δ-Protocadherins partition the zebrafish optic tectum into radial columns of neurons, and the neurons within a column are siblings derived from common neuronal progenitors.
Abstract
Cell–cell recognition guides the assembly of the vertebrate brain during development. δ-Protocadherins comprise a family of neural adhesion molecules that are differentially expressed and have been implicated in a range of neurodevelopmental disorders. Here we show that the expression of δ-protocadherins partitions the zebrafish optic tectum into radial columns of neurons. Using in vivo two-photon imaging of bacterial artificial chromosome transgenic zebrafish, we show that pcdh19 is expressed in discrete columns of neurons, and that these columnar modules are derived from proliferative pcdh19+ neuroepithelial precursors. Elimination of pcdh19 results in both a disruption of columnar organization and defects in visually guided behaviors. These results reveal a fundamental mechanism for organizing the developing nervous system: subdivision of the early neuroepithelium into precursors with distinct molecular identities guides the autonomous development of parallel neuronal units, organizing neural circuit formation and behavior.
Introduction
The vertebrate nervous system becomes progressively regionalized during development (Kiecker and Lumsden, 2005). These compartments may be large morphological subdivisions such as cerebellum, functional specializations such as visual cortex, or local regions such as laminae or nuclei. This partitioning of the nervous system into distinct domains enables each region to undergo a distinct developmental program. Despite the importance to nervous system development, the molecular and cellular mechanisms governing this modular assembly are not well understood.
The optic tectum, the largest subdivision in the zebrafish brain, processes retinal inputs to mediate vision (Portugues and Engert, 2009). Morphological studies in frogs (Lázár, 1973) and fish (Vanegas et al., 1974; Meek and Schellart, 1978) have identified multiple types of tectal neurons and have also revealed a conserved laminated structure. However, neither the cellular architecture of the optic tectum nor the mechanisms governing tectum development are known (Recher et al., 2013). δ-Protocadherins (δ-pcdhs) comprise a family of homophilic cell adhesion molecules (Wolverton and Lalande, 2001; Vanhalst et al., 2005), and prior work has shown that δ-pcdhs are strongly expressed in the zebrafish optic tectum (Biswas and Jontes, 2009; Emond et al., 2009; Liu et al., 2009, 2010; Blevins et al., 2011). Although the detailed function of these molecules is unclear, members of this family can participate in axon guidance (Leung et al., 2013), arborization (Biswas et al., 2014), and fasciculation (Hayashi et al., 2014). δ-Pcdhs are essential for neural development, as several have been implicated in neurodevelopmental disorders (Hirano and Takeichi, 2012; Redies et al., 2012). In particular, mutations in human PCDH19 result in a female-limited form of infant-onset epilepsy (Dibbens et al., 2008; Depienne et al., 2009), making PCDH19 the second most clinically relevant gene in epilepsy (Depienne and LeGuern, 2012). However, it is not known how loss of pcdh19 alters neural development or leads to epileptogenesis.
Here we show that δ-pcdhs are expressed in radial columns of neurons in the developing zebrafish optic tectum and that neurons within a column arise from a common progenitor cell. Elimination of pcdh19 degrades the columnar organization of the tectum because of reduced cell cohesion and increased cell proliferation. Moreover, pcdh19 mutants exhibit defects in visually guided behaviors. These data reveal a previously unknown columnar architecture of the optic tectum, suggesting that the tectum has an organization more similar to that of mammalian cortex than previously realized. In addition, the defects in visual processing suggest that the columnar organization is important for neural function. Thus, our results provide an initial link between δ-pcdhs, the development of neural architecture, and neural function.
Results and discussion
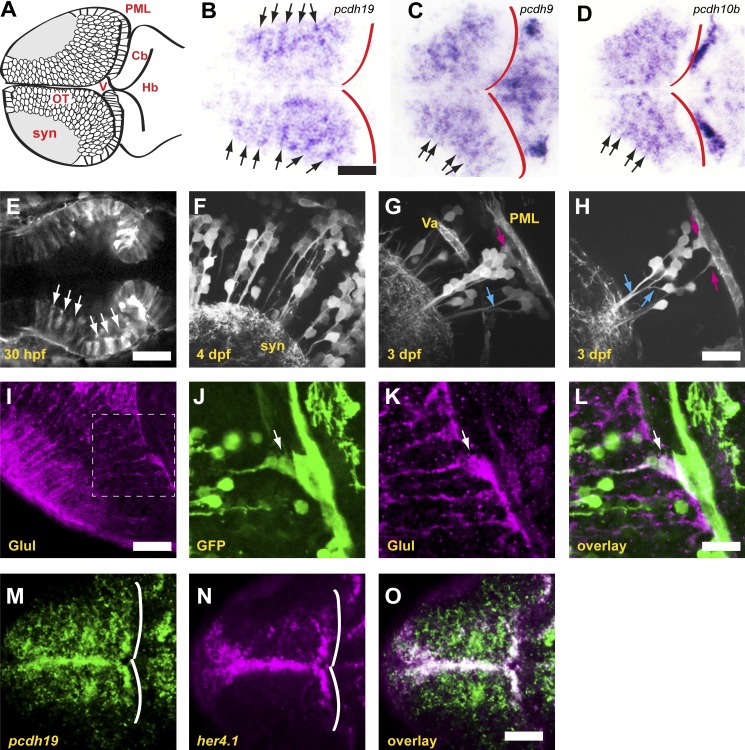
To better understand the expression of δ-pcdhs within the tectum, we imaged horizontal sections of zebrafish larvae at 4 d postfertilization (dpf) that were labeled with riboprobes against pcdh19, pcdh9, and pcdh10b (Fig. 1, B–D). Strikingly, larvae exhibited stripes of expression in the tectum, revealing that neurons expressing a particular δ-pcdh are organized as radial columns. These columns are not apparent in either whole-mount larvae or transverse sections. To investigate this phenomenon in more detail, we identified a bacterial artificial chromosome (BAC) clone harboring the complete pcdh19 gene and generated a BAC transgenic line, TgBAC(pcdh19:Gal4-VP16,UAS:Lifeact-GFP)os49. The doubly modified BAC clone uses the regulatory elements of pcdh19 to express Gal4-VP16, which activates expression of Lifeact-GFP (Riedl et al., 2008) and labels F-actin in pcdh19-expressing cells. At 30 h postfertilization (hpf), the pcdh19 reporter generates a striped pattern in the midbrain neuroepithelium that will give rise to the optic tectum (Fig. 1 E). Consistent with the in situ hybridization data, cells that express pcdh19 in 3- to 4-dpf larvae are organized as radial columns (Fig. 1 F). Individual columns consist of clusters of neurons tightly associated with the radial fibers of one or more radial glia-like cells (Fig. 1, G and H). These cells are likely radial glia, as they express common glial markers including glutamine synthase (Fig. 1, I–L) and Her4.1 (Fig. 1, M–O).
Figure 1.
δ-Protocadherins define neuronal columns in the zebrafish optic tectum. (A) Schematic of the optic tectum of a larval zebrafish. Neurons are organized around a synaptic neuropil that includes both the axons and dendrites of tectal neurons, as well as the axonal arbors of retinal ganglion cells. Cb, cerebellum; Hb, hindbrain; OT, optic tectum; PML, peripheral midbrain layer; syn, synaptic neuropil; V, ventricle. (B) Horizontal section through the optic tectum of a 4-dpf larva labeled with a riboprobe against pcdh19. Labeling for pcdh19 is concentrated into radial stripes (arrows). The posterior boundary of the tectum is shown by red outlines. Bar, 50 µm in B–D. (C and D) Horizontal sections of optic tecta labeled with riboprobes against pcdh9 (C) and pcdh10b (D) showing radial stripes comparable to those observed for pcdh19. (E) Single optical section from a two-photon image stack of a 30-hpf BAC transgenic embryo expressing the F-actin marker, Lifeact-GFP, under the control of pcdh19 regulatory elements. The midbrain neuroepithelium expresses Lifeact-GFP in distinct stripes (arrows). Bar, 50 µm. (F) Maximum-intensity projection of five optical sections (1-µm spacing) of a 4-dpf BAC transgenic larva. Lifeact-GFP is expressed in discrete columns of neurons, as was observed by in situ hybridization for endogenous pcdh19. syn, synaptic neuropil. (G) Maximum-intensity projection of five optical sections (1-µm spacing) of a 3-dpf BAC transgenic larva. Labeled neurons are clustered around a central radial glia (magenta arrow), and most of the primary neurites fasciculate as they project to the synaptic neuropil (blue arrow). Va, vasculature; PML, posterior midbrain layer. (H) Single optical section through a column in a 3-dpf larva. Columns of pcdh19+ neurons are typically associated with one or more pcdh19+ radial glia (magenta arrows). Fasciculation of neuronal processes is highlighted by blue arrows. Neurons arborize in the area immediately adjacent to the column. Bar, 20 µm in F–H. (I–L) Horizontal sections through the optic tectum of a 4-dpf BAC transgenic larva, labeled with antibodies against glutamine synthase (Glul) to reveal radial glia (I and K) or GFP (J). The boxed area in I is highlighted in J–L. (L) Glutamine synthase staining reveals that a subset of radial glia are also pcdh19+ (arrow). Bars: (I) 30 µm; (J–L) 15 µm. (M–O) Single optical section through the dorsal optic tectum of a 3-dpf larva labeled by double FISH with riboprobes against pcdh19 (M) or the neural progenitor marker her4.1 (N). White lines show the posterior boundary of the tectum. An overlay of the signals reveals colocalization along the medial and posterior margins of the tectum (O). Bar, 50 µm.
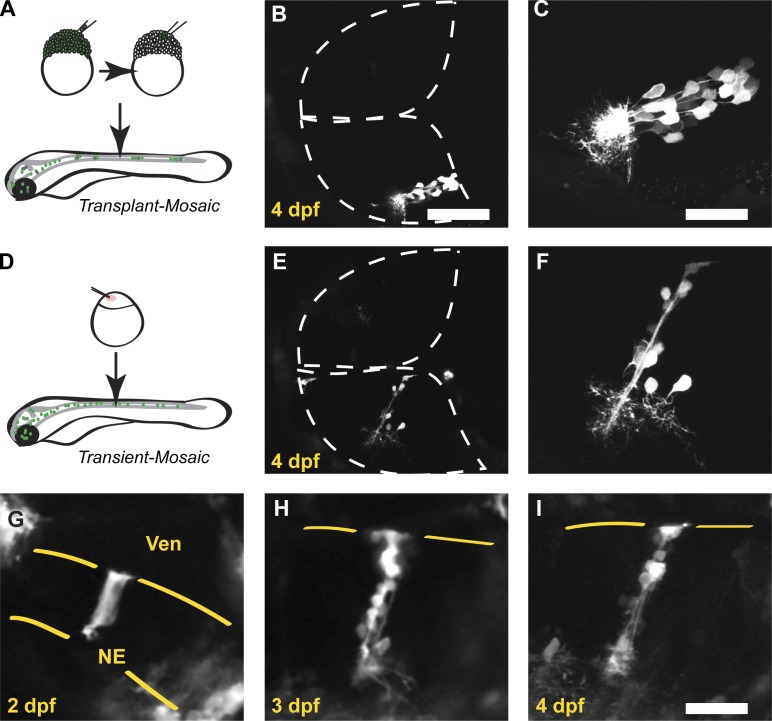
The columnar organization of pcdh19-expressing neurons is reminiscent of the proliferative radial units observed in the developing mammalian cortex (Rakic, 1988; Mountcastle, 1997). To address whether the neurons within a pcdh19+ column are siblings (Noctor et al., 2001), we used two methods to mosaically label cells in the optic tectum. First, we transplanted 20–40 cells from transgenic [TgBAC(pcdh19:Gal4-VP16,UAS:Lifeact-GFP)os49] blastula-stage embryos into unlabeled host embryos (Fig. 2 A). These cells disperse during gastrulation and contribute to different parts of the embryo; those cells fated to express pcdh19 will be labeled with Lifeact-GFP. Labeled tectal cells were predominantly organized in columns, even in the absence of other tectal expression, suggesting that these cells were derived from a common progenitor (Fig. 2, B and C). Additionally, we injected BAC DNA into embryos to transiently express Lifeact-GFP in pcdh19+ cells (Fig. 2 D). Transient expression of fluorescent reporters was largely restricted to radial columns (Fig. 2, E and F), again suggesting that the neurons within these columns arise from an initial pcdh19+ precursor. To further investigate the origins of tectal columns, we imaged individual TgBAC(pcdh19:Gal4-VP16,UAS:Lifeact-GFP)os49 embryos on consecutive days (Fig. 2, G–I). In fortuitous cases, isolated columns identified at 3–4 dpf (Fig. 2, H and I) could be unambiguously traced back to labeled neuroepithelial cells (Fig. 2 G). Our observations demonstrate that pcdh19 is expressed in radial columns within the optic tectum and that these columns assemble from the proliferation of one or a small number of pcdh19+ precursors.
Figure 2.
pcdh19+ neuronal columns are clonal. (A) Schematic outlining our approach to mosaically label zebrafish larva using blastula-stage cell transplantation. Cells were transplanted from BAC transgenic donor embryos into unlabeled WT host embryos. (B) Two-photon image stack of a 4-dpf host embryo with a single pcdh19+ column present in the bottom hemitectum. (C) Higher-magnification image of the labeled cells shown in B. Labeled neurons are organized as a column, and their neurites arborize within an adjacent, restricted region. (D) Schematic outlining approach to mosaically label zebrafish larva by injecting recombinant BAC. BAC DNA is injected at the one-cell stage along with mRNA for Tol2 transposase. (E) Two-photon image stack of a 4-dpf larva injected with recombinant pcdh19 BAC DNA. The only labeled cells in the bottom hemitectum are present in a column reminiscent of those found in the BAC transgenic larvae. (F) Higher-magnification view of the column shown in F. (G–I) Image sequence from a BAC transgenic zebrafish collected on consecutive days. (G) Single optical section collected through the dorsal optic tectum of a 2-dpf embryo, showing one or two strongly labeled neuroepithelial cells. Yellow lines define the boundaries of the neuroepithelium. NE, neuroepithelial cell; Ven, ventricle. At 3 (H) and 4 (I) dpf, columns of neurons are visible at the same position within the tectum. Yellow lines mark the ventricular surface of the tectum. Bars: (B and E) 80 µm; (C and F–I) 40 µm.
To show whether other δ-pcdhs define similar developmental units, as suggested by in situ hybridization (Fig. 1, B–D), we injected BAC clones of zebrafish pcdh18b and pcdh1a, modified as was done for pcdh19. In each case, we observed columns of labeled cells (Fig. S1, A and B), supporting the idea that the zebrafish optic tectum could be partitioned into distinct radial domains based on the expression of δ-pcdhs (Fig. S1 C). To test this, we performed double FISH with riboprobes directed against pcdh7a and pcdh19 (Fig. S1, D–I). Although some overlap of labeling can be observed (Fig. S1, F and I), the columnar distribution of pcdh7a and pcdh19 largely appears to be mutually exclusive, implying that the expression of each δ-pcdh labels distinct sets of columns.
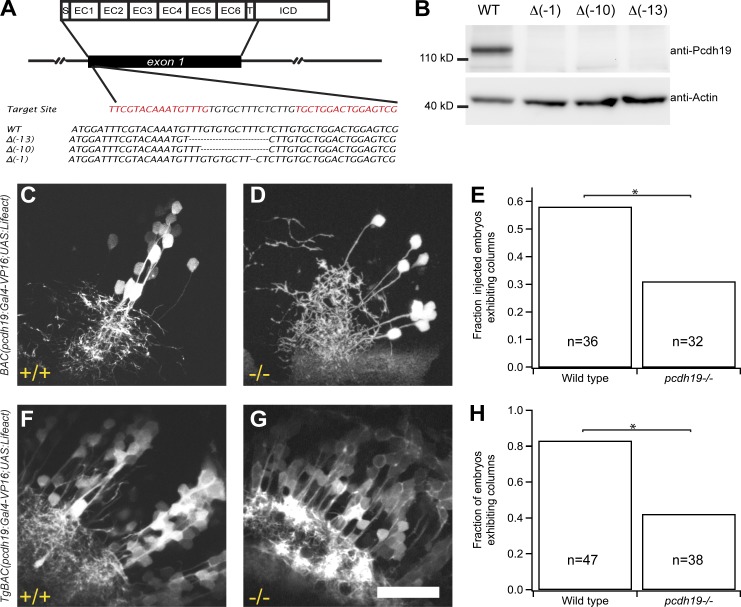
To investigate the role of pcdh19 in the formation of tectal columns, we used transcriptional activator-like effector nucleases (TALENs; Cermak et al., 2011; Bedell et al., 2012; Dahlem et al., 2012) to generate germline lesions in zebrafish pcdh19 (Fig. 3 A). Homozygous mutants completely lack Pcdh19 (Fig. 3 B) but do not exhibit gross defects in neural organization (unpublished data). To investigate the effects of pcdh19 loss in detail, we injected BAC(pcdh19:Gal4-VP16,UAS:Lifeact-GFP) into one-cell–stage wild-type (WT) and mutant embryos (Fig. 3, C and D). At 4 dpf, 58% of labeled WT larvae exhibited columns (Fig. 3, C and E; n = 21/36 larvae). In contrast, only 31% of mutant larvae exhibited evidence of columnar organization (Fig. 3, D and E; n = 10/32 larvae, P = 0.028, Fisher exact test). Labeled neurons in mutants were more dispersed and arborized over a larger area within the synaptic neuropil (Fig. 3 D). To further assess the effects of Pcdh19 loss, we crossed the pcdh19−/− mutants into the TgBAC(pcdh19:Gal4,UAS:Lifeact-GFP)os49 line. Elimination of pcdh19 severely disrupted columnar organization in TgBAC(pcdh19:Gal4,UAS:Lifeact-GFP)os49 larvae (Fig. 3, F–H; WT, 82%, n = 39/47; pcdh19−/−, 41%, n = 16/38; P = 0.0001, Fisher exact test). In addition, cell transplantation experiments indicated that pcdh19 is required in both host and donor cells for normal column development and normal neurite outgrowth and arborization (Fig. S2). Whereas groups of pcdh19+ cells appear less tightly clustered in pcdh19−/− mutants (Fig. 3 D), the loss of apparent columnar organization could also a result of an increased number of labeled cells.
Figure 3.
Loss of pcdh19 disrupts the columnar organization of the optic tectum. (A) Germline lesioning of the pcdh19 gene using TALENs. Exon 1 encodes the entire extracellular domain of Pcdh19, as well as the transmembrane domain and a short segment of the intracellular domain. We designed a TALEN pair against a target site just downstream of the signal peptide encoding sequence within exon 1. We isolated mutants with germline frameshift indel mutations that result in premature stop codons early within EC1. (B) Western blotting with a polyclonal antibody directed against the Pcdh19 intracellular domain confirms the loss of Pcdh19 in the mutants. (C–E) Individual clusters of neurons were labeled by BAC DNA injection in WT (C) or pcdh19−/− (D) larvae at 4 dpf. Distinct columns were observed in 58% (n = 21/36) of WT larvae with expression in the tectum, whereas columns were observed in only 31% (n = 10/32) of pcdh19−/− larvae (E). *, P = 0.028, Fisher exact test. (F–H) pcdh19 mutants were crossed with BAC transgenics. The transgenic:pcdh19−/− mutants exhibited a loss of columnar organization (41%, n = 16/38) compared with transgenic:pcdh19+/+ larvae (82%, n = 39/47; *, P = 0.0001, Fisher exact test). Bar, 50 µm.
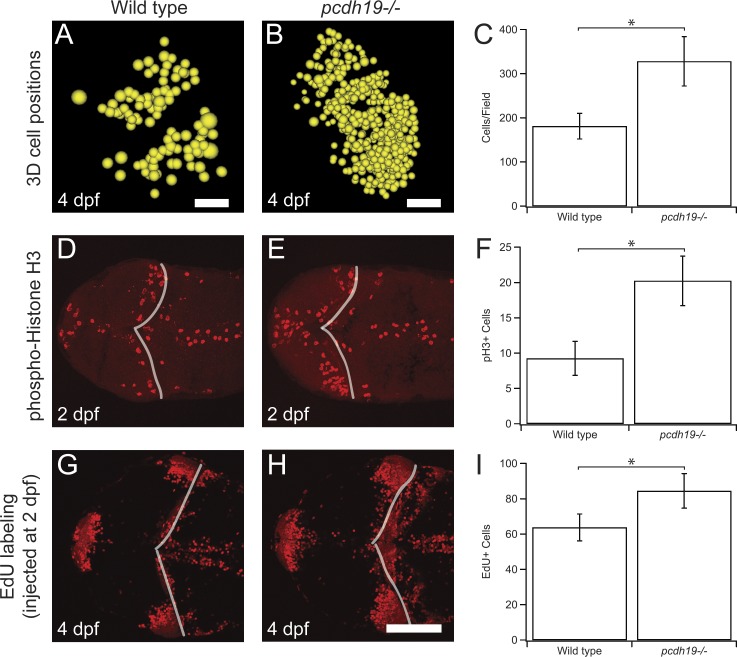
To determine whether there is an increase of cells expressing Lifeact-GFP in mutants, we identified and counted all labeled cells in image stacks collected from the optic tecta of six WT and five mutant larvae (Fig. 4, A–C). We found that there was an approximately twofold increase in the number of labeled cells in the mutants (WT, 181 ± 29, n = 6; pcdh19−/−, 328 ± 56, n = 5; P = 0.036), suggesting an increase of proliferation within this cell population. To directly assess levels of proliferation within the optic tectum, we sectioned and labeled 2-dpf embryos with antibodies against phosphohistone H3 (pH3; Fig. 4, D–F). There was an increase in pH3-labeled nuclei in pcdh19−/− mutants compared with WT (pcdh19−/−, 20.3 ± 3.5, n = 4; WT, 9.3 ± 2.4, n = 3; P = 0.04, Student’s t test). Similarly, we injected 5-ethynyl-2′-deoxyuridine (EdU) into 2-dpf embryos, then fixed them at 4 dpf (Fig. 4, G–I). Again, we found evidence for increased proliferation in pcdh19−/− mutants (pcdh19−/−, 84.5.3 ± 9.8, n = 4; WT, 63.8 ± 7.6, n = 4; P = 0.032, Student’s t test). Our data indicate that the loss of pcdh19 degrades the columnar organization of pcdh19+ neurons through both an increase in neuron production and a reduction in cell cohesion.
Figure 4.
Increased proliferation in the optic tectum of pcdh19−/− mutants. (A–C) Two-photon image stacks were collected in WT (A) or pcdh19−/− (B) larvae. The 3D positions of transgenically labeled cell bodies were modeled as spheres. (C) The number of labeled cells was counted, confirming the visual impression of increased numbers of labeled cells in the pcdh19−/− mutants (WT, 181 ± 29, n = 6; pcdh19−/−, 328 ± 56, n = 5; *, P = 0.036, Student’s t test). Bars: (A) 20 µm; (B) 25 µm. (D–F) To determine whether rates of cell proliferation in the tectum are changed in the mutants, we performed pH3 immunocytochemistry at 2 dpf in both WT (D) and pcdh19−/− (E) larvae. (F) The number of pH3-labeled cells in the tectum was counted, revealing a significant increase in proliferation in the pcdh19−/− mutants (WT, 9.3 ± 2.4, n = 3; pcdh19−/−, 20.3 ± 3.5, n = 4; *, P = 0.04, Student’s t test). (G–I) To confirm our observations with pH3 labeling, we labeled WT (G) or pcdh19−/− (H) larvae with EdU at 2 dpf and fixed the larvae at 4 dpf. (I) The number of EdU-labeled cells in the tectum was counted, confirming a significant increase in proliferation in the pcdh19−/− mutants (WT, 63.8 ± 7.6, n = 4; pcdh19−/−, 84.5 ± 9.8, n = 4; P = 0.032, Student’s t test). Bar, 50 µm.
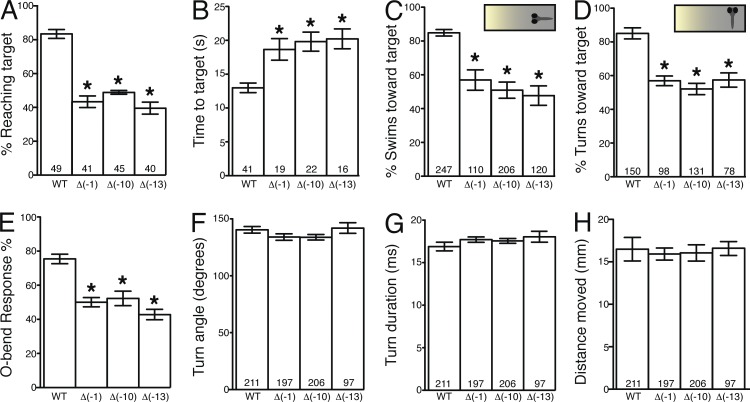
Distinct tectal regions are known to differentially process visual input; therefore, disruption of tectal columnar organization may alter visually guided behaviors. To determine whether pcdh19 loss of function affects visual processing, we tested 6-dpf larvae for positive phototaxis, a behavior in which larvae stereotypically turn and swim toward a target light. Compared with WT larvae, fewer pcdh19−/− larvae reached the target light area, and the pcdh19−/− larvae that did show positive phototaxis required more time to reach the target (Fig. 5, A and B; Burgess et al., 2010). Kinematic analyses of turning and swimming behaviors with millisecond resolution indicated that the reduced phototaxis was not caused by motor impairment (Fig. S3). To determine whether the reduced phototaxis in pcdh19−/− larvae was caused by a visual processing defect, we tested whether pcdh19−/− larvae initiated turns and swims that were biased toward the target light. As expected, WT larvae showed a strong bias to swim or turn toward the target light based on their orientation (Burgess et al., 2010); however, pcdh19−/− larvae showed no directional bias, indicating that a visual impairment underlies the phototaxis defect (Fig. 5, C and D). Consistent with these results, we also found that pcdh19−/− larvae showed a reduced likelihood of initiating the O-bend maneuver (Fig. 5 E), a stereotyped visuomotor behavior to a sudden extinguishment of light (Burgess and Granato, 2007). Again, once initiated, the performance of O-bend maneuvers was indistinguishable between WT and pcdh19−/− larvae (Fig. 5, F–H), suggesting that the behavior initiation defect is caused by a visual, not motor, impairment. In contrast, the acoustic startle response was normal in the pcdh19−/− mutants (unpublished data). Thus, visual processing is impaired in pcdh19−/− larvae, potentially because of disrupted tectal neuronal columns.
Figure 5.
pcdh19−/− mutants exhibit impaired visually guided behaviors. (A) Mean percentage of larvae that reached target light within 30 s during phototaxis. (B) Mean time to reach target area by larvae showing phototaxis. (C and D) Insets represent larvae orientation to target light (insets) at each second before reaching the target. Mean percentage of swims (C) and turns (D) biased toward target. n at bottom of bars indicates larvae (A and B) or number of events based on orientation (C and D). (E) Mean initiation of O-bend response to dark flash stimulus. n = 3 groups of 15 larvae/group. (F–H) Mean turning angle (F), duration (G), and distance moved (H) per O-bend maneuver. n at bottom of bars indicates number of O-bend maneuvers per genotype. Error bars denote SEM. *, P < 0.001 versus WT by analysis of variance.
Retroviral lineage tracing has suggested the radial development of the optic tectum in chickens (Gray and Sanes, 1991, 1992) and medaka fish (Nguyen et al., 1999). Here we show that this radial development results in neuronal columns that may be analogous to the microcolumns found in mammalian cortex (Peters and Walsh, 1972; Peters and Sethares, 1996). However, it remains to be shown if the columns defined by δ-pcdh expression correspond to functional microcircuits. Although our results suggest the existence of a molecular code in which differential expression of δ-pcdhs could partition the optic tectum into columnar modules with distinct identities, further work is required to determine whether this expression is mutually exclusive (one δ-pcdh per column) or combinatorial. In addition, we show that pcdh19+ columns derive from the proliferation of neural progenitors expressing pcdh19. Thus, continued expression of a δ-pcdh could provide continuity between early patterning and differentiation with the final stages of synaptogenesis and circuit assembly. Further investigation is required to dissect the involvement of δ-pcdhs at different stages of column and neuronal development, as pcdh19 mutants exhibit defects in a range of processes that include cell proliferation, fasciculation of primary processes, and neurite arborization. Loss of pcdh19 impairs visually guided behaviors, revealing specific functional defects. It will be instructive to determine whether other δ-pcdhs exhibit comparable defects, and how this family of molecules collaborates to guide both circuit assembly and the development of neural function.
Materials and methods
Fish maintenance
Adult zebrafish (Danio rerio) and embryos of the Tübingen longfin and AB strains were maintained at ∼28.5°C and staged according to Westerfield (1995). Embryos were raised in E3 embryo medium (Westerfield, 1995) with 0.003% phenylthiourea (Sigma-Aldrich) to inhibit pigment formation.
Whole-mount in situ hybridization
Riboprobes directed against δ-pcdhs were amplified by PCR as described previously (Biswas and Jontes, 2009; Emond et al., 2009; Biswas et al., 2014). A T7 RNA polymerase binding site was included in each of the reverse primers, and these PCR products were used as templates for in vitro transcription (Promega). Antisense riboprobes were labeled with digoxygenin-dUTP (Roche). Whole-mount in situ hybridizations were performed using standard methods (Westerfield, 1995). In brief, embryos were fixed at 4°C overnight in 4% paraformaldehyde in PBS, dehydrated in a methanol series, and stored in 100% methanol overnight at −20°C. They were rehydrated in decreasing concentrations of methanol, and embryos ≥24 hpf were permeabilized using proteinase K (10 µg/ml; Roche). Embryos were refixed in 4% paraformaldehyde before hybridization. Digoxygenin-dUTP–labeled riboprobe was added to the hybridization buffer at a final concentration of 200 ng/ml, and hybridization was performed at 65°C overnight. AP-conjugated anti-digoxygenin Fab fragments (Roche) were used at 1:5,000 dilution. Nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate (Roche) was used for the coloration reaction. Labeled embryos were equilibrated in 30% sucrose in PBS overnight. They were then embedded in OCT (Ted Pella), sectioned on a cryostat at 14 µm, and placed on gelatin-coated glass slides. Images were captured on an AxioStar (Carl Zeiss).
For double FISH, 3- and 4-dpf fish were fixed and hybridized with riboprobes against pcdh7a (labeled with digoxygenin-dUTP) and pcdh19 (labeled with fluorescein-dUTP). The digoxygenin-labeled probe was detected using antidigoxygenin Fab–HRP (Roche) and developed using the tetramethylrhodamine substrate from the TSA Plus kit (Perkin-Elmer). Subsequently, the fluorescein probe was detected with an antifluorescein primary antibody (Roche) and a goat anti-mouse HRP secondary antibody (Invitrogen) and developed using the fluorescein substrate from the TSA Plus kit.
The PCR primers used to generate DNA template for riboprobe synthesis are as follows: pcdh19-F: 5′-ATGCATTCCAAGGACATGGATTTCG-3′; pcdh19-T7R: 5′-GTAATACGACTCACTATAGGGCGAGACTGGTCCGATTTGTTCCTGTGC-3′; pcdh10b-F: 5′-ATGTTTGTGTTTTTGCTCCTGCTG-3′; pcdh10b-T7R: 5′-GTAATACGACTCACTATAGGGCGAACGCGTCTTTGGCCTTGGCCGTGC-3′; pcdh9-F: 5′-GCAACATTAGTTGCTTGTTTCTGG-3′; pcdh9-T7R: 5′-CGTAATACGACTCACTATAGGGCGCATCACAGTACCGTTGGTTGG-3′; pcdh7a-F: 5′-ATGGCAGAAAACGCCCCACCTGGC-3′; pcdh7a-T7R: 5′-CGTAATACGACTCACTATAGGGCGGTCCCGAACGAGGATTCGTCC-3′; her4.1-F: 5′-ATGACTCCTACAATCACTGG-3′; and her4.1-T7R: 5′-GTAATACGACTCACTATAGGGCGAATTAAGTCTACCAGGGTCTCC-3′.
Cryosectioning and immunocytochemistry
Zebrafish larvae were fixed for 1 h in 4% paraformaldehyde in PBS, equilibrated in 30% sucrose in PBS overnight, embedded in OCT (Ted Pella, Inc.), sectioned on a cryostat at 16–20 µm, and placed on gelatin-coated glass slides. Sections were permeabilized and blocked in PBS, 0.5% Triton X-100, 1% DMSO, and 10% normal goat serum. For immunocytochemistry, sections were labeled with chicken anti-GFP (1:1,000; Abcam), rabbit antiglutamine synthase (1:100; Genetex), and rabbit anti-pH3 (1:200; Cell Signaling). Rhodamine- and fluorescein-conjugated secondary antibodies were used at 1:500 (Jackson ImmunoResearch). Images were obtained using a TCS-SL confocal microscope (Leica) and either a Leica 20×/NA 0.7 HC Plan Apochromat multi-immersion (pH3 or EdU) or a 40× HCX Plan Apochromat oil NA 0.75–1.25 (glutamine synthase).
To label larvae with EdU, 1 nl of 25 µg/ml EdU (Life Technologies) was injected into the yolk of WT and mutant embryos at 2 dpf. All fish were then fixed at 4 dpf with 4% paraformaldehyde at 4°C overnight. Embryos were transferred into 30% sucrose in PBS at RT for 3 h, then mounted in OCT and frozen on dry ice. Fish were sectioned on a cryostat at a thickness of 25 µm. Staining was performed using the Click-IT EdU Alexa Fluor 555 Imaging kit (Life Technologies) according to the manufacturer’s protocol.
BAC recombineering and transgenesis
BAC clone CH211-156N5 was obtained from BACPAC Resources, Children’s Hospital Oakland. CH211-156N5 includes 21 kb upstream of the start codon and 15 kb downstream of the stop codon. Purified BAC DNA was introduced into EL250 cells (Lee et al., 2001) obtained from N. Copeland (National Institutes of Health, Bethesda, MD). To facilitate transgenesis, we first introduced an inverted Tol2 cassette (iTol2) into the backbone (Suster et al., 2011). The iTol2 reagents were provided by M. Suster (Uni Research AS High Technology Center, Bergen, Norway). We then inserted a Gal4-VP16-FRT-Kanamycin-FRT cassette into exon 1 of the pcdh19 gene. The kanamycin marker was excised by arabinose induction of Flpe recombinase. Finally, a 5xUAS:Lifeact-GFP cassette was introduced into exon 2, which is ∼45 kb downstream of exon 1. The same procedure was performed for BAC clones harboring genes for pcdh1a (DKEY-225G6; Source Bioscience) and pcdh18b (CH211-154P8; BACPAC Resources). The CHORI-211 library (CH211) was built in the pTARBAC2.1 vector, whereas the DanioKey library (DKEY) was built in the pIndigoBac-536 vector. Each of the BAC clones contained a complete protocadherin gene.
To generate germline BAC transgenic zebrafish, we coinjected one-cell–stage embryos with 1 nl of 100 ng/µl purified BAC DNA and 100 ng/µl mRNA encoding the Tol2 transposase. At 2 dpf, embryos were screened for Lifeact-GFP expression, and fluorescent embryos were grown to adulthood. To establish stable lines, founders were outcrossed with WT zebrafish, and fluorescent embryos were pooled and grown to adulthood. For transient expression, injections were performed as described, but embryos were kept in 0.01% propylthiouracil and imaged at 3–4 dpf.
TALEN construction and production of germline lesions
We used the online tool ZiFit (Sander et al., 2010; Zinc Finger Consortium) to search for a TALEN target site in exon 1 of zebrafish pcdh19. We identified an appropriate target site downstream of the signal peptide, 5′-TTTCGTACAAATGTTTGtgtgctttctcttgTGCTGGACTGGAGTCGA-3′ (uppercase indicates TAL left and right binding sites). The TALEN arrays (left, NG-NG-HD-NN-NG-NI-HD-NI-NI-NI-NG-NN-NG-NG-NG-NN; right, HD-NN-NI-HD-NG-HD-NI-NN-NG-HD-HD-NI-NN-HD-NI) were assembled in RCIscript-GoldyTALEN (Bedell et al., 2012) using the TAL Effector kit 1.0 (Addgene; Cermak et al., 2011). Plasmid encoding assembled TALENs was linearized with SacI and used as template for mRNA synthesis with a T3 mMessage Machine kit (Ambion). To generate germline lesions in zebrafish pcdh19, we injected one-cell–stage embryos with 50 pg mRNA encoding the left and right nucleases. Injected embryos were grown to adulthood and screened for germline lesions. For screening, adult F0 fish were outcrossed with WT, and genomic DNA was prepared from eight embryos of each cross. High-resolution melting analysis (HRMA) was used to identify putative founders (Dahlem et al., 2012). PCR products exhibiting aberrant melting curves were cloned and sequenced. F0 adults exhibiting frameshift mutations were outcrossed, and the F1 embryos were grown to adulthood. To screen the adult F1 fish, genomic DNA was prepared from caudal fin clips screened by HRMA. We obtained three mutants: 1-bp deletion (Δ-1), 10-bp deletion (Δ-10), and 13-bp deletion (Δ-13). These heterozygote F1 founders were outcrossed again, and the F2 offspring were raised and screened to establish mutant lines pcdh19os50, pcdh19os51, and pcdh19os52. To obtain homozygous pcdh19 mutants, which are viable and fertile, heterozygotes for each allele were incrossed, and embryos were grown to adulthood and screened by HRMA.
To establish mutant/transgenic lines, each of the three homozygous mutant lines was crossed with TgBAC(pcdh19:Gal4VP16; UAS:Lifeact)os49. Fluorescent embryos were selected and grown to adulthood. These transgenic-pcdh19+/− fish were incrossed, and the offspring were raised to adulthood and screened for homozygous mutants.
Western blotting
Protein lysates were prepared by homogenizing 40 deyolked embryos at 3 dpf in lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0.5% Triton X-100, 1 mM PMSF, and complete protease inhibitor cocktail [Roche]). Lysates were microcentrifuged at 4°C for 10 min, and identical volumes were loaded onto NuPAGE 10% Bis-Tris SDS-PAGE gels (Life Technologies) for each condition. Proteins were subjected to electrophoresis and transferred (Bio-Rad Laboratories) to PVDF (GE Healthcare), blocked with 5% nonfat milk in TBST (10 mM Tris-HCL, pH 7.6, 100 mM NaCl, and 0.1% Tween-20), and incubated overnight with primary antibody (1:1,000 anti-Pcdh19, custom polyclonal [Covance]; 1:5,000 mouse monoclonal anti–β-actin [Santa Cruz Biotechnology]). HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) were used at 1:5,000, and the chemiluminescent signal was amplified using Western Lightning Ultra (PerkinElmer). Blots were imaged on a molecular imaging system (Omega 12iC; UltraLum). Custom polyclonal antibodies against zebrafish Pcdh19 were generated in rabbit, directed against the Pcdh19 intracellular domain (aa 702–1083).
Two-photon imaging
Two-photon imaging of live embryos and larvae was performed at RT on a custom-built microscope controlled by ScanImage (Pologruto et al., 2003). Excitation was provided by a Chameleon-XR Ti:Sapphire laser (Coherent) tuned to 890 nm. Fluorescence was detected using a Multiphoton Detection Unit mounted on a SliceScope (Scientifica). We used water immersion objectives from Olympus, either 60×/NA1.0 (LUMPLFLN60X/W) or 20×/NA1.0 (XLUMPLFLN20XW). For imaging, larvae were embedded in 1% low melting point agarose, covered with embryo medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, and 0.33 mM MgCl2), and imaged at RT.
For the imaging of transgenic/mutant larvae, TgBAC/pcdh19+/− fish were crossed and embryos were kept in individual wells of a 24-well plate. After imaging, fish were genotyped by HRMA to identify homozygous mutants. As homozygous mutants are viable and fertile, some of the experiments were performed on incrossed TgBAC/pcdh19−/− adults.
Image analysis
All image analysis was performed with Fiji (Schindelin et al., 2012). For cell identification and modeling, the TrakEM2 plugin (Cardona et al., 2012) in Fiji was used. Image stacks of ∼100 optical sections were collected from the transgenically labeled tecta of WT or pcdh19−/− larvae. Each detected cell body was modeled as a sphere, and primary projections were traced from cell bodies to the synaptic neuropil when possible. Multipanel images were assembled in Adobe Photoshop, and figures were made in Adobe Illustrator.
Cell transplantation
Transplantations were performed as described previously (Biswas et al., 2014). Host and donor embryos were dechorionated with pronase, and then arranged in an agar injection tray. At 3–3.5 hpf, 20–40 cells from transgenic donor embryos (WT or pcdh19−/−) were transplanted into unlabeled host embryos (WT or pcdh19−/−). Embryos were maintained in embryo medium supplemented with penicillin/streptomycin. At 24 hpf, embryos were screened for health, normal morphology, and fluorescence. At 3–4 dpf, larvae exhibiting fluorescence in the brain were imaged on a two-photon microscope as described in Two-photon imaging.
Visual behavior analysis
Behavioral experiments were performed at 6 dpf and analyzed with the FLOTE software package as previously described (Burgess and Granato, 2007; Burgess et al., 2010). Video was captured at 1,000 frames per second with an IDT MotionPro Y4 camera. 20 larvae of identical genotype were grouped in a 6-cm-wide Petri dish with 9 ml of E3 embryo medium and preadapted to the uniform light intensity for 3 h before testing. 3 dishes of each genotype were tested for positive phototaxis and a response to a sudden removal of preadapted light (termed a “dark flash”). The phototaxis assay was performed as described previously to elicit positive phototaxis (Burgess et al., 2010). Larvae were given 30 s to reach the target light, and only larvae initially positioned on the opposite side of the dish from the target light were included in our analyses. To determine directionality of turns and swims (Fig. 4, C and D), the 30-s video was partitioned into thirty 1-s events. For each event, larvae oriented within 30° of the target light were considered “facing” the target and used for swim direction analyses (Fig. 4 C). Larvae oriented 75°–104° away from the target light were used for turn direction analyses (Fig. 4 D). The visual dark flash assay was performed and analyzed as previously described (Burgess and Granato, 2007). In brief, larvae are adapted to uniform light, and then exposed to ten 1-s periods of darkness separated by 30 s of light. Larvae are expected to perform an O-bend, which has stereotyped kinematic parameters.
Online supplemental material
Fig. S1 shows transgene expression by injection of recombinant BAC clones into zebrafish embryos and double FISH with riboprobes against pcdh7a and pcdh19. In Fig. S2, cell transplantation reveals that pcdh19 function is required in cells and their environment. Fig. S3 shows that the kinematics of swimming behaviors is normal in pcdh19 mutants. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201507108/DC1.
Supplementary Material
Acknowledgments
We thank Min An for technical assistance. We thank Koichi Kawakami and Maximilliano Suster for iTol2 plasmids. The reagents used for BAC recombineering were a gift of Dr. Neal Copeland. The plasmid kit used for the generation of TALENs was a gift of Daniel Voytas and Adam Bogdanove (Addgene kit #1000000016).
This work was supported by a National Institute of Mental Health grant to J.D. Jontes (MH099453), a grant from the National Science Foundation (IOS 1457126), and a Neurosciences Core Grant (P30 NS045758).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- BAC
- bacterial artificial chromosome
- dpf
- days postfertilization
- EdU
- 5-ethynyl-2′-deoxyuridine
- hpf
- hours postfertilization
- HRMA
- high-resolution melting analysis
- pcdh
- protocadherin
- pH3
- phosphohistone H3
- TALEN
- transcriptional activator-like effector nuclease
- WT
- wild type
References
- Bedell V.M., Wang Y., Campbell J.M., Poshusta T.L., Starker C.G., Krug R.G. II, Tan W., Penheiter S.G., Ma A.C., Leung A.Y., et al. 2012. In vivo genome editing using a high-efficiency TALEN system. Nature. 491:114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S., and Jontes J.D.. 2009. Cloning and characterization of zebrafish protocadherin-17. Dev. Genes Evol. 219:265–271. 10.1007/s00427-009-0288-6 [DOI] [PubMed] [Google Scholar]
- Biswas S., Emond M.R., Duy P.Q., Hao T., Beattie C.E., and Jontes J.D.. 2014. Protocadherin-18b interacts with Nap1 to control motor axon growth and arborization in zebrafish. Mol. Biol. Cell. 25:633–642. 10.1091/mbc.E13-08-0475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins C.J., Emond M.R., Biswas S., and Jontes J.D.. 2011. Differential expression, alternative splicing, and adhesive properties of the zebrafish δ1-protocadherins. Neuroscience. 199:523–534. 10.1016/j.neuroscience.2011.09.061 [DOI] [PubMed] [Google Scholar]
- Burgess H.A., and Granato M.. 2007. Modulation of locomotor activity in larval zebrafish during light adaptation. J. Exp. Biol. 210:2526–2539. 10.1242/jeb.003939 [DOI] [PubMed] [Google Scholar]
- Burgess H.A., Schoch H., and Granato M.. 2010. Distinct retinal pathways drive spatial orientation behaviors in zebrafish navigation. Curr. Biol. 20:381–386. 10.1016/j.cub.2010.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona A., Saalfeld S., Schindelin J., Arganda-Carreras I., Preibisch S., Longair M., Tomancak P., Hartenstein V., and Douglas R.J.. 2012. TrakEM2 software for neural circuit reconstruction. PLoS One. 7:e38011 10.1371/journal.pone.0038011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J.A., Somia N.V., Bogdanove A.J., and Voytas D.F.. 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39:e82 10.1093/nar/gkr218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlem T.J., Hoshijima K., Jurynec M.J., Gunther D., Starker C.G., Locke A.S., Weis A.M., Voytas D.F., and Grunwald D.J.. 2012. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8:e1002861 10.1371/journal.pgen.1002861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C., and LeGuern E.. 2012. PCDH19-related infantile epileptic encephalopathy: an unusual X-linked inheritance disorder. Hum. Mutat. 33:627–634. 10.1002/humu.22029 [DOI] [PubMed] [Google Scholar]
- Depienne C., Bouteiller D., Keren B., Cheuret E., Poirier K., Trouillard O., Benyahia B., Quelin C., Carpentier W., Julia S., et al. 2009. Sporadic infantile epileptic encephalopathy caused by mutations in PCDH19 resembles Dravet syndrome but mainly affects females. PLoS Genet. 5:e1000381 10.1371/journal.pgen.1000381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibbens L.M., Tarpey P.S., Hynes K., Bayly M.A., Scheffer I.E., Smith R., Bomar J., Sutton E., Vandeleur L., Shoubridge C., et al. 2008. X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat. Genet. 40:776–781. 10.1038/ng.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emond M.R., Biswas S., and Jontes J.D.. 2009. Protocadherin-19 is essential for early steps in brain morphogenesis. Dev. Biol. 334:72–83. 10.1016/j.ydbio.2009.07.008 [DOI] [PubMed] [Google Scholar]
- Gray G.E., and Sanes J.R.. 1991. Migratory paths and phenotypic choices of clonally related cells in the avian optic tectum. Neuron. 6:211–225. 10.1016/0896-6273(91)90357-6 [DOI] [PubMed] [Google Scholar]
- Gray G.E., and Sanes J.R.. 1992. Lineage of radial glia in the chicken optic tectum. Development. 114:271–283. [DOI] [PubMed] [Google Scholar]
- Hayashi S., Inoue Y., Kiyonari H., Abe T., Misaki K., Moriguchi H., Tanaka Y., and Takeichi M.. 2014. Protocadherin-17 mediates collective axon extension by recruiting actin regulator complexes to interaxonal contacts. Dev. Cell. 30:673–687. 10.1016/j.devcel.2014.07.015 [DOI] [PubMed] [Google Scholar]
- Hirano S., and Takeichi M.. 2012. Cadherins in brain morphogenesis and wiring. Physiol. Rev. 92:597–634. 10.1152/physrev.00014.2011 [DOI] [PubMed] [Google Scholar]
- Kiecker C., and Lumsden A.. 2005. Compartments and their boundaries in vertebrate brain development. Nat. Rev. Neurosci. 6:553–564. 10.1038/nrn1702 [DOI] [PubMed] [Google Scholar]
- Lázár G. 1973. The development of the optic tectum in Xenopus laevis: a Golgi study. J. Anat. 116:347–355. [PMC free article] [PubMed] [Google Scholar]
- Lee E.C., Yu D., Martinez de Velasco J., Tessarollo L., Swing D.A., Court D.L., Jenkins N.A., and Copeland N.G.. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 73:56–65. 10.1006/geno.2000.6451 [DOI] [PubMed] [Google Scholar]
- Leung L.C., Urbančič V., Baudet M.-L., Dwivedy A., Bayley T.G., Lee A.C., Harris W.A., and Holt C.E.. 2013. Coupling of NF-protocadherin signaling to axon guidance by cue-induced translation. Nat. Neurosci. 16:166–173. 10.1038/nn.3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Chen Y., Pan J.J., and Murakami T.. 2009. Expression of protocadherin-9 and protocadherin-17 in the nervous system of the embryonic zebrafish. Gene Expr. Patterns. 9:490–496. 10.1016/j.gep.2009.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Chen Y., Kubota F., Pan J.J., and Murakami T.. 2010. Expression of protocadherin-19 in the nervous system of the embryonic zebrafish. Int. J. Dev. Biol. 54:905–911. 10.1387/ijdb.092882ql [DOI] [PubMed] [Google Scholar]
- Meek J., and Schellart N.A.. 1978. A Golgi study of goldfish optic tectum. J. Comp. Neurol. 182:89–122. 10.1002/cne.901820107 [DOI] [PubMed] [Google Scholar]
- Mountcastle V.B. 1997. The columnar organization of the neocortex. Brain. 120:701–722. 10.1093/brain/120.4.701 [DOI] [PubMed] [Google Scholar]
- Nguyen V., Deschet K., Henrich T., Godet E., Joly J.S., Wittbrodt J., Chourrout D., and Bourrat F.. 1999. Morphogenesis of the optic tectum in the medaka (Oryzias latipes): a morphological and molecular study, with special emphasis on cell proliferation. J. Comp. Neurol. 413:385–404. [DOI] [PubMed] [Google Scholar]
- Noctor S.C., Flint A.C., Weissman T.A., Dammerman R.S., and Kriegstein A.R.. 2001. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 409:714–720. 10.1038/35055553 [DOI] [PubMed] [Google Scholar]
- Peters A., and Sethares C.. 1996. Myelinated axons and the pyramidal cell modules in monkey primary visual cortex. J. Comp. Neurol. 365:232–255. [DOI] [PubMed] [Google Scholar]
- Peters A., and Walsh T.M.. 1972. A study of the organization of apical dendrites in the somatic sensory cortex of the rat. J. Comp. Neurol. 144:253–268. 10.1002/cne.901440302 [DOI] [PubMed] [Google Scholar]
- Pologruto T.A., Sabatini B.L., and Svoboda K.. 2003. ScanImage: flexible software for operating laser scanning microscopes. Biomed. Eng. Online. 2:13 10.1186/1475-925X-2-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portugues R., and Engert F.. 2009. The neural basis of visual behaviors in the larval zebrafish. Curr. Opin. Neurobiol. 19:644–647. 10.1016/j.conb.2009.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. 1988. Specification of cerebral cortical areas. Science. 241:170–176. 10.1126/science.3291116 [DOI] [PubMed] [Google Scholar]
- Recher G., Jouralet J., Brombin A., Heuzé A., Mugniery E., Hermel J.M., Desnoulez S., Savy T., Herbomel P., Bourrat F., et al. 2013. Zebrafish midbrain slow-amplifying progenitors exhibit high levels of transcripts for nucleotide and ribosome biogenesis. Development. 140:4860–4869. 10.1242/dev.099010 [DOI] [PubMed] [Google Scholar]
- Redies C., Hertel N., and Hübner C.A.. 2012. Cadherins and neuropsychiatric disorders. Brain Res. 1470:130–144. 10.1016/j.brainres.2012.06.020 [DOI] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607. 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J.D., Maeder M.L., Reyon D., Voytas D.F., Joung J.K., and Dobbs D.. 2010. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 38(Web Server):W462–W468. 10.1093/nar/gkq319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suster M.L., Abe G., Schouw A., and Kawakami K.. 2011. Transposon-mediated BAC transgenesis in zebrafish. Nat. Protoc. 6:1998–2021. 10.1038/nprot.2011.416 [DOI] [PubMed] [Google Scholar]
- Vanegas H., Laufer M., and Amat J.. 1974. The optic tectum of a perciform teleost. I. General configuration and cytoarchitecture. J. Comp. Neurol. 154:43–60. 10.1002/cne.901540104 [DOI] [PubMed] [Google Scholar]
- Vanhalst K., Kools P., Staes K., van Roy F., and Redies C.. 2005. delta-Protocadherins: a gene family expressed differentially in the mouse brain. Cell. Mol. Life Sci. 62:1247–1259. 10.1007/s00018-005-5021-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. 1995. The zebrafish book. University of Oregon Press, Eugene, OR. [Google Scholar]
- Wolverton T., and Lalande M.. 2001. Identification and characterization of three members of a novel subclass of protocadherins. Genomics. 76:66–72. 10.1006/geno.2001.6592 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.