temperature, lactate, and glucose levels (Fig. S1 G). Mutants also suffer from malocclusion and kyphosis with incomplete penetrance (Fig. S1, H and I).
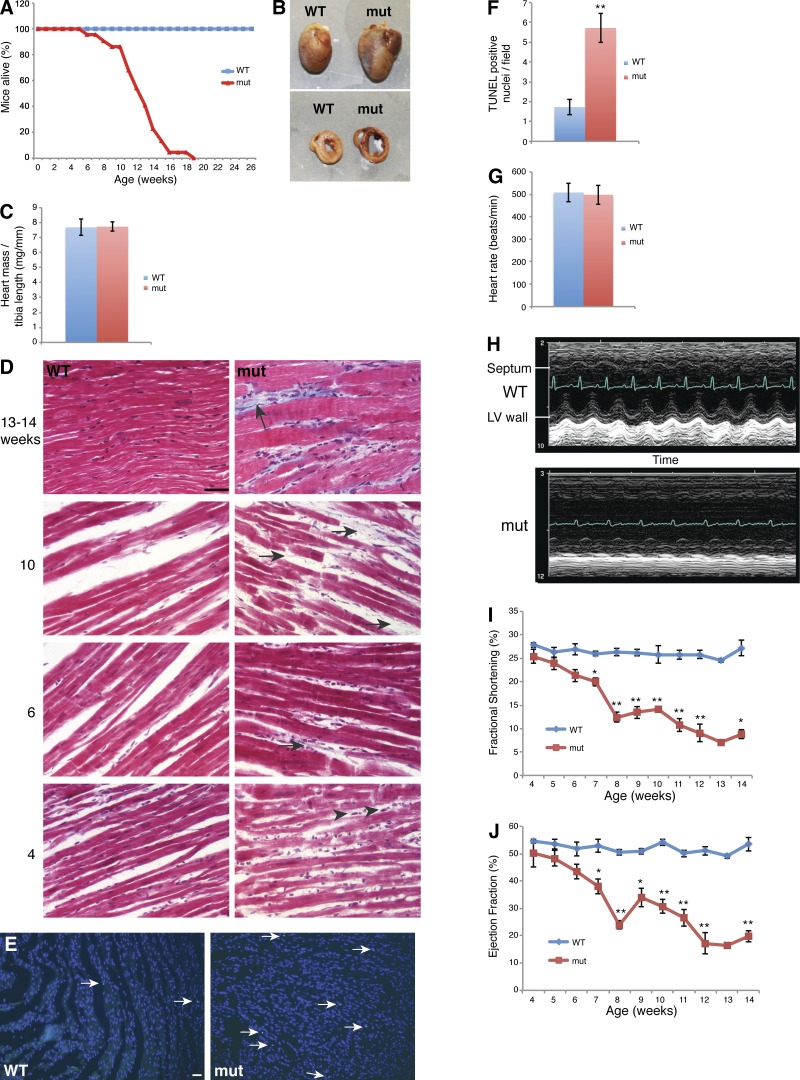
Figure 1.
Cardiomyopathy in Mffgt mice. (A) Survival curve. Mffgt mutant (mut) mice (n = 22) die by 13 wk on average, as opposed to wild-type controls (n = 14). (B) Increased size, dilation of chambers, and thinning of walls in mutant hearts. (C) Heart mass normalized to tibia length. (D) Masson’s trichrome stain of heart sections. Arrowheads indicate fibroblasts. Blue strands (arrows) denote collagen deposition. (E) TUNEL staining of heart sections. Arrows indicate apoptotic nuclei. (F) Quantification of apoptotic nuclei per field (n ≥ 11). (G) Heart rate (n ≥ 6). (H) M-mode echocardiogram of 13–14-wk-old mice. Electrocardiogram is superimposed. (I and J) Longitudinal echocardiography measuring fractional shortening (I) and ejection fraction (J). Study conducted on 5 wild-type and 10 Mffgt mice (five mutants died during the course of the study). Error bars = SEM. *, P ≤ 0.01; **, P ≤ 0.001. Bars, 50 µm. WT, wild type.
Mffgt mice demonstrate prominent neuromuscular defects. Their gait is altered, showing a side-to-side wobble and high step with a lowered tail (Video 1). Mutants also have decreased grasping ability (Fig. S1 J). Fertility of Mffgt mice is decreased in both males and females (Fig. S1 K). In males, this reduced fertility is associated with reduced sperm count (Fig. S1 L).
Finally, Mffgt mice demonstrate an altered immunological profile, with increased amounts of IgG heavy chain in tissues (Fig. S2 A). Whole blood contains normal IgG levels (Fig. S2 B), suggesting that IgG deposition, not production, is increased.
Dilated cardiomyopathy leads to heart failure
The early lethality of Mffgt mice is caused by dilated cardiomyopathy, leading to heart failure. Both atria and ventricles are notably enlarged in 13–14-wk-old mutants (Fig. 1 B). Cross sections show that whereas the lumens are expanded, the heart walls are thinner. As expected in dilated cardiomyopathy, heart weight (normalized to tibia length) does not differ between wild-type and mutant mice despite the larger size of mutant hearts (Fig. 1 C).
End-stage myocardium shows disruption of the myofibril architecture by dramatic interstitial fibrosis (Fig. 1 D). Fibroblast proliferation can be detected by 4 wk of age. Subsequent collagen deposition, detected by Masson's trichrome staining, appears as wisps of blue from 6 to 8 wk of age and broad bands at late stages. Fibrosis is a common hallmark of human cardiomyopathies. The replacement of cardiomyocytes by fibroblasts and collagen disrupts myocardial structure and interferes with heart contractility (Harvey and Leinwand, 2011). Because fibrosis is generally considered a sequela to myocardiocyte death, we performed TUNEL staining to investigate the apoptotic rate. We found a threefold increase in apoptosis in Mffgt hearts (Fig. 1, E and F), but the overall rate remains low and is unlikely to account for the fibrosis.
Although heart rate and rhythm remain largely unaffected, echocardiography confirms severe heart failure in 13–14-wk-old Mffgt mice (Fig. 1, G and H). M-mode readings expose a severe decrease in contraction of the left ventricular wall, resulting in an almost static left ventricle (LV) chamber volume throughout the cardiac cycle. Longitudinal experiments indicate that both fractional shortening and ejection fraction of the LV are fairly normal in 4–5-wk-old Mffgt mice. Thereafter, systolic function steadily deteriorates with age, culminating in heart failure (Fig. 1, I and J).
Deficient respiratory chain function in mitochondria of Mffgt heart
Cellular metabolism of cardiomyocytes predominantly relies on oxidative phosphorylation of fatty acids (Lopaschuk and Jaswal, 2010). Respiration, measured by oxygen consumption rates of isolated heart mitochondria, is severely compromised in Mff mutants. By 6 wk of age, the mutant respiratory control ratio, the most useful indicator of mitochondrial function (Brand and Nicholls, 2011), is 50% that of the wild type (Fig. 2 A). Consistent with reduced respiratory chain function, the steady-state level of ATP in end-stage Mffgt hearts is half that of the wild type (Fig. 2 B). At 8 wk, the difference in ATP is not yet statistically significant, suggesting that heart tissue can maintain overall ATP levels for a time even when respiratory capacity is compromised. It is likely, however, that sporadic cardiomyocytes can have reduced ATP and die, leading to a fibrotic response.
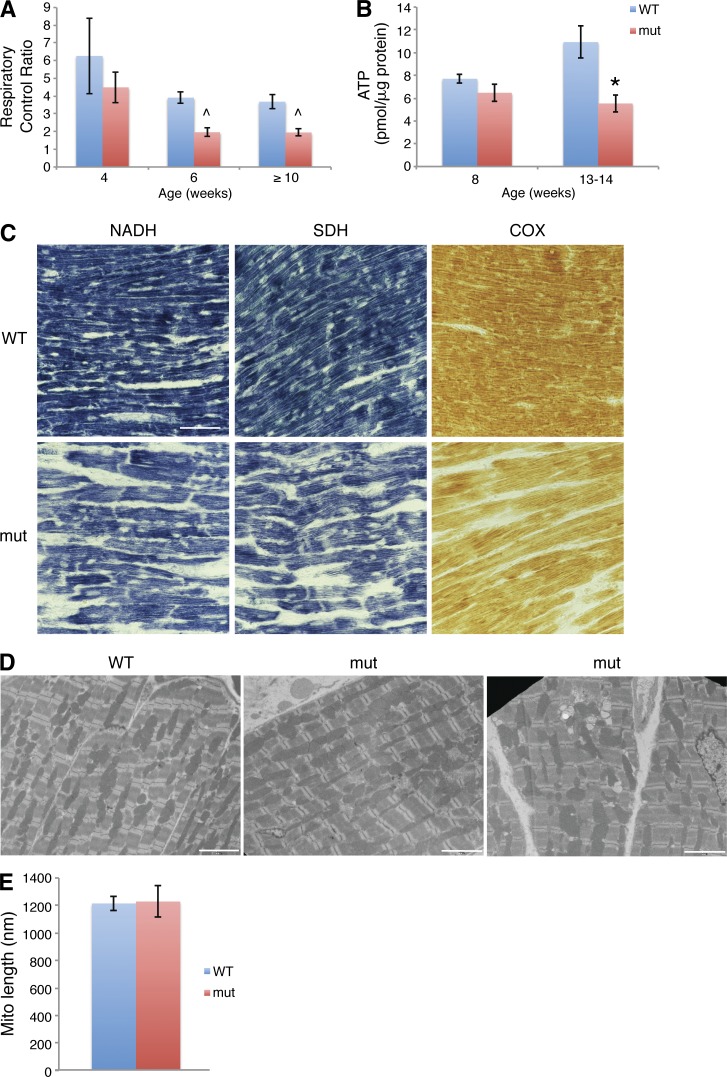
Figure 2.
Mitochondrial function and structure in Mffgt hearts. (A) Respiratory control ratio (ADP stimulated/endogenous) of isolated heart mitochondria. A significant decrease is observed at 6 wk (n = 2 samples; two mice/sample) and ≥10 wk (three mutants; five wild types), but not at 4 wk (n = 3 samples; two mice/sample). (B) ATP levels in heart (n = 6). (C) Histochemical stains of mitochondrial respiration complex activities in hearts. Bar, 50 µm. (D) EM of cardiomyocytes. Bars, 10 µm. (E) Mean mitochondrial length. Error bars = SEM. ^, P ≤ 0.05; *, P ≤ 0.01. WT, wild type.
Histochemical analysis demonstrates reduced activity of respiratory complexes I (NADH dehydrogenase), II (succinate dehydrogenase [SDH]), and IV (cytochrome c oxidase [COX]) in Mff mutants (Fig. 2 C and Fig. S2 C). This profile is distinct from that of fusion-deficient mitochondria, where there is decreased NADH dehydrogenase and COX but increased SDH activity (Chen et al., 2007, 2010). The parallel loss of these activities in Mffgt and Drp1−/− hearts (Kageyama et al., 2014; Ishihara et al., 2015) suggests that the fission-deficient respiration defects are not caused by loss of mitochondrial DNA (mtDNA). Interestingly, metabolite profiling of mutant heart tissue revealed a concurrent decrease in key Krebs cycle components (Fig. S2 D), supporting a generalized deficiency in energy production in Mff-deficient mitochondria.
In contrast to Mffgt fibroblasts, which have dramatically elongated mitochondria (Losón et al., 2013), mutant cardiomyocytes do not show a substantial change in mitochondrial length (Fig. 2, D and E). Instead, they demonstrate increased heterogeneity in mitochondrial shape and abundance (compare the two mutant images in Fig. 2 D).
Decreasing mitochondrial fusion rescues Mffgt lethality
Mffgt mice die at 13 wk of age. Mfn1-deficient mice (Mfn1loxP/loxP [Chen et al., 2007],Sox2-Cre [Hayashi et al., 2002]) die at birth (Fig. S2 E). Remarkably, when these two lethal mutations are combined, Mffgt,Mfn1loxP/−,Sox2-Cre double mutant animals (dm) demonstrate reciprocal rescue (Fig. 3 A and Fig. S2 E). In other words, ablating Mfn1 eliminates the cardiac-related lethality of Mffgt mice, and mutating Mff prevents the perinatal lethality of Mfn1-deficient mice. In fact, a cohort of 10 dm mice followed for over a year never exhibited signs of ill health.
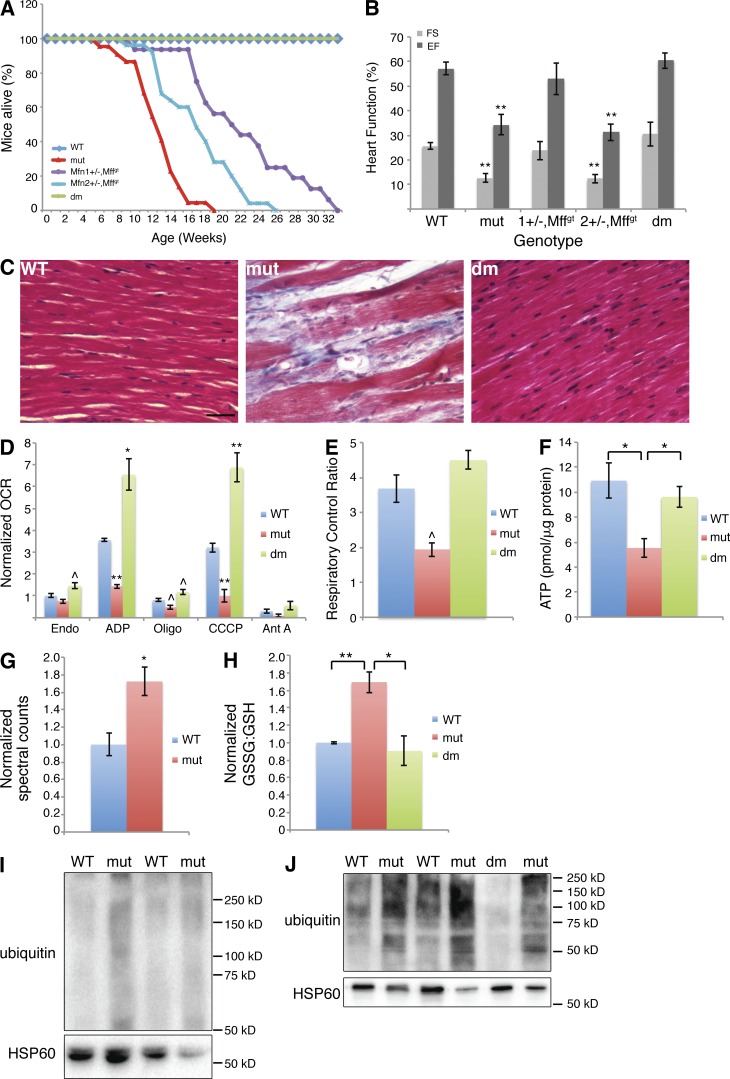
Figure 3.
Rescue of Mffgt phenotypes by deletion of Mfn1. (A) Survival curve demonstrating extension of life with loss of Mfn alleles (n ≥ 14). (B) Echocardiography of different genotypes (n = 3–8). (C) Masson's trichrome stained heart. Bar, 50 µm. (D) Normalized oxygen consumption rates (OCR) of isolated heart mitochondria (≥10 wk; n = 3–5). (E) Respiratory control ratios (n = 3–5). (F) ATP levels in heart (n = 6). (G) Normalized oxidized glutathione levels of 13–14-wk-old hearts (n = 5). (H) Normalized oxidized/reduced glutathione ratio. Increased level in mutants (n = 8) over the wild type (n = 7) is abolished in dm hearts (n = 3). (I and J) Ubiquitin protein blots of 4- (I) and 13–14- (J) wk-old heart mitochondria. Error bars = SEM. P-values reflect comparisons to the wild type unless otherwise noted: ^, P ≤ 0.05; *, P ≤ 0.01; **, P ≤ 0.001. Ant A, complex III inhibitor; CCCP, uncoupler; EF, ejection fraction; Endo, endogenous; FS, fractional shortening; Oligo, ATPase inhibitor; WT, wild type.
The rescue by a mitofusin deletion strongly suggests that mitochondrial, rather than peroxisomal, defects cause the Mffgt cardiomyopathy. Peroxisomal shape was not changed in Mffgt cardiomyocytes (Fig. S2 F). Moreover, in Mffgt mouse embryonic fibroblasts (MEFs), both peroxisomes and mitochondria were highly elongated, and simultaneous removal of Mfn1 restored the morphology of mitochondria, but not peroxisomes (Fig. S2, G and H).
Interestingly, even single mutant alleles of either Mfn1 or Mfn2 have a beneficial effect on the life span of Mffgt mice (Fig. 3 A). Using our original null alleles (Chen et al., 2003), we determined that Mffgt,Mfn2+/− mice live to ∼17 wk, and Mffgt,Mfn1+/− mice live to ∼21 wk (60% life span extension). However, Mffgt,Mfn2loxP/−,Sox2-Cre double mutants die perinatally, as do Mfn2lox/loxP,Sox2-Cre single mutants. Thus, in direct contrast to the Mfn1 mutant, Mff deficiency cannot restore viability to Mfn2-deficient mice.
To determine whether Mfn2 deletion in Mffgt mice can fully rescue the cardiomyopathy, we used the Myh6-Cre transgenic (Agah et al., 1997) to selectively ablate Mfn2 in the heart, thus circumventing the lethality caused by universal deletion of Mfn2. The Mffgt,Mfn2loxP/loxP,Myh6-Cre double mutants had a life span extension identical to that of Mffgt,Mfn2+/− mice (Fig. S2 I). Therefore, deletion of Mfn2 only modestly extends life span, whereas deletion of Mfn1 completely rescues the Mffgt cardiomyopathy. Collectively, these results show that the Mff cardiomyopathy can be rescued to differing degrees, depending on the Mfn gene and number of alleles removed. Therefore, optimal rescue requires precise titration of the levels of fusion.
Restoration of cardiac contractility, mitochondrial function, and cellular integrity in dm hearts
The life span extension observed in combining Mfn and Mff mutations is associated with restoration of cardiac function, as indicated by echocardiography of 13–14-wk-old mice (Fig. 3 B). Mff/Mfn1 mutants, whether Mfn1+/− or dm, exhibit fully normal cardiac contractility. Mffgt,Mfn2+/− mice, however, retain substantial systolic dysfunction, consistent with the shorter life span of these mice. Histological analysis confirms tissue integrity of the myocardium in dm mice (Fig. 3 C). Surprisingly, dm cardiac mitochondria not only have restored respiration, but they also consistently demonstrate oxygen consumption rates exceeding those of wild-type mitochondria (Fig. 3 D). As expected, dm respiratory control ratios and ATP levels are normal (Fig. 3, E and F).
Metabolomics analysis of 13–14-wk-old hearts reveals an increased level of oxidized glutathione in Mff mutants (Fig. 3 G). Fluorometric measurements of the oxidized/reduced glutathione ratio (GSSG/GSH) confirm this shift toward oxidation (Fig. 3 H). Because glutathione is one of the key redox regulators in the heart, this change implies increased oxidative stress (Chen and Zweier, 2014). dm hearts have GSSG/GSH ratios similar to those of wild-type hearts. Ubiquitination, a common response to oxidative stress and first step in both proteasome-mediated protein degradation and mitophagy, is also increased in Mffgt mitochondria (Fig. 3, I and J). Significantly, this difference can be seen at 4 wk of age and therefore precedes myocardial contractile dysfunction.
Multiple lines of evidence suggest that there is a generalized loss of mitochondrial density and integrity in Mffgt hearts. Quantitative EM analysis reveals a reduction in the area occupied by mitochondria to 68% of that in wild-type cells (Fig. 4, A and B). The HSP60 (mitochondrial)/actin (cytoplasmic) protein ratio shows a drop in mitochondrial protein content to 55% of the wild type in 13–14-wk-old Mff mutants (Fig. 4 C). The TOM20/actin ratio confirms this reduction (Fig. S3 A). Quantitative real-time PCR reveals an age-dependent decrease in mtDNA molecules per Mffgt cell (Fig. 4 D and Fig. S3 B). EM images show an increase in structurally compromised mitochondria, with internal vacuolization along with both early and late stage mitophagy intermediates (Fig. 4 E). Revealingly, LC3 and p62 puncta are increased in Mffgt heart and colocalize with mitochondria (Fig. 4 F). Collectively, the findings of reduced mitochondrial density, protein and mtDNA coupled with increased ubiquitination, and LC3 and p62 puncta strongly suggest that mitophagy is elevated in Mffgt cardiomyocytes. Consistent with their normal heart function, dm animals exhibit low levels of ubiquitination, no LC3 or p62 puncta, along with partial normalization of mitochondrial protein (Figs. 3 J; 4, C and F; and S3 A). Interestingly, mitochondrial area and mtDNA levels are actually elevated in dm hearts (Fig. 4, A, B, and D).
Rescue of mutant phenotypes is tissue specific
In Mffgt heart, retuning of mitochondrial dynamics to an alternative but optimal equilibrium dramatically restores tissue integrity and mitochondrial function. However, each tissue or organ has its own metabolic requirements, and we wondered whether there would be tissue-specific effects. The presence of multiple tissue defects in Mff, Mfn1, and Mfn2 mutants allows us to examine this issue.
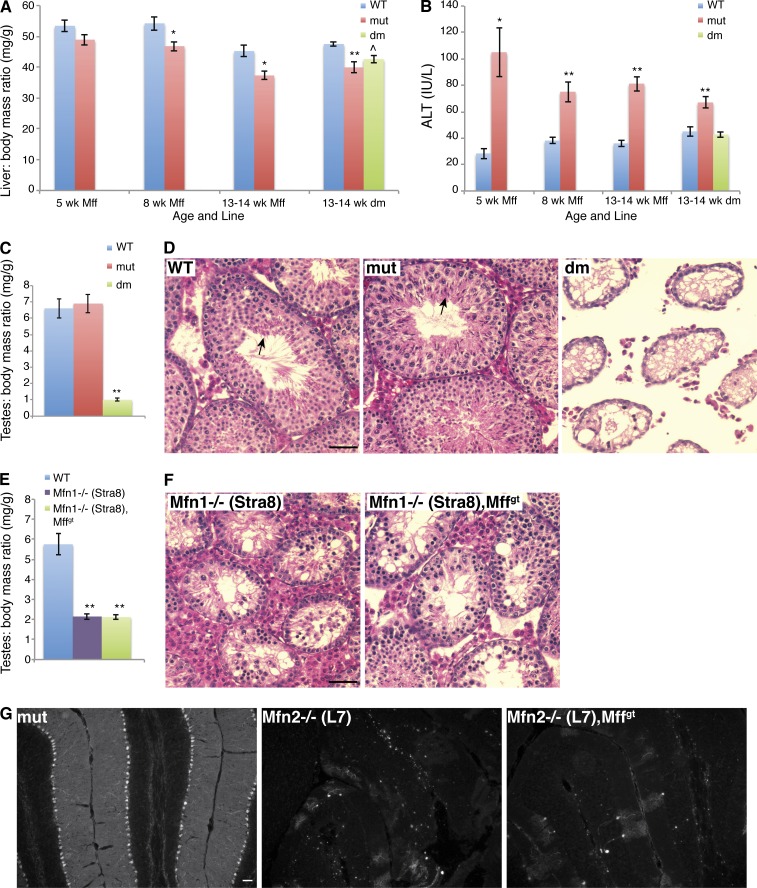
The livers in Mff mutants are consistently smaller than wild-type livers, even when normalized to body weight (Fig. 5 A). The discrepancy in liver mass increases with age and is associated with greatly elevated alanine aminotransferase (ALT) levels, an indicator of hepatocyte injury (Fig. 5 B). dm mice have normal levels of ALT, and their liver size is midway between that of wild-type and mutant animals. Therefore, in liver as in heart, Mfn1 deficiency compensates for the loss of Mff. However, targeted deletion of Mfn2 in Mffgt liver by Alb-Cre (Postic et al., 1999) does not rescue liver size (Fig. S3 C).
Figure 5.
Mffgt and dm phenotypes in liver, testis, and cerebellum. (A) Comparison of liver to body mass ratio (n ≥ 10). (B) ALT levels in serum (n = 5–13). (C and E) Testis to body mass ratio in Mffgt and dm (C; n ≥ 5) and Stra8-Cre lines (E; n ≥ 6). (D and F) Hematoxylin and eosin stain of testicular cross sections in 8–9-wk-old Mffgt and dm (D) and Stra8-Cre lines (F). Note that only control and Mffgt sections contain spermatozoa (arrows). (G) Anticalbindin immunofluorescence illuminating cerebellar PC. Error bars = SEM. P-values as compared with the wild type: ^, P ≤ 0.05; *, P ≤ 0.01; **, P ≤ 0.001. Bars, 50 µm. WT, wild type.
Although mutation of Mff rescues the perinatal lethality of Mfn1loxP/loxP,Sox2-Cre mice (Fig. S2 E), it does not rescue two other mitofusin phenotypes. Whereas wild-type and Mffgt testes are equivalent in size when adjusted for body mass, those of dm mice are much smaller (Fig. 5 C). Histological analysis reveals severe degeneration of the testis in dm mice (Fig. 5 D). Because Mfn1loxP/loxP,Sox2-Cre mice die before puberty, their testes could not be examined. Instead, we used the testis-specific Stra8-Cre (Sadate-Ngatchou et al., 2008) to create Mfn1-deficient testes. Indeed, these testes are also greatly decreased in size (Fig. 5 E). Importantly, Mffgt,Mfn1loxP/−,Stra8-Cre testes are indistinguishable from Mfn1loxP/loxP,Stra8-Cre testes (Fig. 5 F), confirming that the testis defect in dm mice is a result of loss of Mfn1 and cannot be rescued by disruption of Mff.
Elimination of Mfn2 in cerebellar Purkinje cells (PCs) by L7-Cre results in cell death by 3 mo of age (Chen et al., 2007). Genetically combining Mfn2 and Mff mutations did not prevent loss of PCs (Fig. 5 G). Interestingly, Mffgt PCs are viable, in direct contrast to Drp1−/− PCs (Wakabayashi et al., 2009).
Summary
Our results indicate that Mffgt cardiomyocytes have reductions in mitochondrial density, protein and mtDNA associated with increased mitophagy, and decreased respiratory complex activity. These features lead to an eventual reduction in cardiac ATP levels. We suspect that this energy deficiency promotes cardiomyocyte cell death, leading to fibrosis and heart failure.
Moreover, our results show that the balance between mitochondrial fusion and fission is critical for maintaining myocardium integrity and mitochondrial physiology. As one of several Drp1 receptors, Mff disruption, compared with Drp1 deletion, generates a partial fission defect (Losón et al., 2013), a feature that may be necessary for functional rescue. By combining Mffgt with heterozygous or homozygous mutations in Mfn1 or Mfn2, we generated an allelic series in which the level of mitochondrial fusion can be varied, resulting in differing degrees of rescue. In the most optimal genetic combination, we obtained full restoration of myocardial integrity, contractility, and respiratory chain activity. These observations indicate that multiple equilibria for mitochondrial fusion and fission are compatible with tissue and cellular function. As in the heart, Mfn and Mff mutations can work in concert to protect the liver from damage caused by Mff disruption. However, such patterns of phenotypic rescue were not observed in the testis or cerebellum.
These results suggest that pharmacological inhibition of mitochondrial fusion or fission has the potential to restore tissue function in a pathological state caused by reduction of the opposing process. However, our results also caution that the degree of therapeutic benefit is likely to be context dependent and will depend critically on precisely fine-tuning the balance between fusion and fission.
Materials and methods
Mouse experiments
The Mff gene trap embryonic stem cell clone (Wellcome Trust Sanger Institute) was microinjected into mouse blastocysts by the Mutant Mouse Resource and Research Centers. Germline transmission was evaluated by coat color, and the resulting line was confirmed by PCR analysis and DNA sequencing of the gene trap insertion site. Mice were maintained on a mixed 129P2,OlaHsd,C57BL/6J background. Female Mffgt/+,Mfn1loxP/loxP × male Mffgt/+,Mfn1+/−,Sox2Cre matings were used to generate Mffgt,Mfn1loxP/−,Sox2-Cre animals (termed dm for double mutant). Mfn1− is a null allele that contains a deletion of the G1 and G2 motifs of the GTPase domain (Chen et al., 2003). Mfn1loxP is a conditional allele in which loxP sites flank exon 4 (Chen et al., 2007). Sox2-Cre is a transgenic line with Cre under the control of the Sox2 regulatory sequence (Hayashi et al., 2002). Mouse experiments were conducted under protocols approved by the Caltech Institutional Animal Care and Use Committee.
Physiological measurements were performed on 13−14-wk-old mice. Body temperature was measured with a rectal probe, using the Thermalert Monitoring Thermometer (Braintree Scientific). To obtain blood measurements, cardiac puncture was performed on mice anesthetized with isoflurane, and the blood was analyzed with the iSTAT system (Abbott). Mice were exercised by swimming for 20 min at ∼37°C before being anesthetized. The grasp test was performed by having mice grasp onto a wire rack and then inverting the rack over a padded surface. Mice were allowed to move around the rack freely. Time was counted from inversion until the mouse released all four paws or 5 min was reached, whichever came first. Fertility was tested by mating mutants to wild-type mice of the opposite sex and counting days from cohabitation until a litter was born. Controls were Mffgt/+ crosses.
Sperm count
Both caudal epididymes were minced in 3 ml of prewarmed PBS and returned to 37°C for 15 min to allow sperm to swim out. The solution was titurated, and 1 ml was removed to a microfuge tube to sit overnight for sperm to stop swimming before counting on a hemocytometer.
Echocardiography
Animals were anesthetized with isoflurane (1.5% induce and maintenance) and placed on a temperature-controlled table (40°C) while maintaining a heart rate of ∼500 beats per minute. Hair was removed from the chest with a hair removal cream (Veet; Reckitt Benckiser), and preheated ultrasound transmission gel was applied. Echocardiograms were performed with a high-resolution ultrasound instrument (Vevo 2100; VisualSonics, Inc.).
A parasternal long-axis B-mode image was obtained. The maximal long axis of the LV was positioned perpendicular to the ultrasound beam. A 90° rotation of the ultrasound probe at the papillary muscle level was performed to obtain a parasternal short-axis view of the LV. An M-mode image was captured to document LV dimensions. Images were saved and analyzed using the Vevo 2100 cardiac analysis package. Echocardiograms were conducted under the guidelines approved by the University of California, Los Angeles (UCLA), Animal Research Committee.
Antibodies for Western blots
Antibodies to the following proteins were used: HSP60 (goat; sc-1052; Santa Cruz Biotechnology, Inc.), ubiquitin (mouse; 3936P; Cell Signaling Technology), actin (rabbit; A2103; Sigma-Aldrich), histone H2B (rabbit; IMG-359; Imgenex), Mfn1 (raised in chicken against mouse Mfn1 residues 348–579; Chen et al., 2003), Mfn2 (rabbit; D2D10; Cell Signaling Technology), Opa1 (mouse ascites produced in-house against the mouse S1 isoform), Fis1 (rabbit; ALX-210-1037-0100; Enzo Life Sciences), Drp1 (mouse; 611113; BD), and Mff (raised in rabbit against human Mff by A. van der Bliek, UCLA, Los Angeles, CA; Gandre-Babbe and van der Bliek, 2008). All secondary antibodies for Western blots were HRP conjugated and obtained from Jackson ImmunoResearch Laboratories, Inc.
Histology
For hematoxylin and eosin or Masson’s trichrome staining, formalin-fixed tissues were embedded in paraffin and sectioned to 5 µm. Slides were dewaxed and rehydrated. For hematoxylin/eosin, slides were stained with Harris-modified hematoxylin, blued with 0.1% NH4OH, stained with eosin, dehydrated, and cover-slipped in xylene-based mounting medium. Trichrome stain was performed according to instructions from the WebPath Internet Pathology Laboratory (The University of Utah Eccles Health Sciences Library). TUNEL analysis was performed according to the manufacturer’s instructions with the ApopTag Fluorescein In situ Apoptosis Detection kit (EMD Millipore). For NADH/SDH/COX staining, freshly dissected tissue was embedded in OCT compound (Tissue-Tek), frozen on dry ice, and sectioned to 10 µm. Staining protocols were obtained from the Washington University Neuromuscular Disease Center website. Because of the high mitochondrial content of cardiomyocytes, staining times were decreased to 10 min, 15 min, and 10 min, respectively. Quantification was conducted by using ImageJ (National Institutes of Health) to mask areas without myofibers and to measure mean intensity in remaining pixels.
For visualization of PCs, 14-µm midsagittal cryosections of formalin-perfused, OCT-embedded brain were immunostained with anticalbindin (Sigma-Aldrich), followed by Cy3-sheep anti-mouse (Jackson ImmunoResearch Laboratories, Inc.).
Images were acquired using a camera (Spot RT; SPOT Imaging Solutions) and NIS Elements software (Nikon) on a microscope(Eclipse E600; Nikon) at RT. Objectives (Nikon) used were Phase 10× (NA 0.25), Plan Fluor 20× (NA 0.5), and Plan Fluor 40× (NA 0.75). Photoshop (Adobe) was used only to change whole-image brightness/contrast and to crop.
Cell culture and immunofluorescence
MEFs expressing mito-DsRed were grown in DMEM containing 10% bovine calf serum. Peroxisomes were visualized by staining for Pex14 (rabbit; 10594–1-AP; Protein Tech) followed by a secondary antibody labeled with Alexa Fluor 488. Cells with any peroxisomes ≥2 µm long were scored as having tubular peroxisomes. Wild-type and Mff patient fibroblasts were provided by F. Alkuraya (Alfaisal University, Riyadh, Saudi Arabia).
For LC3 (rabbit; 2775; Cell Signaling Technology), P62 (rabbit; PM045; MBL), and PEX14 (rabbit; 10594–1-AP; Protein Tech) immunofluorescence, 10-µm cryosections of formalin-perfused, OCT-embedded heart was immunostained with the relevant antibodies and an Alexa Fluor 546–labeled donkey anti-rabbit (Life Technologies) secondary. HSP60 (sc-1052; Santa Cruz Biotechnology, Inc.) was visualized with Alexa Fluor 488–labeled donkey anti-goat.
Images were acquired using Zen 2009 software (Carl Zeiss) on a confocal microscope (LSM710; Carl Zeiss) at RT. A Plan-Apochromat 63×/1.4 oil objective was used. Photoshop was used only to change whole-image brightness/contrast and to crop.
EM analysis
Mice were perfused with 3% paraformaldehyde, 1.5% glutaraldehyde, 100-mM cacodylate, 5-mM CaCl2, and 2.5% sucrose, pH 7.4. Dissected LV tissue was postfixed for 2 h. The papillary muscle was removed and trimmed into small 1 × 2-mm3 pieces. Samples were postfixed in Palade’s OsO4, en bloc stained in Kellenberger uranyl acetate, dehydrated through a graded series of EtOH, and flat embedded in EMBED 812 (EMS). 80-nm longitudinal sections of papillary muscle were prepared on an ultramicrotome (UCT; Leica), collected onto 400 mesh high-transmission nickel grids, and poststained with lead citrate and uranyl acetate. Images were collected with a transmission electron microscope (Tecnai 12; FEI) operating at 100 kV and equipped with an Olympus Soft Imaging System (OSIS) digital camera (Megaview III; Olympus). 8-bit TIFF images were subsequently exported from the OSIS iTEM database, and figures were assembled in Photoshop with only linear adjustments made in brightness and contrast.
To quantify mitochondrial length, the freehand line tool available in the OSIS iTEM software (version 5.0; Olympus) was used to measure the lengths of hundreds of mitochondria for each genotype. To quantify mitochondrial area, eight random images were captured at a magnification of 4,900. The total cardiac muscle area and total mitochondrial area were determined using the freehand polygon line tool in the OSIS iTEM software (version 5.0). The percent ratio of total mitochondrial area versus total cardiac muscle area was calculated for each genotype.
Cellular respiration
Mitochondria were isolated from heart in MSHE (70-mM sucrose, 220-mM mannitol, 10-mM Hepes, 1-mM EGTA, and 0.5% BSA, pH 7.2). Tissue was minced with a tissue homogenizer (Polytron; VWR) and centrifuged twice at 800 g to remove cellular debris. The mitochondrial fraction was isolated with an 8,500 g spin, and the pellet was resuspended and spun a second time. The final pellet was resuspended in 50 µl MSHE, and protein quantification was performed. Mitochondria were diluted in MAS/PM (70-mM sucrose, 220-mM mannitol, 10-mM Hepes, 1-mM EGTA, 0.2% BSA, 10-mM KH2PO4, 5-mM MgCl2, pH 7.2, 5-mM pyruvate, and 2.5-mM malate), plated at 0.8 µg/well, and analyzed on the XF96 Extracellular Flux Analyzer (Seahorse Bioscience). Drugs added were 4-mM ADP, 5-µM oligomycin, 5-µM CCCP, and 5-µM antimycin A.
Biochemical measurements
The following measurements were performed according to the manufacturer’s instructions: ATP Colorimetric Assay kit (BioVision) with Deproteinizing Sample Preparation kit (BioVision); GSH/GSSG Ratio Detection Assay kit (Fluorometric green; Abcam), and ALT Reagent (Colorimetric, Endpoint Method; BQ kits).
mtDNA quantification
To quantify the amount of mtDNA per nuclear genome, we used the following primers: mtDNA forward primer, 5′-CCTATCACCCTTGCCATCAT-3′; mtDNA reverse primer, 5′-GAGGCTGTTGCTTGTGTGAC-3′; nuclear DNA (Pecam) forward primer, 5′-ATGGAAAGCCTGCCATCATG-3′; and nuclear DNA (Pecam) reverse primer, 5′-TCCTTGTTGTTCAGCATCAC-3′. Quantification of relative copy number differences was performed using the difference in threshold amplification between mtDNA and nuclear DNA (ΔΔC(t) method).
Metabolite profiling
Three distinct liquid chromatography tandem mass spectrometry methods were used to measure polar metabolites and lipids in plasma and heart homogenates. Negative ionization mode polar metabolite profiling data were acquired using an ACQUITY UPLC (Waters Corp.) coupled to a 5500 QTRAP triple quadrupole mass spectrometer (AB SCIEX). Plasma or tissue homogenate samples (30 µl) were extracted using 120 µl of 80% methanol containing 0.05 ng/µl inosine-[15N]4, 0.05 ng/µl thymine-d4, and 0.1 ng/µl glycocholate-d4 as internal standards (Cambridge Isotope Laboratories, Inc.). The samples were centrifuged (10 min at 9,000 g and 4°C), and the supernatants (10 µl) were injected directly onto a 150 × 2.0-mm column (Luna NH2; Phenomenex). The column was eluted at a flow rate of 400 µl/min with initial conditions of 10% mobile phase A (20-mM ammonium acetate and 20-mM ammonium hydroxide in water) and 90% mobile phase B (10-mM ammonium hydroxide in 75:25 vol/vol acetonitrile/methanol), followed by a 10-min linear gradient to 100% mobile phase A. The ion spray voltage was −4.5 kV and the source temperature was 500°C.
Positive ionization mode polar metabolite data were acquired using a 4000 QTRAP triple quadrupole mass spectrometer (AB SCIEX) coupled to a binary HPLC pump (1200 Series; Agilent Technologies) and an autosampler (HTS PAL; Leap Technologies). 10 µl of plasma or tissue homogenate samples was extracted using nine volumes of 74.9:24.9:0.2 (vol/vol/vol) acetonitrile/methanol/formic acid containing stable isotope-labeled internal standards (0.2 ng/µl valine-d8 [Isotec]; and 0.2 ng/µl phenylalanine-d8 [Cambridge Isotope Laboratories, Inc.]). The samples were centrifuged (10 min at 9,000 g and 4°C), and the supernatants (10 µl) were injected onto a 150 × 2.1-mm column (Atlantis HILIC; Waters Corp.). The column was eluted isocratically at a flow rate of 250 µl/min with 5% mobile phase A (10-mM ammonium formate and 0.1% formic acid in water) for 1 min, followed by a linear gradient to 40% mobile phase B (acetonitrile with 0.1% formic acid) over 10 min. The ion spray voltage was 4.5 kV, and the source temperature was 450°C.
Analyses of polar and nonpolar lipids were conducted using an Open Accela 1250 ultra (U)-HPLC and a hybrid quadrupole orbitrap mass spectrometer (Q Exactive; Thermo Fisher Scientific). 10 µl plasma or heart homogenate samples were prepared with the addition of 190 µl isopropanol containing 1-dodecanoyl-2-tridecanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids) as an internal standard, followed by centrifugation at 9,000 g for 15 min. 10-µl samples were injected onto a 150 × 3.0-mm column (Prosphere HP C4; Grace). The column was eluted isocratically with 80% mobile phase A (95:5:0.1 vol/vol/vol 10-mM ammonium acetate/methanol/acetic acid) for 2 min, followed by a linear gradient to 80% mobile phase B (99.9:0.1 vol/vol methanol/acetic acid) over 1 min, a linear gradient to 100% mobile phase B over 12 min, and then 10 min at 100% mobile phase B. Mass spectrometry (MS) data were acquired in the positive ion mode using electrospray ionization and full scan MS over mass-to-charge ratio 400–1,100. Other MS settings were spray voltage, 3 kV; capillary temperature, 300°C; sheath gas, 50 arbitrary units; auxiliary gas, 15 arbitrary units; heater temperature, 300°C; S-lens level, 60; and resolution, 70,000. Raw data were integrated and visually inspected using MultiQuant 1.2 software (AB SCIEX) for polar metabolites and TraceFinder 3.0 software (Thermo Fisher Scientific) for lipids.
Online supplemental material
Fig. S1 shows the gene trap locus, loss of Mff protein in the mutant, and a variety of Mff mutant phenotypes. The phenotypes include small size, low weight, malocclusion, kyphosis, grip weakness, reduced fertility, and reduced sperm count. Fig. S2 shows additional characterization of Mff mutants. Fig. S3 shows the effect of Mfn1 or Mfn2 mutations. Video 1 shows the gait alterations in the Mff mutant. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201507035/DC1.
Supplementary Material
Acknowledgements
We thank Maria Jordan and Dr. Kenneth Roos for performing echocardiography on 13–14-wk-old mice. We are grateful to Dr. Prashant Mishra for many hours of technical instruction and discussion.
This work was supported by grant RO1GM062967 (to D.C. Chan).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ALT
- alanine aminotransferase
- COX
- cytochrome c oxidase
- ES
- embryonic stem cell
- LV
- left ventricle
- MEF
- mouse embryonic fibroblast
- MS
- mass spectrometry
- mtDNA
- mitochondrial DNA
- PC
- Purkinje cell
- SDH
- succinate dehydrogenase
References
- Agah R., Frenkel P.A., French B.A., Michael L.H., Overbeek P.A., and Schneider M.D.. 1997. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J. Clin. Invest. 100:169–179. 10.1172/JCI119509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander C., Votruba M., Pesch U.E., Thiselton D.L., Mayer S., Moore A., Rodriguez M., Kellner U., Leo-Kottler B., Auburger G., et al. 2000. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 26:211–215. 10.1038/79944 [DOI] [PubMed] [Google Scholar]
- Brand M.D., and Nicholls D.G.. 2011. Assessing mitochondrial dysfunction in cells. Biochem. J. 435:297–312. 10.1042/BJ20110162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Stone A., Chipuk J.E., Ingerman E., Song C., Yoo C., Kuwana T., Kurth M.J., Shaw J.T., Hinshaw J.E., Green D.R., and Nunnari J.. 2008. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev. Cell. 14:193–204. 10.1016/j.devcel.2007.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.R., and Zweier J.L.. 2014. Cardiac mitochondria and reactive oxygen species generation. Circ. Res. 114:524–537. 10.1161/CIRCRESAHA.114.300559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E., and Chan D.C.. 2003. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160:189–200. 10.1083/jcb.200211046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., McCaffery J.M., and Chan D.C.. 2007. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 130:548–562. 10.1016/j.cell.2007.06.026 [DOI] [PubMed] [Google Scholar]
- Chen H., Vermulst M., Wang Y.E., Chomyn A., Prolla T.A., McCaffery J.M., and Chan D.C.. 2010. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 141:280–289. 10.1016/j.cell.2010.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Y., and Dorn G.W. II. 2011. Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circ. Res. 109:1327–1331. 10.1161/CIRCRESAHA.111.258723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delettre C., Lenaers G., Griffoin J.M., Gigarel N., Lorenzo C., Belenguer P., Pelloquin L., Grosgeorge J., Turc-Carel C., Perret E., et al. 2000. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 26:207–210. 10.1038/79936 [DOI] [PubMed] [Google Scholar]
- Dorn G.W. II, Song M., and Walsh K.. 2015. Functional implications of mitofusin 2-mediated mitochondrial-SR tethering. J. Mol. Cell. Cardiol. 78:123–128. 10.1016/j.yjmcc.2014.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., and Nunnari J.. 2014. Mitochondrial form and function. Nature. 505:335–343. 10.1038/nature12985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S., and van der Bliek A.M.. 2008. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 19:2402–2412. 10.1091/mbc.E07-12-1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey P.A., and Leinwand L.A.. 2011. The cell biology of disease: Cellular mechanisms of cardiomyopathy. J. Cell Biol. 194:355–365. 10.1083/jcb.201101100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Lewis P., Pevny L., and McMahon A.P.. 2002. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev. 119:S97–S101. 10.1016/S0925-4773(03)00099-6 [DOI] [PubMed] [Google Scholar]
- Ikeda Y., Shirakabe A., Maejima Y., Zhai P., Sciarretta S., Toli J., Nomura M., Mihara K., Egashira K., Ohishi M., et al. 2015. Endogenous Drp1 mediates mitochondrial autophagy and protects the heart against energy stress. Circ. Res. 116:264–278. 10.1161/CIRCRESAHA.116.303356 [DOI] [PubMed] [Google Scholar]
- Ishihara T., Ban-Ishihara R., Maeda M., Matsunaga Y., Ichimura A., Kyogoku S., Aoki H., Katada S., Nakada K., Nomura M., et al. 2015. Dynamics of mitochondrial DNA nucleoids regulated by mitochondrial fission is essential for maintenance of homogeneously active mitochondria during neonatal heart development. Mol. Cell. Biol. 35:211–223. 10.1128/MCB.01054-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y., Hoshijima M., Seo K., Bedja D., Sysa-Shah P., Andrabi S.A., Chen W., Höke A., Dawson V.L., Dawson T.M., et al. 2014. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 33:2798–2813. 10.15252/embj.201488658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara A., Cipolat S., Chen Y., Dorn G.W. II, and Scorrano L.. 2013. Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and Notch signaling. Science. 342:734–737. 10.1126/science.1241359 [DOI] [PubMed] [Google Scholar]
- Lopaschuk G.D., and Jaswal J.S.. 2010. Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J. Cardiovasc. Pharmacol. 56:130–140. 10.1097/FJC.0b013e3181e74a14 [DOI] [PubMed] [Google Scholar]
- Losón O.C., Song Z., Chen H., and Chan D.C.. 2013. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell. 24:659–667. 10.1091/mbc.E12-10-0721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P., and Chan D.C.. 2014. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 15:634–646. 10.1038/nrm3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H., Wang C., Cleland M.M., Setoguchi K., Yokota S., Youle R.J., and Mihara K.. 2010. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 191:1141–1158. 10.1083/jcb.201007152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanicolaou K.N., Kikuchi R., Ngoh G.A., Coughlan K.A., Dominguez I., Stanley W.C., and Walsh K.. 2012. Mitofusins 1 and 2 are essential for postnatal metabolic remodeling in heart. Circ. Res. 111:1012–1026. 10.1161/CIRCRESAHA.112.274142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic C., Shiota M., Niswender K.D., Jetton T.L., Chen Y., Moates J.M., Shelton K.D., Lindner J., Cherrington A.D., and Magnuson M.A.. 1999. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic β cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 274:305–315. 10.1074/jbc.274.1.305 [DOI] [PubMed] [Google Scholar]
- Qi X., Qvit N., Su Y.C., and Mochly-Rosen D.. 2013. A novel Drp1 inhibitor diminishes aberrant mitochondrial fission and neurotoxicity. J. Cell Sci. 126:789–802. 10.1242/jcs.114439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadate-Ngatchou P.I., Payne C.J., Dearth A.T., and Braun R.E.. 2008. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis. 46:738–742. 10.1002/dvg.20437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamseldin H.E., Alshammari M., Al-Sheddi T., Salih M.A., Alkhalidi H., Kentab A., Repetto G.M., Hashem M., and Alkuraya F.S.. 2012. Genomic analysis of mitochondrial diseases in a consanguineous population reveals novel candidate disease genes. J. Med. Genet. 49:234–241. 10.1136/jmedgenet-2012-100836 [DOI] [PubMed] [Google Scholar]
- Song M., Mihara K., Chen Y., Scorrano L., and Dorn G.W. II. 2015. Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell Metab. 21:273–285. 10.1016/j.cmet.2014.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi J., Zhang Z., Wakabayashi N., Tamura Y., Fukaya M., Kensler T.W., Iijima M., and Sesaki H.. 2009. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J. Cell Biol. 186:805–816. 10.1083/jcb.200903065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham H.R., Koster J., van Roermund C.W., Mooyer P.A., Wanders R.J., and Leonard J.V.. 2007. A lethal defect of mitochondrial and peroxisomal fission. N. Engl. J. Med. 356:1736–1741. 10.1056/NEJMoa064436 [DOI] [PubMed] [Google Scholar]
- Züchner S., Mersiyanova I.V., Muglia M., Bissar-Tadmouri N., Rochelle J., Dadali E.L., Zappia M., Nelis E., Patitucci A., Senderek J., et al. 2004. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet. 36:449–451. 10.1038/ng1341 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.