Abstract
The HORMA domain is a multifunctional protein–protein interaction module found in diverse eukaryotic signaling pathways including the spindle assembly checkpoint, numerous DNA recombination/repair pathways, and the initiation of autophagy. In all of these pathways, HORMA domain proteins occupy key signaling junctures and function through the controlled assembly and disassembly of signaling complexes using a stereotypical “safety belt” peptide interaction mechanism. A recent explosion of structural and functional work has shed new light on these proteins, illustrating how strikingly similar structural mechanisms give rise to radically different functional outcomes in each family of HORMA domain proteins.
Introduction
The HORMA domain was first identified through sequence similarity among three functionally unrelated proteins in Saccharomyces cerevisiae: Hop1, Rev7, and Mad2 (Aravind and Koonin, 1998). Hop1 is a member of a conserved family of proteins termed HORMADs, which bind chromosomes in early meiosis and control many aspects of meiotic recombination and chromosome segregation (Muniyappa et al., 2014; Vader and Musacchio, 2014). The multifunctional Rev7 protein (also called Mad2B or Mad2L2; here, we will use Rev7 throughout) is a subunit of the translesion DNA polymerase ζ (Sale, 2013; Makarova and Burgers, 2015), participates in mitotic cell-cycle control (Pfleger et al., 2001; Listovsky and Sale, 2013), and controls recombination pathway choice in DNA double-strand break (DSB) repair (Boersma et al., 2015; Xu et al., 2015). Mad2 is an essential mediator of the spindle assembly checkpoint (SAC) and is the best-characterized HORMA domain protein both structurally and functionally (Mapelli and Musacchio, 2007; Luo and Yu, 2008). Another HORMA domain protein, p31comet (also called MAD2L1BP), also participates in SAC signaling through its interactions with Mad2 and Pch2/TRIP13, a conserved regulator of HORMA domain proteins (Xia et al., 2004; Yang et al., 2007; Tipton et al., 2012; Eytan et al., 2014; Wang et al., 2014; Ye et al., 2015). Recently, two autophagy-signaling proteins, Atg13 and Atg101, were also shown to possess HORMA domains (Jao et al., 2013; Hegedűs et al., 2014; Suzuki et al., 2015a). In all of these different pathways, the role of the HORMA domain is highly conserved, acting as a signal-responsive adaptor mediating protein–protein interactions through a structurally unique mechanism. In this review, we will outline the general structure and interaction mechanisms of the HORMA domain and discuss how these properties uniquely contribute to signaling in each family. Along the way, we will attempt to draw parallels between different HORMA domain protein families, using lessons from well-understood systems such as Mad2 to inform our understanding of the others.
Conserved structural features of the HORMA domain
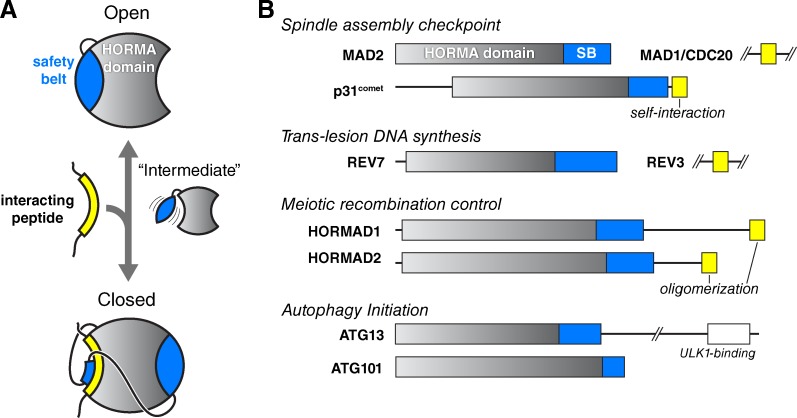
As of this writing, the Protein Data Bank lists 28 nuclear magnetic resonance (NMR) and x-ray crystal structures of HORMA domain proteins (Table S1) that together reveal both common structural features and family-specific variations within the HORMA domain (Fig. 1 and Table 1). The ∼200-aa HORMA domain consists of two functionally distinct regions: the core, comprising the first ∼150 aa, and the C-terminal “safety belt” region. The core (Figs. 1 and S1, gray) comprises three α-helices (αA, αB, and αC) packed against a three-stranded β-sheet (β4, β5, and β6), usually with an additional pair of β-strands (the β2-β3 hairpin) on the “back” of the α-helices (Fig. S1). The structure of the HORMA domain core is stabilized by a buried hydrogen bond network involving an arginine on αA and a glutamate on β4; these are the only two residues conserved among all HORMA domain proteins (Fig. S2; Aravind and Koonin, 1998). The C-terminal safety belt region (Figs. 1 and S1, light blue) can pack against the HORMA domain core in two very different conformations to produce so-called open or closed states. In the open state, the safety belt folds into two β-strands (β7 and β8) that extend over one side of the core β-sheet. In the more commonly observed closed state, the safety belt wraps entirely around the domain and forms two new β-strands (β8′ and β8″) against the opposite side of the HORMA domain core. This change enables a short peptide from a binding partner to interact with the HORMA domain core; the bound peptide is then embraced by the safety belt as it wraps around the domain (Figs. 1 and S1). Because of the topological linkage of the HORMA domain with its binding partner, complex assembly or disassembly likely requires either (a) that the partner protein bind in the process of an open-to-closed conformational conversion of the HORMA domain or (b) that the safety belt transiently disengage from the HORMA domain core to allow partner binding. Mad2 is the only HORMA domain protein known to convert between open and closed states, and Mad2 conformational conversion is intimately linked with partner protein binding and checkpoint signaling (see next section). Rev7 and the meiotic HORMADs are known to bind partner proteins in a manner equivalent to Mad2 but have only been observed in the closed conformation, so the structural mechanisms behind complex assembly and disassembly in these proteins remain unknown.
Figure 1.
Architecture and roles of HORMA domain proteins. (A) Schematic illustrating how conformational changes in the HORMA domain safety belt (blue) are coupled to the binding of interacting peptides (yellow). In the open state, the safety belt occupies the peptide-interaction site. The hypothetical intermediate state would enable an interacting peptide to bind and subsequently become locked into position once the safety belt binds the opposite side of the domain. Both the safety belt and the interacting peptide associate with the HORMA domain core through β-sheet interactions. (B) Domain diagram of human HORMA domain proteins. Proteins containing verified interacting peptides for each protein are shown in yellow. p31comet interacts in cis with its own C-terminal peptide, whereas the meiotic HORMADs’ C-terminal closure motifs are thought to interact in trans to generate oligomeric assemblies. See Fig. S1 for structures and detailed secondary-structure diagrams of each family.
Table 1. Structural features of HORMA domain proteins.
| Protein | Open state? | Closed state? | Partner binding | Dimerization with |
|---|---|---|---|---|
| Mad2 | ✓ | ✓ | Mad1, Cdc20, Sgo2 | Mad2, p31comet |
| p31comet | — | ✓ | Self-bound | Mad2 |
| Rev7 | — | ✓ | Rev3 and othersa | Rev7 |
| HORMADs | — | ✓ | HORMAD C termini | ? |
| Atg13 | — | ✓ | Atg9? | Atg101 |
| Atg101 | ✓ | — | N/A | Atg13 |
N/A, not applicable.
Human ADAM9 and ELK1 have been shown to contain motifs fitting the consensus for Rev7 binding (Hanafusa et al., 2010), but the biological relevance of these interactions is not well established.
Mad2, Rev7, and the meiotic HORMADs each bind multiple different partner proteins through the previously described mechanism. Mad2-binding proteins contain similar Mad2-interacting motifs (MIMs) that fit a consensus sequence of K/RψψxϕxxxP, where K/R is a lysine or arginine; ψ is an aliphatic residue (usually leucine, isoleucine, or methionine), ϕ is a hydrophobic residue (aliphatic or aromatic), and P is proline (Fig. S3 A; Luo et al., 2002; Sironi et al., 2002; Hanafusa et al., 2010). The polar K/R side chain usually forms hydrogen bonds with safety belt residues, and the first two aliphatic side chains are mostly buried by the safety belt as it wraps around the MIM sequence. The downstream aliphatic residue and the proline, whose location varies in different Mad2 binding partners, are usually observed with their side-chains buried in a hydrophobic cleft between αB and β6 (Fig. S3 B). Rev7 has a similar consensus binding sequence of xψψxPxxxpP, where p is a less well-conserved proline residue (Fig. S3 C). As in Mad2, the conserved aliphatic residues are buried by the safety belt, and both conserved proline side chains pack between αB and β6 (Fig. S3 D). The meiotic HORMAD proteins bind short “closure motifs” in their own disordered C-terminal tails to mediate self-assembly (Kim et al., 2014). The best-characterized HORMAD closure motifs are from Caenorhabditis elegans and possess a pair of conserved residues (tyrosine–glycine) that, like the aliphatic residues in Mad2/Rev7, are mostly buried by the safety belt wrapping over the closure motif (Fig. S3, E–H; Kim et al., 2014). Closure motifs from mammalian HORMADs have also been identified (Kim et al., 2014), but comparing these sequences to those from C. elegans reveals no shared consensus sequence for closure motifs across different eukaryotic families.
Another common feature of the HORMA domain is dimerization, through a common interface involving helix αC and the β2-β3 hairpin (Fig. 2, B and C; and Fig. 5 A). Dimerization is usually observed between open and closed HORMA domains and plays distinct roles in different HORMA domain families. In autophagy signaling, the formation of a dimer between Atg13 and Atg101 may simply mediate these proteins’ association as part of an autophagy-initiation signaling complex (Fig. 5 A; Suzuki et al., 2015a). In the SAC, however, the formation of Mad2:Mad2 homodimers and Mad2:p31comet heterodimers (Fig. 2, B and C) is critical for signaling, playing a key role in the conversion of Mad2 between its open and closed states (Mapelli and Musacchio, 2007; Luo and Yu, 2008). There are strong indications that Rev7 can dimerize in at least one of its functional contexts (Hara et al., 2009), though the exact role of dimerization in this family is unknown (see section on Rev7). The only HORMA domain protein family lacking direct evidence for dimerization is the meiotic HORMADs.
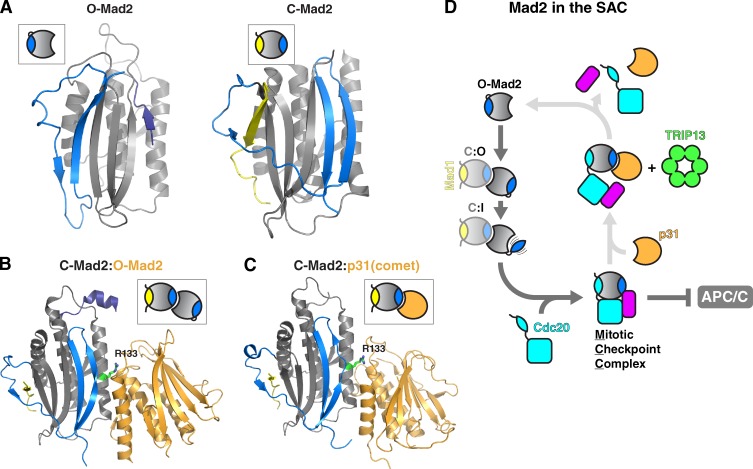
Figure 2.
Structure and function of Mad2 and p31comet in the spindle assembly checkpoint. (A) Structures of Mad2 in the unliganded open state (O-Mad2; right; from PDB ID 1DUJ; Luo et al., 2000) and the Cdc20-bound closed state (C-Mad2; left; from PDB ID 4AEZ; Chao et al., 2012), with structural elements colored as in Fig. 1. (B) Structure of the Homo sapiens C-MAD2:O-MAD2 dimer (PDB ID 2V64; Mapelli et al., 2007). C-MAD2 is colored as in Fig. 1 and O-MAD2 is colored orange. Arginine 133 (R133), which is critical for dimerization (Sironi et al., 2001), is highlighted in green. (C) Structure of the H. sapiens MAD2:p31comet dimer (PDB ID 2QYF; Yang et al., 2007). C-MAD2 is colored as in Fig. 1 and p31comet is colored orange. (D) SAC activation by conversion of O-Mad2 to closed, with Cdc20-bound Mad2 in the MCC. O-Mad2 is recruited to kinetochores by Mad1:Mad2, generating a C:O dimer. O-Mad2 is converted to the proposed intermediate state (I-Mad2), promoting Cdc20 binding and MCC assembly. Once the SAC signal has ceased, p31comet (orange) acts as an adaptor for TRIP13 (green)-mediated MCC disassembly, allowing APC/C activation. This mechanism of SAC inactivation is not conserved in budding yeast: S. cerevisiae lacks p31comet, and its TRIP13 ortholog Pch2 functions only in the disassembly of HORMADs on meiotic chromosomes (see Fig. 3).
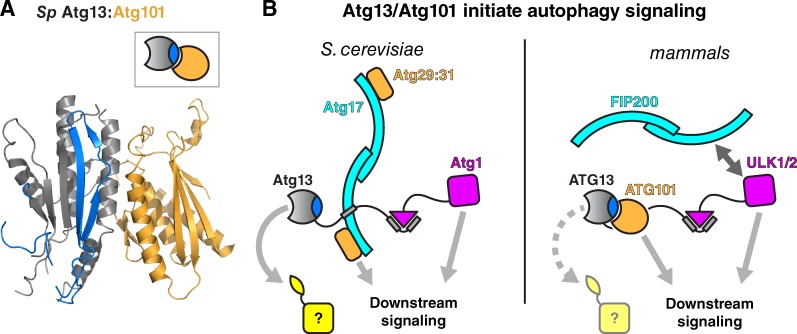
Figure 5.
Structure and function of Atg13 and Atg101. (A) Structure of the S. pombe Atg13:Atg101 dimer (PDB ID 4YK8; Suzuki et al., 2015a). Closed-conformation Atg13 is colored as in Fig. 1 and open-conformation Atg101 is colored orange. (B) Role of Atg13 and Atg101 in autophagy signaling. In S. cerevisiae (left), Atg13 acts as a scaffold for assembly of the Atg1 complex. The Atg13 HORMA domain likely binds a downstream component (yellow) to mediate autophagy initiation; the best candidate protein identified so far is Atg9 (Suzuki et al., 2015b). In mammals (right), ATG13 plays a similar scaffolding role, along with ATG101, but the ATG13 HORMA domain may not be capable of binding partners because of its shortened safety belt region (Qi et al., 2015).
Mad2 and p31comet in the SAC
To ensure accurate chromosome segregation in mitosis, eukaryotic cells monitor kinetochore-microtubule attachment through the SAC (Musacchio and Salmon, 2007; Pines, 2011; Lara-Gonzalez et al., 2012; London and Biggins, 2014). In the SAC, unattached kinetochores catalyze the assembly of a soluble mitotic checkpoint complex (MCC) that inhibits an E3 ubiquitin ligase, the anaphase-promoting complex/cyclosome (APC/C). After all kinetochores become attached to microtubules, assembly of new MCC ceases and existing MCC is disassembled or degraded. The APC/C, directed by its coactivator Cdc20, then promotes the degradation of a defined set of cell-cycle proteins to mediate anaphase onset.
Mad2’s functions in the SAC are intimately linked to the balance between the signaling-inactive open state and the active closed state, and the factors that promote conversion between these two states (Fig. 2). In prometaphase, unattached kinetochores recruit Mad1, bound through a MIM to closed Mad2 (called C-Mad2; Sironi et al., 2002). This Mad1:C-Mad2 complex is thought to stimulate the conversion of open Mad2 (O-Mad2) to the signaling-active closed state and its assembly into the MCC. This stimulation is accomplished by the recruitment of soluble O-Mad2 to the kinetochore-bound Mad1:C-Mad2 complex to form a C-Mad2:O-Mad2 dimer (Fig. 2, B and D; Howell et al., 2004; Shah et al., 2004; De Antoni et al., 2005; Mapelli et al., 2007). Dimerization subtly alters the structure of the O-Mad2 core and may lower the activation energy required to adopt a partially unfolded “intermediate” (I-Mad2) state (Hara et al., 2015). I-Mad2 is thought to then bind a MIM in Cdc20 and refold into the C-Mad2 conformation (De Antoni et al., 2005). The resulting C-Mad2–Cdc20 complex then binds BubR1 and Bub3 to form the full MCC (Sudakin et al., 2001).
Once kinetochores are stably attached to microtubules, the Mad1:C-Mad2 complex is removed, halting assembly of new MCC (Howell et al., 2001; Lara-Gonzalez et al., 2012). The cell now faces a new problem of how to disassemble the existing MCC to inactivate the SAC and allow anaphase onset. Recently, a mechanism for SAC inactivation through direct MCC disassembly has been described, involving p31comet and the AAA+ ATPase TRIP13. p31comet is a structurally unique HORMA domain protein with a stable closed-like conformation (Fig. S1 C) and forms a heterodimer specifically with C-Mad2 (Fig. 2 C; Habu et al., 2002; Mapelli et al., 2006; Yang et al., 2007). Disruption of p31comet in mammalian cells results in prolonged metaphase, implying it has a role in SAC inactivation (Habu et al., 2002; Xia et al., 2004; Hagan et al., 2011), and experiments in cell extracts have suggested that p31comet functions by disassembling the MCC (Reddy et al., 2007; Westhorpe et al., 2011). Recently, p31comet was found to interact with TRIP13, a hexameric AAA+ ATPase related to a large family of protein complex remodelers/unfoldases (Tipton et al., 2012; Wang et al., 2014; Ye et al., 2015). The two proteins together were shown to directly disassemble the MCC in an ATP-dependent manner (Teichner et al., 2011; Eytan et al., 2014). Later work showed that TRIP13 directly converts C-Mad2 to O-Mad2, likely through transient unfolding of the Mad2 safety belt region (Ye et al., 2015). In the context of the MCC, conversion of Cdc20-bound C-Mad2 to the open state likely enables Cdc20 dissociation and MCC disassembly. It is important to note that MCC assembly and disassembly occur together throughout prometaphase, with disassembly dominating only after the SAC is satisfied (Kraft et al., 2003; Vink et al., 2006; Westhorpe et al., 2011). Whether and how MCC disassembly by p31comet and TRIP13 is regulated during the cell cycle is an important open question; recent findings in mammals and Xenopus laevis suggest that this regulation may be achieved at least partially through phosphorylation of p31comet to modulate its affinity for Mad2 (Date et al., 2014; Mo et al., 2015).
Although MCC disassembly is a key activity of p31comet and TRIP13, these proteins may have additional roles that are not yet well understood. Recently, C. elegans p31comet (CMT-1) and TRIP13 (PCH-2) were found to be required for SAC activation when spindle assembly is disrupted (Nelson et al., 2015). This paradoxical finding suggests that p31comet and TRIP13 may maintain the soluble pool of Mad2 in a state conducive to SAC activation (that is, in the less thermodynamically stable open state; Luo et al., 2002; Nelson et al., 2015; Ye et al., 2015). Finally, both p31comet and TRIP13 localize to unattached kinetochores in many organisms (Habu et al., 2002; Hagan et al., 2011; Tipton et al., 2012; Eytan et al., 2014; Wang et al., 2014; Nelson et al., 2015). This localization depends on Mad2, suggesting that p31comet directly recognizes kinetochore-bound Mad1:C-Mad2, but the roles of kinetochore-localized p31comet and TRIP13 remain largely mysterious.
Meiotic HORMADs: Meiotic chromosome organization and recombination
In meiosis, cells must reduce their ploidy by half to generate gametes in preparation for sexual reproduction. Ploidy reduction requires that homologous chromosomes become physically linked by reciprocal DNA recombination events, called crossovers, in meiotic prophase. Crossover formation is controlled by a specialized structure called the chromosome axis, of which the meiotic HORMADs are a key component. In early meiotic prophase, meiotic HORMADs localize along the entire length of chromosomes and promote DNA DSB formation by the Spo11 endonuclease (Mao-Draayer et al., 1996; Woltering et al., 2000; Goodyer et al., 2008; Shin et al., 2010). After DNA breakage, meiotic HORMADs suppress recombination with the sister chromatid, thereby promoting recombination with the homolog to generate crossovers (Fig. 3). This “homolog bias” of meiotic recombination stands in stark contrast to homologous recombination in mitotic cells, where repair via the sister is strongly favored (Humphryes and Hochwagen, 2014). Later in meiotic prophase, assembly of the specialized synaptonemal complex (SC) along each pair of homologs is, in most organisms, coordinated with the removal of meiotic HORMADs from chromosomes (Börner et al., 2008; Wojtasz et al., 2009; Lambing et al., 2015). Meiotic HORMAD removal is thought to shut down further DNA breakage by Spo11 and also alleviate the block to sister-chromosome–mediated repair (Fig. 3; Wojtasz et al., 2009; Kauppi et al., 2013; Thacker et al., 2014). The complex signaling networks governing these activities, and the roles of meiotic HORMADs in these networks, have been nicely summarized in several recent reviews (MacQueen and Hochwagen, 2011; Humphryes and Hochwagen, 2014; Subramanian and Hochwagen, 2014).
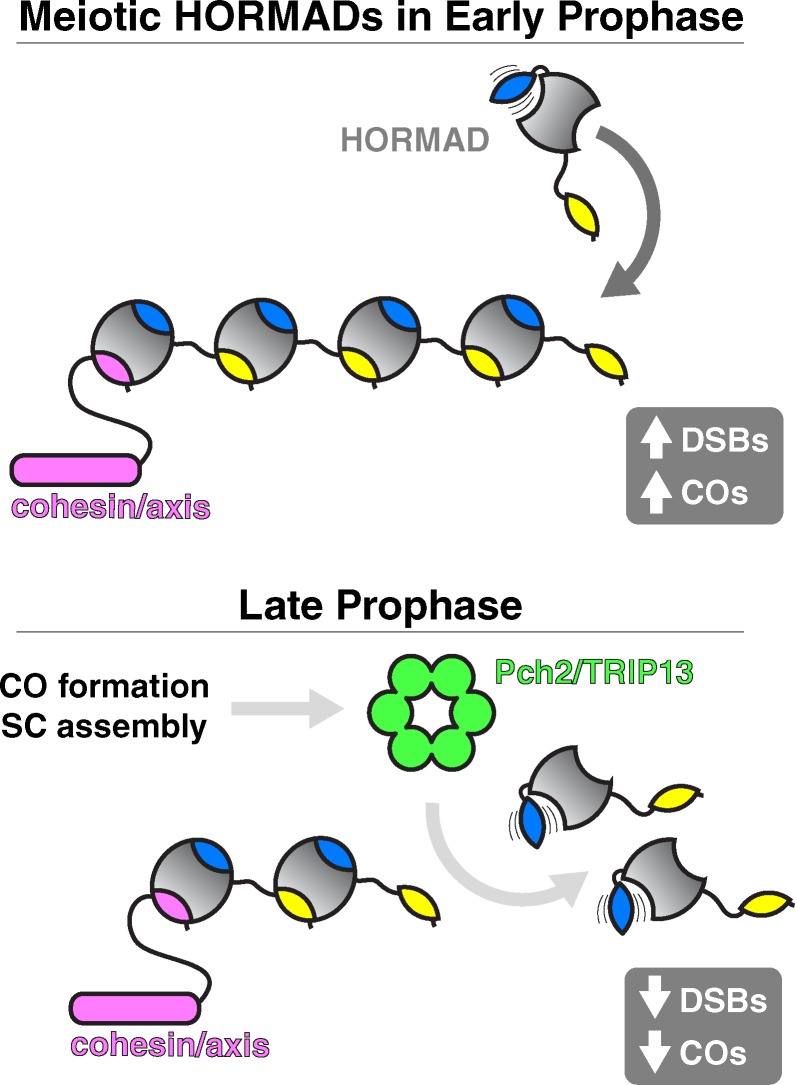
Figure 3.
Biological roles of the meiotic HORMADs. Model for meiotic HORMAD assembly/disassembly at the meiotic chromosome axis. In early meiotic prophase (top), HORMADs are likely recruited to chromosomes through closure motifs in cohesin/SC proteins (pink), then self-assemble through HORMA–closure motif interactions. On chromosomes, HORMADs promote DSB and CO formation through largely unknown mechanisms. Coinciding with the maturation of COs and SC assembly in late prophase (bottom), HORMADs are removed from the chromosomes in a TRIP13-dependent manner, downregulating further DSB and CO formation.
Fungi have a single meiotic HORMAD (Hop1), mammals and plants have two (HORMAD1/2 and ASY1/2), and the nematode C. elegans has four (HIM-3, HTP-1/2/3; Hollingsworth and Johnson, 1993; Caryl et al., 2000; Couteau and Zetka, 2005; Goodyer et al., 2008; Wojtasz et al., 2009). Although meiotic HORMADs from fungi and plants contain additional domains that likely bind DNA or other proteins (Hollingsworth et al., 1990; Kironmai et al., 1998; Aravind and Iyer, 2002), most proteins in this family possess only an N-terminal HORMA domain and a disordered C-terminal tail (Figs. 1 B and S1 E). Recent work with the expanded meiotic HORMAD family in C. elegans showed that these proteins contain closure motifs in their tails (one each in HIM-3, HTP-1, and HTP-2 and six in HTP-3) and form a hierarchical complex through specific HORMA domain–closure motif interactions (Kim et al., 2014; Fig. 3). These interactions are necessary for axis localization of HIM-3, HTP-1, and HTP-2 and as such are required for SC assembly and crossover formation (Kim et al., 2014). Further, this mode of interaction is conserved: mammalian HORMAD1 and HORMAD2 contain closure motifs in their C termini that bind to the HORMA domain of HORMAD1 in vitro (Kim et al., 2014), and highly conserved motifs at the C termini of both fungal and plant meiotic HORMADs (not depicted) suggest that these proteins may also assemble in a similar manner.
Recent work has provided some insight into the mechanisms for initial recruitment of HORMADs to the chromosome axis. Mass spectrometry of meiotic HORMAD complexes in C. elegans suggests a direct interaction between HTP-3 and cohesin complexes; as HTP-3’s HORMA domain does not bind closure motifs in other meiotic HORMADs, it may bind a sequence within the cohesin complex to nucleate assembly on chromosomes (Kim et al., 2014). In other organisms, meiotic HORMAD recruitment requires a second chromosome axis component, which may in turn bind cohesin (Sakuno and Watanabe, 2015); S. cerevisiae Hop1 requires Red1 for chromosome localization (Smith and Roeder, 1997; Woltering et al., 2000), and similar dependencies on chromosome axis proteins, potentially orthologous to S. cerevisiae Red1, have been reported for meiotic HORMADs from both plants and mammals (Fukuda et al., 2010; Wang et al., 2011; Ferdous et al., 2012). Thus, an attractive, though as yet unproven, model for meiotic HORMAD localization is that their HORMA domains bind closure motif sequences within cohesin or cohesin-binding axis proteins to mediate initial recruitment, followed by head-to-tail assembly of larger complexes on chromosomes (Fig. 3). The extent of meiotic HORMAD self-association along chromosome axes in vivo, and crucially how the resulting assemblies promote and control meiotic DSB formation and interhomolog recombination, are not yet well understood.
As mentioned previously, assembly of the SC is coordinated with the removal of meiotic HORMADs from chromosomes in many organisms. In a striking parallel with Mad2, meiotic HORMADs depend on TRIP13 (Pch2 in S. cerevisiae) for this removal. S. cerevisiae Pch2 was first identified as a protein that mediates the removal of Hop1 from chromosomes upon SC assembly, and both mammalian TRIP13 and plant PCH2 also share this function (San-Segundo and Roeder, 1999; Börner et al., 2008; Wojtasz et al., 2009; Chen et al., 2014; Lambing et al., 2015). The recent finding that mammalian TRIP13 can disassemble MAD2-containing complexes, likely through direct manipulation of the safety belt, strongly suggests that meiotic HORMAD removal from chromosomes involves a similar disassembly mechanism (Ye et al., 2015). Although meiotic HORMADs have not been shown to possess a defined open state, transient unfolding of their safety belt region by TRIP13 could nonetheless disrupt closure motif binding and mediate complex disassembly and removal from the chromosome axis (Fig. 3). How recognition of meiotic HORMADs by Pch2/TRIP13 is coordinated with SC assembly remains unknown. S. cerevisiae Pch2 directly interacts with Hop1 in vitro and can remove Hop1 from DNA (Chen et al., 2014), and Arabidopsis thaliana PCH2 was recently shown to copurify with the meiotic HORMAD ASY1 (Lambing et al., 2015). As TRIP13 recognition of Mad2 requires that it form a dimer with p31comet (Ye et al., 2015), it is intriguing to wonder whether meiotic HORMAD recognition requires either homodimerization or binding to an as-yet-unidentified adapter protein.
Rev7: A polymerase adaptor with a secret life (or two)
All cells possess specialized pathways to synthesize DNA past damaged bases using a mechanism known as translesion synthesis (TLS). In eukaryotes, TLS involves the coordinated action of DNA polymerases that can insert bases opposite a lesion (“inserter” polymerase) and then continue synthesis past the lesion (“extender” polymerase; Prakash et al., 2005; Waters et al., 2009; Sale, 2013). The major extender polymerase in eukaryotes is Pol ζ, which consists of the catalytic Rev3 subunit and the HORMA-domain protein Rev7 (Makarova and Burgers, 2015). Rev7 is required for full activity of Pol ζ (Nelson et al., 1996a) and links Pol ζ to Rev1, an inserter polymerase that also coordinates the activity of Pol ζ with other inserter polymerases (Fig. 4 A; Nelson et al., 1996b, 2000; Haracska et al., 2001; Ross et al., 2005). Thus, Rev7 plays a central organizing role in TLS through its interactions with Rev3 and Rev1 (Fig. 4 B).
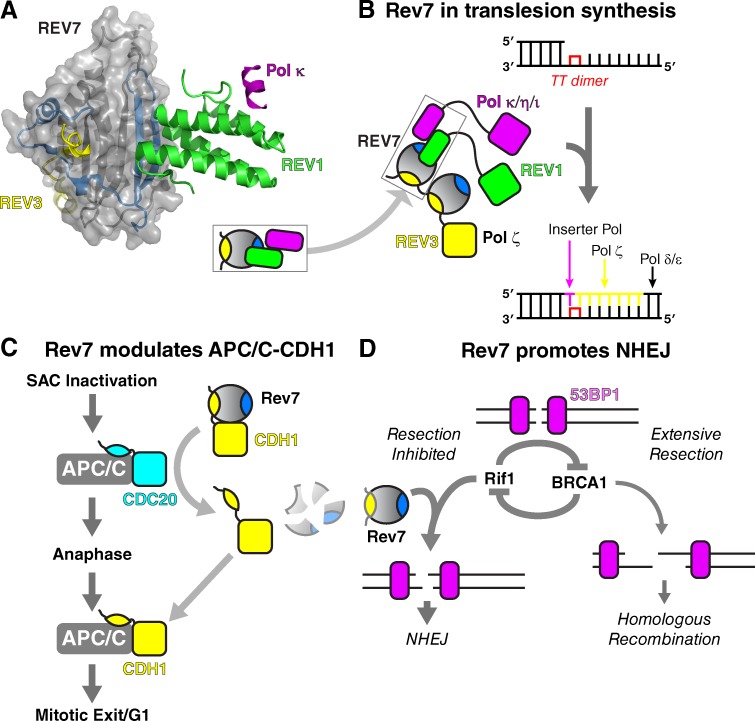
Figure 4.
Biological roles of Rev7. (A) Crystal structure of the complex between Rev7 (shown as gray “surface” representation), Rev3 (yellow), Rev1 (green), and the Rev1-interacting region of Pol κ (purple; Wojtaszek et al., 2012; Xie et al., 2012). (inset) Schematic version of this structure. (B) Diagram outlining the role of Rev7 and associated TLS polymerases during lesion bypass. Rev1 or an associated inserter polymerase inserts the first base opposite the thymine dimer, and Pol ζ performs the following extension step. (C) Proposed role of Rev7 as an inhibitor of APC/C-CDH1 during mitosis. Rev7 binds CDH1/FZR1, potentially through a HORMA domain–binding motif as in Mad2–Cdc20, sequestering it from the APC/C. Once APC/C-CDC20 is activated at anaphase onset, Rev7 is targeted for degradation, resulting in the release of CDH1 and its incorporation into the APC/C. (D) Role of Rev7 during DSB repair. After 53BP1 association with a DSB, Rif1 and BRCA1 play antagonistic roles to either promote end resection and homologous recombination (right branch) or instead inhibit resection, leading to NHEJ (left branch). Rev7 acts downstream of Rif1 through an unknown mechanism to inhibit resection and promote NHEJ.
Human REV3 contains two REV7-binding motifs: (a) residues 1,877–1,898 and (b) residues 1,993–2,003 of 3,130 in human REV3, suggesting that active Pol ζ might incorporate two copies of REV7 (Hara et al., 2009, 2010; Tomida et al., 2015). Interestingly, a reconstituted REV7–REV31,848–1,898 complex forms a heterotetramer in solution, and this tetramer is disrupted by mutation of REV7 arginine 124, located within the canonical HORMA domain dimerization interface on helix αC (Hara et al., 2009). Because mutation of the equivalent residue in Mad2 (Arg133) disrupts Mad2 dimer formation (Sironi et al., 2001), this evidence supports the idea that Rev7 can form canonical HORMA domain dimers. Our own yeast two-hybrid analysis confirms that REV7 can self-associate and that mutation of arginine 124 disrupts this association (not depicted). Thus, an attractive model is that two copies of Rev7 might bind the two motifs in Rev3 and then homodimerize. This Rev7 homodimer could promote a productive conformation of Rev3 or recruit additional proteins to coordinate TLS (Tomida et al., 2015).
Rev7 binds to the inserter polymerase Rev1 through a surface on its β-sheet face that includes β8′ and β8″, close to but not overlapping the putative Rev7 dimerization surface (Fig. 4 A; Kikuchi et al., 2012; Wojtaszek et al., 2012). This interaction surface is unique among HORMA domain proteins and, because of its dependence on β8′ and β8″, likely requires the closed conformation of Rev7. Another potential binding partner is the transcription factor TFII-I, which is proposed to bind Rev7 and recruit Pol ζ to sites of damage through an interaction with PCNA (Fattah et al., 2014). In vitro, TFII-I can bind to a reconstituted Rev3:Rev7:Rev1 complex (Fattah et al., 2014), suggesting that the Rev7 HORMA domain may possess yet another unique protein interaction surface. Finally, one or more Rev7 interactions may be phosphoregulated: the multifunctional Dbf4-dependent protein kinase, which is required for TLS in yeast (Yamada et al., 2013), is proposed to phosphorylate Rev7 to promote Pol ζ localization to sites of repair (Brandão et al., 2014). Overall, although much work remains to tease out the details of its multiple interactions, it is clear that Rev7 is central to the assembly and function of TLS polymerases.
Although early work on Rev7 revealed its highly conserved roles in the TLS pathway, mammalian REV7 also plays a supporting role in the control of cell division. Based on sequence similarity to MAD2, human REV7 was rediscovered in 1999 and named MAD2B/MAD2L2 (Cahill et al., 1999). Later work showed that REV7 inhibits APC/C in vitro through a direct interaction with CDH1/FZR1, a paralog of CDC20 (Chen and Fang, 2001; Pfleger et al., 2001; Listovsky and Sale, 2013). CDH1 directs APC/C activity after CDC20-mediated anaphase onset to promote mitotic exit and the transition to G1 (Primorac and Musacchio, 2013; Sivakumar and Gorbsky, 2015). REV7 binds and sequesters CDH1 during metaphase and is targeted for degradation by APC/C-CDC20 in early anaphase, thereby releasing CDH1 (Fig. 4 C; Listovsky and Sale, 2013). This mechanism complements other pathways for control of CDH1-APC/C interactions (Primorac and Musacchio, 2013) and is required for the proper timing of mitotic exit (Listovsky and Sale, 2013). In an intriguing connection with Rev7’s TLS functions, Rev1 is also a substrate for APC/C-Cdc20–mediated degradation at anaphase, and its ubiquitylation and degradation depend on the presence of Rev7 (Chun et al., 2013).
Recently, Rev7 has been implicated in recombination pathway choice during DNA DSB repair. Eukaryotic cells can repair DNA DSBs by either nonhomologous end joining (NHEJ), which is highly error prone, or homologous recombination. The choice of pathway is heavily influenced by cell cycle stage, and the signaling networks controlling pathway choice have only recently become well understood. A critical step determining which pathway is used for repair is the amount of 5′-to-3′ resection that occurs at a break: the generation of a long 3′ single-stranded overhang favors homologous recombination, whereas limited resection favors NHEJ. A cascade of factors is recruited to DNA break sites, including the proteins 53BP1 and Rif1, which together inhibit end resection to promote NHEJ. Recently, two separate screens for factors governing recombination pathway choice in very different contexts identified Rev7 as another inhibitor of DSB end resection (Boersma et al., 2015; Xu et al., 2015). Rev7 was shown to act downstream of 53BP1 and Rif1 to limit resection and promote NHEJ (Fig. 4 D), but the mechanism for its recruitment to DSB sites, and how it ultimately inhibits end resection, is currently unknown.
Finally, there are hints that Rev7 has even more roles than space here allows us to discuss, including epigenetic reprogramming of germ cells and maintenance of pluripotency (Pirouz et al., 2013, 2015; Sale, 2013; Watanabe et al., 2013), heterochromatin maintenance (Vermeulen et al., 2010), and additional roles in mitosis (Medendorp et al., 2009, 2010) and DNA damage signaling (Zhang et al., 2007). An important avenue for future inquiry will be to determine if these diverse roles of Rev7 are linked, perhaps by a shared cell-cycle dependence or as-yet-unappreciated common regulation in developmental pathways.
HORMA domain proteins in autophagy initiation
Autophagy (literally, “self-eating”) is a conserved starvation-response pathway in eukaryotic cells, in which part of a cell’s contents are engulfed and then degraded (Mizushima, 2007; Xie and Klionsky, 2007). The first step in autophagy is formation of the preautophagosomal structure or phagophore assembly site (PAS), which in S. cerevisiae is initiated by assembly of the Atg1 kinase complex (Mizushima, 2010). This complex is composed of three functional units: the Atg1 kinase, the Atg17/29/31 scaffold complex, and Atg13 (Fig. 5 B). Atg13 possesses an N-terminal HORMA domain (Jao et al., 2013) and an extended C-terminal tail with regulatory phosphorylation sites and binding sites for both Atg1 and the Atg17 scaffold complex (Fujioka et al., 2014; Stjepanovic et al., 2014). Autophagy initiation is regulated by Atg13 tail phosphorylation: in nutrient-rich conditions, the TOR kinase phosphorylates Atg13, inactivating it. In response to starvation, Atg13 is rapidly dephosphorylated, allowing the C terminus to bind both Atg1 (Kabeya et al., 2005; Ragusa et al., 2012) and Atg17 (Kamada et al., 2000), thereby initiating PAS formation.
When the crystal structure of the N-terminal domain of budding yeast Atg13 was determined in 2013, it revealed a previously unidentified HORMA domain in a closed conformation but without a binding partner (Fig. S1 F; Jao et al., 2013), immediately raising the question as to what this domain might bind. Recently, it was found that the Atg13 HORMA domain is required for the recruitment of Atg9-containing vesicles, a critical step in development of the PAS (Suzuki et al., 2015b). This was further demonstrated to be mediated by a direct interaction between the Atg13 HORMA domain and the N-terminal disordered region of Atg9, strongly suggesting that the Atg13 HORMA domain binds a motif within Atg9 in a manner equivalent to other HORMA domain proteins (Suzuki et al., 2015b).
In contrast to the case in fission yeast and animals (see next paragraph), budding yeast Atg13 has not been shown to form homo- or heterodimers. It has been suggested that a budding yeast–specific addition to the HORMA domain, termed the cap (Fig. S1 F), stabilizes the closed monomeric structure of Atg13 (Jao et al., 2013; Suzuki et al., 2015a). On the other hand, S. cerevisiae Atg13 promotes self-interaction and activation of the Atg1 kinase, suggesting that budding yeast Atg13 may in fact self-associate (Yeh et al., 2011). Further work will be required to determine whether the putative self-interaction of Atg13 is mediated by the HORMA domain, through either homodimerization or HORMAD-style head-to-tail association.
Autophagy initiation in animals and the fission yeast Schizosaccharomyces pombe involve orthologues of Atg1 kinase (ULK1/2 in animals) and Atg13, whereas the Atg17 scaffold complex is functionally replaced by other proteins (Mizushima, 2010; Hurley and Schulman, 2014). These proteins include a second HORMA domain protein, Atg101, which folds into the open state and binds directly to Atg13, forming an open–closed heterodimer similar in structure to the open–closed Mad2 dimer (Hosokawa et al., 2009; Mercer et al., 2009; Michel et al., 2015; Qi et al., 2015; Suzuki et al., 2015a; Fig. 5 A). The dimerization interface of Atg101 is highly conserved, and disruption of the interface results in severe autophagy defects in vivo (Suzuki et al., 2015a). The functional significance of Atg13:Atg101 dimerization, and whether this complex is dynamic as in Mad2, is not currently known. The Atg101 safety belt is truncated, containing a β7 strand but no β8 strand, making it unlikely that the protein can adopt the open state and supporting the idea that the Atg13:Atg101 dimer is stable/constitutive (Michel et al., 2015; Qi et al., 2015; Suzuki et al., 2015a). In addition to binding Atg13, Atg101 has several unique structural features that are important for autophagy signaling. In particular, an extended loop connecting β4 and β5, referred to as the “WF finger” motif because of the presence of conserved aromatic residues, is required for autophagy and is proposed to mediate protein–protein interactions important for autophagy initiation (Suzuki et al., 2015a). These interactions may compensate for a proposed loss of interaction potential within the HORMA domain of Atg13 in these species: a recent structure of human ATG13 showed that this protein’s safety belt is shorter than that in budding or fission yeast and may not be capable of binding peptide motifs (Qi et al., 2015). Overall, the roles of the HORMA domains of Atg13 and Atg101 in autophagy signaling are just beginning to be explored, and it will be exciting to see how the common themes of HORMA domain structure and function contribute to signaling in this pathway.
Conclusions
Recent years have seen great progress in defining the mechanisms of HORMA domain function in many different cellular contexts. Although originally identified as a domain shared among three protein families (Hop1, Rev7, and Mad2; Aravind and Koonin, 1998), three additional families with the same fold have since been defined (p31comet, Atg13, and Atg101). Despite the great functional diversity among these families, some common themes can be drawn from the work on HORMA domain proteins to date. With few exceptions, these proteins’ mechanisms are based on two types of interactions: the binding of the safety belt region around short peptide motifs and homo-/heterodimerization. The assembly and disassembly of complexes using these interactions, and the functional interplay between protein interactions and conformational changes within the domain, account for many aspects of these proteins’ functions. Although each family has unique modifications and interaction modes that enable specialized functions, the core role of the HORMA domain remains remarkably constant.
There are many outstanding questions regarding the mechanisms of the known HORMA domain protein families. Chief among them, to us, is the mechanism for binding and exchange of partner proteins in those families that have not been shown to adopt an open state. Of 28 structures of HORMA domain proteins in the Protein Data Bank, 22 depict HORMA domains with their safety belts topologically embracing a binding partner; in the other six, there either is no partner bound or the HORMA domain is in the open state. The consistent observation that partners are topologically linked to the closed HORMA domain strongly supports the idea that these interactions universally involve opening of the safety belt during binding or release, be this a transition to a state like Mad2’s open state or simply a transient unfolding or disengagement of the safety belt from the core. The question of whether and how HORMA domain proteins open has important implications for these proteins’ mechanisms, especially when considering the dynamics of HORMA domain protein complexes during signaling pathway activation and inactivation. Overall, understanding the dynamics and mechanisms of partner protein binding, exchange, and release by each family of HORMA domain proteins will be an important avenue of future research.
Could there be additional HORMA domain proteins waiting to be identified? Our own structure-based searches of several representative genomes reveal no additional examples, but as the saying goes, “The absence of evidence is not evidence of absence.” Given the functional plasticity of the HORMA domain and its consequent central roles in many different pathways, it would not be surprising to find this domain at key signaling junctures in even more pathways in the future.
Supplementary Material
Acknowledgments
The authors thank Juan Wang, Susan Ferro-Novick, Pablo Lara Gonzalez, Dhanya Cheerambathur, Arshad Desai, Yumi Kim, and members of the Corbett laboratory for critical reading and helpful discussions.
K.D. Corbett acknowledges support from the Ludwig Institute for Cancer Research and the National Institutes of Health (R01GM104141).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- APC/C
- anaphase-promoting complex/cyclosome
- DSB
- double-strand break
- MCC
- mitotic checkpoint complex
- MIM
- Mad2-interacting motif
- NHEJ
- nonhomologous end joining
- NMR
- nuclear magnetic resonance
- PAS
- preautophagosomal structure or phagophore assembly site
- SAC
- spindle assembly checkpoint
- SC
- synaptonemal complex
- TLS
- translesion synthesis
References
- Aravind L., and Iyer L.M.. 2002. The SWIRM domain: A conserved module found in chromosomal proteins points to novel chromatin-modifying activities. Genome Biol. 3:research0039–research0039.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., and Koonin E.V.. 1998. The HORMA domain: A common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem. Sci. 23:284–286. 10.1016/S0968-0004(98)01257-2 [DOI] [PubMed] [Google Scholar]
- Boersma V., Moatti N., Segura-Bayona S., Peuscher M.H., van der Torre J., Wevers B.A., Orthwein A., Durocher D., and Jacobs J.J.L.. 2015. MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5′ end resection. Nature. 521:537–540. 10.1038/nature14216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner G.V., Barot A., and Kleckner N.. 2008. Yeast Pch2 promotes domainal axis organization, timely recombination progression, and arrest of defective recombinosomes during meiosis. Proc. Natl. Acad. Sci. USA. 105:3327–3332. 10.1073/pnas.0711864105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandão L.N., Ferguson R., Santoro I., Jinks-Robertson S., and Sclafani R.A.. 2014. The role of Dbf4-dependent protein kinase in DNA polymerase ζ-dependent mutagenesis in Saccharomyces cerevisiae. Genetics. 197:1111–1122. 10.1534/genetics.114.165308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill D.P., da Costa L.T., Carson-Walter E.B., Kinzler K.W., Vogelstein B., and Lengauer C.. 1999. Characterization of MAD2B and other mitotic spindle checkpoint genes. Genomics. 58:181–187. 10.1006/geno.1999.5831 [DOI] [PubMed] [Google Scholar]
- Caryl A.P., Armstrong S.J., Jones G.H., and Franklin F.C.H.. 2000. A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma. 109:62–71. 10.1007/s004120050413 [DOI] [PubMed] [Google Scholar]
- Chao W.C.H., Kulkarni K., Zhang Z., Kong E.H., and Barford D.. 2012. Structure of the mitotic checkpoint complex. Nature. 484:208–213. 10.1038/nature10896 [DOI] [PubMed] [Google Scholar]
- Chen J., and Fang G.. 2001. MAD2B is an inhibitor of the anaphase-promoting complex. Genes Dev. 15:1765–1770. 10.1101/gad.898701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Jomaa A., Ortega J., and Alani E.E.. 2014. Pch2 is a hexameric ring ATPase that remodels the chromosome axis protein Hop1. Proc. Natl. Acad. Sci. USA. 111:E44–E53. 10.1073/pnas.1310755111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun A.C.-S., Kok K.-H., and Jin D.-Y.. 2013. REV7 is required for anaphase-promoting complex-dependent ubiquitination and degradation of translesion DNA polymerase REV1. Cell Cycle. 12:365–378. 10.4161/cc.23214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F., and Zetka M.. 2005. HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes Dev. 19:2744–2756. 10.1101/gad.1348205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date D.A., Burrows A.C., and Summers M.K.. 2014. Phosphorylation regulates the p31Comet-mitotic arrest-deficient 2 (Mad2) interaction to promote spindle assembly checkpoint (SAC) activity. J. Biol. Chem. 289:11367–11373. 10.1074/jbc.M113.520841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A., Pearson C.G., Cimini D., Canman J.C., Sala V., Nezi L., Mapelli M., Sironi L., Faretta M., Salmon E.D., and Musacchio A.. 2005. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 15:214–225. 10.1016/j.cub.2005.01.038 [DOI] [PubMed] [Google Scholar]
- Eytan E., Wang K., Miniowitz-Shemtov S., Sitry-Shevah D., Kaisari S., Yen T.J., Liu S.T., and Hershko A.. 2014. Disassembly of mitotic checkpoint complexes by the joint action of the AAA-ATPase TRIP13 and p31(comet). Proc. Natl. Acad. Sci. USA. 111:12019–12024. 10.1073/pnas.1412901111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattah F.J., Hara K., Fattah K.R., Yang C., Wu N., Warrington R., Chen D.J., Zhou P., Boothman D.A., and Yu H.. 2014. The transcription factor TFII-I promotes DNA translesion synthesis and genomic stability. PLoS Genet. 10:e1004419 10.1371/journal.pgen.1004419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdous M., Higgins J.D., Osman K., Lambing C., Roitinger E., Mechtler K., Armstrong S.J., Perry R., Pradillo M., Cuñado N., and Franklin F.C.H.. 2012. Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLoS Genet. 8:e1002507 10.1371/journal.pgen.1002507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y., Suzuki S.W., Yamamoto H., Kondo-Kakuta C., Kimura Y., Hirano H., Akada R., Inagaki F., Ohsumi Y., and Noda N.N.. 2014. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat. Struct. Mol. Biol. 21:513–521. 10.1038/nsmb.2822 [DOI] [PubMed] [Google Scholar]
- Fukuda T., Daniel K., Wojtasz L., Toth A., and Höög C.. 2010. A novel mammalian HORMA domain-containing protein, HORMAD1, preferentially associates with unsynapsed meiotic chromosomes. Exp. Cell Res. 316:158–171. 10.1016/j.yexcr.2009.08.007 [DOI] [PubMed] [Google Scholar]
- Goodyer W., Kaitna S., Couteau F., Ward J.D., Boulton S.J., and Zetka M.. 2008. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev. Cell. 14:263–274. 10.1016/j.devcel.2007.11.016 [DOI] [PubMed] [Google Scholar]
- Habu T., Kim S.H., Weinstein J., and Matsumoto T.. 2002. Identification of a MAD2-binding protein, CMT2, and its role in mitosis. EMBO J. 21:6419–6428. 10.1093/emboj/cdf659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan R.S., Manak M.S., Buch H.K., Meier M.G., Meraldi P., Shah J.V., Sorger P.K., and Doxsey S.J.. 2011. p31(comet) acts to ensure timely spindle checkpoint silencing subsequent to kinetochore attachment. Mol. Biol. Cell. 22:4236–4246. 10.1091/mbc.E11-03-0216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanafusa T., Habu T., Tomida J., Ohashi E., Murakumo Y., and Ohmori H.. 2010. Overlapping in short motif sequences for binding to human REV7 and MAD2 proteins. Genes Cells. 15:281–296. 10.1111/j.1365-2443.2009.01380.x [DOI] [PubMed] [Google Scholar]
- Hara K., Shimizu T., Unzai S., Akashi S., Sato M., and Hashimoto H.. 2009. Purification, crystallization and initial X-ray diffraction study of human REV7 in complex with a REV3 fragment. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65:1302–1305. 10.1107/S1744309109046181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Hashimoto H., Murakumo Y., Kobayashi S., Kogame T., Unzai S., Akashi S., Takeda S., Shimizu T., and Sato M.. 2010. Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase zeta and REV1. J. Biol. Chem. 285:12299–12307. 10.1074/jbc.M109.092403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M., Özkan E., Sun H., Yu H., and Luo X.. 2015. Structure of an intermediate conformer of the spindle checkpoint protein Mad2. Proc. Natl. Acad. Sci. USA. 112:11252–11257. 10.1073/pnas.1512197112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L., Unk I., Johnson R.E., Johansson E., Burgers P.M., Prakash S., and Prakash L.. 2001. Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes Dev. 15:945–954. 10.1101/gad.882301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedűs K., Nagy P., Gáspári Z., and Juhász G.. 2014. The putative HORMA domain protein Atg101 dimerizes and is required for starvation-induced and selective autophagy in Drosophila. BioMed Res. Int. 2014:470482 10.1155/2014/470482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N.M., and Johnson A.D.. 1993. A conditional allele of the Saccharomyces cerevisiae HOP1 gene is suppressed by overexpression of two other meiosis-specific genes: RED1 and REC104. Genetics. 133:785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N.M., Goetsch L., and Byers B.. 1990. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell. 61:73–84. 10.1016/0092-8674(90)90216-2 [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Sasaki T., Iemura S., Natsume T., Hara T., and Mizushima N.. 2009. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 5:973–979. 10.4161/auto.5.7.9296 [DOI] [PubMed] [Google Scholar]
- Howell B.J., McEwen B.F., Canman J.C., Hoffman D.B., Farrar E.M., Rieder C.L., and Salmon E.D.. 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155:1159–1172. 10.1083/jcb.200105093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B.J., Moree B., Farrar E.M., Stewart S., Fang G., and Salmon E.D.. 2004. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 14:953–964. 10.1016/j.cub.2004.05.053 [DOI] [PubMed] [Google Scholar]
- Humphryes N., and Hochwagen A.. 2014. A non-sister act: Recombination template choice during meiosis. Exp. Cell Res. 329:53–60. 10.1016/j.yexcr.2014.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.H., and Schulman B.A.. 2014. Atomistic autophagy: The structures of cellular self-digestion. Cell. 157:300–311. 10.1016/j.cell.2014.01.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao C.C., Ragusa M.J., Stanley R.E., and Hurley J.H.. 2013. A HORMA domain in Atg13 mediates PI 3-kinase recruitment in autophagy. Proc. Natl. Acad. Sci. USA. 110:5486–5491. 10.1073/pnas.1220306110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Kamada Y., Baba M., Takikawa H., Sasaki M., and Ohsumi Y.. 2005. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell. 16:2544–2553. 10.1091/mbc.E04-08-0669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., and Ohsumi Y.. 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150:1507–1513. 10.1083/jcb.150.6.1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L., Barchi M., Lange J., Baudat F., Jasin M., and Keeney S.. 2013. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev. 27:873–886. 10.1101/gad.213652.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S., Hara K., Shimizu T., Sato M., and Hashimoto H.. 2012. Structural basis of recruitment of DNA polymerase ζ by interaction between REV1 and REV7 proteins. J. Biol. Chem. 287:33847–33852. 10.1074/jbc.M112.396838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Rosenberg S.C., Kugel C.L., Kostow N., Rog O., Davydov V., Su T.Y., Dernburg A.F., and Corbett K.D.. 2014. The chromosome axis controls meiotic events through a hierarchical assembly of HORMA domain proteins. Dev. Cell. 31:487–502. 10.1016/j.devcel.2014.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kironmai K.M., Muniyappa K., Friedman D.B., Hollingsworth N.M., and Byers B.. 1998. DNA-binding activities of Hop1 protein, a synaptonemal complex component from Saccharomyces cerevisiae. Mol. Cell. Biol. 18:1424–1435. 10.1128/MCB.18.3.1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C., Herzog F., Gieffers C., Mechtler K., Hagting A., Pines J., and Peters J.-M.. 2003. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 22:6598–6609. 10.1093/emboj/cdg627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambing C., Osman K., Nuntasoontorn K., West A., Higgins J.D., Copenhaver G.P., Yang J., Armstrong S.J., Mechtler K., Roitinger E., and Franklin F.C.H.. 2015. Arabidopsis PCH2 mediates meiotic chromosome remodeling and maturation of crossovers. PLoS Genet. 11:e1005372 10.1371/journal.pgen.1005372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P., Westhorpe F.G., and Taylor S.S.. 2012. The spindle assembly checkpoint. Curr. Biol. 22:R966–R980. 10.1016/j.cub.2012.10.006 [DOI] [PubMed] [Google Scholar]
- Listovsky T., and Sale J.E.. 2013. Sequestration of CDH1 by MAD2L2 prevents premature APC/C activation prior to anaphase onset. J. Cell Biol. 203:87–100. 10.1083/jcb.201302060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N., and Biggins S.. 2014. Signalling dynamics in the spindle checkpoint response. Nat. Rev. Mol. Cell Biol. 15:736–747. 10.1038/nrm3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., and Yu H.. 2008. Protein metamorphosis: The two-state behavior of Mad2. Structure. 16:1616–1625. 10.1016/j.str.2008.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Fang G., Coldiron M., Lin Y., Yu H., Kirschner M.W., and Wagner G.. 2000. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat. Struct. Biol. 7:224–229. 10.1038/73338 [DOI] [PubMed] [Google Scholar]
- Luo X., Tang Z., Rizo J., and Yu H.. 2002. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell. 9:59–71. 10.1016/S1097-2765(01)00435-X [DOI] [PubMed] [Google Scholar]
- MacQueen A.J., and Hochwagen A.. 2011. Checkpoint mechanisms: The puppet masters of meiotic prophase. Trends Cell Biol. 21:393–400. 10.1016/j.tcb.2011.03.004 [DOI] [PubMed] [Google Scholar]
- Makarova A.V., and Burgers P.M.. 2015. Eukaryotic DNA polymerase ζ. DNA Repair (Amst.). 29:47–55. 10.1016/j.dnarep.2015.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao-Draayer Y., Galbraith A.M., Pittman D.L., Cool M., and Malone R.E.. 1996. Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae. Genetics. 144:71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli M., and Musacchio A.. 2007. MAD contortions: conformational dimerization boosts spindle checkpoint signaling. Curr. Opin. Struct. Biol. 17:716–725. 10.1016/j.sbi.2007.08.011 [DOI] [PubMed] [Google Scholar]
- Mapelli M., Filipp F.V., Rancati G., Massimiliano L., Nezi L., Stier G., Hagan R.S., Confalonieri S., Piatti S., Sattler M., and Musacchio A.. 2006. Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J. 25:1273–1284. 10.1038/sj.emboj.7601033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli M., Massimiliano L., Santaguida S., and Musacchio A.. 2007. The Mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell. 131:730–743. 10.1016/j.cell.2007.08.049 [DOI] [PubMed] [Google Scholar]
- Medendorp K., van Groningen J.J.M., Vreede L., Hetterschijt L., van den Hurk W.H., de Bruijn D.R.H., Brugmans L., and van Kessel A.G.. 2009. The mitotic arrest deficient protein MAD2B interacts with the small GTPase RAN throughout the cell cycle. PLoS One. 4:e7020 10.1371/journal.pone.0007020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medendorp K., Vreede L., van Groningen J.J.M., Hetterschijt L., Brugmans L., Jansen P.A.M., van den Hurk W.H., de Bruijn D.R.H., and van Kessel A.G.. 2010. The mitotic arrest deficient protein MAD2B interacts with the clathrin light chain A during mitosis. PLoS One. 5:e15128 10.1371/journal.pone.0015128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer C.A., Kaliappan A., and Dennis P.B.. 2009. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 5:649–662. 10.4161/auto.5.5.8249 [DOI] [PubMed] [Google Scholar]
- Michel M., Schwarten M., Decker C., Nagel-Steger L., Willbold D., and Weiergräber O.H.. 2015. The mammalian autophagy initiator complex contains two HORMA domain proteins. Autophagy. 0:0 10.1080/15548627.2015.1076605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. 2007. Autophagy: Process and function. Genes Dev. 21:2861–2873. 10.1101/gad.1599207 [DOI] [PubMed] [Google Scholar]
- Mizushima N. 2010. The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 22:132–139. 10.1016/j.ceb.2009.12.004 [DOI] [PubMed] [Google Scholar]
- Mo M., Arnaoutov A., and Dasso M.. 2015. Phosphorylation of Xenopus p31(comet) potentiates mitotic checkpoint exit. Cell Cycle. 0 10.1080/15384101.2015.1033590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniyappa K., Kshirsagar R., and Ghodke I.. 2014. The HORMA domain: An evolutionarily conserved domain discovered in chromatin-associated proteins, has unanticipated diverse functions. Gene. 545:194–197. 10.1016/j.gene.2014.05.020 [DOI] [PubMed] [Google Scholar]
- Musacchio A., and Salmon E.D.. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393. 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Nelson C.R., Hwang T., Chen P.-H., and Bhalla N.. 2015. TRIP13PCH-2 promotes Mad2 localization to unattached kinetochores in the spindle checkpoint response. J. Cell Biol. 211:503–516. 10.1126/science.272.5268.1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J.R., Lawrence C.W., and Hinkle D.C.. 1996a Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science. 272:1646–1649. 10.1126/science.272.5268.1646 [DOI] [PubMed] [Google Scholar]
- Nelson J.R., Lawrence C.W., and Hinkle D.C.. 1996b Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 382:729–731. 10.1038/382729a0 [DOI] [PubMed] [Google Scholar]
- Nelson J.R., Gibbs P.E., Nowicka A.M., Hinkle D.C., and Lawrence C.W.. 2000. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol. Microbiol. 37:549–554. 10.1046/j.1365-2958.2000.01997.x [DOI] [PubMed] [Google Scholar]
- Pfleger C.M., Salic A., Lee E., and Kirschner M.W.. 2001. Inhibition of Cdh1-APC by the MAD2-related protein MAD2L2: A novel mechanism for regulating Cdh1. Genes Dev. 15:1759–1764. 10.1101/gad.897901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J. 2011. Cubism and the cell cycle: The many faces of the APC/C. Nat. Rev. Mol. Cell Biol. 12:427–438. 10.1038/nrm3132 [DOI] [PubMed] [Google Scholar]
- Pirouz M., Pilarski S., and Kessel M.. 2013. A critical function of Mad2l2 in primordial germ cell development of mice. PLoS Genet. 9:e1003712 10.1371/journal.pgen.1003712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirouz M., Rahjouei A., Shamsi F., Eckermann K.N., Salinas-Riester G., Pommerenke C., and Kessel M.. 2015. Destabilization of pluripotency in the absence of Mad2l2. Cell Cycle. 14:1596–1610. 10.1080/15384101.2015.1026485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S., Johnson R.E., and Prakash L.. 2005. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu. Rev. Biochem. 74:317–353. 10.1146/annurev.biochem.74.082803.133250 [DOI] [PubMed] [Google Scholar]
- Primorac I., and Musacchio A.. 2013. Panta rhei: The APC/C at steady state. J. Cell Biol. 201:177–189. 10.1083/jcb.201301130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S., Kim J., Stjepanovic G., and Hurley J.H.. 2015. Structure of the human Atg13-Atg101 HORMA heterodimer: An interaction hub within the ULK1 complex. Structure. 23:1848–1857. 10.1016/j.str.2015.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa M.J., Stanley R.E., and Hurley J.H.. 2012. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 151:1501–1512. 10.1016/j.cell.2012.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S.K., Rape M., Margansky W.A., and Kirschner M.W.. 2007. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 446:921–925. 10.1038/nature05734 [DOI] [PubMed] [Google Scholar]
- Ross A.-L., Simpson L.J., and Sale J.E.. 2005. Vertebrate DNA damage tolerance requires the C-terminus but not BRCT or transferase domains of REV1. Nucleic Acids Res. 33:1280–1289. 10.1093/nar/gki279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuno T., and Watanabe Y.. 2015. Phosphorylation of cohesin Rec11/SA3 by casein kinase 1 promotes homologous recombination by assembling the meiotic chromosome axis. Dev. Cell. 32:220–230. 10.1016/j.devcel.2014.11.033 [DOI] [PubMed] [Google Scholar]
- Sale J.E. 2013. Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb. Perspect. Biol. 5:a012708 10.1101/cshperspect.a012708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Segundo P.A., and Roeder G.S.. 1999. Pch2 links chromatin silencing to meiotic checkpoint control. Cell. 97:313–324. 10.1016/S0092-8674(00)80741-2 [DOI] [PubMed] [Google Scholar]
- Shah J.V., Botvinick E., Bonday Z., Furnari F., Berns M., and Cleveland D.W.. 2004. Dynamics of centromere and kinetochore proteins; Implications for checkpoint signaling and silencing. Curr. Biol. 14:942–952. [DOI] [PubMed] [Google Scholar]
- Shin Y.-H., Choi Y., Erdin S.U., Yatsenko S.A., Kloc M., Yang F., Wang P.J., Meistrich M.L., and Rajkovic A.. 2010. Hormad1 mutation disrupts synaptonemal complex formation, recombination, and chromosome segregation in mammalian meiosis. PLoS Genet. 6:e1001190 (published erratum appears in PLoS Genet. 2011;7) 10.1371/journal.pgen.1001190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L., Melixetian M., Faretta M., Prosperini E., Helin K., and Musacchio A.. 2001. Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J. 20:6371–6382. 10.1093/emboj/20.22.6371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L., Mapelli M., Knapp S., De Antoni A., Jeang K.-T., and Musacchio A.. 2002. Crystal structure of the tetrameric Mad1-Mad2 core complex: Implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 21:2496–2506. 10.1093/emboj/21.10.2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar S., and Gorbsky G.J.. 2015. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat. Rev. Mol. Cell Biol. 16:82–94. 10.1038/nrm3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.V., and Roeder G.S.. 1997. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell Biol. 136:957–967. 10.1083/jcb.136.5.957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stjepanovic G., Davies C.W., Stanley R.E., Ragusa M.J., Kim J., and Hurley J.H.. 2014. Assembly and dynamics of the autophagy-initiating Atg1 complex. Proc. Natl. Acad. Sci. USA. 111:12793–12798. 10.1073/pnas.1407214111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian V.V., and Hochwagen A.. 2014. The meiotic checkpoint network: Step-by-step through meiotic prophase. Cold Spring Harb. Perspect. Biol. 6:a016675 10.1101/cshperspect.a016675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V., Chan G.K., and Yen T.J.. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154:925–936. 10.1083/jcb.200102093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Kaizuka T., Mizushima N., and Noda N.N.. 2015a Structure of the Atg101-Atg13 complex reveals essential roles of Atg101 in autophagy initiation. Nat. Struct. Mol. Biol. 22:572–580. 10.1038/nsmb.3036 [DOI] [PubMed] [Google Scholar]
- Suzuki S.W., Yamamoto H., Oikawa Y., Kondo-Kakuta C., Kimura Y., Hirano H., and Ohsumi Y.. 2015b Atg13 HORMA domain recruits Atg9 vesicles during autophagosome formation. Proc. Natl. Acad. Sci. USA. 112:3350–3355. 10.1073/pnas.1421092112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichner A., Eytan E., Sitry-Shevah D., Miniowitz-Shemtov S., Dumin E., Gromis J., and Hershko A.. 2011. p31comet promotes disassembly of the mitotic checkpoint complex in an ATP-dependent process. Proc. Natl. Acad. Sci. USA. 108:3187–3192. 10.1073/pnas.1100023108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker D., Mohibullah N., Zhu X., and Keeney S.. 2014. Homologue engagement controls meiotic DNA break number and distribution. Nature. 510:241–246. 10.1038/nature13120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton A.R., Wang K., Oladimeji P., Sufi S., Gu Z., and Liu S.-T.. 2012. Identification of novel mitosis regulators through data mining with human centromere/kinetochore proteins as group queries. BMC Cell Biol. 13:15 10.1186/1471-2121-13-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomida J., Takata K., Lange S.S., Schibler A.C., Yousefzadeh M.J., Bhetawal S., Dent S.Y.R., and Wood R.D.. 2015. REV7 is essential for DNA damage tolerance via two REV3L binding sites in mammalian DNA polymerase ζ. Nucleic Acids Res. 43:1000–1011. 10.1093/nar/gku1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader G., and Musacchio A.. 2014. HORMA domains at the heart of meiotic chromosome dynamics. Dev. Cell. 31:389–391. 10.1016/j.devcel.2014.11.009 [DOI] [PubMed] [Google Scholar]
- Vermeulen M., Eberl H.C., Matarese F., Marks H., Denissov S., Butter F., Lee K.K., Olsen J.V., Hyman A.A., Stunnenberg H.G., and Mann M.. 2010. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 142:967–980. 10.1016/j.cell.2010.08.020 [DOI] [PubMed] [Google Scholar]
- Vink M., Simonetta M., Transidico P., Ferrari K., Mapelli M., De Antoni A., Massimiliano L., Ciliberto A., Faretta M., Salmon E.D., and Musacchio A.. 2006. In vitro FRAP identifies the minimal requirements for Mad2 kinetochore dynamics. Curr. Biol. 16:755–766. 10.1016/j.cub.2006.03.057 [DOI] [PubMed] [Google Scholar]
- Wang K., Wang M., Tang D., Shen Y., Qin B., Li M., and Cheng Z.. 2011. PAIR3, an axis-associated protein, is essential for the recruitment of recombination elements onto meiotic chromosomes in rice. Mol. Biol. Cell. 22:12–19. 10.1091/mbc.E10-08-0667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Sturt-Gillespie B., Hittle J.C., Macdonald D., Chan G.K., Yen T.J., and Liu S.-T.. 2014. Thyroid hormone receptor interacting protein 13 (TRIP13) AAA-ATPase is a novel mitotic checkpoint-silencing protein. J. Biol. Chem. 289:23928–23937. 10.1074/jbc.M114.585315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N., Mii S., Asai N., Asai M., Niimi K., Ushida K., Kato T., Enomoto A., Ishii H., Takahashi M., and Murakumo Y.. 2013. The REV7 subunit of DNA polymerase ζ is essential for primordial germ cell maintenance in the mouse. J. Biol. Chem. 288:10459–10471. 10.1074/jbc.M112.421966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters L.S., Minesinger B.K., Wiltrout M.E., D’Souza S., Woodruff R.V., and Walker G.C.. 2009. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 73:134–154. 10.1128/MMBR.00034-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe F.G., Tighe A., Lara-Gonzalez P., and Taylor S.S.. 2011. p31comet-mediated extraction of Mad2 from the MCC promotes efficient mitotic exit. J. Cell Sci. 124:3905–3916. 10.1242/jcs.093286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasz L., Daniel K., Roig I., Bolcun-Filas E., Xu H., Boonsanay V., Eckmann C.R., Cooke H.J., Jasin M., Keeney S., et al. 2009. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 5:e1000702 10.1371/journal.pgen.1000702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek J., Lee C.-J., D’Souza S., Minesinger B., Kim H., D’Andrea A.D., Walker G.C., and Zhou P.. 2012. Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric polymerase (Pol) ζ, and Pol κ. J. Biol. Chem. 287:33836–33846. 10.1074/jbc.M112.394841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltering D., Baumgartner B., Bagchi S., Larkin B., Loidl J., de los Santos T., and Hollingsworth N.M.. 2000. Meiotic segregation, synapsis, and recombination checkpoint functions require physical interaction between the chromosomal proteins Red1p and Hop1p. Mol. Cell. Biol. 20:6646–6658. 10.1128/MCB.20.18.6646-6658.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G., Luo X., Habu T., Rizo J., Matsumoto T., and Yu H.. 2004. Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 23:3133–3143. 10.1038/sj.emboj.7600322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., and Klionsky D.J.. 2007. Autophagosome formation: Core machinery and adaptations. Nat. Cell Biol. 9:1102–1109. 10.1038/ncb1007-1102 [DOI] [PubMed] [Google Scholar]
- Xie W., Yang X., Xu M., and Jiang T.. 2012. Structural insights into the assembly of human translesion polymerase complexes. Protein Cell. 3:864–874. 10.1007/s13238-012-2102-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Chapman J.R., Brandsma I., Yuan J., Mistrik M., Bouwman P., Bartkova J., Gogola E., Warmerdam D., Barazas M., et al. 2015. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature. 521:541–544. 10.1038/nature14328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., Watanabe K., Mistrik M., Vesela E., Protivankova I., Mailand N., Lee M., Masai H., Lukas J., and Bartek J.. 2013. ATR-Chk1-APC/CCdh1-dependent stabilization of Cdc7-ASK (Dbf4) kinase is required for DNA lesion bypass under replication stress. Genes Dev. 27:2459–2472. 10.1101/gad.224568.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Li B., Tomchick D.R., Machius M., Rizo J., Yu H., and Luo X.. 2007. p31comet blocks Mad2 activation through structural mimicry. Cell. 131:744–755. 10.1016/j.cell.2007.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., Rosenberg S.C., Moeller A., Speir J.A., Su T.Y., and Corbett K.D.. 2015. TRIP13 is a protein-remodeling AAA+ ATPase that catalyzes MAD2 conformation switching. eLife. 4:e07367 10.7554/eLife.07367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Y.-Y., Shah K.H., and Herman P.K.. 2011. An Atg13 protein-mediated self-association of the Atg1 protein kinase is important for the induction of autophagy. J. Biol. Chem. 286:28931–28939. 10.1074/jbc.M111.250324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Yang S.-H., and Sharrocks A.D.. 2007. Rev7/MAD2B links c-Jun N-terminal protein kinase pathway signaling to activation of the transcription factor Elk-1. Mol. Cell. Biol. 27:2861–2869. 10.1128/MCB.02276-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.